

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148819.6 - phase: 0
(156 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
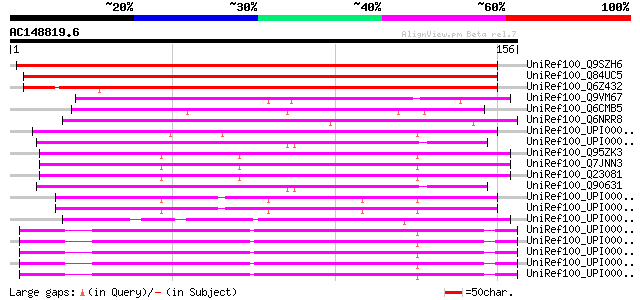
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9SZH6 Hypothetical protein F20B18.130 [Arabidopsis th... 155 4e-37
UniRef100_Q84UC5 4/1 protein [Arabidopsis thaliana] 150 1e-35
UniRef100_Q6Z432 Putative 4/1 protein [Oryza sativa] 122 2e-27
UniRef100_Q9VM67 CG18304-PA [Drosophila melanogaster] 54 9e-07
UniRef100_Q6CMB5 Kluyveromyces lactis strain NRRL Y-1140 chromos... 50 2e-05
UniRef100_Q6NRR8 LOC431812 protein [Xenopus laevis] 49 4e-05
UniRef100_UPI00003C1CAF UPI00003C1CAF UniRef100 entry 48 9e-05
UniRef100_UPI00003AE09E UPI00003AE09E UniRef100 entry 47 1e-04
UniRef100_Q95ZK3 Lin-5 (Five) interacting protein protein 1, iso... 47 1e-04
UniRef100_Q7JNN3 Lin-5 (Five) interacting protein protein 1, iso... 47 1e-04
UniRef100_Q23081 Lin-5 (Five) interacting protein protein 1, iso... 47 1e-04
UniRef100_Q90631 Kinectin [Gallus gallus] 47 1e-04
UniRef100_UPI00003AA72F UPI00003AA72F UniRef100 entry 47 2e-04
UniRef100_UPI00003AA72E UPI00003AA72E UniRef100 entry 47 2e-04
UniRef100_UPI000023D00A UPI000023D00A UniRef100 entry 47 2e-04
UniRef100_UPI000035EEBB UPI000035EEBB UniRef100 entry 46 3e-04
UniRef100_UPI000035EEBA UPI000035EEBA UniRef100 entry 46 3e-04
UniRef100_UPI000035EEB8 UPI000035EEB8 UniRef100 entry 46 3e-04
UniRef100_UPI000035EEB7 UPI000035EEB7 UniRef100 entry 46 3e-04
UniRef100_UPI000035EEB6 UPI000035EEB6 UniRef100 entry 46 3e-04
>UniRef100_Q9SZH6 Hypothetical protein F20B18.130 [Arabidopsis thaliana]
Length = 273
Score = 155 bits (391), Expect = 4e-37
Identities = 75/148 (50%), Positives = 111/148 (74%)
Query: 3 FSEQGHRRALKLLEEEYNEKIARLEAQVKESLHEKASYEATISQLHGDIAAHKNHMQILA 62
F++Q HR AL+ L +++ K+ LE +++ L EKA+ + I +L D+ A+K+H+Q ++
Sbjct: 124 FADQEHRNALESLRQKHVTKVEELEYKIRSLLVEKATNDMVIDRLRQDLTANKSHIQAMS 183
Query: 63 NRLDQVHFEVESKYSSEIRDLKDCLMAEQEEKNDLNRKMQLLEKELLLFKAKMVDQQQEM 122
+LD+V EVE KY EI+DLKDCL+ EQ EKND++ K+Q L+KELL+ + + ++Q++
Sbjct: 184 KKLDRVVTEVECKYELEIQDLKDCLLMEQAEKNDISNKLQSLQKELLISRTSIAEKQRDT 243
Query: 123 TSNWQVETLKQKIMKLRKENEVLKRKFS 150
TSN QVETLKQK+MKLRKENE+LKRK S
Sbjct: 244 TSNRQVETLKQKLMKLRKENEILKRKLS 271
>UniRef100_Q84UC5 4/1 protein [Arabidopsis thaliana]
Length = 242
Score = 150 bits (379), Expect = 1e-35
Identities = 73/146 (50%), Positives = 108/146 (73%)
Query: 5 EQGHRRALKLLEEEYNEKIARLEAQVKESLHEKASYEATISQLHGDIAAHKNHMQILANR 64
E HR AL+ L +++ K+ LE +++ L EKA+ + I +L D+ A+K+H+Q ++ +
Sbjct: 95 EHEHRNALESLRQKHVTKVEELEYKIRSLLVEKATNDMVIDRLRQDLTANKSHIQAMSKK 154
Query: 65 LDQVHFEVESKYSSEIRDLKDCLMAEQEEKNDLNRKMQLLEKELLLFKAKMVDQQQEMTS 124
LD+V EVE KY EI+DLKDCL+ EQ E+ND++ K+Q L+KELL+ + + ++Q++ TS
Sbjct: 155 LDRVVTEVECKYELEIQDLKDCLLMEQAERNDISNKLQSLQKELLISRTSIAEKQRDTTS 214
Query: 125 NWQVETLKQKIMKLRKENEVLKRKFS 150
N QVETLKQK+MKLRKENE+LKRK S
Sbjct: 215 NRQVETLKQKLMKLRKENEILKRKLS 240
>UniRef100_Q6Z432 Putative 4/1 protein [Oryza sativa]
Length = 256
Score = 122 bits (307), Expect = 2e-27
Identities = 68/157 (43%), Positives = 103/157 (65%), Gaps = 12/157 (7%)
Query: 5 EQGHRRALKLLEEEYNEKIARL-----------EAQVKESLHEKASYEATISQLHGDIAA 53
E+ + R L + EEEY +I +L E ++ ++ ++A+ EA I QL+ ++ A
Sbjct: 94 EKANTRLLSM-EEEYKREIEQLKLGSEMNSNDLENKLSCAVVQQATNEAVIKQLNLELEA 152
Query: 54 HKNHMQILANRLDQVHFEVESKYSSEIRDLKDCLMAEQEEKNDLNRKMQLLEKELLLFKA 113
HK H+ +L +RL+QV +V +Y +EI+DLKD ++ EQEEKND++RK+Q E EL + K
Sbjct: 153 HKAHIDMLNSRLEQVTADVHQQYKNEIQDLKDVVIVEQEEKNDMHRKLQNTENELRIMKM 212
Query: 114 KMVDQQQEMTSNWQVETLKQKIMKLRKENEVLKRKFS 150
K +QQ++ S VETLKQK+MK RKENE LKR+ +
Sbjct: 213 KQAEQQRDSISVQHVETLKQKVMKFRKENESLKRRLA 249
>UniRef100_Q9VM67 CG18304-PA [Drosophila melanogaster]
Length = 1833
Score = 54.3 bits (129), Expect = 9e-07
Identities = 40/148 (27%), Positives = 77/148 (52%), Gaps = 16/148 (10%)
Query: 21 EKIARLEAQVKESLHEKASYEATISQLHGDIAAHKNHMQILANRLDQVHFEVESKYSS-- 78
EK++ LE +++ EK EA ISQL ++ + K + + L++ ++++K S
Sbjct: 1028 EKLSSLEKDMEKQAKEKEKLEAKISQLDAELLSAKKSAEKSKSSLEKEIKDLKTKASKSD 1087
Query: 79 --EIRDLKD-------CLMAEQEEKNDLNRKMQLLEKELLLFKAKMVDQQQEMTSNWQVE 129
+++DLK L AEQ+ DLN + L +E +L +A++ ++Q + + ++
Sbjct: 1088 SKQVQDLKKQVEEVQASLSAEQKRYEDLNNHWEKLSEETILMRAQLTTEKQSLQA--ELN 1145
Query: 130 TLKQKIMK---LRKENEVLKRKFSHIEE 154
KQKI + +R E + RK S ++
Sbjct: 1146 ASKQKIAEMDTIRIERTDMARKLSEAQK 1173
>UniRef100_Q6CMB5 Kluyveromyces lactis strain NRRL Y-1140 chromosome E of strain NRRL
Y- 1140 of Kluyveromyces lactis [Kluyveromyces lactis]
Length = 858
Score = 49.7 bits (117), Expect = 2e-05
Identities = 37/154 (24%), Positives = 79/154 (51%), Gaps = 27/154 (17%)
Query: 20 NEKIARLEAQVKESLHEKASYEATISQLHGDIAA-HKNHMQILANRLDQVHFEVESKYSS 78
NE I LEAQ++ + H++ S E +S L ++++ + H +I+ ++ Q+ E+ SS
Sbjct: 315 NEAIRNLEAQLQRNEHQRQSLEREVSLLQEELSSIRETHQKIITHKDQQIKQLTENLSSS 374
Query: 79 EIRDLK--DCLMAEQEEKNDLNRKMQLLEKELLLFKAKMVDQ----QQEMTSNW------ 126
+ +K + L +E+ + N+ ++ EK+L + DQ Q+E+ ++
Sbjct: 375 DSEAVKRLNELSSERLRLLETNKSLKFSEKDLKQTVKSLEDQLNSCQEELHASQDKHKEK 434
Query: 127 --------------QVETLKQKIMKLRKENEVLK 146
+VE ++Q+++++RK N++LK
Sbjct: 435 VSELLRKQLPYTSSEVEAMRQELLEMRKSNDILK 468
Score = 31.2 bits (69), Expect = 8.6
Identities = 34/148 (22%), Positives = 68/148 (44%), Gaps = 35/148 (23%)
Query: 9 RRALKLLEEEYNEKIARLEAQVKESLHEKASYEATISQLHGDIAAHKNHMQILANRLDQV 68
++++++ + N +I L + VK+ HE + +S+L + HK +Q L +++
Sbjct: 488 KKSVEVDIQRKNNEIRALRSTVKDLEHELQDVTSNVSKLKEN---HKEEIQKLRKQVNP- 543
Query: 69 HFEVESKYSSEIRDLKDCLMAEQEEKNDLNRKMQLLEKELLLFKAKMVDQQQEMTSNWQV 128
DL ++ EEK+ L ++ LL+ EL K D +++ + W
Sbjct: 544 -------------DLPTDVL---EEKSKLQNRISLLQMELNSIK----DTKEKELAMW-- 581
Query: 129 ETLKQKIMKLRKENEVLKRKFSHIEEGK 156
++K +R+ NE L ++EGK
Sbjct: 582 ---RRKYETIRRSNEEL------LQEGK 600
>UniRef100_Q6NRR8 LOC431812 protein [Xenopus laevis]
Length = 648
Score = 48.9 bits (115), Expect = 4e-05
Identities = 36/148 (24%), Positives = 73/148 (49%), Gaps = 8/148 (5%)
Query: 17 EEYNEKIARLEAQVKESLHEKASYEATISQLHGDIAAHKNHMQILANRLDQVHFEVESKY 76
E + E++ +++ + +E L K E + + ++ A K ++ + DQ +++ +Y
Sbjct: 71 EAFVEELMQMKQRFQEILLVKEEQEDLLRKRERELTALKGALKEEVSTHDQEIDKLKEQY 130
Query: 77 SSEIRDLKDCLMAEQEEKNDL-NRKMQLLEKELLLFKAKMVDQQQEMTSNWQVETLKQKI 135
E++ ++D L E + +D+ N+K + + + + V Q+ W++E L+ KI
Sbjct: 131 DKELKKIRDNLNEEVKNSSDIANQKNEASQLKAVFENKVKVLLQENEELKWKIENLEDKI 190
Query: 136 MKLRKE-------NEVLKRKFSHIEEGK 156
LR+E NE LK K S E+ K
Sbjct: 191 AGLRQEINDLSEENERLKDKLSRSEKEK 218
Score = 31.6 bits (70), Expect = 6.6
Identities = 35/154 (22%), Positives = 67/154 (42%), Gaps = 20/154 (12%)
Query: 6 QGHRRALK-------LLEEEYNEKIARLEAQVK---ESLHEKASYEATISQLHGDIAAHK 55
Q H+RA LL++ + L+A+ + + + S E ISQL ++ K
Sbjct: 367 QDHQRAQDEALTKNHLLQQTIKDLQYELDAKTHVKDDRIRQIKSMEDKISQLEMELDEEK 426
Query: 56 NHMQILANRLDQVHFEVESKYSSEIRDLKDCLMAEQEEKNDLNRKMQLLEKELLLFKAKM 115
N+ +L R+ + ++E S L E+ K DL L+++ K+++
Sbjct: 427 NNSDLLLERVTRCREQIEQTRSE--------LFQERATKQDLECDKISLDRQNKDLKSRI 478
Query: 116 VDQQQEMTSNWQ--VETLKQKIMKLRKENEVLKR 147
V + SN + V ++ +I +L + E +R
Sbjct: 479 VHLENSHRSNKEGLVAQMETRIAELEERLEYEER 512
>UniRef100_UPI00003C1CAF UPI00003C1CAF UniRef100 entry
Length = 2363
Score = 47.8 bits (112), Expect = 9e-05
Identities = 38/152 (25%), Positives = 75/152 (49%), Gaps = 9/152 (5%)
Query: 8 HRRALKLLEEEYNEKIARLEAQVKESLHEKASYEATISQLH-GDIAAHKNHMQILANR-- 64
H AL ++ + E +A+ + E AS A + H + H+N ++ L
Sbjct: 1016 HASALAESQDRHREAVAKHQNSKSELAAAHASELAALQDGHRAKLDEHRNSIEELVTSHA 1075
Query: 65 -----LDQVHFEVESKYSSEIRDLKDCLMAEQEEKNDLNRKMQLLEKELLLFKAKMVDQQ 119
L+Q H E + ++++ K + A +E N+++++ KEL KA++ D Q
Sbjct: 1076 AALSVLEQSHREALATRDNDLQAAKQEVDARTKELAARNQELEMRTKELGAAKAEIDDLQ 1135
Query: 120 QEMTS-NWQVETLKQKIMKLRKENEVLKRKFS 150
+ +T+ ++E LKQ++ +L+K + KR+ S
Sbjct: 1136 KNITALRAEMEQLKQQLAELQKSHTAAKRELS 1167
>UniRef100_UPI00003AE09E UPI00003AE09E UniRef100 entry
Length = 1365
Score = 47.4 bits (111), Expect = 1e-04
Identities = 36/142 (25%), Positives = 69/142 (48%), Gaps = 5/142 (3%)
Query: 9 RRALKLLEEEYNEKIARLEAQVKESLHEKASYEATISQLHGDIAAHKNHMQILANRLDQV 68
R + LE+E+ +++ +ES K ++ Q+ +I+ K IL + +
Sbjct: 373 RERINALEKEHGTFQSKIHVSYQESQQMKIKFQQRCEQMEAEISHLKQENTILRDAVSTS 432
Query: 69 HFEVESKYSSEIRDLK-DC--LMAEQEEKNDLNRKMQLLEKELLLFKAKMVDQQQEMTSN 125
++ESK ++E+ L+ DC L+ E EKN ++ +L +K A++ QQQE
Sbjct: 433 TNQMESKQAAELNKLRQDCARLVNELGEKNSKLQQEELQKKNAEQAVAQLKVQQQEAERR 492
Query: 126 WQVETLKQKIMKLRKENEVLKR 147
W E ++ + K E+E ++
Sbjct: 493 W--EEIQVYLRKRTAEHEAAQQ 512
Score = 40.8 bits (94), Expect = 0.011
Identities = 34/144 (23%), Positives = 66/144 (45%), Gaps = 12/144 (8%)
Query: 9 RRALKLLEEEYNEKIARLEAQVKESLHEKASYEATISQLHGDIA---AHKNHMQILANRL 65
+R+++ E ++ K+ + ++K+ AS E + +L +I K + L + L
Sbjct: 1162 QRSVEEEESKWKIKVEESQKELKQMRSSVASLEHEVERLKEEIKEVETLKKEREHLESEL 1221
Query: 66 DQVHFEVESKYSSEIRDLKDCLMAEQEEKNDLNRKMQLLEKELLLFKAKM--------VD 117
++ E S Y SE+R+LKD L Q++ +D + +EL L K K+ VD
Sbjct: 1222 EKAEIE-RSTYVSEVRELKDLLTELQKKLDDSYSEAVRQNEELNLLKMKLSETLSKLKVD 1280
Query: 118 QQQEMTSNWQVETLKQKIMKLRKE 141
Q + + ++ + L +E
Sbjct: 1281 QNERQKVAGDLPKAQESLASLERE 1304
Score = 34.3 bits (77), Expect = 1.0
Identities = 32/137 (23%), Positives = 65/137 (47%), Gaps = 19/137 (13%)
Query: 6 QGHRRALKLLEEEYNEKIARLEAQVKESLHEKASYEATISQLHGDIAAHKNHMQILANRL 65
+G +K +E EK E ++ + + + T++QL + ++ L
Sbjct: 875 KGKENQVKTMEALLEEK----EKEIVQKGEQLKGQQDTVAQLTSKV------QELEQQNL 924
Query: 66 DQVHFEVESKYSSEIRDLKDCLMAEQEEKNDLNRKMQLLEKELLLFKAKMVDQQQEMTSN 125
Q+ + +S+++DL+ L E+E+ + L ++ E+E+ A V Q Q M S
Sbjct: 925 QQLQ---QVPAASQVQDLESRLRGEEEQISKLKAVLEEKEREI----ASQVKQLQTMQS- 976
Query: 126 WQVETLKQKIMKLRKEN 142
+ E+ K +I +L++EN
Sbjct: 977 -ENESFKVQIQELKQEN 992
>UniRef100_Q95ZK3 Lin-5 (Five) interacting protein protein 1, isoform b [Caenorhabditis
elegans]
Length = 2315
Score = 47.4 bits (111), Expect = 1e-04
Identities = 40/176 (22%), Positives = 82/176 (45%), Gaps = 31/176 (17%)
Query: 10 RALKLLEEEYNEKIARLEAQVKESLHEKASYEATIS--------------QLHGDIAAHK 55
R+L+ EE+++ +LE +++ES + Y+ I+ +L G +
Sbjct: 1758 RSLRDKEEQWDSSRFQLETKMRESDSDTNKYQLQIASFESERQILTEKIKELDGALRLSD 1817
Query: 56 NHMQILANRLDQVH----------------FEVESKYSSEIRDLKDCLMAEQEEKNDLNR 99
+ +Q + + D++ +++SK S E + LKD L+ Q E N N
Sbjct: 1818 SKVQDMKDDTDKLRRDLTKAESVENELRKTIDIQSKTSHEYQLLKDQLLNTQNELNGANN 1877
Query: 100 KMQLLEKELLLFKAKMVDQQQEMTS-NWQVETLKQKIMKLRKENEVLKRKFSHIEE 154
+ Q LE ELL ++++ D +Q + N +V L++++ E ++ +F +E+
Sbjct: 1878 RKQQLENELLNVRSEVRDYKQRVHDVNNRVSELQRQLQDANTEKNRVEDRFLSVEK 1933
Score = 41.6 bits (96), Expect = 0.006
Identities = 40/147 (27%), Positives = 62/147 (41%), Gaps = 5/147 (3%)
Query: 9 RRALKLLEEEYNEKIARLEAQVKESLHEKASYEATISQLHGDIAAHKNHMQILANRLDQV 68
RR L E NE ++ Q K S HE + + ++ N Q L N L V
Sbjct: 1831 RRDLTKAESVENELRKTIDIQSKTS-HEYQLLKDQLLNTQNELNGANNRKQQLENELLNV 1889
Query: 69 HFEVESKYSSEIRDLKDCLMAEQEEKNDLNRKMQLLEKELLLFKAKMVDQQQ--EMTSNW 126
EV Y + D+ + + Q + D N + +E L + K+V+ + E
Sbjct: 1890 RSEVRD-YKQRVHDVNNRVSELQRQLQDANTEKNRVEDRFLSVE-KVVNTMRTTETDLRQ 1947
Query: 127 QVETLKQKIMKLRKENEVLKRKFSHIE 153
Q+ET K + KE E LKR+ + +E
Sbjct: 1948 QLETAKNEKRVATKELEDLKRRLAQLE 1974
>UniRef100_Q7JNN3 Lin-5 (Five) interacting protein protein 1, isoform d [Caenorhabditis
elegans]
Length = 2350
Score = 47.4 bits (111), Expect = 1e-04
Identities = 40/176 (22%), Positives = 82/176 (45%), Gaps = 31/176 (17%)
Query: 10 RALKLLEEEYNEKIARLEAQVKESLHEKASYEATIS--------------QLHGDIAAHK 55
R+L+ EE+++ +LE +++ES + Y+ I+ +L G +
Sbjct: 1793 RSLRDKEEQWDSSRFQLETKMRESDSDTNKYQLQIASFESERQILTEKIKELDGALRLSD 1852
Query: 56 NHMQILANRLDQVH----------------FEVESKYSSEIRDLKDCLMAEQEEKNDLNR 99
+ +Q + + D++ +++SK S E + LKD L+ Q E N N
Sbjct: 1853 SKVQDMKDDTDKLRRDLTKAESVENELRKTIDIQSKTSHEYQLLKDQLLNTQNELNGANN 1912
Query: 100 KMQLLEKELLLFKAKMVDQQQEMTS-NWQVETLKQKIMKLRKENEVLKRKFSHIEE 154
+ Q LE ELL ++++ D +Q + N +V L++++ E ++ +F +E+
Sbjct: 1913 RKQQLENELLNVRSEVRDYKQRVHDVNNRVSELQRQLQDANTEKNRVEDRFLSVEK 1968
Score = 41.6 bits (96), Expect = 0.006
Identities = 40/147 (27%), Positives = 62/147 (41%), Gaps = 5/147 (3%)
Query: 9 RRALKLLEEEYNEKIARLEAQVKESLHEKASYEATISQLHGDIAAHKNHMQILANRLDQV 68
RR L E NE ++ Q K S HE + + ++ N Q L N L V
Sbjct: 1866 RRDLTKAESVENELRKTIDIQSKTS-HEYQLLKDQLLNTQNELNGANNRKQQLENELLNV 1924
Query: 69 HFEVESKYSSEIRDLKDCLMAEQEEKNDLNRKMQLLEKELLLFKAKMVDQQQ--EMTSNW 126
EV Y + D+ + + Q + D N + +E L + K+V+ + E
Sbjct: 1925 RSEVRD-YKQRVHDVNNRVSELQRQLQDANTEKNRVEDRFLSVE-KVVNTMRTTETDLRQ 1982
Query: 127 QVETLKQKIMKLRKENEVLKRKFSHIE 153
Q+ET K + KE E LKR+ + +E
Sbjct: 1983 QLETAKNEKRVATKELEDLKRRLAQLE 2009
>UniRef100_Q23081 Lin-5 (Five) interacting protein protein 1, isoform a [Caenorhabditis
elegans]
Length = 2396
Score = 47.4 bits (111), Expect = 1e-04
Identities = 40/176 (22%), Positives = 82/176 (45%), Gaps = 31/176 (17%)
Query: 10 RALKLLEEEYNEKIARLEAQVKESLHEKASYEATIS--------------QLHGDIAAHK 55
R+L+ EE+++ +LE +++ES + Y+ I+ +L G +
Sbjct: 1866 RSLRDKEEQWDSSRFQLETKMRESDSDTNKYQLQIASFESERQILTEKIKELDGALRLSD 1925
Query: 56 NHMQILANRLDQVH----------------FEVESKYSSEIRDLKDCLMAEQEEKNDLNR 99
+ +Q + + D++ +++SK S E + LKD L+ Q E N N
Sbjct: 1926 SKVQDMKDDTDKLRRDLTKAESVENELRKTIDIQSKTSHEYQLLKDQLLNTQNELNGANN 1985
Query: 100 KMQLLEKELLLFKAKMVDQQQEMTS-NWQVETLKQKIMKLRKENEVLKRKFSHIEE 154
+ Q LE ELL ++++ D +Q + N +V L++++ E ++ +F +E+
Sbjct: 1986 RKQQLENELLNVRSEVRDYKQRVHDVNNRVSELQRQLQDANTEKNRVEDRFLSVEK 2041
Score = 41.6 bits (96), Expect = 0.006
Identities = 40/147 (27%), Positives = 62/147 (41%), Gaps = 5/147 (3%)
Query: 9 RRALKLLEEEYNEKIARLEAQVKESLHEKASYEATISQLHGDIAAHKNHMQILANRLDQV 68
RR L E NE ++ Q K S HE + + ++ N Q L N L V
Sbjct: 1939 RRDLTKAESVENELRKTIDIQSKTS-HEYQLLKDQLLNTQNELNGANNRKQQLENELLNV 1997
Query: 69 HFEVESKYSSEIRDLKDCLMAEQEEKNDLNRKMQLLEKELLLFKAKMVDQQQ--EMTSNW 126
EV Y + D+ + + Q + D N + +E L + K+V+ + E
Sbjct: 1998 RSEVRD-YKQRVHDVNNRVSELQRQLQDANTEKNRVEDRFLSVE-KVVNTMRTTETDLRQ 2055
Query: 127 QVETLKQKIMKLRKENEVLKRKFSHIE 153
Q+ET K + KE E LKR+ + +E
Sbjct: 2056 QLETAKNEKRVATKELEDLKRRLAQLE 2082
>UniRef100_Q90631 Kinectin [Gallus gallus]
Length = 1364
Score = 47.4 bits (111), Expect = 1e-04
Identities = 36/142 (25%), Positives = 69/142 (48%), Gaps = 5/142 (3%)
Query: 9 RRALKLLEEEYNEKIARLEAQVKESLHEKASYEATISQLHGDIAAHKNHMQILANRLDQV 68
R + LE+E+ +++ +ES K ++ Q+ +I+ K IL + +
Sbjct: 371 RERINALEKEHGTFQSKIHVSYQESQQMKIKFQQRCEQMEAEISHLKQENTILRDAVSTS 430
Query: 69 HFEVESKYSSEIRDLK-DC--LMAEQEEKNDLNRKMQLLEKELLLFKAKMVDQQQEMTSN 125
++ESK ++E+ L+ DC L+ E EKN ++ +L +K A++ QQQE
Sbjct: 431 TNQMESKQAAELNKLRQDCARLVNELGEKNSKLQQEELQKKNAEQAVAQLKVQQQEAERR 490
Query: 126 WQVETLKQKIMKLRKENEVLKR 147
W E ++ + K E+E ++
Sbjct: 491 W--EEIQVYLRKRTAEHEAAQQ 510
Score = 40.8 bits (94), Expect = 0.011
Identities = 34/144 (23%), Positives = 66/144 (45%), Gaps = 12/144 (8%)
Query: 9 RRALKLLEEEYNEKIARLEAQVKESLHEKASYEATISQLHGDIA---AHKNHMQILANRL 65
+R+++ E ++ K+ + ++K+ AS E + +L +I K + L + L
Sbjct: 1160 QRSVEEEESKWKIKVEESQKELKQMRSSVASLEHEVERLKEEIKEVETLKKEREHLESEL 1219
Query: 66 DQVHFEVESKYSSEIRDLKDCLMAEQEEKNDLNRKMQLLEKELLLFKAKM--------VD 117
++ E S Y SE+R+LKD L Q++ +D + +EL L K K+ VD
Sbjct: 1220 EKAEIE-RSTYVSEVRELKDLLTELQKKLDDSYSEAVRQNEELNLLKMKLSETLSKLKVD 1278
Query: 118 QQQEMTSNWQVETLKQKIMKLRKE 141
Q + + ++ + L +E
Sbjct: 1279 QNERQKVAGDLPKAQESLASLERE 1302
Score = 33.5 bits (75), Expect = 1.7
Identities = 32/137 (23%), Positives = 64/137 (46%), Gaps = 19/137 (13%)
Query: 6 QGHRRALKLLEEEYNEKIARLEAQVKESLHEKASYEATISQLHGDIAAHKNHMQILANRL 65
+G +K +E EK E ++ + + T++QL + ++ L
Sbjct: 873 KGKENQVKTMEALLEEK----EKEIVQKGERLKGQQDTVAQLTSKV------QELEQQNL 922
Query: 66 DQVHFEVESKYSSEIRDLKDCLMAEQEEKNDLNRKMQLLEKELLLFKAKMVDQQQEMTSN 125
Q+ + +S+++DL+ L E+E+ + L ++ E+E+ A V Q Q M S
Sbjct: 923 QQLQ---QVPAASQVQDLESRLRGEEEQISKLKAVLEEKEREI----ASQVKQLQTMQS- 974
Query: 126 WQVETLKQKIMKLRKEN 142
+ E+ K +I +L++EN
Sbjct: 975 -ENESFKVQIQELKQEN 990
>UniRef100_UPI00003AA72F UPI00003AA72F UniRef100 entry
Length = 380
Score = 47.0 bits (110), Expect = 2e-04
Identities = 38/142 (26%), Positives = 73/142 (50%), Gaps = 8/142 (5%)
Query: 15 LEEEYNEKIARLEAQVKESLHEKASYEATIS-QLHGDIAAHKNHMQILANRLDQVHFEVE 73
+EE N +I+ L +++ H+ +Y ++ Q I+ K M+ + R + H E E
Sbjct: 183 VEERKNNQISELMKNHEKAFHDFKNYHDDVTFQNLALISLLKEQMEEMKKR--ETHLEKE 240
Query: 74 SKYSS-EIRDLKDCLMAEQEEKNDLNRKMQLLEKE---LLLFKAKMVDQQQEMTS-NWQV 128
+ + + LKD L QE+ +L +K+ +K+ L+ KA + Q+E+ W+
Sbjct: 241 KADALLQNKQLKDSLQQTQEQVFELQKKLAHYDKDKEALMNTKAHLKVTQKELKDLRWEH 300
Query: 129 ETLKQKIMKLRKENEVLKRKFS 150
E L+Q+ K++ E + L +KF+
Sbjct: 301 EVLEQRFSKVQAERDELYQKFT 322
>UniRef100_UPI00003AA72E UPI00003AA72E UniRef100 entry
Length = 485
Score = 47.0 bits (110), Expect = 2e-04
Identities = 38/142 (26%), Positives = 73/142 (50%), Gaps = 8/142 (5%)
Query: 15 LEEEYNEKIARLEAQVKESLHEKASYEATIS-QLHGDIAAHKNHMQILANRLDQVHFEVE 73
+EE N +I+ L +++ H+ +Y ++ Q I+ K M+ + R + H E E
Sbjct: 218 VEERKNNQISELMKNHEKAFHDFKNYHDDVTFQNLALISLLKEQMEEMKKR--ETHLEKE 275
Query: 74 SKYSS-EIRDLKDCLMAEQEEKNDLNRKMQLLEKE---LLLFKAKMVDQQQEMTS-NWQV 128
+ + + LKD L QE+ +L +K+ +K+ L+ KA + Q+E+ W+
Sbjct: 276 KADALLQNKQLKDSLQQTQEQVFELQKKLAHYDKDKEALMNTKAHLKVTQKELKDLRWEH 335
Query: 129 ETLKQKIMKLRKENEVLKRKFS 150
E L+Q+ K++ E + L +KF+
Sbjct: 336 EVLEQRFSKVQAERDELYQKFT 357
>UniRef100_UPI000023D00A UPI000023D00A UniRef100 entry
Length = 774
Score = 46.6 bits (109), Expect = 2e-04
Identities = 39/143 (27%), Positives = 68/143 (47%), Gaps = 12/143 (8%)
Query: 17 EEYNEKIARLEAQVKESLHEKASYEATISQLHGDIAAHKNHMQILANRLDQVHFEVESKY 76
E+ EKI LEAQ +L E A I+ L ++ K + L E K
Sbjct: 56 EDLREKIKELEAQSSLALDET---HARIAILQDEL---KKGGDSTSEELRSTKEAAEQK- 108
Query: 77 SSEIRDLKDCLMAEQEEKNDLNRKMQLLEKELLLFKAKMVDQQQ-----EMTSNWQVETL 131
+ E+ D K L A +E+ L ++ Q + EL KA++V+ ++ E +++TL
Sbjct: 109 AKELEDAKSSLTATEEKLKGLEQERQSIADELATLKAELVEAKEAREALEAALTKEIDTL 168
Query: 132 KQKIMKLRKENEVLKRKFSHIEE 154
K +I + ++++ L + S +EE
Sbjct: 169 KTQISEAEQKHQALTKAHSTLEE 191
>UniRef100_UPI000035EEBB UPI000035EEBB UniRef100 entry
Length = 1585
Score = 46.2 bits (108), Expect = 3e-04
Identities = 37/154 (24%), Positives = 77/154 (49%), Gaps = 13/154 (8%)
Query: 4 SEQGHRRALKLLEEEYNEKIARLEAQVKESLHEKASYEATISQLHGDIAAHKNHMQILAN 63
SE+G +RA L+E L A++ ++ E+ E + +L +I + +
Sbjct: 1204 SERGRKRAESQLQE--------LSARLAQADREREDREERVQKLQSEIETVSSTLSSSET 1255
Query: 64 RLDQVHFEVESKYSSEIRDLKDCLMAEQEEKNDLNRKMQLLEKELLLFKAKMVDQQQEMT 123
+ + EV S S++ D K+ L E +K L +++ LE+E ++ ++++
Sbjct: 1256 KSLRFSKEVNS-LESQLNDAKESLQDETRQKMTLATRVRALEEEKNGLMERLEEEEERSK 1314
Query: 124 S-NWQVETLKQKIMKLRKENEVLKRKFSHIEEGK 156
+ Q++TL Q++ +LRK++E + S +E G+
Sbjct: 1315 ELSRQIQTLSQQLAELRKQSEEVN---SAVETGE 1345
>UniRef100_UPI000035EEBA UPI000035EEBA UniRef100 entry
Length = 1801
Score = 46.2 bits (108), Expect = 3e-04
Identities = 37/154 (24%), Positives = 77/154 (49%), Gaps = 13/154 (8%)
Query: 4 SEQGHRRALKLLEEEYNEKIARLEAQVKESLHEKASYEATISQLHGDIAAHKNHMQILAN 63
SE+G +RA L+E L A++ ++ E+ E + +L +I + +
Sbjct: 1233 SERGRKRAESQLQE--------LSARLAQADREREDREERVQKLQSEIETVSSTLSSSET 1284
Query: 64 RLDQVHFEVESKYSSEIRDLKDCLMAEQEEKNDLNRKMQLLEKELLLFKAKMVDQQQEMT 123
+ + EV S S++ D K+ L E +K L +++ LE+E ++ ++++
Sbjct: 1285 KSLRFSKEVNS-LESQLNDAKESLQDETRQKMTLATRVRALEEEKNGLMERLEEEEERSK 1343
Query: 124 S-NWQVETLKQKIMKLRKENEVLKRKFSHIEEGK 156
+ Q++TL Q++ +LRK++E + S +E G+
Sbjct: 1344 ELSRQIQTLSQQLAELRKQSEEVN---SAVETGE 1374
>UniRef100_UPI000035EEB8 UPI000035EEB8 UniRef100 entry
Length = 1843
Score = 46.2 bits (108), Expect = 3e-04
Identities = 37/154 (24%), Positives = 77/154 (49%), Gaps = 13/154 (8%)
Query: 4 SEQGHRRALKLLEEEYNEKIARLEAQVKESLHEKASYEATISQLHGDIAAHKNHMQILAN 63
SE+G +RA L+E L A++ ++ E+ E + +L +I + +
Sbjct: 1272 SERGRKRAESQLQE--------LSARLAQADREREDREERVQKLQSEIETVSSTLSSSET 1323
Query: 64 RLDQVHFEVESKYSSEIRDLKDCLMAEQEEKNDLNRKMQLLEKELLLFKAKMVDQQQEMT 123
+ + EV S S++ D K+ L E +K L +++ LE+E ++ ++++
Sbjct: 1324 KSLRFSKEVNS-LESQLNDAKESLQDETRQKMTLATRVRALEEEKNGLMERLEEEEERSK 1382
Query: 124 S-NWQVETLKQKIMKLRKENEVLKRKFSHIEEGK 156
+ Q++TL Q++ +LRK++E + S +E G+
Sbjct: 1383 ELSRQIQTLSQQLAELRKQSEEVN---SAVETGE 1413
>UniRef100_UPI000035EEB7 UPI000035EEB7 UniRef100 entry
Length = 1826
Score = 46.2 bits (108), Expect = 3e-04
Identities = 37/154 (24%), Positives = 77/154 (49%), Gaps = 13/154 (8%)
Query: 4 SEQGHRRALKLLEEEYNEKIARLEAQVKESLHEKASYEATISQLHGDIAAHKNHMQILAN 63
SE+G +RA L+E L A++ ++ E+ E + +L +I + +
Sbjct: 1249 SERGRKRAESQLQE--------LSARLAQADREREDREERVQKLQSEIETVSSTLSSSET 1300
Query: 64 RLDQVHFEVESKYSSEIRDLKDCLMAEQEEKNDLNRKMQLLEKELLLFKAKMVDQQQEMT 123
+ + EV S S++ D K+ L E +K L +++ LE+E ++ ++++
Sbjct: 1301 KSLRFSKEVNS-LESQLNDAKESLQDETRQKMTLATRVRALEEEKNGLMERLEEEEERSK 1359
Query: 124 S-NWQVETLKQKIMKLRKENEVLKRKFSHIEEGK 156
+ Q++TL Q++ +LRK++E + S +E G+
Sbjct: 1360 ELSRQIQTLSQQLAELRKQSEEVN---SAVETGE 1390
>UniRef100_UPI000035EEB6 UPI000035EEB6 UniRef100 entry
Length = 1846
Score = 46.2 bits (108), Expect = 3e-04
Identities = 37/154 (24%), Positives = 77/154 (49%), Gaps = 13/154 (8%)
Query: 4 SEQGHRRALKLLEEEYNEKIARLEAQVKESLHEKASYEATISQLHGDIAAHKNHMQILAN 63
SE+G +RA L+E L A++ ++ E+ E + +L +I + +
Sbjct: 1275 SERGRKRAESQLQE--------LSARLAQADREREDREERVQKLQSEIETVSSTLSSSET 1326
Query: 64 RLDQVHFEVESKYSSEIRDLKDCLMAEQEEKNDLNRKMQLLEKELLLFKAKMVDQQQEMT 123
+ + EV S S++ D K+ L E +K L +++ LE+E ++ ++++
Sbjct: 1327 KSLRFSKEVNS-LESQLNDAKESLQDETRQKMTLATRVRALEEEKNGLMERLEEEEERSK 1385
Query: 124 S-NWQVETLKQKIMKLRKENEVLKRKFSHIEEGK 156
+ Q++TL Q++ +LRK++E + S +E G+
Sbjct: 1386 ELSRQIQTLSQQLAELRKQSEEVN---SAVETGE 1416
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.313 0.127 0.334
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 226,001,666
Number of Sequences: 2790947
Number of extensions: 8100646
Number of successful extensions: 65274
Number of sequences better than 10.0: 4287
Number of HSP's better than 10.0 without gapping: 302
Number of HSP's successfully gapped in prelim test: 4045
Number of HSP's that attempted gapping in prelim test: 58296
Number of HSP's gapped (non-prelim): 10152
length of query: 156
length of database: 848,049,833
effective HSP length: 116
effective length of query: 40
effective length of database: 524,299,981
effective search space: 20971999240
effective search space used: 20971999240
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 69 (31.2 bits)
Medicago: description of AC148819.6


