

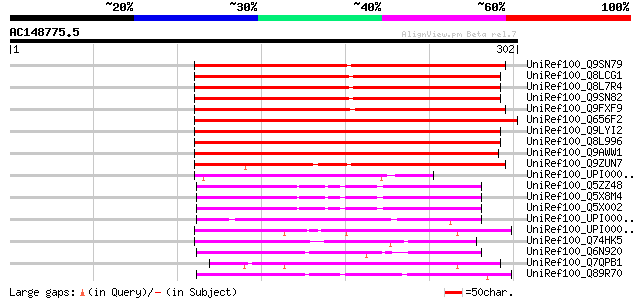
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148775.5 + phase: 0 /pseudo
(302 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9SN79 Hypothetical protein F1P2.140 [Arabidopsis thal... 266 4e-70
UniRef100_Q8LCG1 Putative esterase [Arabidopsis thaliana] 260 3e-68
UniRef100_Q8L7R4 Hypothetical protein At3g47560 [Arabidopsis tha... 260 3e-68
UniRef100_Q9SN82 Hypothetical protein F1P2.110 [Arabidopsis thal... 260 3e-68
UniRef100_Q9FXF9 F1N18.12 protein [Arabidopsis thaliana] 257 2e-67
UniRef100_Q656F2 Esterase/lipase/thioesterase family protein-lik... 250 4e-65
UniRef100_Q9LYI2 Putative esterase-like protein [Arabidopsis tha... 246 5e-64
UniRef100_Q8L996 Putative esterase-like protein [Arabidopsis tha... 244 2e-63
UniRef100_Q9AWW1 Esterase/lipase/thioesterase family protein-lik... 221 1e-56
UniRef100_Q9ZUN7 Putative esterase [Arabidopsis thaliana] 214 3e-54
UniRef100_UPI000042DDBD UPI000042DDBD UniRef100 entry 67 4e-10
UniRef100_Q5ZZ48 Hydrolases of the alpha/beta superfamily [Legio... 65 2e-09
UniRef100_Q5X8M4 Hypothetical protein [Legionella pneumophila st... 64 5e-09
UniRef100_Q5X002 Hypothetical protein [Legionella pneumophila st... 63 8e-09
UniRef100_UPI00002C6CBA UPI00002C6CBA UniRef100 entry 61 3e-08
UniRef100_UPI00003C1ACF UPI00003C1ACF UniRef100 entry 59 1e-07
UniRef100_Q74HK5 Hypothetical protein [Lactobacillus johnsonii] 58 3e-07
UniRef100_Q6N920 Possible hydrolase [Rhodopseudomonas palustris] 58 4e-07
UniRef100_Q7QPB1 GLP_122_2606_3436 [Giardia lamblia ATCC 50803] 57 5e-07
UniRef100_Q89R70 Bll2902 protein [Bradyrhizobium japonicum] 57 8e-07
>UniRef100_Q9SN79 Hypothetical protein F1P2.140 [Arabidopsis thaliana]
Length = 309
Score = 266 bits (681), Expect = 4e-70
Identities = 126/185 (68%), Positives = 150/185 (80%), Gaps = 2/185 (1%)
Query: 111 ESEGSFEYGNYWKEVDDLHAVAQHFRESNRVIRAIVGHSKGGDVVLLYASKYHEIKTVVN 170
ESEGSF YGNY E DDLH+V Q+F NRV+ I+GHSKGGDVVLLYASKYH+++ V+N
Sbjct: 123 ESEGSFYYGNYNHEADDLHSVVQYFSNKNRVVPIILGHSKGGDVVLLYASKYHDVRNVIN 182
Query: 171 LSGRYDLKAGIEERLGKDYLERIRKDGFFDVKRSSGKLDYRVTEESLMDRLGTNMHEACL 230
LSGRYDLK GI ERLG+D+LERI++ GF DV GK YRVTE+SLMDRL T++HEACL
Sbjct: 183 LSGRYDLKKGIRERLGEDFLERIKQQGFIDV--GDGKSGYRVTEKSLMDRLSTDIHEACL 240
Query: 231 QIDKDCRILTIHGSSDEIIPVQDAHEFAKIIPNHKLHIIEGADHAYNNHQDELSSVFMSF 290
+IDK+CR+LT+HGS DE+IPV+DA EFAKIIPNHKL I+EGA+H Y HQ +L S M F
Sbjct: 241 KIDKECRVLTVHGSEDEVIPVEDAKEFAKIIPNHKLEIVEGANHGYTEHQSQLVSTVMEF 300
Query: 291 IKETI 295
IK I
Sbjct: 301 IKTVI 305
>UniRef100_Q8LCG1 Putative esterase [Arabidopsis thaliana]
Length = 265
Score = 260 bits (665), Expect = 3e-68
Identities = 125/182 (68%), Positives = 147/182 (80%), Gaps = 2/182 (1%)
Query: 111 ESEGSFEYGNYWKEVDDLHAVAQHFRESNRVIRAIVGHSKGGDVVLLYASKYHEIKTVVN 170
ESEGSF YGNY E DDLH+V Q+F NRV+ I+GHSKGGDVVLLYASKYH+I V+N
Sbjct: 74 ESEGSFYYGNYNYEADDLHSVIQYFSNLNRVVTIILGHSKGGDVVLLYASKYHDIPNVIN 133
Query: 171 LSGRYDLKAGIEERLGKDYLERIRKDGFFDVKRSSGKLDYRVTEESLMDRLGTNMHEACL 230
LSGRYDLK GI ERLG+D+LERI++ G+ DVK G YRVTEESLMDRL T+MHEACL
Sbjct: 134 LSGRYDLKKGIGERLGEDFLERIKQQGYIDVK--DGDSGYRVTEESLMDRLNTDMHEACL 191
Query: 231 QIDKDCRILTIHGSSDEIIPVQDAHEFAKIIPNHKLHIIEGADHAYNNHQDELSSVFMSF 290
+IDK+CR+LT+HGS DE +PV+DA EFAKIIPNH+L I+EGADH Y N+ +L S M F
Sbjct: 192 KIDKECRVLTVHGSGDETVPVEDAKEFAKIIPNHELQIVEGADHCYTNYXSQLVSTVMEF 251
Query: 291 IK 292
IK
Sbjct: 252 IK 253
>UniRef100_Q8L7R4 Hypothetical protein At3g47560 [Arabidopsis thaliana]
Length = 265
Score = 260 bits (665), Expect = 3e-68
Identities = 125/182 (68%), Positives = 147/182 (80%), Gaps = 2/182 (1%)
Query: 111 ESEGSFEYGNYWKEVDDLHAVAQHFRESNRVIRAIVGHSKGGDVVLLYASKYHEIKTVVN 170
ESEGSF YGNY E DDLH+V Q+F NRV+ I+GHSKGGDVVLLYASKYH+I V+N
Sbjct: 74 ESEGSFYYGNYNYEADDLHSVIQYFSNLNRVVTIILGHSKGGDVVLLYASKYHDIPNVIN 133
Query: 171 LSGRYDLKAGIEERLGKDYLERIRKDGFFDVKRSSGKLDYRVTEESLMDRLGTNMHEACL 230
LSGRYDLK GI ERLG+D+LERI++ G+ DVK G YRVTEESLMDRL T+MHEACL
Sbjct: 134 LSGRYDLKKGIGERLGEDFLERIKQQGYIDVK--DGDSGYRVTEESLMDRLNTDMHEACL 191
Query: 231 QIDKDCRILTIHGSSDEIIPVQDAHEFAKIIPNHKLHIIEGADHAYNNHQDELSSVFMSF 290
+IDK+CR+LT+HGS DE +PV+DA EFAKIIPNH+L I+EGADH Y N+Q +L M F
Sbjct: 192 KIDKECRVLTVHGSGDETVPVEDAKEFAKIIPNHELQIVEGADHCYTNYQSQLVLTVMEF 251
Query: 291 IK 292
IK
Sbjct: 252 IK 253
>UniRef100_Q9SN82 Hypothetical protein F1P2.110 [Arabidopsis thaliana]
Length = 265
Score = 260 bits (665), Expect = 3e-68
Identities = 125/182 (68%), Positives = 147/182 (80%), Gaps = 2/182 (1%)
Query: 111 ESEGSFEYGNYWKEVDDLHAVAQHFRESNRVIRAIVGHSKGGDVVLLYASKYHEIKTVVN 170
ESEGSF YGNY E DDLH+V Q+F NRV+ I+GHSKGGDVVLLYASKYH+I V+N
Sbjct: 74 ESEGSFYYGNYNYEADDLHSVIQYFSNLNRVVTIILGHSKGGDVVLLYASKYHDIPNVIN 133
Query: 171 LSGRYDLKAGIEERLGKDYLERIRKDGFFDVKRSSGKLDYRVTEESLMDRLGTNMHEACL 230
LSGRYDLK GI ERLG+D+LERI++ G+ DVK G YRVTEESLMDRL T+MHEACL
Sbjct: 134 LSGRYDLKKGIGERLGEDFLERIKQQGYIDVK--DGDSGYRVTEESLMDRLNTDMHEACL 191
Query: 231 QIDKDCRILTIHGSSDEIIPVQDAHEFAKIIPNHKLHIIEGADHAYNNHQDELSSVFMSF 290
+IDK+CR+LT+HGS DE +PV+DA EFAKIIPNH+L I+EGADH Y N+Q +L M F
Sbjct: 192 KIDKECRVLTVHGSGDETVPVEDAKEFAKIIPNHELQIVEGADHCYTNYQSQLVLTVMEF 251
Query: 291 IK 292
IK
Sbjct: 252 IK 253
>UniRef100_Q9FXF9 F1N18.12 protein [Arabidopsis thaliana]
Length = 263
Score = 257 bits (657), Expect = 2e-67
Identities = 121/185 (65%), Positives = 150/185 (80%), Gaps = 2/185 (1%)
Query: 111 ESEGSFEYGNYWKEVDDLHAVAQHFRESNRVIRAIVGHSKGGDVVLLYASKYHEIKTVVN 170
ES+GSF +GNY E DDLH+V ++F NRV+ I+GHSKGGDVVL+YASKY +I+ V+N
Sbjct: 77 ESKGSFYFGNYNYEADDLHSVIRYFTNMNRVVPIIIGHSKGGDVVLVYASKYQDIRNVIN 136
Query: 171 LSGRYDLKAGIEERLGKDYLERIRKDGFFDVKRSSGKLDYRVTEESLMDRLGTNMHEACL 230
LSGRYDLK GI ERLG+DYLERI++ GF D+K G +RVTEESLM+RL T+MHEACL
Sbjct: 137 LSGRYDLKRGIGERLGEDYLERIKQQGFIDIKE--GNAGFRVTEESLMERLNTDMHEACL 194
Query: 231 QIDKDCRILTIHGSSDEIIPVQDAHEFAKIIPNHKLHIIEGADHAYNNHQDELSSVFMSF 290
+IDK+CR+LT+HGS+DE+IP++DA EFAKIIPNHKL I+EGADH Y HQ +L + M F
Sbjct: 195 KIDKECRVLTVHGSADEVIPLEDAKEFAKIIPNHKLEIVEGADHCYTKHQSQLITNVMEF 254
Query: 291 IKETI 295
IK I
Sbjct: 255 IKTVI 259
>UniRef100_Q656F2 Esterase/lipase/thioesterase family protein-like [Oryza sativa]
Length = 275
Score = 250 bits (638), Expect = 4e-65
Identities = 117/192 (60%), Positives = 151/192 (77%)
Query: 111 ESEGSFEYGNYWKEVDDLHAVAQHFRESNRVIRAIVGHSKGGDVVLLYASKYHEIKTVVN 170
ESEG F+YGNY KE DDLH+V H + ++AIVGHSKGGDVV+LYAS Y +++TVVN
Sbjct: 84 ESEGEFQYGNYRKEADDLHSVISHLNQEKYDVKAIVGHSKGGDVVVLYASIYDDVRTVVN 143
Query: 171 LSGRYDLKAGIEERLGKDYLERIRKDGFFDVKRSSGKLDYRVTEESLMDRLGTNMHEACL 230
LSGR+ L+ GIEERLGK+++ I K+G+ DVK +SGK+ Y+VT+ESLM+RL T+MH+ACL
Sbjct: 144 LSGRFHLEKGIEERLGKEFMNIIDKEGYIDVKTNSGKVLYKVTKESLMERLTTDMHKACL 203
Query: 231 QIDKDCRILTIHGSSDEIIPVQDAHEFAKIIPNHKLHIIEGADHAYNNHQDELSSVFMSF 290
I K+CR T+HGS+DEIIPV+DA+EFAK IPNHKLH+IEGA+H Y H+ ELS + F
Sbjct: 204 SISKECRFFTVHGSADEIIPVEDAYEFAKHIPNHKLHVIEGANHCYTAHRKELSDAVVDF 263
Query: 291 IKETIDHSKSTA 302
I + D S+A
Sbjct: 264 ITSSEDGDNSSA 275
>UniRef100_Q9LYI2 Putative esterase-like protein [Arabidopsis thaliana]
Length = 339
Score = 246 bits (628), Expect = 5e-64
Identities = 115/182 (63%), Positives = 148/182 (81%)
Query: 111 ESEGSFEYGNYWKEVDDLHAVAQHFRESNRVIRAIVGHSKGGDVVLLYASKYHEIKTVVN 170
ES+GSF+YGNY +EV+DL +V QH R NRVI AI+GHSKGG+VVLLYA+KY++++TVVN
Sbjct: 126 ESQGSFQYGNYRREVEDLRSVLQHLRGVNRVISAIIGHSKGGNVVLLYAAKYNDVQTVVN 185
Query: 171 LSGRYDLKAGIEERLGKDYLERIRKDGFFDVKRSSGKLDYRVTEESLMDRLGTNMHEACL 230
+SGR+ L GIE RLGKDY +RI+ +GF DV GK +YRVTEESLMDRL TN HEACL
Sbjct: 186 ISGRFFLDRGIEFRLGKDYFKRIKDNGFIDVSNRKGKFEYRVTEESLMDRLTTNAHEACL 245
Query: 231 QIDKDCRILTIHGSSDEIIPVQDAHEFAKIIPNHKLHIIEGADHAYNNHQDELSSVFMSF 290
I ++CR+LT+HGS+D I+ V +A EFAK I NHKL++IEGADH + +HQ +L+S+ +SF
Sbjct: 246 SIRENCRVLTVHGSNDRIVHVTEASEFAKQIKNHKLYVIEGADHEFTSHQHQLASIVLSF 305
Query: 291 IK 292
K
Sbjct: 306 FK 307
>UniRef100_Q8L996 Putative esterase-like protein [Arabidopsis thaliana]
Length = 297
Score = 244 bits (623), Expect = 2e-63
Identities = 114/182 (62%), Positives = 148/182 (80%)
Query: 111 ESEGSFEYGNYWKEVDDLHAVAQHFRESNRVIRAIVGHSKGGDVVLLYASKYHEIKTVVN 170
ES+GSF+YGNY +E++DL +V QH R NRVI AI+GHSKGG+VVLLYA+KY++++TVVN
Sbjct: 86 ESQGSFQYGNYRRELEDLRSVLQHLRGVNRVISAIIGHSKGGNVVLLYAAKYNDVQTVVN 145
Query: 171 LSGRYDLKAGIEERLGKDYLERIRKDGFFDVKRSSGKLDYRVTEESLMDRLGTNMHEACL 230
+SGR+ L GIE RLGKDY +RI+ +GF DV GK +YRVTEESLMDRL TN HEACL
Sbjct: 146 ISGRFFLDRGIEFRLGKDYFKRIKDNGFIDVGNRKGKFEYRVTEESLMDRLTTNAHEACL 205
Query: 231 QIDKDCRILTIHGSSDEIIPVQDAHEFAKIIPNHKLHIIEGADHAYNNHQDELSSVFMSF 290
I ++CR+LT+HGS+D I+ V +A EFAK I NHKL++IEGADH + +HQ +L+S+ +SF
Sbjct: 206 SIRENCRVLTVHGSNDRIVHVTEASEFAKQIKNHKLYVIEGADHEFTSHQHQLASIVLSF 265
Query: 291 IK 292
K
Sbjct: 266 FK 267
>UniRef100_Q9AWW1 Esterase/lipase/thioesterase family protein-like [Oryza sativa]
Length = 324
Score = 221 bits (564), Expect = 1e-56
Identities = 102/181 (56%), Positives = 137/181 (75%)
Query: 111 ESEGSFEYGNYWKEVDDLHAVAQHFRESNRVIRAIVGHSKGGDVVLLYASKYHEIKTVVN 170
ESEG FEYGNY KE DDLH+V + + + AIVGHSKGGDVV LYAS Y +++ V+N
Sbjct: 141 ESEGEFEYGNYRKEADDLHSVVSYLCKEKYDVTAIVGHSKGGDVVTLYASIYDDVRLVIN 200
Query: 171 LSGRYDLKAGIEERLGKDYLERIRKDGFFDVKRSSGKLDYRVTEESLMDRLGTNMHEACL 230
+SGR+DL+ GIEER+G+ ++RI K+G+ DVK SG + YRVT+ESLM+RL T++ +
Sbjct: 201 VSGRFDLEKGIEERIGEGSIDRINKEGYLDVKDKSGNVQYRVTKESLMERLNTDIRAVSM 260
Query: 231 QIDKDCRILTIHGSSDEIIPVQDAHEFAKIIPNHKLHIIEGADHAYNNHQDELSSVFMSF 290
I K+CR T+HGS+DE IPV+DA++FAK IPNHKL +IEGA+H Y H++EL+ + F
Sbjct: 261 SITKECRFFTVHGSADETIPVEDAYKFAKHIPNHKLQVIEGANHNYTAHREELADAVVDF 320
Query: 291 I 291
I
Sbjct: 321 I 321
>UniRef100_Q9ZUN7 Putative esterase [Arabidopsis thaliana]
Length = 332
Score = 214 bits (544), Expect = 3e-54
Identities = 108/190 (56%), Positives = 140/190 (72%), Gaps = 9/190 (4%)
Query: 111 ESEGSFEYGNYWKEV-DDLHAVAQHFRESN---RVIRAIVGHSKGGDVVLLYASKYHE-I 165
+SEG+F YGN+ E DDLH V QH SN R++ I+GHSKGGDVVLLYASK+ + I
Sbjct: 68 DSEGTFYYGNFNSEAEDDLHYVIQHLSSSNIMNRLVPVILGHSKGGDVVLLYASKFPDYI 127
Query: 166 KTVVNLSGRYDLKAGIEERLGKDYLERIRKDGFFDVKRSSGKLDYRVTEESLMDRLGTNM 225
+ VVN+SGR+DLK + RLG Y+E+I++ GF D + GK +RVT+ESLMDRL T+M
Sbjct: 128 RNVVNISGRFDLKNDV--RLGDGYIEKIKEQGFIDA--TEGKSCFRVTQESLMDRLNTDM 183
Query: 226 HEACLQIDKDCRILTIHGSSDEIIPVQDAHEFAKIIPNHKLHIIEGADHAYNNHQDELSS 285
H+ACL IDK C++LT+HGS D ++P +DA EFAK+IPNHKL I+EGA+H Y HQ EL S
Sbjct: 184 HQACLNIDKQCKVLTVHGSDDTVVPGEDAKEFAKVIPNHKLEIVEGANHGYTKHQKELVS 243
Query: 286 VFMSFIKETI 295
+ + F K I
Sbjct: 244 IAVEFTKTAI 253
>UniRef100_UPI000042DDBD UPI000042DDBD UniRef100 entry
Length = 220
Score = 67.4 bits (163), Expect = 4e-10
Identities = 48/150 (32%), Positives = 83/150 (55%), Gaps = 12/150 (8%)
Query: 111 ESEG--SFEYGNYWKEV-DDLHAVAQHFRESNRVIRAIVGHSKGGDVVLLYASKYHEIKT 167
ES+G S+ +G+Y EV DDL V + R I+AI+GHS+GG L+Y+ Y ++
Sbjct: 71 ESQGADSWSFGDYHGEVNDDLRKVVEFLRNKGLEIKAIIGHSRGGVETLMYSWMYDDVDI 130
Query: 168 VVNLSGRYDLKAGIEER-LGKDYLERIRKDGFFDVK-RSSGKLDYRVTEESLMDR--LGT 223
++++S R+DL I R + + E+++ + V+ + ++T E + R +
Sbjct: 131 IISISARFDLANSIITRFISDEQYEKLKNNELESVEIIPRDNIPRKITLECINKRNLVDY 190
Query: 224 NMHEACLQIDKDCR-ILTIHGSSDEIIPVQ 252
NM L+ K+ + L IHG+ D+I+PVQ
Sbjct: 191 NM----LKTVKNTKYFLLIHGTKDDIVPVQ 216
>UniRef100_Q5ZZ48 Hydrolases of the alpha/beta superfamily [Legionella pneumophila
subsp. pneumophila str. Philadelphia 1]
Length = 273
Score = 65.5 bits (158), Expect = 2e-09
Identities = 46/171 (26%), Positives = 83/171 (47%), Gaps = 8/171 (4%)
Query: 112 SEGSFEYGNYWKEVDDLHAVAQHFRESNRVIRAIVGHSKGGDVVLLYASKYHEIKTVVNL 171
SEGSF N+ V+DL A A + R + ++GHS GG VLL A K E+K + +
Sbjct: 84 SEGSFSETNFSSNVEDLVAAADYLRTHYQAPVLLIGHSLGGAAVLLAAKKITEVKAIATI 143
Query: 172 SGRYDLKAGIEERLGKDYLERIRKDGFFDVKRSSGKLDYRVTEESLMDRLGTNMHEACLQ 231
G ++ D L +I DG + + G + + ++ L D + ++ ++
Sbjct: 144 -GAPASAHHVKHHFSAD-LSKIESDG--EAHVTLGPRSFTIKKQFLQD---IDRYQETIK 196
Query: 232 IDKDCRILTIHGSSDEIIPVQDAHEFAKIIPNHKLHI-IEGADHAYNNHQD 281
D +L +H D+++ +++A + K + K I ++ ADH +N +D
Sbjct: 197 SDAGKALLIMHSPIDKVVSIKEAEKIYKAAQHPKSFISLDKADHLLSNKRD 247
>UniRef100_Q5X8M4 Hypothetical protein [Legionella pneumophila str. Paris]
Length = 257
Score = 63.9 bits (154), Expect = 5e-09
Identities = 45/171 (26%), Positives = 82/171 (47%), Gaps = 8/171 (4%)
Query: 112 SEGSFEYGNYWKEVDDLHAVAQHFRESNRVIRAIVGHSKGGDVVLLYASKYHEIKTVVNL 171
SEGSF N+ V+DL A A + R R ++GHS GG VLL A E+K + +
Sbjct: 69 SEGSFSETNFSSNVEDLVAAADYLRAHYRAPVLLIGHSLGGAAVLLAAKNISEVKAIATI 128
Query: 172 SGRYDLKAGIEERLGKDYLERIRKDGFFDVKRSSGKLDYRVTEESLMDRLGTNMHEACLQ 231
G ++ D L +I DG + + + G + + ++ L D + ++ +
Sbjct: 129 -GAPASAHHVKHHFSAD-LSKIESDG--EAQVTLGPRSFTIKKQFLQD---IDRYQDTIN 181
Query: 232 IDKDCRILTIHGSSDEIIPVQDAHEFAKIIPNHKLHI-IEGADHAYNNHQD 281
D +L +H D+++ +++A + K + K I ++ ADH ++ +D
Sbjct: 182 SDAGKALLILHSPIDKVVSIKEAEKIYKAAKHPKSFISLDKADHLLSDKRD 232
>UniRef100_Q5X002 Hypothetical protein [Legionella pneumophila str. Lens]
Length = 257
Score = 63.2 bits (152), Expect = 8e-09
Identities = 46/171 (26%), Positives = 82/171 (47%), Gaps = 8/171 (4%)
Query: 112 SEGSFEYGNYWKEVDDLHAVAQHFRESNRVIRAIVGHSKGGDVVLLYASKYHEIKTVVNL 171
SEGSF N+ V+DL A A + R R ++GHS GG VLL A K E+K + +
Sbjct: 69 SEGSFAETNFSSNVEDLVAAADYLRTHYRAPVLLIGHSLGGAAVLLAAKKISEVKAIATI 128
Query: 172 SGRYDLKAGIEERLGKDYLERIRKDGFFDVKRSSGKLDYRVTEESLMDRLGTNMHEACLQ 231
G ++ D L +I DG + + + G + ++ L D + ++ +
Sbjct: 129 -GAPASAHHVKHHFSAD-LSKIESDG--EAQVTLGPRSVTIKKQFLED---IDRYQEAIT 181
Query: 232 IDKDCRILTIHGSSDEIIPVQDAHEFAKIIPNHKLHI-IEGADHAYNNHQD 281
D +L +H D+++ +++A + K + K I ++ ADH ++ +D
Sbjct: 182 SDAGKALLIMHSPIDKVVSIKEAEKIYKAAQHPKSFISLDKADHLLSDKRD 232
>UniRef100_UPI00002C6CBA UPI00002C6CBA UniRef100 entry
Length = 231
Score = 61.2 bits (147), Expect = 3e-08
Identities = 49/174 (28%), Positives = 78/174 (44%), Gaps = 10/174 (5%)
Query: 112 SEGSFEYGNYWKEVDDLHAVAQHFRESNRVIRAIVGHSKGGDVVLLYASKYH-EIKTVVN 170
S G FE G K ++ H + F I I+G S GG + L+ A + I ++
Sbjct: 51 SAGEFEDGGISKWSEEAHEI---FNLCKSKINIIIGSSMGGWISLIVAKRNPLTINGLIG 107
Query: 171 LSGRYDLKAGIEERLGKDYLERIRKDGFFDVKRSSGKLDYRVTEESLMD-RLGTNMHEAC 229
++ D G RL K+ +I+KDG + DY +T + L D + +HE
Sbjct: 108 IASAPDFVVGEWNRLSKEQKTQIKKDGKIIINWDKYSEDYTITYKFLEDGKKNMILHE-- 165
Query: 230 LQIDKDCRILTIHGSSDEIIPVQDAHEFAKII--PNHKLHIIEGADHAYNNHQD 281
I C I +HG D+++ + + + I N +L IIE DH+ + QD
Sbjct: 166 -PIKIKCPIRLLHGRLDQVVSFETSEKIINKIESENKQLKIIEDGDHSLSRDQD 218
>UniRef100_UPI00003C1ACF UPI00003C1ACF UniRef100 entry
Length = 562
Score = 59.3 bits (142), Expect = 1e-07
Identities = 50/207 (24%), Positives = 92/207 (44%), Gaps = 20/207 (9%)
Query: 111 ESEGSFEYGNYWKEVDDLHAVAQHFR-ESNRVIRAIVGHSKGGDVVLLYASKY------- 162
E+ G++ GN +V+DL AV + R + + I+GHS+GG + +K+
Sbjct: 161 ETPGTWSMGNLADDVEDLVAVVDYLRTKLEYTVEIIIGHSRGGLDGFAWFAKHCPDALPP 220
Query: 163 -HEIKTVVNLSGRYDLKAGIEERLGKDYLERIRKDGFF--DVKRSSGKLDYRVTEESLMD 219
+ V LS R+++ A I ER YL K+GFF + + + V E +
Sbjct: 221 SLRVPFFVALSARFNM-ANIHER-DPVYLSAFAKEGFFRWQARVAGQDKELHVYPEQVEQ 278
Query: 220 RLGTNMHEACLQIDKDCRILTIHGSSDEIIPVQDAHEFAKIIPN-HK------LHIIEGA 272
E L + +L IHG++D+ +P D + I+ H+ + +I+ A
Sbjct: 279 FAAWPTREIALAFPYNTDVLLIHGTADKSVPASDVTSYGNILSGIHRRPGSCSVKLIDHA 338
Query: 273 DHAYNNHQDELSSVFMSFIKETIDHSK 299
DH + ++ + ++ E + +K
Sbjct: 339 DHLFRGFYPQVVEAIVEWLAERSELTK 365
>UniRef100_Q74HK5 Hypothetical protein [Lactobacillus johnsonii]
Length = 249
Score = 58.2 bits (139), Expect = 3e-07
Identities = 47/173 (27%), Positives = 83/173 (47%), Gaps = 15/173 (8%)
Query: 111 ESEGSFEYGNYWKEVDDLHAVAQHFRESNRVIRA-IVGHSKGGDVVLLYASKYHE-IKTV 168
+S+G FE E++D +A+ + + V +VGHS+GG V + A Y + IK V
Sbjct: 67 DSDGKFENMTVLNEIEDANAILNYVKTDPHVRNIYLVGHSQGGVVASMLAGLYPDLIKKV 126
Query: 169 VNLSGRYDLKAGIEERLGKDYLERIRKDGFFDVKRSSGKLDYR-VTEESLMDRLGTNM-- 225
V L+ LK+ D LE + ++ +L ++ +T R+ +
Sbjct: 127 VLLAPAATLKS--------DALEGNTQGVTYNPDHIPDRLPFKDLTLGGFYLRIAQQLPI 178
Query: 226 HEACLQIDKDCRILTIHGSSDEIIPVQDAHEFAKIIPNHKLHIIEGADHAYNN 278
+E Q K + IHG+ D ++ + ++ +I N LH+IEGADH +++
Sbjct: 179 YEVSAQFTKP--VCLIHGTDDTVVSPNASKKYDQIYQNSTLHLIEGADHCFSD 229
>UniRef100_Q6N920 Possible hydrolase [Rhodopseudomonas palustris]
Length = 407
Score = 57.8 bits (138), Expect = 4e-07
Identities = 45/177 (25%), Positives = 80/177 (44%), Gaps = 19/177 (10%)
Query: 112 SEGSFEYGNYWKEVDDLHAVAQHFRESNRVIRAIVGHSKGGDVVLLYASKYHEIKTVVNL 171
SEG FE + V DL A H R ++R ++GHS GG VL A++ E K + +
Sbjct: 71 SEGEFENATFSSNVADLVLAADHLRATHRAPTLLIGHSLGGAAVLAAAAQIPEAKAIATI 130
Query: 172 SGRYDLKAGIEERLGKDYLERIRKDGFFDVKRSSGKLDYR------VTEESLMDRLGTNM 225
+ D L D +E IR +G +V + + + E +LM +G +
Sbjct: 131 AAPSD--PSHVTGLFADDIETIRTEGRVNVSLAGRPFTIKREFLDDIAEHNLMAEIG-KL 187
Query: 226 HEACLQIDKDCRILTIHGSSDEIIPVQDAHEFAKIIPNHKLHI-IEGADHAYNNHQD 281
H+A +L +H +D+ + + +A + + K + ++ ADH ++ +D
Sbjct: 188 HKA---------LLILHAPTDDTVGIDNATKIFLAAKHPKSFVSLDHADHLLSDRRD 235
>UniRef100_Q7QPB1 GLP_122_2606_3436 [Giardia lamblia ATCC 50803]
Length = 276
Score = 57.4 bits (137), Expect = 5e-07
Identities = 44/191 (23%), Positives = 86/191 (44%), Gaps = 20/191 (10%)
Query: 120 NYWKEVDDLHAVAQHFRES------NRVIRAIVGHSKGGDVVLLYASKY-------HEIK 166
+Y EVD L +V H +E +R + +VGHS+G ++ LLY + E+
Sbjct: 79 SYADEVDVLSSVLDHLKEYIQEWGLSRTV--LVGHSRGANISLLYIQRLVLQCPEQPELP 136
Query: 167 TVVNLSGRYDLKAGIEE-RLGKDYLERIRKDGFFDVKRSSGKLDYRVTEESLMDRLGTNM 225
VV + GR+DL + + + G + + ++ F + VT +++R +
Sbjct: 137 YVVVVRGRFDLSGTLNQSKRGPEEVLQLENGHEFTKAFPGCRCPLLVTPAFILERRSIQL 196
Query: 226 HEACLQIDKDCRILTIHGSSDEIIPVQDAHEFAKIIPNHK----LHIIEGADHAYNNHQD 281
+AC + + R+ +HG++D +P Q++ + K L +++ A H + +
Sbjct: 197 GDACRVLSQTDRLAVLHGTNDRAVPPQESSLMVQFYSEEKKRSRLRLVDEATHNWKYKLE 256
Query: 282 ELSSVFMSFIK 292
E F I+
Sbjct: 257 EGYHAFCGLIE 267
>UniRef100_Q89R70 Bll2902 protein [Bradyrhizobium japonicum]
Length = 407
Score = 56.6 bits (135), Expect = 8e-07
Identities = 49/191 (25%), Positives = 84/191 (43%), Gaps = 9/191 (4%)
Query: 112 SEGSFEYGNYWKEVDDLHAVAQHFRESNRVIRAIVGHSKGGDVVLLYASKYHEIKTVVNL 171
SEG F + V DL A H R + + ++GHS GG +L ASK E K VV +
Sbjct: 71 SEGDFANSTFSSNVADLVRAADHLRSTRKAPSILIGHSLGGAAILAAASKVPEAKAVVTI 130
Query: 172 SGRYDLKAGIEERLGKDYLERIRKDGFFDVKRSSGKLDYRVTEESLMDRLGTNMHEACLQ 231
+ D L +++++ IR G +V+ S +R+ E L D + +
Sbjct: 131 AAPSD--PTHVTGLFREHVDAIRAQG--EVEVSLAGRPFRIKREFLEDIAEHELMKDVTG 186
Query: 232 IDKDCRILTIHGSSDEIIPVQDAHEFAKIIPNHKLHI-IEGADHAYNNHQDEL--SSVFM 288
+ K +L +H D+ + + +A + + K + ++ ADH D L + V
Sbjct: 187 LHK--ALLVMHSPLDDTVGIDNATKLFVAAKHPKSFVSLDHADHLLTRPADALYAADVIA 244
Query: 289 SFIKETIDHSK 299
++ ID +K
Sbjct: 245 AWASRYIDAAK 255
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.337 0.147 0.492
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 480,446,868
Number of Sequences: 2790947
Number of extensions: 19870710
Number of successful extensions: 71837
Number of sequences better than 10.0: 501
Number of HSP's better than 10.0 without gapping: 117
Number of HSP's successfully gapped in prelim test: 384
Number of HSP's that attempted gapping in prelim test: 71542
Number of HSP's gapped (non-prelim): 569
length of query: 302
length of database: 848,049,833
effective HSP length: 127
effective length of query: 175
effective length of database: 493,599,564
effective search space: 86379923700
effective search space used: 86379923700
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 74 (33.1 bits)
Medicago: description of AC148775.5


