

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148965.10 + phase: 0
(636 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
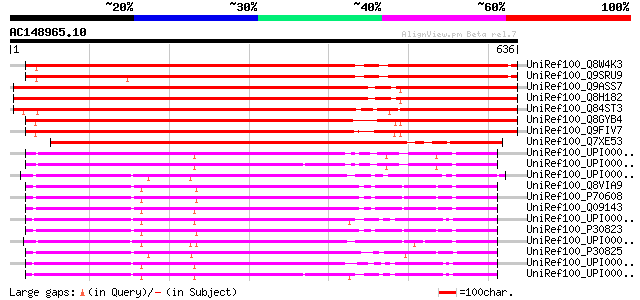
UniRef100_Q8W4K3 Putative cationic amino acid transporter [Arabi... 808 0.0
UniRef100_Q9SRU9 Putative cationic amino acid transporter [Arabi... 798 0.0
UniRef100_Q9ASS7 AT5g36940/MLF18_60 [Arabidopsis thaliana] 797 0.0
UniRef100_Q8H182 Hypothetical protein [Arabidopsis thaliana] 795 0.0
UniRef100_Q84ST3 Putative cationic amino acid transporter [Oryza... 784 0.0
UniRef100_Q8GYB4 Putative cationic amino acid transporter [Arabi... 761 0.0
UniRef100_Q9FIV7 Cationic amino acid transporter-like protein [A... 756 0.0
UniRef100_Q7XE53 Putative cationic amino acid transporter-like p... 615 e-174
UniRef100_UPI000043683B UPI000043683B UniRef100 entry 378 e-103
UniRef100_UPI000043683A UPI000043683A UniRef100 entry 376 e-103
UniRef100_UPI00003AA095 UPI00003AA095 UniRef100 entry 376 e-103
UniRef100_Q8VIA9 Ecotropic retrovirus receptor [Rattus norvegicus] 376 e-103
UniRef100_P70608 Cationic amino acid transporter-1 [Rattus norve... 376 e-103
UniRef100_Q09143 High-affinity cationic amino acid transporter-1... 375 e-102
UniRef100_UPI00003654CA UPI00003654CA UniRef100 entry 372 e-101
UniRef100_P30823 High-affinity cationic amino acid transporter-1... 371 e-101
UniRef100_UPI000024805A UPI000024805A UniRef100 entry 369 e-100
UniRef100_P30825 High-affinity cationic amino acid transporter-1... 369 e-100
UniRef100_UPI00003654C9 UPI00003654C9 UniRef100 entry 368 e-100
UniRef100_UPI00003654CB UPI00003654CB UniRef100 entry 368 e-100
>UniRef100_Q8W4K3 Putative cationic amino acid transporter [Arabidopsis thaliana]
Length = 600
Score = 808 bits (2087), Expect = 0.0
Identities = 401/620 (64%), Positives = 485/620 (77%), Gaps = 25/620 (4%)
Query: 20 SLIRRKQVDSIHY---RGHPQLARKLSVVDLVGIGVGATIGAGVYILIGTVAREQAGPAL 76
SL+RRKQVDS+H G QLA+KLS VDLV IGVG TIGAGVYIL+GTVARE GPAL
Sbjct: 3 SLVRRKQVDSVHLIKNDGPHQLAKKLSAVDLVAIGVGTTIGAGVYILVGTVAREHTGPAL 62
Query: 77 VISLFIAGIAAALSALCYAELACRCPSAGSAYHYTYICIGEGVAWLVGWSLILEYTIGAS 136
+S FIAG+AAALSA CYAELA RCPSAGSAYHY YIC+GEG+AWLVGW+L+L+YTIG S
Sbjct: 63 AVSFFIAGVAAALSACCYAELASRCPSAGSAYHYAYICLGEGIAWLVGWALVLDYTIGGS 122
Query: 137 AVARGITPNLALFFGGQDNLPSFLARHTLPGLGIVVDPCAAVLIVLITLLLCLGIKESST 196
A+ARGITPNLA FFGG DNLP FLAR T+PG+GIVVDPCAA+LI+++T+LLC GIKESST
Sbjct: 123 AIARGITPNLASFFGGLDNLPVFLARQTIPGVGIVVDPCAALLIMIVTILLCFGIKESST 182
Query: 197 VQSIVTTINVSVMLFIIIVGGYLGFKAGWVGYELPSGYFPYGVNGMFAGSAIVFFSYIGF 256
VQ+IVT++NV ++FII+VGGYL K GWVGY+LPSGYFP+G+NG+ AGSA+VFFSYIGF
Sbjct: 183 VQAIVTSVNVCTLVFIIVVGGYLACKTGWVGYDLPSGYFPFGLNGILAGSAVVFFSYIGF 242
Query: 257 DSVTSTAEEVKNPQRDLPIGISTALAICCVLYMLVSAVIVGLVPYYELNPDTPISSAFSS 316
D+VTSTAEEVKNPQRDLP+GI AL ICC+LYML+S VIVGLVPYY LNPDTPISSAF
Sbjct: 243 DTVTSTAEEVKNPQRDLPLGIGIALLICCILYMLLSVVIVGLVPYYSLNPDTPISSAFGD 302
Query: 317 YGMEWAVYIITTGAVTALFSSLLGSVLPQPRVFMAMARDGLLPTFFSDIHRRTQIPLKST 376
GM+WA YI+TTGA+TAL +SLLGS+L QPR+FMAMARDGLLP FFS+I RTQ+P+KST
Sbjct: 303 SGMQWAAYILTTGAITALCASLLGSLLAQPRIFMAMARDGLLPAFFSEISPRTQVPVKST 362
Query: 377 IVTGLFAAVLAFFMDVSQLAGMVSVGTLLAFTTVAVSVLIIRYVPPDEIPIPASLLTSVD 436
I G+ AA LAFFMDV+QL+ MVSVGTL+AFT VAV VL++RYVPPD +P+ +S T
Sbjct: 363 IAIGVLAAALAFFMDVAQLSEMVSVGTLMAFTAVAVCVLVLRYVPPDGVPLSSSSQTL-- 420
Query: 437 PLLRHSGDDIEEDRTVSPVDLASYSDNSHLHDKSDVLLEHPLIIKEVTKEQHNEKTRRKL 496
D +E R + L ++S + PL+ E +++ RRK+
Sbjct: 421 -------SDTDESRAETENFLVDAIESS----------DSPLLGNETARDEKYFGKRRKI 463
Query: 497 AAWTIALLCIGILIVSGSASAERCPRILRVTLFGAGVVIFLCSIIVLACIKQDDTRHTFG 556
AAW+IAL+CIG+L ++ +ASAER P R T+ G VI L S+I L I +D+ RH FG
Sbjct: 464 AAWSIALVCIGVLGLASAASAERLPSFPRFTICGVSAVILLGSLITLGYIDEDEERHNFG 523
Query: 557 HSGGFACPFVPFLPAACILINTYLLIDLGVDTWLRVSVWLLIGVLIYLFYGRTHSSLLNA 616
H GGF CPFVP+LP CILINTYL+I++G TW+RV +WLLIG +IY+FYGR+HS L NA
Sbjct: 524 HKGGFLCPFVPYLPVLCILINTYLIINIGAGTWIRVLIWLLIGSMIYIFYGRSHSLLNNA 583
Query: 617 IYVPSARADEIHRSQANHLA 636
+YVP+ R +HLA
Sbjct: 584 VYVPTMTCT---RKTTDHLA 600
>UniRef100_Q9SRU9 Putative cationic amino acid transporter [Arabidopsis thaliana]
Length = 614
Score = 798 bits (2062), Expect = 0.0
Identities = 401/634 (63%), Positives = 485/634 (76%), Gaps = 39/634 (6%)
Query: 20 SLIRRKQVDSIHY---RGHPQLARKLSVVDLVGIGVGATIGAGVYILIGTVAREQAGPAL 76
SL+RRKQVDS+H G QLA+KLS VDLV IGVG TIGAGVYIL+GTVARE GPAL
Sbjct: 3 SLVRRKQVDSVHLIKNDGPHQLAKKLSAVDLVAIGVGTTIGAGVYILVGTVAREHTGPAL 62
Query: 77 VISLFIAGIAAALSALCYAELACRCPSAGSAYHYTYICIGEGVAWLVGWSLILEYTIGAS 136
+S FIAG+AAALSA CYAELA RCPSAGSAYHY YIC+GEG+AWLVGW+L+L+YTIG S
Sbjct: 63 AVSFFIAGVAAALSACCYAELASRCPSAGSAYHYAYICLGEGIAWLVGWALVLDYTIGGS 122
Query: 137 AVARGITPNL--------------ALFFGGQDNLPSFLARHTLPGLGIVVDPCAAVLIVL 182
A+ARGITPNL A FFGG DNLP FLAR T+PG+GIVVDPCAA+LI++
Sbjct: 123 AIARGITPNLVFAFELYVFGFSQEASFFGGLDNLPVFLARQTIPGVGIVVDPCAALLIMI 182
Query: 183 ITLLLCLGIKESSTVQSIVTTINVSVMLFIIIVGGYLGFKAGWVGYELPSGYFPYGVNGM 242
+T+LLC GIKESSTVQ+IVT++NV ++FII+VGGYL K GWVGY+LPSGYFP+G+NG+
Sbjct: 183 VTILLCFGIKESSTVQAIVTSVNVCTLVFIIVVGGYLACKTGWVGYDLPSGYFPFGLNGI 242
Query: 243 FAGSAIVFFSYIGFDSVTSTAEEVKNPQRDLPIGISTALAICCVLYMLVSAVIVGLVPYY 302
AGSA+VFFSYIGFD+VTSTAEEVKNPQRDLP+GI AL ICC+LYML+S VIVGLVPYY
Sbjct: 243 LAGSAVVFFSYIGFDTVTSTAEEVKNPQRDLPLGIGIALLICCILYMLLSVVIVGLVPYY 302
Query: 303 ELNPDTPISSAFSSYGMEWAVYIITTGAVTALFSSLLGSVLPQPRVFMAMARDGLLPTFF 362
LNPDTPISSAF GM+WA YI+TTGA+TAL +SLLGS+L QPR+FMAMARDGLLP FF
Sbjct: 303 SLNPDTPISSAFGDSGMQWAAYILTTGAITALCASLLGSLLAQPRIFMAMARDGLLPAFF 362
Query: 363 SDIHRRTQIPLKSTIVTGLFAAVLAFFMDVSQLAGMVSVGTLLAFTTVAVSVLIIRYVPP 422
S+I RTQ+P+KSTI G+ AA LAFFMDV+QL+ MVSVGTL+AFT VAV VL++RYVPP
Sbjct: 363 SEISPRTQVPVKSTIAIGVLAAALAFFMDVAQLSEMVSVGTLMAFTAVAVCVLVLRYVPP 422
Query: 423 DEIPIPASLLTSVDPLLRHSGDDIEEDRTVSPVDLASYSDNSHLHDKSDVLLEHPLIIKE 482
D +P+ +S T D +E R + L ++S + PL+ E
Sbjct: 423 DGVPLSSSSQTL---------SDTDESRAETENFLVDAIESS----------DSPLLGNE 463
Query: 483 VTKEQHNEKTRRKLAAWTIALLCIGILIVSGSASAERCPRILRVTLFGAGVVIFLCSIIV 542
+++ RRK+AAW+IAL+CIG+L ++ +ASAER P R T+ G VI L S+I
Sbjct: 464 TARDEKYFGKRRKIAAWSIALVCIGVLGLASAASAERLPSFPRFTICGVSAVILLGSLIT 523
Query: 543 LACIKQDDTRHTFGHSGGFACPFVPFLPAACILINTYLLIDLGVDTWLRVSVWLLIGVLI 602
L I +D+ RH FGH GGF CPFVP+LP CILINTYL+I++G TW+RV +WLLIG +I
Sbjct: 524 LGYIDEDEERHNFGHKGGFLCPFVPYLPVLCILINTYLIINIGAGTWIRVLIWLLIGSMI 583
Query: 603 YLFYGRTHSSLLNAIYVPSARADEIHRSQANHLA 636
Y+FYGR+HS L NA+YVP+ R +HLA
Sbjct: 584 YIFYGRSHSLLNNAVYVPTMTCT---RKTTDHLA 614
>UniRef100_Q9ASS7 AT5g36940/MLF18_60 [Arabidopsis thaliana]
Length = 635
Score = 797 bits (2058), Expect = 0.0
Identities = 402/643 (62%), Positives = 496/643 (76%), Gaps = 23/643 (3%)
Query: 5 VGNGEEGGGRFRGF-GSLIRRKQVDSIHYRGHP-QLARKLSVVDLVGIGVGATIGAGVYI 62
V +EGGG G+ SL+RRKQVDS + + H QLAR L+V LV IGVGATIGAGVYI
Sbjct: 5 VDTQKEGGGHSWGYVRSLVRRKQVDSANGQSHGHQLARALTVPHLVAIGVGATIGAGVYI 64
Query: 63 LIGTVAREQAGPALVISLFIAGIAAALSALCYAELACRCPSAGSAYHYTYICIGEGVAWL 122
L+GTVARE +GP+L +S IAGIAA LSA CYAEL+ RCPSAGSAYHY+YIC+GEGVAW+
Sbjct: 65 LVGTVAREHSGPSLALSFLIAGIAAGLSAFCYAELSSRCPSAGSAYHYSYICVGEGVAWI 124
Query: 123 VGWSLILEYTIGASAVARGITPNLALFFGGQDNLPSFLARHTLPGLGIVVDPCAAVLIVL 182
+GW+LILEYTIG SAVARGI+PNLAL FGG+D LP+ LARH +PGL IVVDPCAA+L+ +
Sbjct: 125 IGWALILEYTIGGSAVARGISPNLALIFGGEDGLPAILARHQIPGLDIVVDPCAAILVFV 184
Query: 183 ITLLLCLGIKESSTVQSIVTTINVSVMLFIIIVGGYLGFKAGWVGYELPSGYFPYGVNGM 242
+T LLC+GIKES+ Q IVT +NV V+LF+I+ G YLGFK GW GYELP+G+FP+GV+GM
Sbjct: 185 VTGLLCMGIKESTFAQGIVTAVNVCVLLFVIVAGSYLGFKTGWPGYELPTGFFPFGVDGM 244
Query: 243 FAGSAIVFFSYIGFDSVTSTAEEVKNPQRDLPIGISTALAICCVLYMLVSAVIVGLVPYY 302
FAGSA VFF++IGFDSV STAEEV+NPQRDLPIGI AL +CC LYM+VS VIVGL+PYY
Sbjct: 245 FAGSATVFFAFIGFDSVASTAEEVRNPQRDLPIGIGLALLLCCSLYMMVSIVIVGLIPYY 304
Query: 303 ELNPDTPISSAFSSYGMEWAVYIITTGAVTALFSSLLGSVLPQPRVFMAMARDGLLPTFF 362
++PDTPISSAF+S+ M+WAVY+IT GAV AL S+L+G++LPQPR+ MAMARDGLLP+ F
Sbjct: 305 AMDPDTPISSAFASHDMQWAVYLITLGAVMALCSALMGALLPQPRILMAMARDGLLPSIF 364
Query: 363 SDIHRRTQIPLKSTIVTGLFAAVLAFFMDVSQLAGMVSVGTLLAFTTVAVSVLIIRYVPP 422
SDI++RTQ+P+K+T+ TGL AA LAFFMDVSQLAGMVSVGTLLAFT VA+SVLI+RYVPP
Sbjct: 365 SDINKRTQVPVKATVATGLCAATLAFFMDVSQLAGMVSVGTLLAFTMVAISVLILRYVPP 424
Query: 423 DEIPIPASLLTSVDPLLRHSGDDIEEDRTVSPVDLASYSDNSH--LHDKSDVLLEHPLII 480
DE P+P+SL +D + G+ SD+SH L +D L++ PLI
Sbjct: 425 DEQPLPSSLQERIDSVSFICGETTSSGH-------VGTSDSSHQPLIVNNDALVDVPLI- 476
Query: 481 KEVTKEQH-------NEKTRRKLAAWTIALLCIGILIVSGSASAERCPRILRVTLFGAGV 533
K Q +E+TRR +A W+I C+G ++S +AS+ P ++R L G G
Sbjct: 477 ----KNQEALGCLVLSEETRRIVAGWSIMFTCVGAFLLSYAASSLSFPGLIRYPLCGVGG 532
Query: 534 VIFLCSIIVLACIKQDDTRHTFGHSGGFACPFVPFLPAACILINTYLLIDLGVDTWLRVS 593
+ L +I L+ I QDD RHTFGHSGG+ CPFVP LP CILIN YLL++LG TW RVS
Sbjct: 533 CLLLAGLIALSSIDQDDARHTFGHSGGYMCPFVPLLPIICILINMYLLVNLGSATWARVS 592
Query: 594 VWLLIGVLIYLFYGRTHSSLLNAIYVPSARADEIHRSQANHLA 636
VWLLIGV++Y+FYGR +SSL NA+YV +A A+EI+R LA
Sbjct: 593 VWLLIGVIVYVFYGRKNSSLANAVYVTTAHAEEIYREHEGSLA 635
>UniRef100_Q8H182 Hypothetical protein [Arabidopsis thaliana]
Length = 635
Score = 795 bits (2052), Expect = 0.0
Identities = 401/643 (62%), Positives = 495/643 (76%), Gaps = 23/643 (3%)
Query: 5 VGNGEEGGGRFRGF-GSLIRRKQVDSIHYRGHP-QLARKLSVVDLVGIGVGATIGAGVYI 62
V +EGGG G+ SL+RRKQVDS + + H QLAR L+V LV IGVGATIGAGVYI
Sbjct: 5 VDTQKEGGGHSWGYVRSLVRRKQVDSANGQSHGHQLARALTVPHLVAIGVGATIGAGVYI 64
Query: 63 LIGTVAREQAGPALVISLFIAGIAAALSALCYAELACRCPSAGSAYHYTYICIGEGVAWL 122
L+GTVARE +GP+L +S IAGIAA LSA CYAEL+ RCPSAGSAYHY+YIC+GEGVAW+
Sbjct: 65 LVGTVAREHSGPSLALSFLIAGIAAGLSAFCYAELSSRCPSAGSAYHYSYICVGEGVAWI 124
Query: 123 VGWSLILEYTIGASAVARGITPNLALFFGGQDNLPSFLARHTLPGLGIVVDPCAAVLIVL 182
+GW+LILEYTIG SAVARGI+PNLAL FGG+D LP+ LARH +PGL IVVDPCAA+L+ +
Sbjct: 125 IGWALILEYTIGGSAVARGISPNLALIFGGEDGLPAILARHQIPGLDIVVDPCAAILVFV 184
Query: 183 ITLLLCLGIKESSTVQSIVTTINVSVMLFIIIVGGYLGFKAGWVGYELPSGYFPYGVNGM 242
+T LLC+GIKES+ Q IVT +NV V+LF+I+ G YLGFK GW GYELP+G+FP+GV+GM
Sbjct: 185 VTGLLCMGIKESTFAQGIVTAVNVCVLLFVIVAGSYLGFKTGWPGYELPTGFFPFGVDGM 244
Query: 243 FAGSAIVFFSYIGFDSVTSTAEEVKNPQRDLPIGISTALAICCVLYMLVSAVIVGLVPYY 302
FAGSA VFF++IGFDSV STAEEV+NPQRDLPIGI AL +CC LYM+VS VIVGL+PYY
Sbjct: 245 FAGSATVFFAFIGFDSVASTAEEVRNPQRDLPIGIGLALLLCCSLYMMVSIVIVGLIPYY 304
Query: 303 ELNPDTPISSAFSSYGMEWAVYIITTGAVTALFSSLLGSVLPQPRVFMAMARDGLLPTFF 362
++PDTPISSAF+S+ M+WAVY+IT GAV AL S+L+G++LPQPR+ MAMARDGLLP+ F
Sbjct: 305 AMDPDTPISSAFASHDMQWAVYLITLGAVMALCSALMGALLPQPRILMAMARDGLLPSIF 364
Query: 363 SDIHRRTQIPLKSTIVTGLFAAVLAFFMDVSQLAGMVSVGTLLAFTTVAVSVLIIRYVPP 422
SDI++RTQ+P+K+T+ TGL AA LAF MDVSQLAGMVSVGTLLAFT VA+SVLI+RYVPP
Sbjct: 365 SDINKRTQVPVKATVATGLCAATLAFIMDVSQLAGMVSVGTLLAFTMVAISVLILRYVPP 424
Query: 423 DEIPIPASLLTSVDPLLRHSGDDIEEDRTVSPVDLASYSDNSH--LHDKSDVLLEHPLII 480
DE P+P+SL +D + G+ SD+SH L +D L++ PLI
Sbjct: 425 DEQPLPSSLQERIDSVSFICGETTSSGH-------VGTSDSSHQPLIVNNDALVDVPLI- 476
Query: 481 KEVTKEQH-------NEKTRRKLAAWTIALLCIGILIVSGSASAERCPRILRVTLFGAGV 533
K Q +E+TRR +A W+I C+G ++S +AS+ P ++R L G G
Sbjct: 477 ----KNQEALGCLVLSEETRRIVAGWSIMFTCVGAFLLSYAASSLSFPGLIRYPLCGVGG 532
Query: 534 VIFLCSIIVLACIKQDDTRHTFGHSGGFACPFVPFLPAACILINTYLLIDLGVDTWLRVS 593
+ L +I L+ I QDD RHTFGHSGG+ CPFVP LP CILIN YLL++LG TW RVS
Sbjct: 533 CLLLAGLIALSSIDQDDARHTFGHSGGYMCPFVPLLPIICILINMYLLVNLGSATWARVS 592
Query: 594 VWLLIGVLIYLFYGRTHSSLLNAIYVPSARADEIHRSQANHLA 636
VWLLIGV++Y+FYGR +SSL NA+YV +A A+EI+R LA
Sbjct: 593 VWLLIGVIVYVFYGRKNSSLANAVYVTTAHAEEIYREHEGSLA 635
>UniRef100_Q84ST3 Putative cationic amino acid transporter [Oryza sativa]
Length = 639
Score = 784 bits (2024), Expect = 0.0
Identities = 387/645 (60%), Positives = 501/645 (77%), Gaps = 24/645 (3%)
Query: 6 GNGEEGGGRFR-----GFGSLIRRKQVDSIHYR-----GHPQLARKLSVVDLVGIGVGAT 55
G GE G R R GF SL+RRKQVDS R G PQLA++L++ LV IGVG+T
Sbjct: 5 GGGEVGRRRRRRGWGGGFPSLMRRKQVDSDRVRAAEGEGQPQLAKELNIPALVAIGVGST 64
Query: 56 IGAGVYILIGTVAREQAGPALVISLFIAGIAAALSALCYAELACRCPSAGSAYHYTYICI 115
IGAGVY+L+GTVARE AGPAL IS IAGIA+ALSA CYAELA RCPSAGSAYHY+YIC+
Sbjct: 65 IGAGVYVLVGTVAREHAGPALTISFLIAGIASALSAFCYAELASRCPSAGSAYHYSYICV 124
Query: 116 GEGVAWLVGWSLILEYTIGASAVARGITPNLALFFGGQDNLPSFLARHTLPGLGIVVDPC 175
GEGVAWL+GW+L+LEYTIG SAVARGI+PNLALFFGG D+LP L+RH LP ++VDPC
Sbjct: 125 GEGVAWLIGWALVLEYTIGGSAVARGISPNLALFFGGPDSLPWILSRHQLPWFDVIVDPC 184
Query: 176 AAVLIVLITLLLCLGIKESSTVQSIVTTINVSVMLFIIIVGGYLGFKAGWVGYELPSGYF 235
AA L+ ++T+LLC+GIKESS VQ ++T +N VM+F+I+ G Y+GF+ GWVGY++ GYF
Sbjct: 185 AAALVFVVTVLLCVGIKESSAVQELITVLNACVMIFVIVAGSYIGFQIGWVGYKVTDGYF 244
Query: 236 PYGVNGMFAGSAIVFFSYIGFDSVTSTAEEVKNPQRDLPIGISTALAICCVLYMLVSAVI 295
P+G+NGM AGSA VFF+YIGFD+V STAEEVKNPQRDLP+GI AL+ICC LYM+VS VI
Sbjct: 245 PHGINGMLAGSATVFFAYIGFDTVASTAEEVKNPQRDLPLGIGAALSICCCLYMMVSVVI 304
Query: 296 VGLVPYYELNPDTPISSAFSSYGMEWAVYIITTGAVTALFSSLLGSVLPQPRVFMAMARD 355
VGLVPY+ ++PDTPISS F+ +GM+WA+YI+T+GAV AL S+LLGS+LPQPR+ MAMARD
Sbjct: 305 VGLVPYFAMDPDTPISSVFAKHGMQWAMYIVTSGAVLALCSTLLGSLLPQPRILMAMARD 364
Query: 356 GLLPTFFSDIHRRTQIPLKSTIVTGLFAAVLAFFMDVSQLAGMVSVGTLLAFTTVAVSVL 415
GLLP+FF+D+++RTQ+P+KST+VTGL AA LAFFMDVSQLAGMVSVGTLLAFT VAVS+L
Sbjct: 365 GLLPSFFADVNKRTQVPVKSTVVTGLCAAALAFFMDVSQLAGMVSVGTLLAFTIVAVSIL 424
Query: 416 IIRYVPPDEIPIPASLLTSVDPLLRHSGDDIEEDRTVSPVDLASYSDNSHLHDKSDVLL- 474
I+RY+PPDE+P+P+SL + + +E+R + + + DV+L
Sbjct: 425 ILRYIPPDEVPLPSSLQETF-----CLSQEYDEERVSGILG----DERCKTSETKDVILA 475
Query: 475 ---EHPLIIKEVTKEQHNEKTRRKLAAWTIALLCIGILIVSGSASAERCPRILRVTLFGA 531
E PLI K++T++ +E RRK+AA++I +C+G+++++ +ASA P +
Sbjct: 476 ESMEDPLIEKKITRKM-DEMKRRKVAAFSIGSVCVGVMVLTSAASATWLPFLPMCIGCIV 534
Query: 532 GVVIFLCSIIVLACIKQDDTRHTFGHSGGFACPFVPFLPAACILINTYLLIDLGVDTWLR 591
G ++ + + +L I QDD RH+FG SGGF CPFVP LP IL+NTYLLI+LG + W+R
Sbjct: 535 GALLLVAGLGLLCWIDQDDGRHSFGQSGGFTCPFVPLLPVLSILVNTYLLINLGGEAWMR 594
Query: 592 VSVWLLIGVLIYLFYGRTHSSLLNAIYVPSARADEIHRSQANHLA 636
V +WLLIGVL+Y+ YGRT+SSL + IYVP A+ADEI++S + +++
Sbjct: 595 VGIWLLIGVLVYILYGRTNSSLKDVIYVPVAQADEIYKSSSGYVS 639
>UniRef100_Q8GYB4 Putative cationic amino acid transporter [Arabidopsis thaliana]
Length = 609
Score = 761 bits (1966), Expect = 0.0
Identities = 382/633 (60%), Positives = 475/633 (74%), Gaps = 45/633 (7%)
Query: 20 SLIRRKQVDSIH-----YRGHPQLARKLSVVDLVGIGVGATIGAGVYILIGTVAREQAGP 74
SL+RRKQ DS + + H QLA+ L+ L+ IGVG+TIGAGVYIL+GTVARE +GP
Sbjct: 6 SLVRRKQFDSSNGKAETHHHHQQLAKALTFPHLIAIGVGSTIGAGVYILVGTVAREHSGP 65
Query: 75 ALVISLFIAGIAAALSALCYAELACRCPSAGSAYHYTYICIGEGVAWLVGWSLILEYTIG 134
AL +S IAGI+AALSA CYAEL+ R PSAGSAYHY+YICIGEGVAWL+GW+LILEYTIG
Sbjct: 66 ALALSFLIAGISAALSAFCYAELSSRFPSAGSAYHYSYICIGEGVAWLIGWALILEYTIG 125
Query: 135 ASAVARGITPNLALFFGGQDNLPSFLARHTLPGLGIVVDPCAAVLIVLITLLLCLGIKES 194
S VARGI+PNLA+ FGG+D LP+ LARH +PGL IVVDPCAAVL+ ++T L CLG+KES
Sbjct: 126 GSTVARGISPNLAMIFGGEDCLPTILARHQIPGLDIVVDPCAAVLVFIVTGLCCLGVKES 185
Query: 195 STVQSIVTTINVSVMLFIIIVGGYLGFKAGWVGYELPSGYFPYGVNGMFAGSAIVFFSYI 254
+ Q IVTT NV VM+F+I+ G YL FK GWVGYELP+GYFPYGV+GM GSA VFF+YI
Sbjct: 186 TFAQGIVTTANVFVMIFVIVAGSYLCFKTGWVGYELPTGYFPYGVDGMLTGSATVFFAYI 245
Query: 255 GFDSVTSTAEEVKNPQRDLPIGISTALAICCVLYMLVSAVIVGLVPYYELNPDTPISSAF 314
GFD+V S AEEVKNP+RDLP+GI +L +CC+LYM+VS VIVGLVPYY ++PDTPISSAF
Sbjct: 246 GFDTVASMAEEVKNPRRDLPLGIGISLLLCCLLYMMVSVVIVGLVPYYAMDPDTPISSAF 305
Query: 315 SSYGMEWAVYIITTGAVTALFSSLLGSVLPQPRVFMAMARDGLLPTFFSDIHRRTQIPLK 374
SS+G++WA Y+I GAV AL S+L+GS+LPQPR+ MAMARDGLLP++FS +++RTQ+P+
Sbjct: 306 SSHGIQWAAYLINLGAVMALCSALMGSILPQPRILMAMARDGLLPSYFSYVNQRTQVPIN 365
Query: 375 STIVTGLFAAVLAFFMDVSQLAGMVSVGTLLAFTTVAVSVLIIRYV-PPDEIPIPASLLT 433
TI TG+ AA+LAFFMDVSQLAGMVSVGTL+AFT VA+S+LI+RYV PPDE+P+P+SL
Sbjct: 366 GTITTGVCAAILAFFMDVSQLAGMVSVGTLVAFTMVAISLLIVRYVVPPDEVPLPSSL-- 423
Query: 434 SVDPLLRHSGDDIEEDRTVSPVDLASYSDNSHLHDKSDVLLEHPLIIKE----VTKEQH- 488
+NS H + + + PL+ K V KE
Sbjct: 424 ---------------------------QENSSSHVGTSIRSKQPLLGKVDDSVVDKENAP 456
Query: 489 -----NEKTRRKLAAWTIALLCIGILIVSGSASAERCPRILRVTLFGAGVVIFLCSIIVL 543
N+K RRK A W+I CIG ++S +AS+ P +LR +L G G + L +IVL
Sbjct: 457 GSWVLNKKNRRKFAGWSIMFTCIGNFLLSYAASSFLLPGLLRYSLCGVGGLFLLVGLIVL 516
Query: 544 ACIKQDDTRHTFGHSGGFACPFVPFLPAACILINTYLLIDLGVDTWLRVSVWLLIGVLIY 603
CI QDD RH+FGHSGGF CPFVP LP CILIN YLL++LG TW+RVSVWL +GV++Y
Sbjct: 517 ICIDQDDARHSFGHSGGFICPFVPLLPIVCILINMYLLVNLGAATWVRVSVWLFLGVVVY 576
Query: 604 LFYGRTHSSLLNAIYVPSARADEIHRSQANHLA 636
+FYGR +SSL+NA+YV +A EI R+ + LA
Sbjct: 577 IFYGRRNSSLVNAVYVSTAHLQEIRRTSGHSLA 609
>UniRef100_Q9FIV7 Cationic amino acid transporter-like protein [Arabidopsis thaliana]
Length = 616
Score = 756 bits (1952), Expect = 0.0
Identities = 383/633 (60%), Positives = 476/633 (74%), Gaps = 38/633 (6%)
Query: 20 SLIRRKQVDSIH-----YRGHPQLARKLSVVDLVGIGVGATIGAGVYILIGTVAREQAGP 74
SL+RRKQ DS + + H QLA+ L+ L+ IGVG+TIGAGVYIL+GTVARE +GP
Sbjct: 6 SLVRRKQFDSSNGKAETHHHHQQLAKALTFPHLIAIGVGSTIGAGVYILVGTVAREHSGP 65
Query: 75 ALVISLFIAGIAAALSALCYAELACRCPSAGSAYHYTYICIGEGVAWLVGWSLILEYTIG 134
AL +S IAGI+AALSA CYAEL+ R PSAGSAYHY+YICIGEGVAWL+GW+LILEYTIG
Sbjct: 66 ALALSFLIAGISAALSAFCYAELSSRFPSAGSAYHYSYICIGEGVAWLIGWALILEYTIG 125
Query: 135 ASAVARGITPNLALFFGGQDNLPSFLARHTLPGLGIVVDPCAAVLIVLITLLLCLGIKES 194
S VARGI+PNLA+ FGG+D LP+ LARH +PGL IVVDPCAAVL+ ++T L CLG+KES
Sbjct: 126 GSTVARGISPNLAMIFGGEDCLPTILARHQIPGLDIVVDPCAAVLVFIVTGLCCLGVKES 185
Query: 195 STVQSIVTTINVSVMLFIIIVGGYLGFKAGWVGYELPSGYFPYGVNGMFAGSAIVFFSYI 254
+ Q IVTT NV VM+F+I+ G YL FK GWVGYELP+GYFPYGV+GM GSA VFF+YI
Sbjct: 186 TFAQGIVTTANVFVMIFVIVAGSYLCFKTGWVGYELPTGYFPYGVDGMLTGSATVFFAYI 245
Query: 255 GFDSVTSTAEEVKNPQRDLPIGISTALAICCVLYMLVSAVIVGLVPYYELNPDTPISSAF 314
GFD+V S AEEVKNP+RDLP+GI +L +CC+LYM+VS VIVGLVPYY ++PDTPISSAF
Sbjct: 246 GFDTVASMAEEVKNPRRDLPLGIGISLLLCCLLYMMVSVVIVGLVPYYAMDPDTPISSAF 305
Query: 315 SSYGMEWAVYIITTGAVTALFSSLLGSVLPQPRVFMAMARDGLLPTFFSDIHRRTQIPLK 374
SS+G++WA Y+I GAV AL S+L+GS+LPQPR+ MAMARDGLLP++FS +++RTQ+P+
Sbjct: 306 SSHGIQWAAYLINLGAVMALCSALMGSILPQPRILMAMARDGLLPSYFSYVNQRTQVPIN 365
Query: 375 STIVTGLFAAVLAFFMDVSQLAGMVSVGTLLAFTTVAVSVLIIRY-VPPDEIPIPASLLT 433
TI TG+ AA+LAFFMDVSQLAGMVSVGTL+AFT VA+S+LI+RY VPPDE+ I L+
Sbjct: 366 GTITTGVCAAILAFFMDVSQLAGMVSVGTLVAFTMVAISLLIVRYVVPPDEVLIRLELV- 424
Query: 434 SVDPLLRHSGDDIEEDRTVSPVDLASYSDNSHLHDKSDVLLEHPLIIK----EVTKEQH- 488
PL +S +NS H + + + PL+ K V KE
Sbjct: 425 ---PL------------------PSSLQENSSSHVGTSIRSKQPLLGKVDDSVVDKENAP 463
Query: 489 -----NEKTRRKLAAWTIALLCIGILIVSGSASAERCPRILRVTLFGAGVVIFLCSIIVL 543
N+K RRK A W+I CIG ++S +AS+ P +LR +L G G + L +IVL
Sbjct: 464 GSWVLNKKNRRKFAGWSIMFTCIGNFLLSYAASSFLLPGLLRYSLCGVGGLFLLVGLIVL 523
Query: 544 ACIKQDDTRHTFGHSGGFACPFVPFLPAACILINTYLLIDLGVDTWLRVSVWLLIGVLIY 603
CI QDD RH+FGHSGGF CPFVP LP CILIN YLL++LG TW+RVSVWL +GV++Y
Sbjct: 524 ICIDQDDARHSFGHSGGFICPFVPLLPIVCILINMYLLVNLGAATWVRVSVWLFLGVVVY 583
Query: 604 LFYGRTHSSLLNAIYVPSARADEIHRSQANHLA 636
+FYGR +SSL+NA+YV +A EI R+ + LA
Sbjct: 584 IFYGRRNSSLVNAVYVSTAHLQEIRRTSGHSLA 616
>UniRef100_Q7XE53 Putative cationic amino acid transporter-like protein [Oryza
sativa]
Length = 673
Score = 615 bits (1585), Expect = e-174
Identities = 317/568 (55%), Positives = 402/568 (69%), Gaps = 21/568 (3%)
Query: 52 VGATIGAGVYILIGTVAREQAGPALVISLFIAGIAAALSALCYAELACRCPSAGSAYHYT 111
VG+TIGAG+Y+L+GTVARE AGPAL +S IAGIAAALSALCYAEL+CR P AGSAYHY+
Sbjct: 120 VGSTIGAGIYVLVGTVAREHAGPALTLSFLIAGIAAALSALCYAELSCRFPLAGSAYHYS 179
Query: 112 YICIGEGVAWLVGWSLILEYTIGASAVARGITPNLALFFGGQDNLPSFLARHTLPGLGIV 171
YICIGE VAWL+GW+LILEYTIG S+VARGI+PNLALFFGG + LP FL + +
Sbjct: 180 YICIGESVAWLIGWALILEYTIGGSSVARGISPNLALFFGGHEKLPFFLTQIHVKWFETP 239
Query: 172 VDPCAAVLIVLITLLLCLGIKESSTVQSIVTTINVSVMLFIIIVGGYLGFKAGWVGYELP 231
+DPCAA+L++++T LLCLGIKESS V+ I+T NV VMLF+I GGYL F+ GW GY
Sbjct: 240 LDPCAAILVLIVTALLCLGIKESSFVEGIITIANVIVMLFVICAGGYLAFQNGWSGYNDE 299
Query: 232 SGYFPYGVNGMFAGSAIVFFSYIGFDSVTSTAEEVKNPQRDLPIGISTALAICCVLYMLV 291
GYFP GV G+ +GSA +FF+YIGFD+V STAEEVKNPQRDLP G+ L++CC LYM+V
Sbjct: 300 QGYFPKGVAGVLSGSATLFFAYIGFDAVASTAEEVKNPQRDLPWGMCLTLSLCCFLYMMV 359
Query: 292 SAVIVGLVPYYELNPDTPISSAFSSYGMEWAVYIITTGAVTALFSSLLGSVLPQPRVFMA 351
S VIVGLVPYY L+P+TPISSAF+ YGM+WAVYII+TGAV AL +SL+G++LPQPR+ MA
Sbjct: 360 SIVIVGLVPYYALDPNTPISSAFAKYGMQWAVYIISTGAVFALIASLIGAILPQPRIVMA 419
Query: 352 MARDGLLPTFFSDIHRRTQIPLKSTIVTGLFAAVLAFFMDVSQLAGMVSVGTLLAFTTVA 411
MARDGLLP FS + TQ+P STI++G+ AA+LA FMDVS+LAGMVSVGTLLAFT VA
Sbjct: 420 MARDGLLPPLFSAVDPTTQVPTLSTILSGICAAILALFMDVSELAGMVSVGTLLAFTMVA 479
Query: 412 VSVLIIRYVPPDEIPIPASLLTSVDPLLRHSG-DDIEEDRTVSPVDLASYSDNSHLHDKS 470
+SVLI+RY PP+EI +L S + L SG + +E+ + + + S
Sbjct: 480 ISVLIVRYAPPNEIATKVALPGSSESLTSDSGYSEPDEENSEDLLGNEGAAIRVRQASTS 539
Query: 471 DVLLEHPLIIKEVTKEQHNEKTRRKLAAWTIALLCIGILIVSGSASAERCPRILRVTLFG 530
V+ + + KE + +L W L++ + RC
Sbjct: 540 KVVHRPASVESDQPKEVMTSRRGIRLPDW---------LVLEFLSPFFRC---------A 581
Query: 531 AGVVIFLCSIIVLACIKQDDTRHTFGHSGGFACPFVPFLPAACILINTYLLIDLGVDTWL 590
G ++ + + I L I QD + + +GGF CPFVP LP CILIN YLL++LG+ TW+
Sbjct: 582 FGSLLLVSATIALWFIGQD--KSSLRQTGGFMCPFVPILPVCCILINVYLLMNLGIHTWI 639
Query: 591 RVSVWLLIGVLIYLFYGRTHSSLLNAIY 618
RVS+WL +G +IY+FYGR +SSL Y
Sbjct: 640 RVSMWLAVGAIIYVFYGRKYSSLTGVAY 667
>UniRef100_UPI000043683B UPI000043683B UniRef100 entry
Length = 600
Score = 378 bits (971), Expect = e-103
Identities = 231/625 (36%), Positives = 336/625 (52%), Gaps = 65/625 (10%)
Query: 20 SLIRRKQVDSIHYRGHPQLARKLSVVDLVGIGVGATIGAGVYILIGTVAREQAGPALVIS 79
SL+RRK VD +L R LS VDL+ +GVG+T+GAGVY+L G VA+ +GP++V+S
Sbjct: 6 SLVRRKNVDQGCLE-ESKLCRCLSTVDLIALGVGSTLGAGVYVLAGEVAKGSSGPSIVVS 64
Query: 80 LFIAGIAAALSALCYAELACRCPSAGSAYHYTYICIGEGVAWLVGWSLILEYTIGASAVA 139
IA +A+ ++ LCYAE R P GSAY Y+Y+ +GE A++ GW+LIL Y IG S+VA
Sbjct: 65 FLIAALASVMAGLCYAEFGARVPKTGSAYLYSYVTVGELWAFITGWNLILSYVIGTSSVA 124
Query: 140 RGITPNLALFFGGQ-DNLPSFLARHTLPGLGIVVDPCAAVLIVLITLLLCLGIKESSTVQ 198
R + GG + + +LPGL D A LI+L++ LL G+KES+ V
Sbjct: 125 RAWSGTFDEIIGGHIEKFCKMYFKMSLPGLAEYPDFFAVCLILLLSGLLSFGVKESAWVN 184
Query: 199 SIVTTINVSVMLFIIIVGGYLGFKAGWVGYEL-----------------------PSGYF 235
I T +NV V++F+II G G W E G+F
Sbjct: 185 KIFTAVNVLVLMFVIISGFVKGDSLNWNISEESLVNITVVKRNISGAANVTSDYGAGGFF 244
Query: 236 PYGVNGMFAGSAIVFFSYIGFDSVTSTAEEVKNPQRDLPIGISTALAICCVLYMLVSAVI 295
PYG G AG+A F++++GFD + +T EEVKNPQR +PIGI +L +C + Y VSA +
Sbjct: 245 PYGFGGTLAGAATCFYAFVGFDCIATTGEEVKNPQRAIPIGIVVSLLVCFLAYFGVSAAL 304
Query: 296 VGLVPYYELNPDTPISSAFSSYGMEWAVYIITTGAVTALFSSLLGSVLPQPRVFMAMARD 355
++PYY L+ +P+ AF G A Y++ G++ AL +SLLGS+ P PRV AMA+D
Sbjct: 305 TLMMPYYLLDEKSPLPLAFEYVGWGPAKYVVAAGSLCALSTSLLGSIFPMPRVIYAMAQD 364
Query: 356 GLLPTFFSDIHRRTQIPLKSTIVTGLFAAVLAFFMDVSQLAGMVSVGTLLAFTTVAVSVL 415
G+L + I+ +T+ PL +T+ +G+ AA++AF D+ L M+S+GTLLA++ VA VL
Sbjct: 365 GVLFKVLAQINPKTKTPLIATMSSGVVAAIMAFLFDLKALVDMMSIGTLLAYSLVAACVL 424
Query: 416 IIRYVPPDEIPIPASLLTSVDPLLRHSGDDIEEDRTVSPVDLASYSDNSHLHDKSD---- 471
I+RY P AS S + +++ E SHL+ D
Sbjct: 425 ILRYQP------DASFERS---RISEGKEEVGESELT--------ESESHLNMLKDGGVT 467
Query: 472 --VLLEHPLIIKEVTKEQHNEKTRRKLAAWTIALLCIGILIVSGSASAERCPRILRVTLF 529
LL PL+ E T N ++ + ++ V + + I+ + L+
Sbjct: 468 LRSLLHPPLLPTENTSSVVN----------VSVIITVLVVCVVSTLNTYYGQAIIAMELW 517
Query: 530 GAGVV---IFLCSIIVLACIKQDDTRHTFGHSGGFACPFVPFLPAACILINTYLLIDLGV 586
GV+ +F+ I V +Q TR F P +PFLP I +N YL++ L
Sbjct: 518 ALGVLAASLFIFIICVFLICRQPQTRKKV----SFMVPLLPFLPILSIFVNVYLMVQLSG 573
Query: 587 DTWLRVSVWLLIGVLIYLFYGRTHS 611
DTW+R S+W+ IG LIY YG HS
Sbjct: 574 DTWIRFSIWMAIGFLIYFGYGMWHS 598
>UniRef100_UPI000043683A UPI000043683A UniRef100 entry
Length = 616
Score = 376 bits (966), Expect = e-103
Identities = 232/625 (37%), Positives = 334/625 (53%), Gaps = 66/625 (10%)
Query: 20 SLIRRKQVDSIHYRGHPQLARKLSVVDLVGIGVGATIGAGVYILIGTVAREQAGPALVIS 79
SL+RRK VD +L R LS VDL+ +GVG+T+GAGVY+L G VA+ +GP++V+S
Sbjct: 6 SLVRRKNVDQGCLE-ESKLCRCLSTVDLIALGVGSTLGAGVYVLAGEVAKGSSGPSIVVS 64
Query: 80 LFIAGIAAALSALCYAELACRCPSAGSAYHYTYICIGEGVAWLVGWSLILEYTIGASAVA 139
IA +A+ ++ LCYAE R P GSAY Y+Y+ +GE A++ GW+LIL Y IG S+VA
Sbjct: 65 FLIAALASVMAGLCYAEFGARVPKTGSAYLYSYVTVGELWAFITGWNLILSYVIGTSSVA 124
Query: 140 RGITPNLALFFGGQ-DNLPSFLARHTLPGLGIVVDPCAAVLIVLITLLLCLGIKESSTVQ 198
R + GG + + +LPGL D A LI+L++ LL G+KES+ V
Sbjct: 125 RAWSGTFDEIIGGHIEKFCKMYFKMSLPGLAEYPDFFAVCLILLLSGLLSFGVKESAWVN 184
Query: 199 SIVTTINVSVMLFIIIVGGYLGFKAGWVGYEL-----------------------PSGYF 235
I T +NV V++F+II G G W E G+F
Sbjct: 185 KIFTAVNVLVLMFVIISGFVKGDSLNWNISEESLVNITVVKRNISGAANVTSDYGAGGFF 244
Query: 236 PYGVNGMFAGSAIVFFSYIGFDSVTSTAEEVKNPQRDLPIGISTALAICCVLYMLVSAVI 295
PYG G AG+A F++++GFD + +T EEVKNPQR +PIGI +L +C + Y VSA +
Sbjct: 245 PYGFGGTLAGAATCFYAFVGFDCIATTGEEVKNPQRAIPIGIVVSLLVCFLAYFGVSAAL 304
Query: 296 VGLVPYYELNPDTPISSAFSSYGMEWAVYIITTGAVTALFSSLLGSVLPQPRVFMAMARD 355
++PYY L+ +P+ AF G A Y++ G++ AL +SLLGS+ P PR+ AMARD
Sbjct: 305 TLMMPYYLLDEKSPLPLAFEYVGWGPAKYVVAAGSLCALSTSLLGSMFPLPRILFAMARD 364
Query: 356 GLLPTFFSDIHRRTQIPLKSTIVTGLFAAVLAFFMDVSQLAGMVSVGTLLAFTTVAVSVL 415
G+L F S + +R Q P+ +T+ G AA++AF D+ L M+S+GTLLA++ VA VL
Sbjct: 365 GVLFRFLSKLSKR-QSPVAATMAAGTTAAIMAFLFDLKALVDMMSIGTLLAYSLVAACVL 423
Query: 416 IIRYVPPDEIPIPASLLTSVDPLLRHSGDDIEEDRTVSPVDLASYSDNSHLHDKSD---- 471
I+RY P AS S + +++ E SHL+ D
Sbjct: 424 ILRYQP------DASFERS---RISEGKEEVGESELT--------ESESHLNMLKDGGVT 466
Query: 472 --VLLEHPLIIKEVTKEQHNEKTRRKLAAWTIALLCIGILIVSGSASAERCPRILRVTLF 529
LL PL+ E T N ++ + ++ V + + I+ + L+
Sbjct: 467 LRSLLHPPLLPTENTSSVVN----------VSVIITVLVVCVVSTLNTYYGQAIIAMELW 516
Query: 530 GAGVV---IFLCSIIVLACIKQDDTRHTFGHSGGFACPFVPFLPAACILINTYLLIDLGV 586
GV+ +F+ I V +Q TR F P +PFLP I +N YL++ L
Sbjct: 517 ALGVLAASLFIFIICVFLICRQPQTRKKV----SFMVPLLPFLPILSIFVNVYLMVQLSG 572
Query: 587 DTWLRVSVWLLIGVLIYLFYGRTHS 611
DTW+R S+W+ IG LIY YG HS
Sbjct: 573 DTWIRFSIWMAIGFLIYFGYGMWHS 597
>UniRef100_UPI00003AA095 UPI00003AA095 UniRef100 entry
Length = 624
Score = 376 bits (966), Expect = e-103
Identities = 226/637 (35%), Positives = 355/637 (55%), Gaps = 59/637 (9%)
Query: 14 RFRGFGS-LIRRKQVDSIHYRGHPQLARKLSVVDLVGIGVGATIGAGVYILIGTVAREQA 72
+F FG+ L+RRK VD R +L+R L+ DLV +GVG+T+GAGVY+L G VAR A
Sbjct: 5 KFIKFGNQLLRRKNVDCT--REDSRLSRCLNTFDLVALGVGSTLGAGVYVLAGAVARVNA 62
Query: 73 GPALVISLFIAGIAAALSALCYAELACRCPSAGSAYHYTYICIGEGVAWLVGWSLILEYT 132
GPA+VIS IA +A+ L+ LCY E R P GSAY Y+Y+ +GE A++ GW+LIL Y
Sbjct: 63 GPAIVISFLIAALASVLAGLCYGEFGARVPKTGSAYLYSYVTVGELWAFITGWNLILSYV 122
Query: 133 IGASAVARGITPNLALFFGGQDNLPSFLARH-TLPGLGIVV---DPCAAVLIVLITLLLC 188
IG S+VAR + GG ++ F R+ T+ G++ D A V+I+++T LL
Sbjct: 123 IGTSSVARAWSATFDEIIGG--HIEDFCKRYMTMNAPGVLAKYPDIFAVVIIIILTGLLT 180
Query: 189 LGIKESSTVQSIVTTINVSVMLFIIIVGGYLGFKAGW--------------------VGY 228
G+KES+ V + T IN+ V+ F+++ G G W G
Sbjct: 181 FGVKESALVNKVFTCINILVIGFVVVSGFVKGSVKNWQLTERDIYNTSPGIHGDNQTQGE 240
Query: 229 EL--PSGYFPYGVNGMFAGSAIVFFSYIGFDSVTSTAEEVKNPQRDLPIGISTALAICCV 286
+L G+ PYG+ G+ +G+A F++++GFD + +T EEVKNPQ+ +PIGI +L IC V
Sbjct: 241 KLYGVGGFMPYGLKGVLSGAATCFYAFVGFDCIATTGEEVKNPQKAIPIGIVASLLICFV 300
Query: 287 LYMLVSAVIVGLVPYYELNPDTPISSAFSSYGMEWAVYIITTGAVTALFSSLLGSVLPQP 346
Y VSA + ++PYY+L+ ++P+ +AF G + A Y + G++ AL +SLLGS+ P P
Sbjct: 301 AYFGVSAALTLMMPYYQLDTNSPLPNAFKYVGWDGANYAVAVGSLCALSTSLLGSMFPMP 360
Query: 347 RVFMAMARDGLLPTFFSDIHRRTQIPLKSTIVTGLFAAVLAFFMDVSQLAGMVSVGTLLA 406
R+ AMA DGLL F + ++ + + P+ +T+ +G AA++AF D+ L ++S+GTLLA
Sbjct: 361 RIIYAMAEDGLLFKFLAKVNDKRKTPVIATVTSGAVAAIMAFLFDLKDLVDLMSIGTLLA 420
Query: 407 FTTVAVSVLIIRYVPPDEIPIPASLLTSVDPLLRHSGDDIEEDRTVSPVDLASYSDNSHL 466
++ VA VL++RY P ++ + + ++ + + +VS S S L
Sbjct: 421 YSLVAACVLVLRYQPEQ---------PNLAYQMARTTEETDNNESVS----TSESQTGFL 467
Query: 467 HDKSDVLLEHPLIIKEVTKEQHNEKTRRKLAAWTIALLCIGILIVSGSASAERCP-RILR 525
++ E +K + +++ ++ I+ IG LIV P +++
Sbjct: 468 PEE-----EEKCSLKAILCPPNSDPSKFSGLVVNISTCIIGFLIVGSCVLTTLKPSTLIK 522
Query: 526 VTLFGAGVVIFLCSIIVLACIKQDDTRHTFGHSGGFACPFVPFLPAACILINTYLLIDLG 585
A +++ + S IV KQ +++ F P +P LP I +N YL++ L
Sbjct: 523 AVWIIAAILVLIISFIVW---KQPESKTKL----SFKVPLLPLLPIVSIFVNVYLMMQLD 575
Query: 586 VDTWLRVSVWLLIGVLIYLFYGRTHSSLLNAIYVPSA 622
+ TW+R +VW+LIG +IY YG HS + A Y SA
Sbjct: 576 LGTWIRFAVWMLIGFIIYFSYGIWHS--VEAAYAASA 610
>UniRef100_Q8VIA9 Ecotropic retrovirus receptor [Rattus norvegicus]
Length = 624
Score = 376 bits (966), Expect = e-103
Identities = 230/611 (37%), Positives = 341/611 (55%), Gaps = 40/611 (6%)
Query: 21 LIRRKQVDSIHYRGHPQLARKLSVVDLVGIGVGATIGAGVYILIGTVAREQAGPALVISL 80
++RRK VD R +L+R L+ DLV +GVG+T+GAGVY+L G VARE AGPA+VIS
Sbjct: 13 MLRRKVVDCS--REESRLSRCLNTYDLVALGVGSTLGAGVYVLAGAVARENAGPAIVISF 70
Query: 81 FIAGIAAALSALCYAELACRCPSAGSAYHYTYICIGEGVAWLVGWSLILEYTIGASAVAR 140
IA +A+ L+ LCY E R P GSAY Y+Y+ +GE A++ GW+LIL Y IG S+VAR
Sbjct: 71 LIAALASVLAGLCYGEFGARVPKTGSAYLYSYVTVGELWAFITGWNLILSYIIGTSSVAR 130
Query: 141 GITPNLALFFGGQDNLPSFLARH---TLPG-LGIVVDPCAAVLIVLITLLLCLGIKESST 196
+ G + F +H PG L D A ++I+++T LL LG+KES+
Sbjct: 131 AWSATFDELIGKP--IGEFSRQHMALNAPGVLAQTPDIFAVIIIIILTGLLTLGVKESAM 188
Query: 197 VQSIVTTINVSVMLFIIIVGGYLGFKAGWVGYELPS----------------GYFPYGVN 240
V I T INV V+ FI++ G G W E S G+ P+G +
Sbjct: 189 VNKIFTCINVLVLCFIMVSGFVKGSIENWQLTENKSSPLCGNDDTNVKYGEGGFMPFGFS 248
Query: 241 GMFAGSAIVFFSYIGFDSVTSTAEEVKNPQRDLPIGISTALAICCVLYMLVSAVIVGLVP 300
G+ +G+A F++++GFD + +T EEVKNPQ+ +P+GI +L IC + Y VSA + ++P
Sbjct: 249 GVLSGAATCFYAFVGFDCIATTGEEVKNPQKAIPVGIVASLLICFIAYFGVSAALTLMMP 308
Query: 301 YYELNPDTPISSAFSSYGMEWAVYIITTGAVTALFSSLLGSVLPQPRVFMAMARDGLLPT 360
Y+ L+ D+P+ AF G E A Y + G++ AL +SLLGS+ P PRV AMA DGLL
Sbjct: 309 YFCLDTDSPLPGAFKYRGWEEAKYAVAVGSLCALSTSLLGSMFPMPRVIYAMAEDGLLFK 368
Query: 361 FFSDIHRRTQIPLKSTIVTGLFAAVLAFFMDVSQLAGMVSVGTLLAFTTVAVSVLIIRYV 420
F + I+ RT+ P+ +T+ +G AAV+AF ++ L ++S+GTLLA++ VA VL++RY
Sbjct: 369 FLAKINDRTKTPIIATVTSGAIAAVMAFLFELKDLVDLMSIGTLLAYSLVAACVLVLRY- 427
Query: 421 PPDEIPIPASLLTSVDPLLRHSGDDIEEDRTVSPVDLASYSDNSHLHDKSDVLLEHPLII 480
P++ + + + D L D ++++ VS AS S L L+ L
Sbjct: 428 QPEQPNLVYQMARTTDEL-----DQVDQNEMVS----ASESQTGFLPAAEKFSLKTILSP 478
Query: 481 KEVTKEQHNEKTRRKLAAWTIALLCIGILIVSGSASAERCPRILRVTLFGAGVVIFLCSI 540
K + + + ++A +A+L I + IV+ L G V+ LC +
Sbjct: 479 KNMEPSKFSGLI-VNISAGLLAVLIITVCIVAVLGREALAEGTLWAVFVMTGSVL-LCML 536
Query: 541 IVLACIKQDDTRHTFGHSGGFACPFVPFLPAACILINTYLLIDLGVDTWLRVSVWLLIGV 600
+ +Q +++ F PFVP LP I +N YL++ L TW+R +VW+LIG
Sbjct: 537 VTGIIWRQPESKTKL----SFKVPFVPVLPVLSIFVNIYLMMQLDQGTWVRFAVWMLIGF 592
Query: 601 LIYLFYGRTHS 611
IY YG HS
Sbjct: 593 AIYFGYGVWHS 603
>UniRef100_P70608 Cationic amino acid transporter-1 [Rattus norvegicus]
Length = 624
Score = 376 bits (966), Expect = e-103
Identities = 230/611 (37%), Positives = 341/611 (55%), Gaps = 40/611 (6%)
Query: 21 LIRRKQVDSIHYRGHPQLARKLSVVDLVGIGVGATIGAGVYILIGTVAREQAGPALVISL 80
++RRK VD R +L+R L+ DLV +GVG+T+GAGVY+L G VARE AGPA+VIS
Sbjct: 13 MLRRKVVDCS--REESRLSRCLNTYDLVALGVGSTLGAGVYVLAGAVARENAGPAIVISF 70
Query: 81 FIAGIAAALSALCYAELACRCPSAGSAYHYTYICIGEGVAWLVGWSLILEYTIGASAVAR 140
IA +A+ L+ LCY E R P GSAY Y+Y+ +GE A++ GW+LIL Y IG S+VAR
Sbjct: 71 LIAALASVLAGLCYGEFGARVPKTGSAYLYSYVTVGELWAFITGWNLILSYIIGTSSVAR 130
Query: 141 GITPNLALFFGGQDNLPSFLARH---TLPG-LGIVVDPCAAVLIVLITLLLCLGIKESST 196
+ G + F +H PG L D A ++I+++T LL LG+KES+
Sbjct: 131 AWSATFDELIGKP--IGEFSRQHMALNAPGVLAQTPDIFAVIIIIILTGLLTLGVKESAM 188
Query: 197 VQSIVTTINVSVMLFIIIVGGYLGFKAGWVGYELPS----------------GYFPYGVN 240
V I T INV V+ FI++ G G W E S G+ P+G +
Sbjct: 189 VNKIFTCINVLVLCFIMVSGFVKGSIENWQLTENKSSPLCGNNDTNVKYGEGGFMPFGFS 248
Query: 241 GMFAGSAIVFFSYIGFDSVTSTAEEVKNPQRDLPIGISTALAICCVLYMLVSAVIVGLVP 300
G+ +G+A F++++GFD + +T EEVKNPQ+ +P+GI +L IC + Y VSA + ++P
Sbjct: 249 GVLSGAATCFYAFVGFDCIATTGEEVKNPQKAIPVGIVASLLICFIAYFGVSAALTLMMP 308
Query: 301 YYELNPDTPISSAFSSYGMEWAVYIITTGAVTALFSSLLGSVLPQPRVFMAMARDGLLPT 360
Y+ L+ D+P+ AF G E A Y + G++ AL +SLLGS+ P PRV AMA DGLL
Sbjct: 309 YFCLDTDSPLPGAFKYRGWEEAKYAVAVGSLCALSTSLLGSMFPMPRVIYAMAEDGLLFK 368
Query: 361 FFSDIHRRTQIPLKSTIVTGLFAAVLAFFMDVSQLAGMVSVGTLLAFTTVAVSVLIIRYV 420
F + I+ RT+ P+ +T+ +G AAV+AF ++ L ++S+GTLLA++ VA VL++RY
Sbjct: 369 FLAKINDRTKTPIIATVTSGAIAAVMAFLFELKDLVDLMSIGTLLAYSLVAACVLVLRY- 427
Query: 421 PPDEIPIPASLLTSVDPLLRHSGDDIEEDRTVSPVDLASYSDNSHLHDKSDVLLEHPLII 480
P++ + + + D L D ++++ VS AS S L L+ L
Sbjct: 428 QPEQPNLVYQMARTTDEL-----DQVDQNEMVS----ASESQTGFLPAAEKFSLKTILSP 478
Query: 481 KEVTKEQHNEKTRRKLAAWTIALLCIGILIVSGSASAERCPRILRVTLFGAGVVIFLCSI 540
K + + + ++A +A+L I + IV+ L G V+ LC +
Sbjct: 479 KNMEPSKFSGLI-VNISAGLLAVLIITVCIVAVLGREALAEGTLWAVFVMTGSVL-LCML 536
Query: 541 IVLACIKQDDTRHTFGHSGGFACPFVPFLPAACILINTYLLIDLGVDTWLRVSVWLLIGV 600
+ +Q +++ F PFVP LP I +N YL++ L TW+R +VW+LIG
Sbjct: 537 VTGIIWRQPESKTKL----SFKVPFVPVLPVLSIFVNIYLMMQLDQGTWVRFAVWMLIGF 592
Query: 601 LIYLFYGRTHS 611
IY YG HS
Sbjct: 593 AIYFGYGVWHS 603
>UniRef100_Q09143 High-affinity cationic amino acid transporter-1 [Mus musculus]
Length = 622
Score = 375 bits (962), Expect = e-102
Identities = 229/609 (37%), Positives = 339/609 (55%), Gaps = 38/609 (6%)
Query: 21 LIRRKQVDSIHYRGHPQLARKLSVVDLVGIGVGATIGAGVYILIGTVAREQAGPALVISL 80
++RRK VD R +L+R L+ DLV +GVG+T+GAGVY+L G VARE AGPA+VIS
Sbjct: 13 MLRRKVVDCS--REESRLSRCLNTYDLVALGVGSTLGAGVYVLAGAVARENAGPAIVISF 70
Query: 81 FIAGIAAALSALCYAELACRCPSAGSAYHYTYICIGEGVAWLVGWSLILEYTIGASAVAR 140
IA +A+ L+ LCY E R P GSAY Y+Y+ +GE A++ GW+LIL Y IG S+VAR
Sbjct: 71 LIAALASVLAGLCYGEFGARVPKTGSAYLYSYVTVGELWAFITGWNLILSYIIGTSSVAR 130
Query: 141 GITPNLALFFGGQDNLPSFLARH---TLPG-LGIVVDPCAAVLIVLITLLLCLGIKESST 196
+ G + F +H PG L D A ++I+++T LL LG+KES+
Sbjct: 131 AWSATFDELIGKP--IGEFSRQHMALNAPGVLAQTPDIFAVIIIIILTGLLTLGVKESAM 188
Query: 197 VQSIVTTINVSVMLFIIIVGGYLGFKAGWVGYEL--------------PSGYFPYGVNGM 242
V I T INV V+ FI++ G G W E G+ P+G +G+
Sbjct: 189 VNKIFTCINVLVLCFIVVSGFVKGSIKNWQLTEKNFSCNNNDTNVKYGEGGFMPFGFSGV 248
Query: 243 FAGSAIVFFSYIGFDSVTSTAEEVKNPQRDLPIGISTALAICCVLYMLVSAVIVGLVPYY 302
+G+A F++++GFD + +T EEVKNPQ+ +P+GI +L IC + Y VSA + ++PY+
Sbjct: 249 LSGAATCFYAFVGFDCIATTGEEVKNPQKAIPVGIVASLLICFIAYFGVSAALTLMMPYF 308
Query: 303 ELNPDTPISSAFSSYGMEWAVYIITTGAVTALFSSLLGSVLPQPRVFMAMARDGLLPTFF 362
L+ D+P+ AF G E A Y + G++ AL +SLLGS+ P PRV AMA DGLL F
Sbjct: 309 CLDIDSPLPGAFKHQGWEEAKYAVAIGSLCALSTSLLGSMFPMPRVIYAMAEDGLLFKFL 368
Query: 363 SDIHRRTQIPLKSTIVTGLFAAVLAFFMDVSQLAGMVSVGTLLAFTTVAVSVLIIRYVPP 422
+ I+ RT+ P+ +T+ +G AAV+AF ++ L ++S+GTLLA++ VA VL++RY P
Sbjct: 369 AKINNRTKTPVIATVTSGAIAAVMAFLFELKDLVDLMSIGTLLAYSLVAACVLVLRY-QP 427
Query: 423 DEIPIPASLLTSVDPLLRHSGDDIEEDRTVSPVDLASYSDNSHLHDKSDVLLEHPLIIKE 482
++ + + + + L D ++++ VS AS S L L+ L K
Sbjct: 428 EQPNLVYQMARTTEEL-----DRVDQNELVS----ASESQTGFLPVAEKFSLKSILSPKN 478
Query: 483 VTKEQHNEKTRRKLAAWTIALLCIGILIVSGSASAERCPRILRVTLFGAGVVIFLCSIIV 542
V + + ++A +A L I + IV+ L G V+ LC ++
Sbjct: 479 VEPSKFSGLI-VNISAGLLAALIITVCIVAVLGREALAEGTLWAVFVMTGSVL-LCMLVT 536
Query: 543 LACIKQDDTRHTFGHSGGFACPFVPFLPAACILINTYLLIDLGVDTWLRVSVWLLIGVLI 602
+Q +++ F PFVP LP I +N YL++ L TW+R +VW+LIG I
Sbjct: 537 GIIWRQPESKTKL----SFKVPFVPVLPVLSIFVNIYLMMQLDQGTWVRFAVWMLIGFTI 592
Query: 603 YLFYGRTHS 611
Y YG HS
Sbjct: 593 YFGYGIWHS 601
>UniRef100_UPI00003654CA UPI00003654CA UniRef100 entry
Length = 616
Score = 372 bits (956), Expect = e-101
Identities = 226/623 (36%), Positives = 336/623 (53%), Gaps = 57/623 (9%)
Query: 20 SLIRRKQVDSIHYRGHPQLARKLSVVDLVGIGVGATIGAGVYILIGTVAREQAGPALVIS 79
SL+RRK VD + +L R L VDL+ +GVG+T+GAGVY+L G VA+ +GP++VIS
Sbjct: 7 SLVRRKVVD-LSSLEDSKLCRCLGTVDLIALGVGSTLGAGVYVLAGEVAKGDSGPSIVIS 65
Query: 80 LFIAGIAAALSALCYAELACRCPSAGSAYHYTYICIGEGVAWLVGWSLILEYTIGASAVA 139
IA +A+ ++ LCYAE R P GSAY Y+Y+ +GE A++ GW+LIL Y IG S+VA
Sbjct: 66 FLIAALASVMAGLCYAEFGARVPKTGSAYLYSYVTVGEIWAFITGWNLILSYVIGTSSVA 125
Query: 140 RGITPNLALFFGGQDNLPSFLARH---TLPGLGIVVDPCAAVLIVLITLLLCLGIKESST 196
R + GG ++ F + PGL D A LI+L++ LL G+KES++
Sbjct: 126 RAWSGTFDEMIGG--HIEKFCKTYFSMNWPGLAQYPDFFAVCLILLLSGLLSFGVKESAS 183
Query: 197 VQSIVTTINVSVMLFIIIVGGYLGFKAGWVGYEL-----------------------PSG 233
V + T++NV V+LF+II G G W E G
Sbjct: 184 VNKVFTSVNVLVLLFVIISGFVKGDIENWTITEESLINVTRETRNLSALTNVSSDYGAGG 243
Query: 234 YFPYGVNGMFAGSAIVFFSYIGFDSVTSTAEEVKNPQRDLPIGISTALAICCVLYMLVSA 293
+ PYG +G AG+A F++++GFD + +T EEVKNPQ+ +PIGI +L +C + Y VSA
Sbjct: 244 FMPYGFSGTLAGAATCFYAFVGFDCIATTGEEVKNPQKAIPIGIVVSLTMCFLAYFGVSA 303
Query: 294 VIVGLVPYYELNPDTPISSAFSSYGMEWAVYIITTGAVTALFSSLLGSVLPQPRVFMAMA 353
+ ++PYY L+ +P+ AF G A Y++ G++ AL +SLLGS+ P PRV AMA
Sbjct: 304 ALTLMMPYYLLDEKSPLPMAFEYVGWGPAKYVVAAGSLCALSASLLGSIFPMPRVIFAMA 363
Query: 354 RDGLLPTFFSDIHRRTQIPLKSTIVTGLFAAVLAFFMDVSQLAGMVSVGTLLAFTTVAVS 413
DG+L + I+ +T+ PL +T+ +GL AA++AF D+ L M+S+GTLLA++ VAV
Sbjct: 364 EDGVLFKALAQINPKTKTPLIATMTSGLVAAIMAFLFDLKALVDMMSIGTLLAYSLVAVC 423
Query: 414 VLIIRYVPPDEI-----PIPASLLTSVDPLLRHSGDDIEEDRTVSPVDLASYSDNSHLHD 468
VLI+RY P + P LTS + E D T S L S
Sbjct: 424 VLILRYQPDGALEQRAGPGEREYLTS---------EGGESDLTESESHLNMLKGGS---S 471
Query: 469 KSDVLLEHPLIIKEVTKEQHNEKTRRKLAAWTIALLCIGILIVSGSASAERCPRILRVTL 528
+L+ P + +H+ + + ++C + + SA + + L
Sbjct: 472 TLQAVLQPP-----SSPSEHSSSVVKMSICVLVVIVCALSYLTTYHISAILGAQAWALVL 526
Query: 529 FGAGVVIFLCSIIVLACIKQDDTRHTFGHSGGFACPFVPFLPAACILINTYLLIDLGVDT 588
+++F C I+++ +Q T F P +P+LP +L+N YL+ L DT
Sbjct: 527 LSLCLLLFSCCILLIC--RQPQT----SKKVSFMVPLLPYLPILSVLVNIYLMFQLSGDT 580
Query: 589 WLRVSVWLLIGVLIYLFYGRTHS 611
W+R SVW+ +G LIY YG HS
Sbjct: 581 WIRFSVWMAVGFLIYFGYGMWHS 603
>UniRef100_P30823 High-affinity cationic amino acid transporter-1 [Rattus norvegicus]
Length = 624
Score = 371 bits (953), Expect = e-101
Identities = 228/611 (37%), Positives = 339/611 (55%), Gaps = 40/611 (6%)
Query: 21 LIRRKQVDSIHYRGHPQLARKLSVVDLVGIGVGATIGAGVYILIGTVAREQAGPALVISL 80
++RRK VD R +L+R L+ DLV +GVG+T+GAGVY+L G VARE AGPA+VIS
Sbjct: 13 MLRRKVVDCS--REESRLSRCLNTYDLVALGVGSTLGAGVYVLAGAVARENAGPAIVISF 70
Query: 81 FIAGIAAALSALCYAELACRCPSAGSAYHYTYICIGEGVAWLVGWSLILEYTIGASAVAR 140
IA +A+ L+ LCY E R P GSAY Y+Y+ +GE A++ GW+LIL Y IG S+VAR
Sbjct: 71 LIAALASVLAGLCYGEFGARVPKTGSAYLYSYVTVGELWAFITGWNLILSYIIGTSSVAR 130
Query: 141 GITPNLALFFGGQDNLPSFLARH---TLPG-LGIVVDPCAAVLIVLITLLLCLGIKESST 196
+ G + F +H PG L D A ++I+++T LL LG+KES+
Sbjct: 131 AWSATFDELIGKP--IGEFSRQHMALNAPGVLAQTPDIFAVIIIIILTGLLTLGVKESAM 188
Query: 197 VQSIVTTINVSVMLFIIIVGGYLGFKAGWVGYELPS----------------GYFPYGVN 240
V I T INV V+ FI++ G G W E S G+ P+G +
Sbjct: 189 VNKIFTCINVLVLCFIMVSGFVKGSIENWQLTENKSSPLCGNNDTNVKYGEGGFMPFGFS 248
Query: 241 GMFAGSAIVFFSYIGFDSVTSTAEEVKNPQRDLPIGISTALAICCVLYMLVSAVIVGLVP 300
G+ +G+A F++++GFD + +T EEVKNPQ+ +P+GI +L IC + Y VSA + ++P
Sbjct: 249 GVLSGAATCFYAFVGFDCIATTGEEVKNPQKAIPVGIVASLLICFIAYFGVSAALTLMMP 308
Query: 301 YYELNPDTPISSAFSSYGMEWAVYIITTGAVTALFSSLLGSVLPQPRVFMAMARDGLLPT 360
Y+ L+ D+P+ AF G E A Y + G++ AL +S LGS+ P PRV AMA DGLL
Sbjct: 309 YFCLDTDSPLPGAFKYRGWEEAKYAVAVGSLCALSTSPLGSMFPMPRVIYAMAEDGLLFK 368
Query: 361 FFSDIHRRTQIPLKSTIVTGLFAAVLAFFMDVSQLAGMVSVGTLLAFTTVAVSVLIIRYV 420
F + I+ RT+ P+ +T+ +G AAV+AF ++ L ++S+GTLLA++ VA VL++RY
Sbjct: 369 FLAKINDRTKTPIIATVTSGAIAAVMAFLFELKDLVDLMSIGTLLAYSLVAACVLVLRY- 427
Query: 421 PPDEIPIPASLLTSVDPLLRHSGDDIEEDRTVSPVDLASYSDNSHLHDKSDVLLEHPLII 480
P++ + + + D L D ++++ VS AS S L L+ L
Sbjct: 428 QPEQPNLVYQMARTTDEL-----DQVDQNEMVS----ASESQTGFLPAAEKFSLKTILSP 478
Query: 481 KEVTKEQHNEKTRRKLAAWTIALLCIGILIVSGSASAERCPRILRVTLFGAGVVIFLCSI 540
K + + + ++A +A+L I + IV+ L G V+ LC +
Sbjct: 479 KNMEPSKFSGLI-VNISAGLLAVLIITVCIVAVLGREALAEGTLWAVFVMTGSVL-LCML 536
Query: 541 IVLACIKQDDTRHTFGHSGGFACPFVPFLPAACILINTYLLIDLGVDTWLRVSVWLLIGV 600
+ +Q +++ F PFVP LP I +N YL++ L TW+R +VW+LI
Sbjct: 537 VTGIIWRQPESKTKL----SFKVPFVPVLPVLSIFVNIYLMMQLDQGTWVRFAVWMLIAF 592
Query: 601 LIYLFYGRTHS 611
IY YG HS
Sbjct: 593 AIYFGYGVWHS 603
>UniRef100_UPI000024805A UPI000024805A UniRef100 entry
Length = 624
Score = 369 bits (946), Expect = e-100
Identities = 224/625 (35%), Positives = 342/625 (53%), Gaps = 48/625 (7%)
Query: 18 FGSLIRRKQVDSIHYRGHPQLARKLSVVDLVGIGVGATIGAGVYILIGTVAREQAGPALV 77
FG + R +V + + +L+R L+ DLV +GVG+T+GAGVY+L G VARE AGPA+V
Sbjct: 9 FGKQLLRVKVVNCNSE-ESRLSRCLNTFDLVALGVGSTLGAGVYVLAGAVARENAGPAIV 67
Query: 78 ISLFIAGIAAALSALCYAELACRCPSAGSAYHYTYICIGEGVAWLVGWSLILEYTIGASA 137
+S IA +A+ L+ LCYAE R P GSAY Y+Y+ +GE A++ GW+LIL Y IG S+
Sbjct: 68 LSFLIAALASVLAGLCYAEFGARVPKTGSAYLYSYVTVGELWAFITGWNLILSYVIGTSS 127
Query: 138 VARGITPNLALFFGGQDNLPSFLARH---TLPG-LGIVVDPCAAVLIVLITLLLCLGIKE 193
VAR + G ++ F ++ PG L D + +I+ +T LL G+KE
Sbjct: 128 VARAWSATFDELIG--KHIEHFCRQYMSMNAPGVLAEYPDMFSVFIILTLTGLLAFGVKE 185
Query: 194 SSTVQSIVTTINVSVMLFIIIVGGYLGFKAGW----------------VGYELPS----- 232
S+ V + T IN+ V+LF+++ G G W LPS
Sbjct: 186 SAMVNKVFTCINILVLLFMVVSGLVKGTLKNWHLDPDEILNATNSTLNATQPLPSEEMLG 245
Query: 233 --GYFPYGVNGMFAGSAIVFFSYIGFDSVTSTAEEVKNPQRDLPIGISTALAICCVLYML 290
G+ P+G G+ +G+A F++++GFD + +T EEVKNPQR +PIGI ++L IC V Y
Sbjct: 246 QGGFMPFGFTGVLSGAATCFYAFVGFDCIATTGEEVKNPQRAIPIGIVSSLLICFVAYFG 305
Query: 291 VSAVIVGLVPYYELNPDTPISSAFSSYGMEWAVYIITTGAVTALFSSLLGSVLPQPRVFM 350
VSA + ++PYY L+ ++P+ AF G E A Y + G++ AL +SLLG++ P PRV
Sbjct: 306 VSAALTMMMPYYMLDKNSPLPVAFKYVGWEGATYAVAVGSLCALSTSLLGAMFPMPRVLW 365
Query: 351 AMARDGLLPTFFSDIHRRTQIPLKSTIVTGLFAAVLAFFMDVSQLAGMVSVGTLLAFTTV 410
AMA DGLL F + I RT+ P+K+TI++G AA++AF D+ L ++S+GTLLA+T V
Sbjct: 366 AMADDGLLFKFMAGISERTKTPIKATIMSGFLAAIMAFLFDLKDLVDLMSIGTLLAYTLV 425
Query: 411 AVSVLIIRYVPPDEIPIPASLLTSVDPLLRHSGDDIEEDRTVSPVDLASYSDNSHLHDKS 470
A VL++RY P S + ++ +D+E T+S +
Sbjct: 426 AACVLVLRYQPEQ---------FSQTYHIANTHEDMEMSETISTPSMGILPGVEERFSFK 476
Query: 471 DVLLEHPLIIKEVTKEQHNEKTRRKLAAWTIALLCI---GILIVSGSASAERCPRILRVT 527
++L + ++ N T L + L C+ G+LI+S S A R I
Sbjct: 477 NLLFPDIIEPSNLSGFTVNICT-SLLVVFLTFLNCVFFPGLLILSFSLLAVR-GGIASWN 534
Query: 528 LFGAGVVIFLCSIIVLACIKQDDTRHTFGHSGGFACPFVPFLPAACILINTYLLIDLGVD 587
+ V+ LC I+ +Q +++ F P +PF+P + +N YL++ L
Sbjct: 535 IITLAVLFGLCVIVTFIIWRQPESKTKL----SFKVPCLPFIPVVSMFVNVYLMMQLDRG 590
Query: 588 TWLRVSVWLLIGVLIYLFYGRTHSS 612
TW+R ++W+ IG++IY YG HS+
Sbjct: 591 TWIRFAIWMSIGLVIYFGYGIWHST 615
>UniRef100_P30825 High-affinity cationic amino acid transporter-1 [Homo sapiens]
Length = 629
Score = 369 bits (946), Expect = e-100
Identities = 228/624 (36%), Positives = 346/624 (54%), Gaps = 61/624 (9%)
Query: 21 LIRRKQVDSIHYRGHPQLARKLSVVDLVGIGVGATIGAGVYILIGTVAREQAGPALVISL 80
++RRK VD R +L+R L+ DLV +GVG+T+GAGVY+L G VARE AGPA+VIS
Sbjct: 13 MLRRKVVDCS--REETRLSRCLNTFDLVALGVGSTLGAGVYVLAGAVARENAGPAIVISF 70
Query: 81 FIAGIAAALSALCYAELACRCPSAGSAYHYTYICIGEGVAWLVGWSLILEYTIGASAVAR 140
IA +A+ L+ LCY E R P GSAY Y+Y+ +GE A++ GW+LIL Y IG S+VAR
Sbjct: 71 LIAALASVLAGLCYGEFGARVPKTGSAYLYSYVTVGELWAFITGWNLILSYIIGTSSVAR 130
Query: 141 GITPNLALFFGGQDNLPSFLARH-TLPGLGIVV---DPCAAVLIVLITLLLCLGIKESST 196
+ G + F H TL G++ D A ++I+++T LL LG+KES+
Sbjct: 131 AWSATFDELIGRP--IGEFSRTHMTLNAPGVLAENPDIFAVIIILILTGLLTLGVKESAM 188
Query: 197 VQSIVTTINVSVMLFIIIVGGYLGFKAGWV---------------------GYELPSGYF 235
V I T INV V+ FI++ G G W G G+
Sbjct: 189 VNKIFTCINVLVLGFIMVSGFVKGSVKNWQLTEEDFGNTSGRLCLNNDTKEGKPGVGGFM 248
Query: 236 PYGVNGMFAGSAIVFFSYIGFDSVTSTAEEVKNPQRDLPIGISTALAICCVLYMLVSAVI 295
P+G +G+ +G+A F++++GFD + +T EEVKNPQ+ +P+GI +L IC + Y VSA +
Sbjct: 249 PFGFSGVLSGAATCFYAFVGFDCIATTGEEVKNPQKAIPVGIVASLLICFIAYFGVSAAL 308
Query: 296 VGLVPYYELNPDTPISSAFSSYGMEWAVYIITTGAVTALFSSLLGSVLPQPRVFMAMARD 355
++PY+ L+ ++P+ AF G E A Y + G++ AL +SLLGS+ P PRV AMA D
Sbjct: 309 TLMMPYFCLDNNSPLPDAFKHVGWEGAKYAVAVGSLCALSASLLGSMFPMPRVIYAMAED 368
Query: 356 GLLPTFFSDIHRRTQIPLKSTIVTGLFAAVLAFFMDVSQLAGMVSVGTLLAFTTVAVSVL 415
GLL F ++++ RT+ P+ +T+ +G AAV+AF D+ L ++S+GTLLA++ VA VL
Sbjct: 369 GLLFKFLANVNDRTKTPIIATLASGAVAAVMAFLFDLKDLVDLMSIGTLLAYSLVAACVL 428
Query: 416 IIRYVP--PDEIPIPASLLTSVDPLLRHSGDDIEEDRTVSPVDLASYSDNSHLHDKSDVL 473
++RY P P+ + AS +DP ++ +LAS +D+ + L
Sbjct: 429 VLRYQPEQPNLVYQMASTSDELDPADQN--------------ELASTNDS-----QLGFL 469
Query: 474 LEHPLI-IKEVTKEQHNEKTRRK-----LAAWTIALLCIGILIVSGSASAERCPRILRVT 527
E + +K + ++ E ++ ++ IA+L I IV+ L
Sbjct: 470 PEAEMFSLKTILSPKNMEPSKISGLIVNISTSLIAVLIITFCIVTVLGREALTKGALWAV 529
Query: 528 LFGAGVVIFLCSIIVLACIKQDDTRHTFGHSGGFACPFVPFLPAACILINTYLLIDLGVD 587
AG + LC+++ +Q +++ F PF+P LP I +N YL++ L
Sbjct: 530 FLLAGSAL-LCAVVTGVIWRQPESKTKLS----FKVPFLPVLPILSIFVNVYLMMQLDQG 584
Query: 588 TWLRVSVWLLIGVLIYLFYGRTHS 611
TW+R +VW+LIG +IY YG HS
Sbjct: 585 TWVRFAVWMLIGFIIYFGYGLWHS 608
>UniRef100_UPI00003654C9 UPI00003654C9 UniRef100 entry
Length = 599
Score = 368 bits (945), Expect = e-100
Identities = 223/618 (36%), Positives = 333/618 (53%), Gaps = 64/618 (10%)
Query: 20 SLIRRKQVDSIHYRGHPQLARKLSVVDLVGIGVGATIGAGVYILIGTVAREQAGPALVIS 79
SL+RRK VD + +L R L VDL+ +GVG+T+GAGVY+L G VA+ +GP++VIS
Sbjct: 4 SLVRRKVVD-LSSLEDSKLCRCLGTVDLIALGVGSTLGAGVYVLAGEVAKGDSGPSIVIS 62
Query: 80 LFIAGIAAALSALCYAELACRCPSAGSAYHYTYICIGEGVAWLVGWSLILEYTIGASAVA 139
IA +A+ ++ LCYAE R P GSAY Y+Y+ +GE A++ GW+LIL Y IG S+VA
Sbjct: 63 FLIAALASVMAGLCYAEFGARVPKTGSAYLYSYVTVGEIWAFITGWNLILSYVIGTSSVA 122
Query: 140 RGITPNLALFFGGQDNLPSFLARH---TLPGLGIVVDPCAAVLIVLITLLLCLGIKESST 196
R + GG ++ F + PGL D A LI+L++ LL G+KES++
Sbjct: 123 RAWSGTFDEMIGG--HIEKFCKTYFSMNWPGLAQYPDFFAVCLILLLSGLLSFGVKESAS 180
Query: 197 VQSIVTTINVSVMLFIIIVGGYLGFKAGWVGYEL-----------------------PSG 233
V + T++NV V+LF+II G G W E G
Sbjct: 181 VNKVFTSVNVLVLLFVIISGFVKGDIENWTITEESLINVTRETRNLSALTNVSSDYGAGG 240
Query: 234 YFPYGVNGMFAGSAIVFFSYIGFDSVTSTAEEVKNPQRDLPIGISTALAICCVLYMLVSA 293
+ PYG +G AG+A F++++GFD + +T EEVKNPQ+ +PIGI +L +C + Y VSA
Sbjct: 241 FMPYGFSGTLAGAATCFYAFVGFDCIATTGEEVKNPQKAIPIGIVVSLTMCFLAYFGVSA 300
Query: 294 VIVGLVPYYELNPDTPISSAFSSYGMEWAVYIITTGAVTALFSSLLGSVLPQPRVFMAMA 353
+ ++PYY L+ +P+ AF G A Y++ G++ AL +SLLGS+ P PRV AMA
Sbjct: 301 ALTLMMPYYLLDEKSPLPMAFEYVGWGPAKYVVAAGSLCALSASLLGSIFPMPRVIFAMA 360
Query: 354 RDGLLPTFFSDIHRRTQIPLKSTIVTGLFAAVLAFFMDVSQLAGMVSVGTLLAFTTVAVS 413
DG+L + I+ +T+ PL +T+ +GL AA++AF D+ L M+S+GTLLA++ VAV
Sbjct: 361 EDGVLFKALAQINPKTKTPLIATMTSGLVAAIMAFLFDLKALVDMMSIGTLLAYSLVAVC 420
Query: 414 VLIIRYVPPDEIPIPASLLTSVDPLLRHSGDDIEEDRTVSPVDLASYSDNSHLHDKSDVL 473
VLI+RY P D L E + L S +S L +
Sbjct: 421 VLILRYQP--------------DGALEQRAGPGEREY------LTSEGGDSTL----QAV 456
Query: 474 LEHPLIIKEVTKEQHNEKTRRKLAAWTIALLCIGILIVSGSASAERCPRILRVTLFGAGV 533
L+ P + +H+ + + ++C + + SA + + L +
Sbjct: 457 LQPP-----SSPSEHSSSVVKMSICVLVVIVCALSYLTTYHISAILGAQAWALVLLSLCL 511
Query: 534 VIFLCSIIVLACIKQDDTRHTFGHSGGFACPFVPFLPAACILINTYLLIDLGVDTWLRVS 593
++F C I+++ +Q T F P +P+LP +L+N YL+ L DTW+R S
Sbjct: 512 LLFSCCILLIC--RQPQT----SKKVSFMVPLLPYLPILSVLVNIYLMFQLSGDTWIRFS 565
Query: 594 VWLLIGVLIYLFYGRTHS 611
VW+ +G LIY YG HS
Sbjct: 566 VWMAVGFLIYFGYGMWHS 583
>UniRef100_UPI00003654CB UPI00003654CB UniRef100 entry
Length = 604
Score = 368 bits (944), Expect = e-100
Identities = 225/623 (36%), Positives = 334/623 (53%), Gaps = 58/623 (9%)
Query: 20 SLIRRKQVDSIHYRGHPQLARKLSVVDLVGIGVGATIGAGVYILIGTVAREQAGPALVIS 79
SL+RRK VD + +L R L VDL+ +GVG+T+GAGVY+L G VA+ +GP++VIS
Sbjct: 7 SLVRRKVVD-LSSLEDSKLCRCLGTVDLIALGVGSTLGAGVYVLAGEVAKGDSGPSIVIS 65
Query: 80 LFIAGIAAALSALCYAELACRCPSAGSAYHYTYICIGEGVAWLVGWSLILEYTIGASAVA 139
IA +A+ ++ LCYAE R P GSAY Y+Y+ +GE A++ GW+LIL Y IG S+VA
Sbjct: 66 FLIAALASVMAGLCYAEFGARVPKTGSAYLYSYVTVGEIWAFITGWNLILSYVIGTSSVA 125
Query: 140 RGITPNLALFFGGQDNLPSFLARH---TLPGLGIVVDPCAAVLIVLITLLLCLGIKESST 196
R + GG ++ F + PGL D A LI+L++ LL G+KES++
Sbjct: 126 RAWSGTFDEMIGG--HIEKFCKTYFSMNWPGLAQYPDFFAVCLILLLSGLLSFGVKESAS 183
Query: 197 VQSIVTTINVSVMLFIIIVGGYLGFKAGWVGYEL-----------------------PSG 233
V + T++NV V+LF+II G G W E G
Sbjct: 184 VNKVFTSVNVLVLLFVIISGFVKGDIENWTITEESLINVTRETRNLSALTNVSSDYGAGG 243
Query: 234 YFPYGVNGMFAGSAIVFFSYIGFDSVTSTAEEVKNPQRDLPIGISTALAICCVLYMLVSA 293
+ PYG +G AG+A F++++GFD + +T EEVKNPQ+ +PIGI +L +C + Y VSA
Sbjct: 244 FMPYGFSGTLAGAATCFYAFVGFDCIATTGEEVKNPQKAIPIGIVVSLTMCFLAYFGVSA 303
Query: 294 VIVGLVPYYELNPDTPISSAFSSYGMEWAVYIITTGAVTALFSSLLGSVLPQPRVFMAMA 353
+ ++PYY L+ +P+ AF G A Y++ G++ AL +SLLG + P PR+ AMA
Sbjct: 304 ALTLMMPYYLLDEKSPLPMAFEYVGWGPAKYVVAAGSLCALSASLLGCMFPLPRILFAMA 363
Query: 354 RDGLLPTFFSDIHRRTQIPLKSTIVTGLFAAVLAFFMDVSQLAGMVSVGTLLAFTTVAVS 413
RDG+L F S + +R Q P+ +T+ G AA++AF D+ L M+S+GTLLA++ VAV
Sbjct: 364 RDGILFKFMSKVSKR-QSPVAATMAAGTTAAIMAFLFDLKALVDMMSIGTLLAYSLVAVC 422
Query: 414 VLIIRYVPPDEI-----PIPASLLTSVDPLLRHSGDDIEEDRTVSPVDLASYSDNSHLHD 468
VLI+RY P + P LTS + E D T S L S
Sbjct: 423 VLILRYQPDGALEQRAGPGEREYLTS---------EGGESDLTESESHLNMLKGGS---S 470
Query: 469 KSDVLLEHPLIIKEVTKEQHNEKTRRKLAAWTIALLCIGILIVSGSASAERCPRILRVTL 528
+L+ P + +H+ + + ++C + + SA + + L
Sbjct: 471 TLQAVLQPP-----SSPSEHSSSVVKMSICVLVVIVCALSYLTTYHISAILGAQAWALVL 525
Query: 529 FGAGVVIFLCSIIVLACIKQDDTRHTFGHSGGFACPFVPFLPAACILINTYLLIDLGVDT 588
+++F C I+++ +Q T F P +P+LP +L+N YL+ L DT
Sbjct: 526 LSLCLLLFSCCILLIC--RQPQT----SKKVSFMVPLLPYLPILSVLVNIYLMFQLSGDT 579
Query: 589 WLRVSVWLLIGVLIYLFYGRTHS 611
W+R SVW+ +G LIY YG HS
Sbjct: 580 WIRFSVWMAVGFLIYFGYGMWHS 602
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.326 0.142 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,056,022,649
Number of Sequences: 2790947
Number of extensions: 46090423
Number of successful extensions: 177390
Number of sequences better than 10.0: 2346
Number of HSP's better than 10.0 without gapping: 1434
Number of HSP's successfully gapped in prelim test: 912
Number of HSP's that attempted gapping in prelim test: 171611
Number of HSP's gapped (non-prelim): 3440
length of query: 636
length of database: 848,049,833
effective HSP length: 134
effective length of query: 502
effective length of database: 474,062,935
effective search space: 237979593370
effective search space used: 237979593370
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 78 (34.7 bits)
Medicago: description of AC148965.10


