

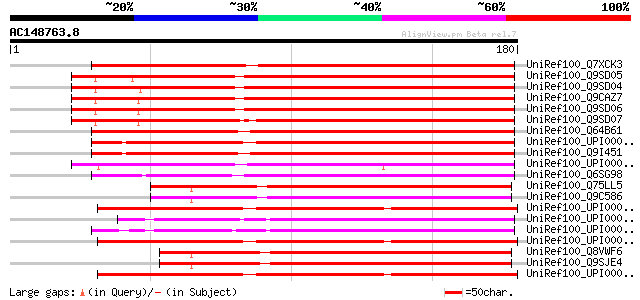
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148763.8 - phase: 1 /pseudo
(180 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q7XCK3 Mucin-like protein [Oryza sativa] 175 6e-43
UniRef100_Q9SD05 Putative mucin protein [Arabidopsis thaliana] 172 4e-42
UniRef100_Q9SD04 Mucin-like protein [Arabidopsis thaliana] 168 5e-41
UniRef100_Q9CAZ7 Strictosidine synthase-like protein [Arabidopsi... 167 9e-41
UniRef100_Q9SD06 Mucin-like protein [Arabidopsis thaliana] 167 9e-41
UniRef100_Q9SD07 Mucin-like protein [Arabidopsis thaliana] 166 3e-40
UniRef100_Q64B61 Hypothetical protein [uncultured archaeon GZfos... 130 1e-29
UniRef100_UPI00002FF7CC UPI00002FF7CC UniRef100 entry 126 2e-28
UniRef100_Q9I451 Hypothetical protein [Pseudomonas aeruginosa] 123 2e-27
UniRef100_UPI000033AA29 UPI000033AA29 UniRef100 entry 114 1e-24
UniRef100_Q6SG98 Strictosidine synthase family protein [uncultur... 105 4e-22
UniRef100_Q75LL5 Putative strictosidine synthase [Oryza sativa] 103 2e-21
UniRef100_Q9C586 Putative strictosidine synthase-like [Arabidops... 101 1e-20
UniRef100_UPI000030DF18 UPI000030DF18 UniRef100 entry 100 1e-20
UniRef100_UPI000025AAFC UPI000025AAFC UniRef100 entry 100 1e-20
UniRef100_UPI00002AD1B3 UPI00002AD1B3 UniRef100 entry 100 3e-20
UniRef100_UPI0000321BB3 UPI0000321BB3 UniRef100 entry 99 4e-20
UniRef100_Q8VWF6 Hypothetical protein At1g08470 [Arabidopsis tha... 99 4e-20
UniRef100_Q9SJE4 T27G7.16 [Arabidopsis thaliana] 99 4e-20
UniRef100_UPI000030FF39 UPI000030FF39 UniRef100 entry 99 7e-20
>UniRef100_Q7XCK3 Mucin-like protein [Oryza sativa]
Length = 369
Score = 175 bits (443), Expect = 6e-43
Identities = 80/150 (53%), Positives = 114/150 (75%), Gaps = 4/150 (2%)
Query: 30 GCEDGWIKRITVADSVVENWVHSSGRPLGLALEKTGELIVADAHLGLLRVTQKEGKEPKV 89
GC DGW++R++V+ VE+W + GRPLG+AL G L+VADA +GLL+V+ + V
Sbjct: 84 GCGDGWVRRVSVSSGDVEDWARTGGRPLGVALTADGGLVVADADIGLLKVSPDKA----V 139
Query: 90 EILANEHDGLKFKLTDGVDVGEDGTIYFTEATYKYNLYDFYNDILEGEPHGRFMSYNPAT 149
E+L +E +G+KF LTDGVDV DG IYFT+A++K++L +F D+LE PHGR MS++P+T
Sbjct: 140 ELLTDEAEGVKFALTDGVDVAGDGVIYFTDASHKHSLAEFMVDVLEARPHGRLMSFDPST 199
Query: 150 KKVTLLARNLYFANRVAIAPDQKFVVYCET 179
++ T+LAR LYFAN VA++PDQ +V+CET
Sbjct: 200 RRTTVLARGLYFANGVAVSPDQDSLVFCET 229
>UniRef100_Q9SD05 Putative mucin protein [Arabidopsis thaliana]
Length = 371
Score = 172 bits (436), Expect = 4e-42
Identities = 85/162 (52%), Positives = 118/162 (72%), Gaps = 8/162 (4%)
Query: 23 EEASYIY-GCEDGWIKRITVA----DSVVENWVHSSGRPLGLALEKTGELIVADAHLGLL 77
E++ +IY GC DGW+KR+ VA DS+VE+ V++ GRPLG+A GE+IVADA+ GLL
Sbjct: 77 EDSGFIYTGCVDGWVKRVKVAESVNDSLVEDLVNTGGRPLGIAFGIHGEVIVADAYKGLL 136
Query: 78 RVTQKEGKEPKVEILANEHDGLKFKLTDGVDVGEDGTIYFTEATYKYNLYDFYNDILEGE 137
++ G K E+L E DG++FKL D V V ++G +YFT+ +YKYNL+ F DILEG+
Sbjct: 137 NIS---GDGKKTELLTEEADGVRFKLPDAVTVADNGVLYFTDGSYKYNLHQFSFDILEGK 193
Query: 138 PHGRFMSYNPATKKVTLLARNLYFANRVAIAPDQKFVVYCET 179
PHGR MS++P TK +L R+LYFAN V+++PDQ +V+CET
Sbjct: 194 PHGRLMSFDPTTKVTRVLLRDLYFANGVSLSPDQTHLVFCET 235
>UniRef100_Q9SD04 Mucin-like protein [Arabidopsis thaliana]
Length = 371
Score = 168 bits (426), Expect = 5e-41
Identities = 82/162 (50%), Positives = 116/162 (70%), Gaps = 8/162 (4%)
Query: 23 EEASYIY-GCEDGWIKRITVADSV----VENWVHSSGRPLGLALEKTGELIVADAHLGLL 77
+E++ IY GC DGW+KR+ VADSV VE+WV++ GRPLG+A GE+IVAD H GLL
Sbjct: 77 KESNLIYTGCVDGWVKRVKVADSVNDSVVEDWVNTGGRPLGIAFGIHGEVIVADVHKGLL 136
Query: 78 RVTQKEGKEPKVEILANEHDGLKFKLTDGVDVGEDGTIYFTEATYKYNLYDFYNDILEGE 137
++ G K E+L +E DG+KFKLTD V V ++G +YFT+A+YKY L D+LEG+
Sbjct: 137 NIS---GDGKKTELLTDEADGVKFKLTDAVTVADNGVLYFTDASYKYTLNQLSLDMLEGK 193
Query: 138 PHGRFMSYNPATKKVTLLARNLYFANRVAIAPDQKFVVYCET 179
P GR +S++P T+ +L ++LYFAN + I+PDQ +++CET
Sbjct: 194 PFGRLLSFDPTTRVTKVLLKDLYFANGITISPDQTHLIFCET 235
>UniRef100_Q9CAZ7 Strictosidine synthase-like protein [Arabidopsis thaliana]
Length = 371
Score = 167 bits (424), Expect = 9e-41
Identities = 81/162 (50%), Positives = 120/162 (74%), Gaps = 8/162 (4%)
Query: 23 EEASYIY-GCEDGWIKRITVADS----VVENWVHSSGRPLGLALEKTGELIVADAHLGLL 77
++++ IY GC DGW+KR++V DS VVE+WV++ GRPLG+A GE+IVADA+ GLL
Sbjct: 77 QDSNLIYTGCIDGWVKRVSVHDSANDSVVEDWVNTGGRPLGIAFGVHGEVIVADAYKGLL 136
Query: 78 RVTQKEGKEPKVEILANEHDGLKFKLTDGVDVGEDGTIYFTEATYKYNLYDFYNDILEGE 137
++ G K E+L ++ +G+KFKLTD V V ++G +YFT+A+YKY L+ DILEG+
Sbjct: 137 NIS---GDGKKTELLTDQAEGVKFKLTDVVAVADNGVLYFTDASYKYTLHQVKFDILEGK 193
Query: 138 PHGRFMSYNPATKKVTLLARNLYFANRVAIAPDQKFVVYCET 179
PHGR MS++P T+ +L ++LYFAN V+++PDQ +++CET
Sbjct: 194 PHGRLMSFDPTTRVTRVLLKDLYFANGVSMSPDQTHLIFCET 235
>UniRef100_Q9SD06 Mucin-like protein [Arabidopsis thaliana]
Length = 367
Score = 167 bits (424), Expect = 9e-41
Identities = 81/162 (50%), Positives = 120/162 (74%), Gaps = 8/162 (4%)
Query: 23 EEASYIY-GCEDGWIKRITVADS----VVENWVHSSGRPLGLALEKTGELIVADAHLGLL 77
++++ IY GC DGW+KR++V DS VVE+WV++ GRPLG+A GE+IVADA+ GLL
Sbjct: 77 QDSNLIYTGCIDGWVKRVSVHDSANDSVVEDWVNTGGRPLGIAFGVHGEVIVADAYKGLL 136
Query: 78 RVTQKEGKEPKVEILANEHDGLKFKLTDGVDVGEDGTIYFTEATYKYNLYDFYNDILEGE 137
++ G K E+L ++ +G+KFKLTD V V ++G +YFT+A+YKY L+ DILEG+
Sbjct: 137 NIS---GDGKKTELLTDQAEGVKFKLTDVVAVADNGVLYFTDASYKYTLHQVKFDILEGK 193
Query: 138 PHGRFMSYNPATKKVTLLARNLYFANRVAIAPDQKFVVYCET 179
PHGR MS++P T+ +L ++LYFAN V+++PDQ +++CET
Sbjct: 194 PHGRLMSFDPTTRVTRVLLKDLYFANGVSMSPDQTHLIFCET 235
>UniRef100_Q9SD07 Mucin-like protein [Arabidopsis thaliana]
Length = 370
Score = 166 bits (420), Expect = 3e-40
Identities = 81/162 (50%), Positives = 119/162 (73%), Gaps = 8/162 (4%)
Query: 23 EEASYIY-GCEDGWIKRITVADS----VVENWVHSSGRPLGLALEKTGELIVADAHLGLL 77
++++ IY GC DGW+KR++V DS +VE+WV++ GRPLG+A GE+IVADA+ GLL
Sbjct: 77 KDSNLIYTGCVDGWVKRVSVHDSANDSIVEDWVNTGGRPLGIAFGLHGEVIVADANKGLL 136
Query: 78 RVTQKEGKEPKVEILANEHDGLKFKLTDGVDVGEDGTIYFTEATYKYNLYDFYNDILEGE 137
++ GK K E+L +E DG++FKLTD V V ++G +YFT+A+ KY+ Y F D LEG+
Sbjct: 137 SISDG-GK--KTELLTDEADGVRFKLTDAVTVADNGVLYFTDASSKYDFYQFIFDFLEGK 193
Query: 138 PHGRFMSYNPATKKVTLLARNLYFANRVAIAPDQKFVVYCET 179
PHGR MS++P T+ +L ++LYFAN ++++PDQ V+CET
Sbjct: 194 PHGRVMSFDPTTRATRVLLKDLYFANGISMSPDQTHFVFCET 235
>UniRef100_Q64B61 Hypothetical protein [uncultured archaeon GZfos27E7]
Length = 352
Score = 130 bits (328), Expect = 1e-29
Identities = 65/150 (43%), Positives = 94/150 (62%), Gaps = 4/150 (2%)
Query: 30 GCEDGWIKRITVADSVVENWVHSSGRPLGLALEKTGELIVADAHLGLLRVTQKEGKEPKV 89
G EDG I R S E + + GRPLGL + G L+V DA+ GLL +T + +
Sbjct: 69 GMEDGRIMRFQADGSQHEVFADTEGRPLGLHFDAAGNLVVCDAYKGLLSITP----DGSI 124
Query: 90 EILANEHDGLKFKLTDGVDVGEDGTIYFTEATYKYNLYDFYNDILEGEPHGRFMSYNPAT 149
+L+ E G+ F+LTD VD+ DG IYF++A++K+ + D++E P+GR ++YNP+T
Sbjct: 125 IVLSTEQGGVPFRLTDDVDIAADGIIYFSDASFKFTEAEHMADLMEHRPNGRLLAYNPST 184
Query: 150 KKVTLLARNLYFANRVAIAPDQKFVVYCET 179
K L+ NLYFAN VA++PDQ FV+ ET
Sbjct: 185 KTTRLVLNNLYFANGVAVSPDQSFVLVVET 214
>UniRef100_UPI00002FF7CC UPI00002FF7CC UniRef100 entry
Length = 307
Score = 126 bits (317), Expect = 2e-28
Identities = 63/150 (42%), Positives = 100/150 (66%), Gaps = 5/150 (3%)
Query: 30 GCEDGWIKRITVADSVVENWVHSSGRPLGLALEKTGELIVADAHLGLLRVTQKEGKEPKV 89
G +DG I RI DS VE+W+ + GRPLGL + +G LIV DA+LGLL ++ +V
Sbjct: 64 GTQDGKIIRIH-PDSTVEDWITTGGRPLGLHFDSSGNLIVCDAYLGLLSISPNA----EV 118
Query: 90 EILANEHDGLKFKLTDGVDVGEDGTIYFTEATYKYNLYDFYNDILEGEPHGRFMSYNPAT 149
+L +E DG+ D +D+ DG I+FT+A++++N ++ D+LEG P+GRF+ Y+P+T
Sbjct: 119 TVLLDEVDGVPLGFADDLDIAVDGRIFFTDASFRWNQANYMLDLLEGRPYGRFIVYDPST 178
Query: 150 KKVTLLARNLYFANRVAIAPDQKFVVYCET 179
+ +L ++LYF N VA++ ++FV+ ET
Sbjct: 179 DEAEVLLKDLYFPNGVALSSSEEFVLINET 208
>UniRef100_Q9I451 Hypothetical protein [Pseudomonas aeruginosa]
Length = 353
Score = 123 bits (309), Expect = 2e-27
Identities = 65/150 (43%), Positives = 95/150 (63%), Gaps = 5/150 (3%)
Query: 30 GCEDGWIKRITVADSVVENWVHSSGRPLGLALEKTGELIVADAHLGLLRVTQKEGKEPKV 89
G DG + R+ A VE +V + GRPLG+ + G LI+ADA GLLR+ + KV
Sbjct: 77 GLADGRVVRLD-ASGKVETFVDTGGRPLGMDFDAAGNLILADAWKGLLRIDP----QGKV 131
Query: 90 EILANEHDGLKFKLTDGVDVGEDGTIYFTEATYKYNLYDFYNDILEGEPHGRFMSYNPAT 149
E LA E D + F TD +D+ DG IYF++A+ K++ D+ D+LE PHGR + Y+P+T
Sbjct: 132 ETLATEADSVPFAFTDDLDIASDGRIYFSDASSKFHQPDYILDLLEARPHGRLLRYDPST 191
Query: 150 KKVTLLARNLYFANRVAIAPDQKFVVYCET 179
K +L ++LYFAN VA++ ++ FV+ ET
Sbjct: 192 GKTEVLLKDLYFANGVALSANEDFVLVNET 221
>UniRef100_UPI000033AA29 UPI000033AA29 UniRef100 entry
Length = 361
Score = 114 bits (285), Expect = 1e-24
Identities = 67/165 (40%), Positives = 95/165 (56%), Gaps = 12/165 (7%)
Query: 23 EEASYIYG-CEDGWIKRITVADSVVENWVHSSGRPLGLALEKTGELIVADAHLGLLRVTQ 81
++ IYG +DG I R ++ + + GRPLGL + G LIVADA GLL +
Sbjct: 68 DQQGIIYGGFDDGKIIRFRSNGDIIGVFADTKGRPLGLDFDYAGNLIVADAKKGLLSID- 126
Query: 82 KEGKEPKVEILANEHDGLKFKLTDGVDVGEDGTIYFTEATYKYNLYDFYN-------DIL 134
K + LA E +G TD VDVG DG IYFT+A+ K+++ DF D+
Sbjct: 127 ---KNGSIITLATEINGTPLGFTDDVDVGPDGMIYFTDASEKFSVIDFPTSMSASRADLW 183
Query: 135 EGEPHGRFMSYNPATKKVTLLARNLYFANRVAIAPDQKFVVYCET 179
E P+G+ + YNP TK+ TLL NLYFAN +A++ D +F+++ ET
Sbjct: 184 EHRPYGKIIRYNPYTKESTLLLDNLYFANGIAVSNDGQFLLFNET 228
>UniRef100_Q6SG98 Strictosidine synthase family protein [uncultured bacterium 561]
Length = 358
Score = 105 bits (263), Expect = 4e-22
Identities = 55/150 (36%), Positives = 88/150 (58%), Gaps = 5/150 (3%)
Query: 30 GCEDGWIKRITVADSVVENWVHSSGRPLGLALEKTGELIVADAHLGLLRVTQKEGKEPKV 89
G +DG I R + + GRPLG+ ++ G LIVADA+ GLL + G++ +V
Sbjct: 79 GLDDGRIVRFNDQGETAL-FAQTEGRPLGMIFDQQGALIVADAYKGLLTI----GRDGQV 133
Query: 90 EILANEHDGLKFKLTDGVDVGEDGTIYFTEATYKYNLYDFYNDILEGEPHGRFMSYNPAT 149
E L ++++G + + D VD+ +GTIYF++A+ + ++ D EG GR +Y+P T
Sbjct: 134 ETLVDQYEGRRLRFVDDVDIASNGTIYFSDASMGFEFHNNLLDFYEGSMTGRLFAYSPQT 193
Query: 150 KKVTLLARNLYFANRVAIAPDQKFVVYCET 179
+ + LL L+FAN VA+ PD +V+ ET
Sbjct: 194 QSIELLLDGLFFANGVALGPDDAYVLINET 223
>UniRef100_Q75LL5 Putative strictosidine synthase [Oryza sativa]
Length = 480
Score = 103 bits (257), Expect = 2e-21
Identities = 52/129 (40%), Positives = 79/129 (60%), Gaps = 4/129 (3%)
Query: 51 HSSGRPLGLALEK-TGELIVADAHLGLLRVTQKEGKEPKVEILANEHDGLKFKLTDGVDV 109
H GR LGL ++ TG+L +ADA+ GLL+V G LA E +G++F T+ +D+
Sbjct: 214 HICGRALGLRFDRRTGDLYIADAYFGLLKVGPDGGLATP---LATEAEGVRFNFTNDLDL 270
Query: 110 GEDGTIYFTEATYKYNLYDFYNDILEGEPHGRFMSYNPATKKVTLLARNLYFANRVAIAP 169
+DG +YFT+++ Y F + G+P GR + Y+P TKK T+L RN+ F N V+++
Sbjct: 271 DDDGNVYFTDSSIHYQRRHFMQLVFSGDPSGRLLKYDPNTKKATVLHRNIQFPNGVSMSK 330
Query: 170 DQKFVVYCE 178
D F V+CE
Sbjct: 331 DGLFFVFCE 339
>UniRef100_Q9C586 Putative strictosidine synthase-like [Arabidopsis thaliana]
Length = 395
Score = 101 bits (251), Expect = 1e-20
Identities = 51/129 (39%), Positives = 77/129 (59%), Gaps = 4/129 (3%)
Query: 51 HSSGRPLGLALEK-TGELIVADAHLGLLRVTQKEGKEPKVEILANEHDGLKFKLTDGVDV 109
H GRPLGL +K TG+L +ADA++GLL+V + G L E +G+ T+ +D+
Sbjct: 133 HICGRPLGLRFDKRTGDLYIADAYMGLLKVGPEGGLATP---LVTEAEGVPLGFTNDLDI 189
Query: 110 GEDGTIYFTEATYKYNLYDFYNDILEGEPHGRFMSYNPATKKVTLLARNLYFANRVAIAP 169
+DGT+YFT+++ Y +F + G+ GR + Y+P KK +L NL F N V+I+
Sbjct: 190 ADDGTVYFTDSSISYQRRNFLQLVFSGDNTGRVLKYDPVAKKAVVLVSNLQFPNGVSISR 249
Query: 170 DQKFVVYCE 178
D F V+CE
Sbjct: 250 DGSFFVFCE 258
>UniRef100_UPI000030DF18 UPI000030DF18 UniRef100 entry
Length = 199
Score = 100 bits (250), Expect = 1e-20
Identities = 58/149 (38%), Positives = 92/149 (60%), Gaps = 6/149 (4%)
Query: 32 EDGWIKRITVADSVVENWVHSSGRPLGLALEKTGELIVADAHLGLLRVTQKEGKEPKVEI 91
EDG I ++ + ++ + GRPLGL + G LIVADA+ GLL + ++
Sbjct: 21 EDGRIIVLSRDGTNPYDFAQTGGRPLGLKFDSQGNLIVADAYEGLLSIDTTG----EITT 76
Query: 92 LANEHDGLKFKLTDGVDVGEDGTIYFTEATYKYNLYDFYNDILEGEPHGRFMSYNPATKK 151
L E + L F LTD +D+ +G IYF++A+ NL + +D GEP+GR ++Y+P T++
Sbjct: 77 LTTECNSLPFLLTDDLDISSNGIIYFSDASSVNNLANNGDDW--GEPNGRLLAYDPQTEE 134
Query: 152 VTLLARNLYFANRVAIAPDQKFVVYCETA 180
+LL +LYFAN VA+APD+ +V+ E++
Sbjct: 135 TSLLLDSLYFANGVALAPDESYVLVNESS 163
>UniRef100_UPI000025AAFC UPI000025AAFC UniRef100 entry
Length = 361
Score = 100 bits (250), Expect = 1e-20
Identities = 57/141 (40%), Positives = 83/141 (58%), Gaps = 5/141 (3%)
Query: 39 ITVADSVVENWVHSSGRPLGLALEKTGELIVADAHLGLLRVTQKEGKEPKVEILANEHDG 98
I +D + N + GRPLG+A + LIVADA GLL + G + I ++ D
Sbjct: 93 IMSSDGIFSN---TEGRPLGMAFDLEDNLIVADAIQGLLLIDMS-GTVSTLSI-KSDSDN 147
Query: 99 LKFKLTDGVDVGEDGTIYFTEATYKYNLYDFYNDILEGEPHGRFMSYNPATKKVTLLARN 158
+ FK D +D+ DG IYF++A+ KY ++ E P+GR + Y+PATKK T L ++
Sbjct: 148 IPFKFVDDLDIASDGKIYFSDASSKYGYGSDRLELFEHSPNGRLLVYDPATKKTTTLLKD 207
Query: 159 LYFANRVAIAPDQKFVVYCET 179
LYFAN VA++ D+ FV+ ET
Sbjct: 208 LYFANGVALSSDESFVLVNET 228
>UniRef100_UPI00002AD1B3 UPI00002AD1B3 UniRef100 entry
Length = 235
Score = 99.8 bits (247), Expect = 3e-20
Identities = 57/150 (38%), Positives = 89/150 (59%), Gaps = 8/150 (5%)
Query: 30 GCEDGWIKRITVADSVVENWVHSSGRPLGLALEKTGELIVADAHLGLLRVTQKEGKEPKV 89
G +DG I + D + + + GRPLG+A + LIVADA GL+ + G+ +
Sbjct: 87 GYDDGVIMNV---DGIFSD---TKGRPLGMAFDSNDNLIVADAIQGLISI-DGSGEVTSL 139
Query: 90 EILANEHDGLKFKLTDGVDVGEDGTIYFTEATYKYNLYDFYNDILEGEPHGRFMSYNPAT 149
I ++ D + FK D +D+ DG IYF++A+ KY + ++LE +GR + Y+PAT
Sbjct: 140 SI-KSDSDNIPFKFVDDLDIASDGKIYFSDASSKYGYGNDRLELLEHSANGRLLVYDPAT 198
Query: 150 KKVTLLARNLYFANRVAIAPDQKFVVYCET 179
+K T L ++LYFAN VA++ D+ FV+ ET
Sbjct: 199 QKTTTLLKDLYFANGVALSSDESFVLVNET 228
>UniRef100_UPI0000321BB3 UPI0000321BB3 UniRef100 entry
Length = 379
Score = 99.4 bits (246), Expect = 4e-20
Identities = 57/149 (38%), Positives = 91/149 (60%), Gaps = 6/149 (4%)
Query: 32 EDGWIKRITVADSVVENWVHSSGRPLGLALEKTGELIVADAHLGLLRVTQKEGKEPKVEI 91
EDG I ++ + ++ + GRPLGL + G LIVADA+ GLL + ++
Sbjct: 27 EDGRIIVLSRDGTNPYDFAQTGGRPLGLKFDSQGNLIVADAYEGLLSIDTTG----EITT 82
Query: 92 LANEHDGLKFKLTDGVDVGEDGTIYFTEATYKYNLYDFYNDILEGEPHGRFMSYNPATKK 151
L E + L F LTD +D+ +G IYF++A+ NL + +D GEP+GR ++Y+P T++
Sbjct: 83 LTTECNSLPFLLTDDLDISSNGIIYFSDASSVNNLANNGDDW--GEPNGRLLAYDPQTEE 140
Query: 152 VTLLARNLYFANRVAIAPDQKFVVYCETA 180
+LL +LYFAN V +APD+ +V+ E++
Sbjct: 141 TSLLLDSLYFANGVVLAPDESYVLVNESS 169
>UniRef100_Q8VWF6 Hypothetical protein At1g08470 [Arabidopsis thaliana]
Length = 390
Score = 99.4 bits (246), Expect = 4e-20
Identities = 49/126 (38%), Positives = 77/126 (60%), Gaps = 4/126 (3%)
Query: 54 GRPLGLALEK-TGELIVADAHLGLLRVTQKEGKEPKVEILANEHDGLKFKLTDGVDVGED 112
GRPLGL +K G+L +ADA+LG+++V + G V NE DG+ + T+ +D+ ++
Sbjct: 130 GRPLGLRFDKKNGDLYIADAYLGIMKVGPEGGLATSV---TNEADGVPLRFTNDLDIDDE 186
Query: 113 GTIYFTEATYKYNLYDFYNDILEGEPHGRFMSYNPATKKVTLLARNLYFANRVAIAPDQK 172
G +YFT+++ + F I+ GE GR + YNP TK+ T L RNL F N +++ D
Sbjct: 187 GNVYFTDSSSFFQRRKFMLLIVSGEDSGRVLKYNPKTKETTTLVRNLQFPNGLSLGKDGS 246
Query: 173 FVVYCE 178
F ++CE
Sbjct: 247 FFIFCE 252
>UniRef100_Q9SJE4 T27G7.16 [Arabidopsis thaliana]
Length = 421
Score = 99.4 bits (246), Expect = 4e-20
Identities = 49/126 (38%), Positives = 77/126 (60%), Gaps = 4/126 (3%)
Query: 54 GRPLGLALEK-TGELIVADAHLGLLRVTQKEGKEPKVEILANEHDGLKFKLTDGVDVGED 112
GRPLGL +K G+L +ADA+LG+++V + G V NE DG+ + T+ +D+ ++
Sbjct: 130 GRPLGLRFDKKNGDLYIADAYLGIMKVGPEGGLATSV---TNEADGVPLRFTNDLDIDDE 186
Query: 113 GTIYFTEATYKYNLYDFYNDILEGEPHGRFMSYNPATKKVTLLARNLYFANRVAIAPDQK 172
G +YFT+++ + F I+ GE GR + YNP TK+ T L RNL F N +++ D
Sbjct: 187 GNVYFTDSSSFFQRRKFMLLIVSGEDSGRVLKYNPKTKETTTLVRNLQFPNGLSLGKDGS 246
Query: 173 FVVYCE 178
F ++CE
Sbjct: 247 FFIFCE 252
>UniRef100_UPI000030FF39 UPI000030FF39 UniRef100 entry
Length = 218
Score = 98.6 bits (244), Expect = 7e-20
Identities = 57/149 (38%), Positives = 90/149 (60%), Gaps = 6/149 (4%)
Query: 32 EDGWIKRITVADSVVENWVHSSGRPLGLALEKTGELIVADAHLGLLRVTQKEGKEPKVEI 91
EDG I ++ + ++ + GRPLGL + G LIVADA+ GLL + ++
Sbjct: 21 EDGRIIVLSRDGTNPYDFAQTGGRPLGLKFDSQGNLIVADAYEGLLSIDTTG----EITT 76
Query: 92 LANEHDGLKFKLTDGVDVGEDGTIYFTEATYKYNLYDFYNDILEGEPHGRFMSYNPATKK 151
L E + L F LTD +D+ +G IYF++A+ NL + +D GEP+GR ++Y+P T +
Sbjct: 77 LTTECNSLPFLLTDDLDISSNGIIYFSDASSVNNLANNGDDW--GEPNGRLLAYDPQTDE 134
Query: 152 VTLLARNLYFANRVAIAPDQKFVVYCETA 180
+LL +LYFAN V +APD+ +V+ E++
Sbjct: 135 TSLLLDSLYFANGVVLAPDESYVLVNESS 163
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.330 0.148 0.475
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 303,727,385
Number of Sequences: 2790947
Number of extensions: 12400076
Number of successful extensions: 36018
Number of sequences better than 10.0: 216
Number of HSP's better than 10.0 without gapping: 97
Number of HSP's successfully gapped in prelim test: 119
Number of HSP's that attempted gapping in prelim test: 35741
Number of HSP's gapped (non-prelim): 229
length of query: 180
length of database: 848,049,833
effective HSP length: 119
effective length of query: 61
effective length of database: 515,927,140
effective search space: 31471555540
effective search space used: 31471555540
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.8 bits)
S2: 70 (31.6 bits)
Medicago: description of AC148763.8


