

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148775.1 - phase: 0
(207 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
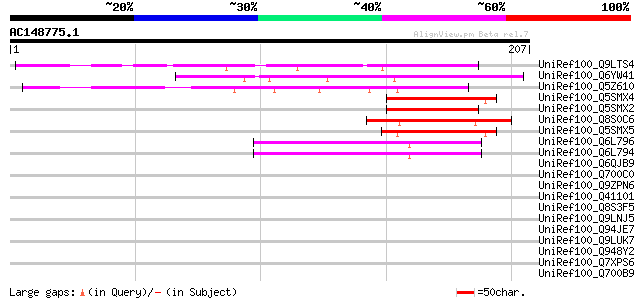
UniRef100_Q9LTS4 Gb|AAC78547.1 [Arabidopsis thaliana] 91 1e-17
UniRef100_Q6YW41 Basic helix-loop-helix (BHLH) family protein-li... 66 6e-10
UniRef100_Q5Z610 Basic helix-loop-helix protein-like [Oryza sativa] 61 2e-08
UniRef100_Q5SMX4 Basic helix-loop-helix protein-like [Oryza sativa] 58 2e-07
UniRef100_Q5SMX2 Basic helix-loop-helix protein-like [Oryza sativa] 57 4e-07
UniRef100_Q8S0C6 Basic helix-loop-helix (BHLH) family protein-li... 52 1e-05
UniRef100_Q5SMX5 Basic helix-loop-helix protein-like [Oryza sativa] 50 4e-05
UniRef100_Q6L796 BHLH transcription activator Ivory seed [Ipomoe... 50 5e-05
UniRef100_Q6L794 BHLH transcription activator Ivory seed [Ipomoe... 47 4e-04
UniRef100_Q6QJB9 MYC protein [Oryza sativa] 45 0.001
UniRef100_Q700C0 MYC transcription factor [Solanum tuberosum] 45 0.001
UniRef100_Q9ZPN6 Transcription factor MYC7E [Zea mays] 45 0.001
UniRef100_Q41101 Phaseolin G-box binding protein PG1 [Phaseolus ... 45 0.001
UniRef100_Q8S3F5 Putative bHLH transcription factor [Arabidopsis... 45 0.001
UniRef100_Q9LNJ5 F6F3.7 protein [Arabidopsis thaliana] 45 0.001
UniRef100_Q94JE7 P0684B02.21 protein [Oryza sativa] 45 0.002
UniRef100_Q9LUK7 BHLH transcription factor [Arabidopsis thaliana] 45 0.002
UniRef100_Q948Y2 R-type basic helix-loop-helix protein [Oryza sa... 44 0.002
UniRef100_Q7XPS6 OSJNBa0065O17.5 protein [Oryza sativa] 44 0.002
UniRef100_Q700B9 MYC transcription factor [Solanum tuberosum] 44 0.002
>UniRef100_Q9LTS4 Gb|AAC78547.1 [Arabidopsis thaliana]
Length = 466
Score = 91.3 bits (225), Expect = 1e-17
Identities = 76/211 (36%), Positives = 103/211 (48%), Gaps = 45/211 (21%)
Query: 3 SLSTAGSPEYSSLIFNIPPGTSPPQFSPDLTQLGGVNIPPMRPVSNTLLPLQLQQLPQIT 62
SLS EYSSL+F + P S T VN+P + P L P+ +
Sbjct: 126 SLSPNNISEYSSLLFPLIPKPS--------TTTEAVNVPVLPP----LAPINMIHPQHQE 173
Query: 63 PTQLYPTTQIENDVIMRAIQNVL---STPPSHQHQPQQNYVAHPEASAFGRYRH---DKS 116
P L+ Q E + + +AI VL S+PPS PQ+ A+AF RY D+
Sbjct: 174 P--LFRNRQREEEAMTQAILAVLTGPSSPPSTSSSPQRK----GRATAFKRYYSMISDRG 227
Query: 117 PNIGSNFRRQSLMKRSFAFFRSLNLMRMRDR--------------------NQAARPSSN 156
+ R+QS+M R+ +F+ LN+ R+R + PS+
Sbjct: 228 RAPLPSVRKQSMMTRAMSFYNRLNI-NQRERFTRENATTHGEGSGGSGGGGRYTSGPSAT 286
Query: 157 QLHHMISERRRREKLNENFQALRALLPQGTK 187
QL HMISER+RREKLNE+FQALR+LLP GTK
Sbjct: 287 QLQHMISERKRREKLNESFQALRSLLPPGTK 317
>UniRef100_Q6YW41 Basic helix-loop-helix (BHLH) family protein-like [Oryza sativa]
Length = 745
Score = 65.9 bits (159), Expect = 6e-10
Identities = 50/187 (26%), Positives = 85/187 (44%), Gaps = 49/187 (26%)
Query: 67 YPTTQIENDVIMRAIQNVLSTPPSHQ-----HQPQQNYVAH------------PEASAFG 109
+P+ + ++ + +A+ +V+S+P + H P + V H P +AF
Sbjct: 375 FPSAEADDAAMAQAMLDVISSPSTSSSAAALHAPWSS-VKHRAQIIRSPRRGTPTTTAFR 433
Query: 110 RYRHDKSPNIGSNFRR-----QSLMKRSFAFFRSLNLMRMRDRNQAAR------------ 152
Y +P ++ R Q ++K F+ R ++++R AA
Sbjct: 434 AYNAALAPRAAASRRPPGAPGQRMIKMGFSILRRMHMVRCSQERAAAAAAAASAAAAQRS 493
Query: 153 --------------PSSNQLHHMISERRRREKLNENFQALRALLPQGTKVRTKLLISSCC 198
P+S+QLHHMISERRRRE+LNE+F+ LR LLP G+K +++
Sbjct: 494 GGGDDEDATAAPPPPTSSQLHHMISERRRRERLNESFEHLRGLLPPGSKKDKATVLAKTL 553
Query: 199 FIINALL 205
+N L+
Sbjct: 554 EYMNLLI 560
>UniRef100_Q5Z610 Basic helix-loop-helix protein-like [Oryza sativa]
Length = 539
Score = 60.8 bits (146), Expect = 2e-08
Identities = 60/202 (29%), Positives = 88/202 (42%), Gaps = 46/202 (22%)
Query: 6 TAGSPEYSSLIFNIPPGTSPPQFSPDLTQLGGVNIPPMRPVSNTLLPLQLQQLPQITPTQ 65
+ GSPEYSSL+ ++ T +G PV LL + P
Sbjct: 185 SVGSPEYSSLVRSMA------------TSVGASAAADPSPVHGGLLAPVYGEFPGSD--- 229
Query: 66 LYPTTQIENDVIMRAIQNVLSTP---PSHQHQPQQNYVAHPE---ASAFGRYRHDKSPNI 119
++ + +A+ V+STP P P++ + A+AF Y SP
Sbjct: 230 -------DDAAMAQAMLAVISTPAPPPPLWRPPRRRARSSSSPRRATAFKAYNAALSPRA 282
Query: 120 GSN--FRRQSLMKRSFAFFRSLNLM---------RMRDRNQAARP-------SSNQLHHM 161
Q ++K + S+++ R RD + A P SS+QLHHM
Sbjct: 283 RPRPGAPGQRMIKTGISLLASVHMQTRSRELAAARQRDTHAAPPPPPPPPPPSSSQLHHM 342
Query: 162 ISERRRREKLNENFQALRALLP 183
ISERRRRE++N++FQ LRALLP
Sbjct: 343 ISERRRRERINDSFQTLRALLP 364
>UniRef100_Q5SMX4 Basic helix-loop-helix protein-like [Oryza sativa]
Length = 439
Score = 57.8 bits (138), Expect = 2e-07
Identities = 29/45 (64%), Positives = 37/45 (81%), Gaps = 1/45 (2%)
Query: 151 ARPSSNQLHHMISERRRREKLNENFQALRALLPQGTKV-RTKLLI 194
A PS NQL HMISER+RREKLN++F AL+A+LP G+K +T +LI
Sbjct: 246 APPSGNQLQHMISERKRREKLNDSFLALKAVLPPGSKKDKTSILI 290
>UniRef100_Q5SMX2 Basic helix-loop-helix protein-like [Oryza sativa]
Length = 412
Score = 56.6 bits (135), Expect = 4e-07
Identities = 26/37 (70%), Positives = 32/37 (86%)
Query: 151 ARPSSNQLHHMISERRRREKLNENFQALRALLPQGTK 187
A PS NQL HMISER+RREKLN++F AL+A+LP G+K
Sbjct: 176 APPSDNQLQHMISERKRREKLNDSFVALKAVLPTGSK 212
>UniRef100_Q8S0C6 Basic helix-loop-helix (BHLH) family protein-like [Oryza sativa]
Length = 454
Score = 52.0 bits (123), Expect = 1e-05
Identities = 29/62 (46%), Positives = 44/62 (70%), Gaps = 4/62 (6%)
Query: 143 RMRDRNQAARPS---SNQLHHMISERRRREKLNENFQALRALLPQ-GTKVRTKLLISSCC 198
R R + +AA S S+QL+HM+SER+RREKLN++F LR+LLP K +T +LI++
Sbjct: 247 RGRQQEEAAAASATNSSQLYHMMSERKRREKLNDSFHTLRSLLPPCSKKDKTTVLINAAK 306
Query: 199 FI 200
++
Sbjct: 307 YL 308
>UniRef100_Q5SMX5 Basic helix-loop-helix protein-like [Oryza sativa]
Length = 338
Score = 50.1 bits (118), Expect = 4e-05
Identities = 27/50 (54%), Positives = 38/50 (76%), Gaps = 4/50 (8%)
Query: 149 QAARP---SSNQLHHMISERRRREKLNENFQALRALLPQGTKV-RTKLLI 194
+AA P +NQL H +SER+RREKLN++F AL+A+LP G+K +T +LI
Sbjct: 221 EAAPPPPSGNNQLQHTMSERKRREKLNDSFVALKAVLPPGSKKDKTSILI 270
>UniRef100_Q6L796 BHLH transcription activator Ivory seed [Ipomoea purpurea]
Length = 665
Score = 49.7 bits (117), Expect = 5e-05
Identities = 34/102 (33%), Positives = 52/102 (50%), Gaps = 11/102 (10%)
Query: 98 NYVAHPEASAFGRYRHDKSPNIGSNFRRQSLMKRSFAFFRSLNLMRMRDRNQAARPSSNQ 157
+YV H +SAF R+ S S+ Q ++K + L+ + AA S++
Sbjct: 398 DYVVHSASSAFSRWTTAASSTCSSHRSAQWVLKYTLLTVPFLHAKNSNGGDGAATILSSK 457
Query: 158 L-----------HHMISERRRREKLNENFQALRALLPQGTKV 188
L +H+++ERRRREKLNE F LR+L+P TK+
Sbjct: 458 LCKAAPQEEPNVNHVLAERRRREKLNERFIILRSLVPFVTKM 499
>UniRef100_Q6L794 BHLH transcription activator Ivory seed [Ipomoea tricolor]
Length = 670
Score = 46.6 bits (109), Expect = 4e-04
Identities = 33/102 (32%), Positives = 48/102 (46%), Gaps = 11/102 (10%)
Query: 98 NYVAHPEASAFGRYRHDKSPNIGSNFRRQSLMKRSFAFFRSLNLMRMRDRNQAARPSSNQ 157
+YV H +SAF R+ S S+ Q +K + L A S++
Sbjct: 400 DYVVHSASSAFSRWTTAASSTCSSHRSAQWALKYTLLTVPFLQAKNSHGGGAADTIPSSK 459
Query: 158 L-----------HHMISERRRREKLNENFQALRALLPQGTKV 188
L +H+++ERRRREKLNE F LR+L+P TK+
Sbjct: 460 LCKAAPQEEPNVNHVLAERRRREKLNERFIILRSLVPFVTKM 501
>UniRef100_Q6QJB9 MYC protein [Oryza sativa]
Length = 699
Score = 45.4 bits (106), Expect = 0.001
Identities = 24/63 (38%), Positives = 39/63 (61%), Gaps = 3/63 (4%)
Query: 145 RDRNQAARPSSNQ---LHHMISERRRREKLNENFQALRALLPQGTKVRTKLLISSCCFII 201
R R + +P++ + L+H+ +ER+RREKLN+ F ALRA++P +K+ L+ I
Sbjct: 507 RPRKRGRKPANGREEPLNHVEAERQRREKLNQRFYALRAVVPNVSKMDKASLLGDAISYI 566
Query: 202 NAL 204
N L
Sbjct: 567 NEL 569
>UniRef100_Q700C0 MYC transcription factor [Solanum tuberosum]
Length = 692
Score = 45.4 bits (106), Expect = 0.001
Identities = 24/63 (38%), Positives = 39/63 (61%), Gaps = 3/63 (4%)
Query: 145 RDRNQAARPSSNQ---LHHMISERRRREKLNENFQALRALLPQGTKVRTKLLISSCCFII 201
R R + +P++ + L+H+ +ER+RREKLN+ F ALRA++P +K+ L+ I
Sbjct: 500 RPRKRGRKPANGREEPLNHVEAERQRREKLNQRFYALRAVVPNVSKMDKASLLGDAISYI 559
Query: 202 NAL 204
N L
Sbjct: 560 NEL 562
>UniRef100_Q9ZPN6 Transcription factor MYC7E [Zea mays]
Length = 702
Score = 45.4 bits (106), Expect = 0.001
Identities = 24/63 (38%), Positives = 39/63 (61%), Gaps = 3/63 (4%)
Query: 145 RDRNQAARPSSNQ---LHHMISERRRREKLNENFQALRALLPQGTKVRTKLLISSCCFII 201
R R + +P++ + L+H+ +ER+RREKLN+ F ALRA++P +K+ L+ I
Sbjct: 507 RPRKRGRKPANGREEPLNHVEAERQRREKLNQRFYALRAVVPNVSKMDKASLLGDAISYI 566
Query: 202 NAL 204
N L
Sbjct: 567 NEL 569
>UniRef100_Q41101 Phaseolin G-box binding protein PG1 [Phaseolus vulgaris]
Length = 642
Score = 45.1 bits (105), Expect = 0.001
Identities = 24/63 (38%), Positives = 38/63 (60%), Gaps = 3/63 (4%)
Query: 145 RDRNQAARPSSNQ---LHHMISERRRREKLNENFQALRALLPQGTKVRTKLLISSCCFII 201
R R + +P + + L+H+ +ER+RREKLN+ F ALRA++P +K+ L+ I
Sbjct: 445 RPRKRGRKPGNGREEPLNHVEAERQRREKLNQRFYALRAVVPNVSKMDKASLLGDAISYI 504
Query: 202 NAL 204
N L
Sbjct: 505 NEL 507
>UniRef100_Q8S3F5 Putative bHLH transcription factor [Arabidopsis thaliana]
Length = 590
Score = 45.1 bits (105), Expect = 0.001
Identities = 24/63 (38%), Positives = 39/63 (61%), Gaps = 3/63 (4%)
Query: 145 RDRNQAARPSSNQ---LHHMISERRRREKLNENFQALRALLPQGTKVRTKLLISSCCFII 201
R R + RP++ + L+H+ +ER+RREKLN+ F ALR+++P +K+ L+ I
Sbjct: 416 RPRKRGRRPANGRAEALNHVEAERQRREKLNQRFYALRSVVPNISKMDKASLLGDAVSYI 475
Query: 202 NAL 204
N L
Sbjct: 476 NEL 478
>UniRef100_Q9LNJ5 F6F3.7 protein [Arabidopsis thaliana]
Length = 590
Score = 45.1 bits (105), Expect = 0.001
Identities = 24/63 (38%), Positives = 39/63 (61%), Gaps = 3/63 (4%)
Query: 145 RDRNQAARPSSNQ---LHHMISERRRREKLNENFQALRALLPQGTKVRTKLLISSCCFII 201
R R + RP++ + L+H+ +ER+RREKLN+ F ALR+++P +K+ L+ I
Sbjct: 416 RPRKRGRRPANGRAEALNHVEAERQRREKLNQRFYALRSVVPNISKMDKASLLGDAVSYI 475
Query: 202 NAL 204
N L
Sbjct: 476 NEL 478
>UniRef100_Q94JE7 P0684B02.21 protein [Oryza sativa]
Length = 685
Score = 44.7 bits (104), Expect = 0.002
Identities = 24/63 (38%), Positives = 40/63 (63%), Gaps = 3/63 (4%)
Query: 145 RDRNQAARPSSNQ---LHHMISERRRREKLNENFQALRALLPQGTKVRTKLLISSCCFII 201
R R + +P++ + L+H+ +ER+RREKLN+ F ALRA++P+ +K+ L+S I
Sbjct: 310 RPRKRGRKPANGREEPLNHVEAERQRREKLNQRFYALRAVVPKISKMDKASLLSDAIAYI 369
Query: 202 NAL 204
L
Sbjct: 370 QEL 372
>UniRef100_Q9LUK7 BHLH transcription factor [Arabidopsis thaliana]
Length = 511
Score = 44.7 bits (104), Expect = 0.002
Identities = 24/64 (37%), Positives = 41/64 (63%), Gaps = 4/64 (6%)
Query: 141 LMRMRDRNQAARPSSNQ---LHHMISERRRREKLNENFQALRALLPQGTKV-RTKLLISS 196
L + + + + +P+ + L+H+ +ER RREKLN F ALRA++P +K+ +T LL +
Sbjct: 322 LEKKKGKKRGRKPAHGRDKPLNHVEAERMRREKLNHRFYALRAVVPNVSKMDKTSLLEDA 381
Query: 197 CCFI 200
C+I
Sbjct: 382 VCYI 385
>UniRef100_Q948Y2 R-type basic helix-loop-helix protein [Oryza sativa]
Length = 451
Score = 44.3 bits (103), Expect = 0.002
Identities = 22/36 (61%), Positives = 26/36 (72%)
Query: 153 PSSNQLHHMISERRRREKLNENFQALRALLPQGTKV 188
P SN H++SERRRREKLNE F L++LLP KV
Sbjct: 372 PGSNIKSHVMSERRRREKLNEMFLILKSLLPSVRKV 407
>UniRef100_Q7XPS6 OSJNBa0065O17.5 protein [Oryza sativa]
Length = 265
Score = 44.3 bits (103), Expect = 0.002
Identities = 22/36 (61%), Positives = 26/36 (72%)
Query: 153 PSSNQLHHMISERRRREKLNENFQALRALLPQGTKV 188
P SN H++SERRRREKLNE F L++LLP KV
Sbjct: 186 PGSNIKSHVMSERRRREKLNEMFLILKSLLPSVRKV 221
>UniRef100_Q700B9 MYC transcription factor [Solanum tuberosum]
Length = 646
Score = 44.3 bits (103), Expect = 0.002
Identities = 23/63 (36%), Positives = 39/63 (61%), Gaps = 3/63 (4%)
Query: 145 RDRNQAARPSSNQ---LHHMISERRRREKLNENFQALRALLPQGTKVRTKLLISSCCFII 201
+ R + +P++ + L+H+ +ER+RREKLN+ F ALRA++P +K+ L+ I
Sbjct: 456 KPRKRGRKPANGREEPLNHVEAERQRREKLNQRFYALRAVVPNVSKMDKASLLGDAIAYI 515
Query: 202 NAL 204
N L
Sbjct: 516 NEL 518
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.320 0.133 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 337,455,760
Number of Sequences: 2790947
Number of extensions: 13399461
Number of successful extensions: 40793
Number of sequences better than 10.0: 320
Number of HSP's better than 10.0 without gapping: 200
Number of HSP's successfully gapped in prelim test: 121
Number of HSP's that attempted gapping in prelim test: 40532
Number of HSP's gapped (non-prelim): 374
length of query: 207
length of database: 848,049,833
effective HSP length: 122
effective length of query: 85
effective length of database: 507,554,299
effective search space: 43142115415
effective search space used: 43142115415
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 72 (32.3 bits)
Medicago: description of AC148775.1


