

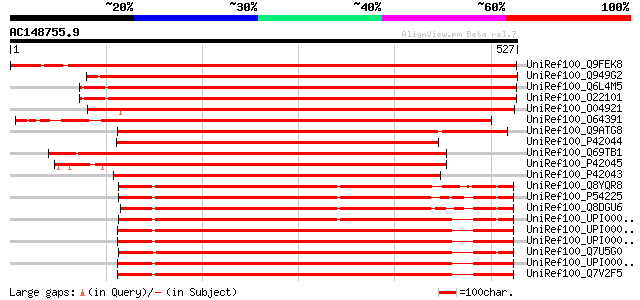
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148755.9 + phase: 0
(527 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9FEK8 Ferrochelatase [Cucumis sativus] 827 0.0
UniRef100_Q949G2 Ferrochelatase [Nicotiana tabacum] 780 0.0
UniRef100_Q6L4M5 Putative ferrochelatase II [Oryza sativa] 773 0.0
UniRef100_O22101 Ferrochelatase II, chloroplast precursor [Oryza... 773 0.0
UniRef100_O04921 Ferrochelatase II, chloroplast precursor [Arabi... 755 0.0
UniRef100_O64391 Ferrochelatase [Solanum tuberosum] 729 0.0
UniRef100_Q9ATG8 Ferrochelatase [Chlamydomonas reinhardtii] 576 e-163
UniRef100_P42044 Ferrochelatase II, chloroplast precursor [Cucum... 522 e-147
UniRef100_Q69TB1 Putative ferrochelatase [Oryza sativa] 518 e-145
UniRef100_P42045 Ferrochelatase II, chloroplast precursor [Horde... 513 e-144
UniRef100_P42043 Ferrochelatase I, chloroplast/mitochondrial pre... 512 e-143
UniRef100_Q8YQR8 Ferrochelatase [Anabaena sp.] 446 e-124
UniRef100_P54225 Ferrochelatase [Synechocystis sp.] 433 e-120
UniRef100_Q8DGU6 Ferrochelatase [Synechococcus elongatus] 427 e-118
UniRef100_UPI0000283E7C UPI0000283E7C UniRef100 entry 417 e-115
UniRef100_UPI00002FD15D UPI00002FD15D UniRef100 entry 414 e-114
UniRef100_UPI000031F6DF UPI000031F6DF UniRef100 entry 412 e-114
UniRef100_Q7U5G0 Ferrochelatase [Synechococcus sp.] 411 e-113
UniRef100_UPI00002B771B UPI00002B771B UniRef100 entry 410 e-113
UniRef100_Q7V2F5 Ferrochelatase [Prochlorococcus marinus subsp. ... 409 e-113
>UniRef100_Q9FEK8 Ferrochelatase [Cucumis sativus]
Length = 522
Score = 827 bits (2135), Expect = 0.0
Identities = 416/526 (79%), Positives = 462/526 (87%), Gaps = 6/526 (1%)
Query: 1 MNSPIRAPSTSSCSYIRPHSCRTCASRNFKLPLLLPQAICTAQKLHRCSGGHTEASSNVN 60
M+S I+A S+ + S C T AS+N K PL P + K HR H +A
Sbjct: 1 MDSAIQASSSPASSSAFRSPCLTSASQNCKFPL--PTSRVVGSKRHRAFRLHMDAC---- 54
Query: 61 PLKTCIVGKFSPGWSEAQPLVSKQSLNRHLLPVEALVTSTTQDVSDTPLIGDDKIGVLLL 120
P K +V ++S ++Q + SK+S+N+ P ALV S TQ+ S PLIG+DK+GVLLL
Sbjct: 55 PTKCHVVSRYSFELPDSQSIFSKKSINKFFPPPRALVASNTQNTSAAPLIGEDKVGVLLL 114
Query: 121 NLGGPETLDDVQPFLFNLFADPDIIRLPRLFSFLQKPLAQFVSVLRAPKSKEGYASIGGG 180
NLGGPETLDDVQPFLFNLFADPDIIRLPRLF FLQ+PLA+F+SVLR+PKS+EGYASIGGG
Sbjct: 115 NLGGPETLDDVQPFLFNLFADPDIIRLPRLFRFLQRPLARFISVLRSPKSREGYASIGGG 174
Query: 181 SPLRRMTDAQAEELRKSLFEKNVPANVYVGMRYWHPFTEEAIELIKRDGITKLVVLPLYP 240
SPLR++TDAQAEEL+K+L++K+VPA VYVGMRYWHPFTEEAIE IK+DGI+KLVVLPLYP
Sbjct: 175 SPLRKITDAQAEELKKALWQKDVPAEVYVGMRYWHPFTEEAIEQIKKDGISKLVVLPLYP 234
Query: 241 QFSISTSGSSLRLLESIFREDEYLVNMQHTVIPSWYQREGYIKAMANLIEKELKGFDLPE 300
QFSISTSGSSLRLLE IFREDEYLVNMQHTVIPSWYQREGYIKAMA+LIEKELK FD PE
Sbjct: 235 QFSISTSGSSLRLLEGIFREDEYLVNMQHTVIPSWYQREGYIKAMADLIEKELKTFDFPE 294
Query: 301 KVMIFFSAHGVPVAYVEEAGDPYKAEMEECVDLIMEELEKRKITNAYTLAYQSRVGPVEW 360
+VM+FFSAHGVP+AYVEEAGDPYKAEMEECVDLIMEELEKR+ITN+YTLAYQSRVGPVEW
Sbjct: 295 QVMVFFSAHGVPLAYVEEAGDPYKAEMEECVDLIMEELEKRRITNSYTLAYQSRVGPVEW 354
Query: 361 LKPYTDETIIELGKKGVKSLLAVPISFVSEHIETLEEIDVEYKELALESGIENWGRVPAL 420
LKPYTDETIIELG+KGVKSLLAVPISFVSEHIETLEEIDVEYKELAL+SGIE WGRVPAL
Sbjct: 355 LKPYTDETIIELGQKGVKSLLAVPISFVSEHIETLEEIDVEYKELALKSGIEKWGRVPAL 414
Query: 421 GCEPTFISDLADAVIESLPYVGAMAVSNLEARQSLVPLGSVEELLAAYDSQRRELPPPIL 480
GCEPTFI+DLADAVIESLPYVGAMAVSNLEARQ LVPLGSVEELLAAYDSQRR+LPPP+
Sbjct: 415 GCEPTFITDLADAVIESLPYVGAMAVSNLEARQPLVPLGSVEELLAAYDSQRRQLPPPVT 474
Query: 481 VWEWGWTKSAETWNGRAAMIAVLLLLFLEVTTGEGFLHQWGILPLF 526
VWEWGWTKSAETWNGRAAM+AVL+LL LEVTTGEGFLHQWGI PLF
Sbjct: 475 VWEWGWTKSAETWNGRAAMLAVLVLLVLEVTTGEGFLHQWGIFPLF 520
>UniRef100_Q949G2 Ferrochelatase [Nicotiana tabacum]
Length = 497
Score = 780 bits (2014), Expect = 0.0
Identities = 393/450 (87%), Positives = 419/450 (92%), Gaps = 3/450 (0%)
Query: 80 LVSKQSLNRHLLPVEALVTSTTQDVSDTPL--IGDDKIGVLLLNLGGPETLDDVQPFLFN 137
+ ++ SL ++P ALVTS Q VS P+ + D KIGVLLLNLGGPE+L+DVQPFLFN
Sbjct: 49 VAARCSLKPSMMP-SALVTSRPQQVSTDPVNVVCDGKIGVLLLNLGGPESLEDVQPFLFN 107
Query: 138 LFADPDIIRLPRLFSFLQKPLAQFVSVLRAPKSKEGYASIGGGSPLRRMTDAQAEELRKS 197
LFADPDIIRLPRLF FLQ+PLAQF+SV RAPKSKEGYASIGGGSPLRR+TDAQAE LRK+
Sbjct: 108 LFADPDIIRLPRLFRFLQRPLAQFISVARAPKSKEGYASIGGGSPLRRITDAQAEALRKA 167
Query: 198 LFEKNVPANVYVGMRYWHPFTEEAIELIKRDGITKLVVLPLYPQFSISTSGSSLRLLESI 257
L E+NVPA VYVGMRYWHPFTEEAIELIKRDGITKLVVLPLYPQFSISTSGSSLRLLESI
Sbjct: 168 LRERNVPAKVYVGMRYWHPFTEEAIELIKRDGITKLVVLPLYPQFSISTSGSSLRLLESI 227
Query: 258 FREDEYLVNMQHTVIPSWYQREGYIKAMANLIEKELKGFDLPEKVMIFFSAHGVPVAYVE 317
FREDEYLVNMQHTVIPSWYQREGYIKAMA+L+EKELK F+ P +VMIFFSAHGVP+AYVE
Sbjct: 228 FREDEYLVNMQHTVIPSWYQREGYIKAMADLMEKELKSFERPGEVMIFFSAHGVPLAYVE 287
Query: 318 EAGDPYKAEMEECVDLIMEELEKRKITNAYTLAYQSRVGPVEWLKPYTDETIIELGKKGV 377
EAGDPYKAEMEECVDLIMEELEKR++ NAYTLAYQSRVGPVEWLKPYTDETIIELGKKGV
Sbjct: 288 EAGDPYKAEMEECVDLIMEELEKRRVYNAYTLAYQSRVGPVEWLKPYTDETIIELGKKGV 347
Query: 378 KSLLAVPISFVSEHIETLEEIDVEYKELALESGIENWGRVPALGCEPTFISDLADAVIES 437
KS+LAVPISFVSEHIETLEEIDVEYKELALESGI+NW RVPALG EPTFISDLADAVIES
Sbjct: 348 KSILAVPISFVSEHIETLEEIDVEYKELALESGIQNWARVPALGVEPTFISDLADAVIES 407
Query: 438 LPYVGAMAVSNLEARQSLVPLGSVEELLAAYDSQRRELPPPILVWEWGWTKSAETWNGRA 497
LPYVGAMAVSNLEARQSLVPLGSVEELLAAYDS RELPPP+ VWEWGWTKSAETWNGRA
Sbjct: 408 LPYVGAMAVSNLEARQSLVPLGSVEELLAAYDSHGRELPPPVTVWEWGWTKSAETWNGRA 467
Query: 498 AMIAVLLLLFLEVTTGEGFLHQWGILPLFR 527
AM+AVL+LL LEVTTGEGFLHQWGILPLFR
Sbjct: 468 AMLAVLVLLVLEVTTGEGFLHQWGILPLFR 497
>UniRef100_Q6L4M5 Putative ferrochelatase II [Oryza sativa]
Length = 526
Score = 773 bits (1995), Expect = 0.0
Identities = 392/455 (86%), Positives = 419/455 (91%), Gaps = 4/455 (0%)
Query: 73 GWSEAQPLVS-KQSLNRHLLPVEALVTSTTQDVSDTPLIGDDKIGVLLLNLGGPETLDDV 131
GWS +QPL S + L H EA++TS Q L+G++KIGVLLLNLGGPETLDDV
Sbjct: 74 GWS-SQPLSSLRHHLRVHSSASEAVLTS--QSDFTKLLVGNEKIGVLLLNLGGPETLDDV 130
Query: 132 QPFLFNLFADPDIIRLPRLFSFLQKPLAQFVSVLRAPKSKEGYASIGGGSPLRRMTDAQA 191
QPFLFNLFADPDIIRLPRLF FLQKPLAQF+SV+RAPKSKEGYASIGGGSPLR++TDAQA
Sbjct: 131 QPFLFNLFADPDIIRLPRLFRFLQKPLAQFISVVRAPKSKEGYASIGGGSPLRQITDAQA 190
Query: 192 EELRKSLFEKNVPANVYVGMRYWHPFTEEAIELIKRDGITKLVVLPLYPQFSISTSGSSL 251
E LRK+L +K++PA VYVGMRYWHPFTEEAIE IKRDGITKLVVLPLYPQFSISTSGSSL
Sbjct: 191 EALRKALCDKDIPAKVYVGMRYWHPFTEEAIEQIKRDGITKLVVLPLYPQFSISTSGSSL 250
Query: 252 RLLESIFREDEYLVNMQHTVIPSWYQREGYIKAMANLIEKELKGFDLPEKVMIFFSAHGV 311
RLLE IFREDEYLVNMQHTVIPSWYQREGYIKAMA LIEKEL+ F P+KVMIFFSAHGV
Sbjct: 251 RLLEGIFREDEYLVNMQHTVIPSWYQREGYIKAMATLIEKELRTFSEPQKVMIFFSAHGV 310
Query: 312 PVAYVEEAGDPYKAEMEECVDLIMEELEKRKITNAYTLAYQSRVGPVEWLKPYTDETIIE 371
P+AYVEEAGDPYKAEMEECVDLIMEELEKR ITN+ TLAYQSRVGPVEWL+PYTDETIIE
Sbjct: 311 PLAYVEEAGDPYKAEMEECVDLIMEELEKRGITNSCTLAYQSRVGPVEWLRPYTDETIIE 370
Query: 372 LGKKGVKSLLAVPISFVSEHIETLEEIDVEYKELALESGIENWGRVPALGCEPTFISDLA 431
LG+KGVKSLLAVPISFVSEHIETLEEIDVEYKELALESGI++WGRVPALGCEPTFI+DLA
Sbjct: 371 LGQKGVKSLLAVPISFVSEHIETLEEIDVEYKELALESGIKHWGRVPALGCEPTFITDLA 430
Query: 432 DAVIESLPYVGAMAVSNLEARQSLVPLGSVEELLAAYDSQRRELPPPILVWEWGWTKSAE 491
DAVIESLPYVGAMAVSNLEARQ LVPLGSVEELLAAYDS+R ELPPP+ VWEWGWTKSAE
Sbjct: 431 DAVIESLPYVGAMAVSNLEARQPLVPLGSVEELLAAYDSKRDELPPPVTVWEWGWTKSAE 490
Query: 492 TWNGRAAMIAVLLLLFLEVTTGEGFLHQWGILPLF 526
TWNGRAAM+AVL LL LEVTTGEGFLHQWGILPLF
Sbjct: 491 TWNGRAAMLAVLALLVLEVTTGEGFLHQWGILPLF 525
>UniRef100_O22101 Ferrochelatase II, chloroplast precursor [Oryza sativa]
Length = 494
Score = 773 bits (1995), Expect = 0.0
Identities = 392/455 (86%), Positives = 419/455 (91%), Gaps = 4/455 (0%)
Query: 73 GWSEAQPLVS-KQSLNRHLLPVEALVTSTTQDVSDTPLIGDDKIGVLLLNLGGPETLDDV 131
GWS +QPL S + L H EA++TS Q L+G++KIGVLLLNLGGPETLDDV
Sbjct: 42 GWS-SQPLSSLRHHLRVHSSASEAVLTS--QSDFTKLLVGNEKIGVLLLNLGGPETLDDV 98
Query: 132 QPFLFNLFADPDIIRLPRLFSFLQKPLAQFVSVLRAPKSKEGYASIGGGSPLRRMTDAQA 191
QPFLFNLFADPDIIRLPRLF FLQKPLAQF+SV+RAPKSKEGYASIGGGSPLR++TDAQA
Sbjct: 99 QPFLFNLFADPDIIRLPRLFRFLQKPLAQFISVVRAPKSKEGYASIGGGSPLRQITDAQA 158
Query: 192 EELRKSLFEKNVPANVYVGMRYWHPFTEEAIELIKRDGITKLVVLPLYPQFSISTSGSSL 251
E LRK+L +K++PA VYVGMRYWHPFTEEAIE IKRDGITKLVVLPLYPQFSISTSGSSL
Sbjct: 159 EALRKALCDKDIPAKVYVGMRYWHPFTEEAIEQIKRDGITKLVVLPLYPQFSISTSGSSL 218
Query: 252 RLLESIFREDEYLVNMQHTVIPSWYQREGYIKAMANLIEKELKGFDLPEKVMIFFSAHGV 311
RLLE IFREDEYLVNMQHTVIPSWYQREGYIKAMA LIEKEL+ F P+KVMIFFSAHGV
Sbjct: 219 RLLEGIFREDEYLVNMQHTVIPSWYQREGYIKAMATLIEKELRTFSEPQKVMIFFSAHGV 278
Query: 312 PVAYVEEAGDPYKAEMEECVDLIMEELEKRKITNAYTLAYQSRVGPVEWLKPYTDETIIE 371
P+AYVEEAGDPYKAEMEECVDLIMEELEKR ITN+ TLAYQSRVGPVEWL+PYTDETIIE
Sbjct: 279 PLAYVEEAGDPYKAEMEECVDLIMEELEKRGITNSCTLAYQSRVGPVEWLRPYTDETIIE 338
Query: 372 LGKKGVKSLLAVPISFVSEHIETLEEIDVEYKELALESGIENWGRVPALGCEPTFISDLA 431
LG+KGVKSLLAVPISFVSEHIETLEEIDVEYKELALESGI++WGRVPALGCEPTFI+DLA
Sbjct: 339 LGQKGVKSLLAVPISFVSEHIETLEEIDVEYKELALESGIKHWGRVPALGCEPTFITDLA 398
Query: 432 DAVIESLPYVGAMAVSNLEARQSLVPLGSVEELLAAYDSQRRELPPPILVWEWGWTKSAE 491
DAVIESLPYVGAMAVSNLEARQ LVPLGSVEELLAAYDS+R ELPPP+ VWEWGWTKSAE
Sbjct: 399 DAVIESLPYVGAMAVSNLEARQPLVPLGSVEELLAAYDSKRDELPPPVTVWEWGWTKSAE 458
Query: 492 TWNGRAAMIAVLLLLFLEVTTGEGFLHQWGILPLF 526
TWNGRAAM+AVL LL LEVTTGEGFLHQWGILPLF
Sbjct: 459 TWNGRAAMLAVLALLVLEVTTGEGFLHQWGILPLF 493
>UniRef100_O04921 Ferrochelatase II, chloroplast precursor [Arabidopsis thaliana]
Length = 512
Score = 755 bits (1949), Expect = 0.0
Identities = 382/455 (83%), Positives = 408/455 (88%), Gaps = 12/455 (2%)
Query: 82 SKQSLNRHLLPVEA-LVTSTTQDVSDTPLIGDD-----------KIGVLLLNLGGPETLD 129
S + L +H LP+ A LVTS ++S + +I D KIGVLLLNLGGPETLD
Sbjct: 56 SNRLLGKHSLPLRAALVTSNPLNISSSSVISDAISSSSVITDDAKIGVLLLNLGGPETLD 115
Query: 130 DVQPFLFNLFADPDIIRLPRLFSFLQKPLAQFVSVLRAPKSKEGYASIGGGSPLRRMTDA 189
DVQPFLFNLFADPDIIRLP +F FLQKPLAQF+SV RAPKSKEGYASIGGGSPLR +TDA
Sbjct: 116 DVQPFLFNLFADPDIIRLPPVFQFLQKPLAQFISVARAPKSKEGYASIGGGSPLRHITDA 175
Query: 190 QAEELRKSLFEKNVPANVYVGMRYWHPFTEEAIELIKRDGITKLVVLPLYPQFSISTSGS 249
QAEELRK L+EKNVPA VYVGMRYWHPFTEEAIE IKRDGITKLVVLPLYPQFSISTSGS
Sbjct: 176 QAEELRKCLWEKNVPAKVYVGMRYWHPFTEEAIEQIKRDGITKLVVLPLYPQFSISTSGS 235
Query: 250 SLRLLESIFREDEYLVNMQHTVIPSWYQREGYIKAMANLIEKELKGFDLPEKVMIFFSAH 309
SLRLLE IFREDEYLVNMQHTVIPSWYQREGYIKAMANLI+ EL F P +V+IFFSAH
Sbjct: 236 SLRLLERIFREDEYLVNMQHTVIPSWYQREGYIKAMANLIQSELGKFGSPNQVVIFFSAH 295
Query: 310 GVPVAYVEEAGDPYKAEMEECVDLIMEELEKRKITNAYTLAYQSRVGPVEWLKPYTDETI 369
GVP+AYVEEAGDPYKAEMEECVDLIMEEL+KRKITNAYTLAYQSRVGPVEWLKPYT+E I
Sbjct: 296 GVPLAYVEEAGDPYKAEMEECVDLIMEELDKRKITNAYTLAYQSRVGPVEWLKPYTEEAI 355
Query: 370 IELGKKGVKSLLAVPISFVSEHIETLEEIDVEYKELALESGIENWGRVPALGCEPTFISD 429
ELGKKGV++LLAVPISFVSEHIETLEEIDVEYKELAL+SGI+NWGRVPALG EP FISD
Sbjct: 356 TELGKKGVENLLAVPISFVSEHIETLEEIDVEYKELALKSGIKNWGRVPALGTEPMFISD 415
Query: 430 LADAVIESLPYVGAMAVSNLEARQSLVPLGSVEELLAAYDSQRRELPPPILVWEWGWTKS 489
LADAV+ESLPYVGAMAVSNLEARQSLVPLGSVEELLA YDSQRRELP P+ +WEWGWTKS
Sbjct: 416 LADAVVESLPYVGAMAVSNLEARQSLVPLGSVEELLATYDSQRRELPAPVTMWEWGWTKS 475
Query: 490 AETWNGRAAMIAVLLLLFLEVTTGEGFLHQWGILP 524
AETWNGRAAM+AVL LL LEVTTG+GFLHQWGILP
Sbjct: 476 AETWNGRAAMLAVLALLVLEVTTGKGFLHQWGILP 510
>UniRef100_O64391 Ferrochelatase [Solanum tuberosum]
Length = 543
Score = 729 bits (1882), Expect = 0.0
Identities = 383/496 (77%), Positives = 415/496 (83%), Gaps = 27/496 (5%)
Query: 7 APSTSSCSYIRPHSCRTCASRNFKLPLLLPQAICTAQKLHRCSGGHTEASSNVNPLKTC- 65
AP C I P S C S + P L CT+ + A+S + K C
Sbjct: 2 APPVIFCREISP-SPANCTSYS---PFLQLDTQCTS----------SFAASCLRLPKNCS 47
Query: 66 IVGKFSPGWSEAQPLVSKQSLNRHLLPVEALVTSTTQDVSDTPLIGDDKIGVLLLNLGGP 125
+V + S WSE +V AL+TS Q +S P+ G+ KIGVLLLNLGGP
Sbjct: 48 LVARCSLKWSEKPSIVPG-----------ALLTSNPQQLSTNPVFGEGKIGVLLLNLGGP 96
Query: 126 ETLDDVQPFLFNLFADPDIIRLPRLFSFLQKPLAQFVSVLRAPKSKEGYASIGGGSPLRR 185
ETL+DVQPFLFNLFADPDIIRLPRLF FLQ+PLAQF+SV RAPKSKEGYASIGGGSPLRR
Sbjct: 97 ETLEDVQPFLFNLFADPDIIRLPRLFRFLQRPLAQFISVARAPKSKEGYASIGGGSPLRR 156
Query: 186 MTDAQAEELRKSLFEKNVPANVYVGMRYWHPFTEEAIELIKRDGITKLVVLPLYPQFSIS 245
+TDAQAE LRK+L+EKNVPA VYVGMRYWHPFTEEAIE IKRDGITKLVVLPLYPQFSIS
Sbjct: 157 ITDAQAEALRKALWEKNVPAKVYVGMRYWHPFTEEAIEQIKRDGITKLVVLPLYPQFSIS 216
Query: 246 TSGSSLRLLESIFREDEYLVNMQHTVIPSWYQREGYIKAMANLIEKELKGFDLPEKVMIF 305
TSGSSLRLLESIFREDEYLVNMQHTVIPSWYQREGYIKAMA+L+EKELK F+ PE+VMIF
Sbjct: 217 TSGSSLRLLESIFREDEYLVNMQHTVIPSWYQREGYIKAMADLMEKELKSFEHPEEVMIF 276
Query: 306 FSAHGVPVAYVEEAGDPYKAEMEECVDLIMEELEKRKITNAYTLAYQSRVGPVEWLKPYT 365
FSAHGVP+AYVEEAGDPYKAEMEECVDLIMEELEKR++ NAYTLAYQSRVGPVEWLKPYT
Sbjct: 277 FSAHGVPLAYVEEAGDPYKAEMEECVDLIMEELEKRRVHNAYTLAYQSRVGPVEWLKPYT 336
Query: 366 DETIIELGKKGVKSLLAVP-ISFVSEHIETLEEIDVEYKELALESGIENWGRVPALGCEP 424
DETIIELGKKGVKS+LAVP ISFVSEHIETLEEIDVEYKELALESGI+NW RVPALG EP
Sbjct: 337 DETIIELGKKGVKSILAVPIISFVSEHIETLEEIDVEYKELALESGIQNWARVPALGVEP 396
Query: 425 TFISDLADAVIESLPYVGAMAVSNLEARQSLVPLGSVEELLAAYDSQRRELPPPILVWEW 484
TFISDLADAVIESLPYVGAMAVSNLEARQSLVPLGSVEELLAAYDSQRRELPPP+ VWEW
Sbjct: 397 TFISDLADAVIESLPYVGAMAVSNLEARQSLVPLGSVEELLAAYDSQRRELPPPVTVWEW 456
Query: 485 GWTKSAETWNGRAAMI 500
GWTKSAETW +++ +
Sbjct: 457 GWTKSAETWKWKSSYV 472
>UniRef100_Q9ATG8 Ferrochelatase [Chlamydomonas reinhardtii]
Length = 493
Score = 576 bits (1485), Expect = e-163
Identities = 285/405 (70%), Positives = 331/405 (81%), Gaps = 4/405 (0%)
Query: 113 DKIGVLLLNLGGPETLDDVQPFLFNLFADPDIIRLPRLFSFLQKPLAQFVSVLRAPKSKE 172
DK+GVLLLNLGGPE LDDV+PFL+NLFADP+IIRLP FLQ LA +S LRAPKS E
Sbjct: 86 DKVGVLLLNLGGPEKLDDVKPFLYNLFADPEIIRLPAAAQFLQPLLATIISTLRAPKSAE 145
Query: 173 GYASIGGGSPLRRMTDAQAEELRKSLFEKNVPANVYVGMRYWHPFTEEAIELIKRDGITK 232
GY +IGGGSPLRR+TD QAE L +SL K PANVYVGMRYWHP+TEEA+E IK DG+T+
Sbjct: 146 GYEAIGGGSPLRRITDEQAEALAESLRAKGQPANVYVGMRYWHPYTEEALEHIKADGVTR 205
Query: 233 LVVLPLYPQFSISTSGSSLRLLESIFREDEYLVNMQHTVIPSWYQREGYIKAMANLIEKE 292
LV+LPLYPQFSISTSGSSLRLLES+F+ D L +++HTVIPSWYQR GY+ AMA+LI +E
Sbjct: 206 LVILPLYPQFSISTSGSSLRLLESLFKSDIALKSLRHTVIPSWYQRRGYVSAMADLIVEE 265
Query: 293 LKGFDLPEKVMIFFSAHGVPVAYVEEAGDPYKAEMEECVDLIMEELEKRKITNAYTLAYQ 352
LK F V +FFSAHGVP +YVEEAGDPYK EMEECV LI +E+++R N +TLAYQ
Sbjct: 266 LKKFRDVPSVELFFSAHGVPKSYVEEAGDPYKEEMEECVRLITDEVKRRGFANTHTLAYQ 325
Query: 353 SRVGPVEWLKPYTDETIIELGKKGVKSLLAVPISFVSEHIETLEEIDVEYKELALESGIE 412
SRVGP EWLKPYTDE+I ELGK+GVKSLLAVPISFVSEHIETLEEID+EY+ELA ESGI
Sbjct: 326 SRVGPAEWLKPYTDESIKELGKRGVKSLLAVPISFVSEHIETLEEIDMEYRELAEESGIR 385
Query: 413 NWGRVPALGCEPTFISDLADAVIESLPYVGAMAVSNLEARQSLVPLGSVEELLAAYDSQR 472
NWGRVPAL FI DLADAV+E+LPYVG +A SLVPLG +E LL AYD +R
Sbjct: 386 NWGRVPALNTNAAFIDDLADAVMEALPYVGCLA----GPTDSLVPLGDLEMLLQAYDRER 441
Query: 473 RELPPPILVWEWGWTKSAETWNGRAAMIAVLLLLFLEVTTGEGFL 517
R LP P+++WEWGWTKSAETWNGR AMIA++++L LE +G+ L
Sbjct: 442 RTLPSPVVMWEWGWTKSAETWNGRIAMIAIIIILALEAASGQSIL 486
>UniRef100_P42044 Ferrochelatase II, chloroplast precursor [Cucumis sativus]
Length = 514
Score = 522 bits (1345), Expect = e-147
Identities = 251/334 (75%), Positives = 298/334 (89%)
Query: 112 DDKIGVLLLNLGGPETLDDVQPFLFNLFADPDIIRLPRLFSFLQKPLAQFVSVLRAPKSK 171
DDK+GVLLLNLGGPETLDDVQPFL+NLFADPDIIRLPRLF FLQ+PLA+ +S RAPKSK
Sbjct: 112 DDKVGVLLLNLGGPETLDDVQPFLYNLFADPDIIRLPRLFRFLQEPLAKLISTYRAPKSK 171
Query: 172 EGYASIGGGSPLRRMTDAQAEELRKSLFEKNVPANVYVGMRYWHPFTEEAIELIKRDGIT 231
EGYASIGGGSPLR++TD QA+ L+ +L EKN+ NVYVGMRYW+PFTEEAI+ IKRDGIT
Sbjct: 172 EGYASIGGGSPLRKITDEQAQALKMALAEKNMSTNVYVGMRYWYPFTEEAIQQIKRDGIT 231
Query: 232 KLVVLPLYPQFSISTSGSSLRLLESIFREDEYLVNMQHTVIPSWYQREGYIKAMANLIEK 291
+LVVLPLYPQ+SIST+GSS+R+L+ +FRED YL ++ ++I SWYQREGYIK+MA+L++
Sbjct: 232 RLVVLPLYPQYSISTTGSSIRVLQKMFREDAYLSSLPVSIIKSWYQREGYIKSMADLMQA 291
Query: 292 ELKGFDLPEKVMIFFSAHGVPVAYVEEAGDPYKAEMEECVDLIMEELEKRKITNAYTLAY 351
ELK F P++VMIFFSAHGVPV+YVE AGDPYK +MEEC+ LIM+EL+ R I N +TLAY
Sbjct: 292 ELKNFANPQEVMIFFSAHGVPVSYVENAGDPYKDQMEECICLIMQELKARGIGNEHTLAY 351
Query: 352 QSRVGPVEWLKPYTDETIIELGKKGVKSLLAVPISFVSEHIETLEEIDVEYKELALESGI 411
QSRVGPV+WLKPYTDE ++ELG+KG+KSLLAVP+SFVSEHIETLEEID+EYK LALESGI
Sbjct: 352 QSRVGPVQWLKPYTDEVLVELGQKGIKSLLAVPVSFVSEHIETLEEIDMEYKHLALESGI 411
Query: 412 ENWGRVPALGCEPTFISDLADAVIESLPYVGAMA 445
+NWGRVPAL C +FISDLADAVIE+LP A+A
Sbjct: 412 QNWGRVPALNCNSSFISDLADAVIEALPSATALA 445
>UniRef100_Q69TB1 Putative ferrochelatase [Oryza sativa]
Length = 482
Score = 518 bits (1334), Expect = e-145
Identities = 258/414 (62%), Positives = 320/414 (76%), Gaps = 1/414 (0%)
Query: 41 TAQKLHRCSGGHTEASSNVNPLKTCIVGKFSPGWSEAQPLVSKQSLNRHLLPVEALVTST 100
T+ L C+ NV P + G S + L +Q+L+ + T
Sbjct: 34 TSTNLAICTKHEQNLHGNVKPSQLAASGS-SYSVHRSPVLKQRQNLSARSTSADVYTTFD 92
Query: 101 TQDVSDTPLIGDDKIGVLLLNLGGPETLDDVQPFLFNLFADPDIIRLPRLFSFLQKPLAQ 160
+ + ++K+GVLLLNLGGPETLDDVQPFLFNLFADPDIIRLPRLF FLQ+PLA+
Sbjct: 93 ENVRAVSSHAAEEKVGVLLLNLGGPETLDDVQPFLFNLFADPDIIRLPRLFRFLQRPLAK 152
Query: 161 FVSVLRAPKSKEGYASIGGGSPLRRMTDAQAEELRKSLFEKNVPANVYVGMRYWHPFTEE 220
+S RAPKSKEGYASIGGGSPLR++TD QA L+ +L +KN+ AN+YVGMRYW+PFTEE
Sbjct: 153 LISTFRAPKSKEGYASIGGGSPLRKITDEQANALKVALKKKNLNANIYVGMRYWYPFTEE 212
Query: 221 AIELIKRDGITKLVVLPLYPQFSISTSGSSLRLLESIFREDEYLVNMQHTVIPSWYQREG 280
AI+ IK+D ITKLVVLPLYPQ+SISTSGSS+R+L++I +ED Y + ++I SWYQR+G
Sbjct: 213 AIDQIKKDKITKLVVLPLYPQYSISTSGSSIRVLQNIVKEDSYFAGLPISIIESWYQRDG 272
Query: 281 YIKAMANLIEKELKGFDLPEKVMIFFSAHGVPVAYVEEAGDPYKAEMEECVDLIMEELEK 340
Y+K+MA+LIEKEL F PE+VMIFFSAHGVP+ YV +AGDPY+ +ME+C+ LIM EL+
Sbjct: 273 YVKSMADLIEKELSIFSNPEEVMIFFSAHGVPLTYVTDAGDPYRDQMEDCIALIMGELKS 332
Query: 341 RKITNAYTLAYQSRVGPVEWLKPYTDETIIELGKKGVKSLLAVPISFVSEHIETLEEIDV 400
R I N++TLAYQSRVGPV+WLKPYTDE ++ELG++GVKSLLAVP+SFVSEHIETLEEID+
Sbjct: 333 RGILNSHTLAYQSRVGPVQWLKPYTDEVLVELGQQGVKSLLAVPVSFVSEHIETLEEIDM 392
Query: 401 EYKELALESGIENWGRVPALGCEPTFISDLADAVIESLPYVGAMAVSNLEARQS 454
EYKELALESGIENWGRVPALGC +FISDLADAV+E+LP A+ ++ S
Sbjct: 393 EYKELALESGIENWGRVPALGCTSSFISDLADAVVEALPSASALVTKKVDESDS 446
>UniRef100_P42045 Ferrochelatase II, chloroplast precursor [Hordeum vulgare]
Length = 484
Score = 513 bits (1321), Expect = e-144
Identities = 265/422 (62%), Positives = 326/422 (76%), Gaps = 18/422 (4%)
Query: 47 RCS---GGHTEASSNVN----PLKTCIVGKF--SPGWSEAQPLVSKQSLNRHLLPVEA-- 95
RCS G T+ N++ PL GK + G P++ Q H L V +
Sbjct: 31 RCSTNLAGSTKCEQNLHGKAKPLLLSASGKARGTSGLVHRSPVLKHQ----HHLSVRSTS 86
Query: 96 --LVTSTTQDVSDTPLIG-DDKIGVLLLNLGGPETLDDVQPFLFNLFADPDIIRLPRLFS 152
+ T+ +DV ++K+GVLLLNLGGPETL+DVQPFLFNLFADPDIIRLPRLF
Sbjct: 87 TDVCTTFDEDVKGVSSHAVEEKVGVLLLNLGGPETLNDVQPFLFNLFADPDIIRLPRLFR 146
Query: 153 FLQKPLAQFVSVLRAPKSKEGYASIGGGSPLRRMTDAQAEELRKSLFEKNVPANVYVGMR 212
FLQ+PLA+ +S RAPKS EGYASIGGGSPLR++TD QA L+ +L KN+ A++YVGMR
Sbjct: 147 FLQRPLAKLISTFRAPKSNEGYASIGGGSPLRKITDEQANALKVALKSKNLEADIYVGMR 206
Query: 213 YWHPFTEEAIELIKRDGITKLVVLPLYPQFSISTSGSSLRLLESIFREDEYLVNMQHTVI 272
YW+PFTEEAI+ IK+D ITKLVVLPLYPQ+SISTSGSS+R+L++I +ED Y + ++I
Sbjct: 207 YWYPFTEEAIDQIKKDKITKLVVLPLYPQYSISTSGSSIRVLQNIVKEDPYFAGLPISII 266
Query: 273 PSWYQREGYIKAMANLIEKELKGFDLPEKVMIFFSAHGVPVAYVEEAGDPYKAEMEECVD 332
SWYQREGY+K+MA+LIEKEL F PE+VMIFFSAHGVP+ YV++AGDPY+ +ME+C+
Sbjct: 267 ESWYQREGYVKSMADLIEKELSVFSNPEEVMIFFSAHGVPLTYVKDAGDPYRDQMEDCIA 326
Query: 333 LIMEELEKRKITNAYTLAYQSRVGPVEWLKPYTDETIIELGKKGVKSLLAVPISFVSEHI 392
LIMEEL+ R N +TLAYQSRVGPV+WLKPYTDE ++ELG+KGVKSLLAVP+SFVSEHI
Sbjct: 327 LIMEELKSRGTLNDHTLAYQSRVGPVQWLKPYTDEVLVELGQKGVKSLLAVPVSFVSEHI 386
Query: 393 ETLEEIDVEYKELALESGIENWGRVPALGCEPTFISDLADAVIESLPYVGAMAVSNLEAR 452
ETLEEID+EY+ELALESGIENWGRVPALGC +FISDLADAV+E+LP AMA ++
Sbjct: 387 ETLEEIDMEYRELALESGIENWGRVPALGCTSSFISDLADAVVEALPSASAMATRKVKDT 446
Query: 453 QS 454
S
Sbjct: 447 DS 448
>UniRef100_P42043 Ferrochelatase I, chloroplast/mitochondrial precursor [Arabidopsis
thaliana]
Length = 466
Score = 512 bits (1318), Expect = e-143
Identities = 244/340 (71%), Positives = 298/340 (86%)
Query: 109 LIGDDKIGVLLLNLGGPETLDDVQPFLFNLFADPDIIRLPRLFSFLQKPLAQFVSVLRAP 168
++ +DKIGVLLLNLGGPETL+DVQPFL+NLFADPDIIRLPR F FLQ +A+F+SV+RAP
Sbjct: 84 VVAEDKIGVLLLNLGGPETLNDVQPFLYNLFADPDIIRLPRPFQFLQGTIAKFISVVRAP 143
Query: 169 KSKEGYASIGGGSPLRRMTDAQAEELRKSLFEKNVPANVYVGMRYWHPFTEEAIELIKRD 228
KSKEGYA+IGGGSPLR++TD QA+ ++ SL KN+ ANVYVGMRYW+PFTEEA++ IK+D
Sbjct: 144 KSKEGYAAIGGGSPLRKITDEQADAIKMSLQAKNIAANVYVGMRYWYPFTEEAVQQIKKD 203
Query: 229 GITKLVVLPLYPQFSISTSGSSLRLLESIFREDEYLVNMQHTVIPSWYQREGYIKAMANL 288
IT+LVVLPLYPQ+SIST+GSS+R+L+ +FR+D YL + +I SWYQR GY+ +MA+L
Sbjct: 204 KITRLVVLPLYPQYSISTTGSSIRVLQDLFRKDPYLAGVPVAIIKSWYQRRGYVNSMADL 263
Query: 289 IEKELKGFDLPEKVMIFFSAHGVPVAYVEEAGDPYKAEMEECVDLIMEELEKRKITNAYT 348
IEKEL+ F P++VMIFFSAHGVPV+YVE AGDPY+ +MEEC+DLIMEEL+ R + N +
Sbjct: 264 IEKELQTFSDPKEVMIFFSAHGVPVSYVENAGDPYQKQMEECIDLIMEELKARGVLNDHK 323
Query: 349 LAYQSRVGPVEWLKPYTDETIIELGKKGVKSLLAVPISFVSEHIETLEEIDVEYKELALE 408
LAYQSRVGPV+WLKPYTDE +++LGK GVKSLLAVP+SFVSEHIETLEEID+EY+ELALE
Sbjct: 324 LAYQSRVGPVQWLKPYTDEVLVDLGKSGVKSLLAVPVSFVSEHIETLEEIDMEYRELALE 383
Query: 409 SGIENWGRVPALGCEPTFISDLADAVIESLPYVGAMAVSN 448
SG+ENWGRVPALG P+FI+DLADAVIESLP AM+ N
Sbjct: 384 SGVENWGRVPALGLTPSFITDLADAVIESLPSAEAMSNPN 423
>UniRef100_Q8YQR8 Ferrochelatase [Anabaena sp.]
Length = 388
Score = 446 bits (1148), Expect = e-124
Identities = 228/410 (55%), Positives = 296/410 (71%), Gaps = 25/410 (6%)
Query: 114 KIGVLLLNLGGPETLDDVQPFLFNLFADPDIIRLPRLFSFLQKPLAQFVSVLRAPKSKEG 173
++GVLLLNLGGP+ L+DV PFLFNLF+DP+IIRLP F +LQKPLA F++ R S+E
Sbjct: 3 RVGVLLLNLGGPDKLEDVAPFLFNLFSDPEIIRLP--FRWLQKPLAWFIASRRTKTSQEN 60
Query: 174 YASIGGGSPLRRMTDAQAEELRKSLFEKNVPANVYVGMRYWHPFTEEAIELIKRDGITKL 233
Y IGGGSPLRR+T+AQ E L++ L AN+YVGMRYWHP+TEEAI L+ +D + L
Sbjct: 61 YKQIGGGSPLRRITEAQGEALKEQLHYLGQEANIYVGMRYWHPYTEEAIALLTQDNLDNL 120
Query: 234 VVLPLYPQFSISTSGSSLRLLESIFREDEYLVNMQHTVIPSWYQREGYIKAMANLIEKEL 293
V+LPLYPQFSISTSGSS RLLE +++ED L +++TVIPSWY+ GY++AMA LI +E+
Sbjct: 121 VILPLYPQFSISTSGSSFRLLERLWQEDPKLQRLEYTVIPSWYKEPGYLQAMAELIRQEI 180
Query: 294 KGFDLPEKVMIFFSAHGVPVAYVEEAGDPYKAEMEECVDLIMEELEKRKITNAYTLAYQS 353
+ F P++V +FFSAHGVP +YVEEAGDPY+ E+EEC LIM+ L + N +TLAYQS
Sbjct: 181 EQFPHPDQVHVFFSAHGVPKSYVEEAGDPYQQEIEECTALIMQTLNR---PNPHTLAYQS 237
Query: 354 RVGPVEWLKPYTDETIIELGKKGVKSLLAVPISFVSEHIETLEEIDVEYKELALESGIEN 413
RVGPVEWL+PYT++ + ELG +GVK L+ VPISFVSEHIETL+EID+EY+E+A E+GI N
Sbjct: 238 RVGPVEWLQPYTEDALKELGAQGVKDLVVVPISFVSEHIETLQEIDIEYREIAEEAGIHN 297
Query: 414 WGRVPALGCEPTFISDLADAVIESLPYVGAMAVSNLEARQSLVPLGSVEELLAAYDSQRR 473
+ RVPA P FI LAD VI++L + L +++++ Y
Sbjct: 298 FRRVPAPNTHPVFIRALADLVIDALN----------KPSFKLSQAAQIKKMVKMY----- 342
Query: 474 ELPPPILVWEWGWTKSAETWNGRAAMIAVLLLLFLEVTTGEGFLHQWGIL 523
PP WEWG T SAE WNGR AM+ + L+ +E+ TG+G LH G+L
Sbjct: 343 --PPE--SWEWGMTSSAEVWNGRIAMLGFIALI-IELVTGQGLLHMIGLL 387
>UniRef100_P54225 Ferrochelatase [Synechocystis sp.]
Length = 387
Score = 433 bits (1114), Expect = e-120
Identities = 228/410 (55%), Positives = 291/410 (70%), Gaps = 25/410 (6%)
Query: 114 KIGVLLLNLGGPETLDDVQPFLFNLFADPDIIRLPRLFSFLQKPLAQFVSVLRAPKSKEG 173
++GVLLLNLGGPE L+DV+PFLFNLFADP+IIRLP F +LQKPLA +S LRA KS+
Sbjct: 3 RVGVLLLNLGGPEKLEDVRPFLFNLFADPEIIRLP--FPWLQKPLAWLISTLRAKKSQAN 60
Query: 174 YASIGGGSPLRRMTDAQAEELRKSLFEKNVPANVYVGMRYWHPFTEEAIELIKRDGITKL 233
YA IGGGSPL ++T+AQA L L A VY+GMRYWHPFTEEA+E IK D + +L
Sbjct: 61 YAEIGGGSPLLQITEAQASALTTRLERLGQDAKVYIGMRYWHPFTEEAVEKIKGDRLQRL 120
Query: 234 VVLPLYPQFSISTSGSSLRLLESIFREDEYLVNMQHTVIPSWYQREGYIKAMANLIEKEL 293
V+LPLYP FSISTSGSS R+LE ++ D L + +++IPSWY GY++AMA+LI +EL
Sbjct: 121 VILPLYPHFSISTSGSSFRVLEEMWHNDPSLRQLDYSLIPSWYDHPGYLQAMADLIAQEL 180
Query: 294 KGFDLPEKVMIFFSAHGVPVAYVEEAGDPYKAEMEECVDLIMEELEKRKITNAYTLAYQS 353
K F P++ IFFSAHGVP +YV+EAGDPY+AE+E C LIM L++ N YTLAYQS
Sbjct: 181 KKFPNPDQAHIFFSAHGVPQSYVDEAGDPYQAEIEACTRLIMRTLDR---PNQYTLAYQS 237
Query: 354 RVGPVEWLKPYTDETIIELGKKGVKSLLAVPISFVSEHIETLEEIDVEYKELALESGIEN 413
RVGPVEWLKPYT+E + +LG +G+ LL VPISFVSEHIETL+EID+EY+E+A E+GI+N
Sbjct: 238 RVGPVEWLKPYTEEALQKLGAEGIDDLLVVPISFVSEHIETLQEIDIEYREIAEEAGIDN 297
Query: 414 WGRVPALGCEPTFISDLADAVIESLPYVGAMAVSNLEARQSLVPLGSVEELLAAYDSQRR 473
+ RVPAL P FI LA V++SL ++ VP ++ + Y +R
Sbjct: 298 FQRVPALNTHPVFIDALAQMVMDSL--------NDPPCTFETVP--HPKKNMKMYPQER- 346
Query: 474 ELPPPILVWEWGWTKSAETWNGRAAMIAVLLLLFLEVTTGEGFLHQWGIL 523
WEWG T +AE WNGR AM+ + LL +E+ +G+G LH G+L
Sbjct: 347 --------WEWGLTTAAEVWNGRLAMLGFIALL-VELISGQGPLHFVGLL 387
>UniRef100_Q8DGU6 Ferrochelatase [Synechococcus elongatus]
Length = 388
Score = 427 bits (1098), Expect = e-118
Identities = 228/408 (55%), Positives = 291/408 (70%), Gaps = 25/408 (6%)
Query: 116 GVLLLNLGGPETLDDVQPFLFNLFADPDIIRLPRLFSFLQKPLAQFVSVLRAPKSKEGYA 175
GVLLLNLGGP+ +DV+PFL+NLF+DP+IIRLP F +LQKPLA F+S RA +S+ YA
Sbjct: 6 GVLLLNLGGPDRPEDVRPFLYNLFSDPEIIRLP--FRWLQKPLAWFISTSRARRSQANYA 63
Query: 176 SIGGGSPLRRMTDAQAEELRKSLFEKNVPANVYVGMRYWHPFTEEAIELIKRDGITKLVV 235
IGGGSPLRR+T+ QA L+ +L + AN+Y+GMRYWHPFTEEAI IK D I +LV+
Sbjct: 64 QIGGGSPLRRITEQQARALKDALEGIGIEANLYIGMRYWHPFTEEAIAQIKADQIRELVI 123
Query: 236 LPLYPQFSISTSGSSLRLLESIFREDEYLVNMQHTVIPSWYQREGYIKAMANLIEKELKG 295
LPLYPQFSISTSGSS RLLES++ +D L +++T+IPSWY GY+ AMA+LI +EL
Sbjct: 124 LPLYPQFSISTSGSSFRLLESLWNQDPELQKIRYTLIPSWYNHPGYVAAMADLIRQELDR 183
Query: 296 FDLPEKVMIFFSAHGVPVAYVEEAGDPYKAEMEECVDLIMEELEKRKITNAYTLAYQSRV 355
P++ +IFFSAHGVP +YV EAGDPY+ E+E CV LIM L + NA+ LAYQSRV
Sbjct: 184 CPNPDEAVIFFSAHGVPKSYVTEAGDPYQEEIEACVRLIMAALNR---PNAHVLAYQSRV 240
Query: 356 GPVEWLKPYTDETIIELGKKGVKSLLAVPISFVSEHIETLEEIDVEYKELALESGIENWG 415
GPVEWL+PYT++ I+EL +GVK+L+ VPISFVSEHIETL+EID+EY+E+A E+GIE +
Sbjct: 241 GPVEWLQPYTEDVILELAAQGVKTLVVVPISFVSEHIETLQEIDIEYREIAAEAGIEVFR 300
Query: 416 RVPALGCEPTFISDLADAVIESLPYVGAMAVSNLEARQSLVPLGSVEELLAAYDSQRREL 475
RVPAL FIS LA V E+L A + E QS + + Y +R
Sbjct: 301 RVPALNDHNGFISALAQLVKEAL---AAPPRTFAEVNQS-------RKRVKLYPQER--- 347
Query: 476 PPPILVWEWGWTKSAETWNGRAAMIAVLLLLFLEVTTGEGFLHQWGIL 523
WEWG T +AE WNGR AM+ L L+ +E+ +G+G LH G+L
Sbjct: 348 ------WEWGMTSAAERWNGRLAMLGFLALM-IELISGQGPLHMLGLL 388
>UniRef100_UPI0000283E7C UPI0000283E7C UniRef100 entry
Length = 387
Score = 417 bits (1071), Expect = e-115
Identities = 218/412 (52%), Positives = 277/412 (66%), Gaps = 29/412 (7%)
Query: 114 KIGVLLLNLGGPETLDDVQPFLFNLFADPDIIRLPRLFSFLQKPLAQFVSVLRAPKSKEG 173
++G++LLNLGGPE + DV PFL+NLF+DP+IIRLP QKPLA +S LR+ KS+E
Sbjct: 3 RVGIVLLNLGGPERIQDVGPFLYNLFSDPEIIRLP--IPAFQKPLAWLISTLRSNKSQEA 60
Query: 174 YASIGGGSPLRRMTDAQAEELRKSLFEKNVPANVYVGMRYWHPFTEEAIELIKRDGITKL 233
Y SIGGGSPLRR+TD QA EL+ L ++ V A YV MRYWHPFTE A+ +K DGI ++
Sbjct: 61 YRSIGGGSPLRRITDQQARELQSLLRQREVDATTYVAMRYWHPFTESAVADMKADGIEQV 120
Query: 234 VVLPLYPQFSISTSGSSLRLLESIFREDEYLVNMQHTVIPSWYQREGYIKAMANLIEKEL 293
VVLPLYP FSISTSGSS R L+ + + DE+ + I SW+ GY++AMA L+ +E+
Sbjct: 121 VVLPLYPHFSISTSGSSFRELQRLRQGDEHFAQLPLRAIRSWHDHPGYLQAMAQLMAREI 180
Query: 294 KGFDLPEKVMIFFSAHGVPVAYVEEAGDPYKAEMEECVDLIMEELEKRKITNAYTLAYQS 353
P +FFSAHGVP +YVEEAGDPY+ E+E C +LIM+ L + N +TLAYQS
Sbjct: 181 DACVDPSSAHVFFSAHGVPKSYVEEAGDPYQKEIESCAELIMKTLGR---DNPWTLAYQS 237
Query: 354 RVGPVEWLKPYTDETIIELGKKGVKSLLAVPISFVSEHIETLEEIDVEYKELALESGIEN 413
RVGPVEWL+PYT+E + ELG++GVK L+ VPISFVSEHIETLEEID+EY+E+A E+G+ N
Sbjct: 238 RVGPVEWLQPYTEEALEELGEQGVKELVVVPISFVSEHIETLEEIDIEYREIATEAGVSN 297
Query: 414 WGRVPALGCEPTFISDLADAVIESL--PYVGAMAVSNLEARQSLVPLGSVEELLAAYDSQ 471
+ RVPAL +PTFI+ LAD V SL P V + L AR L P
Sbjct: 298 FRRVPALDTDPTFIASLADLVETSLAGPEVDLEEAAALPARTKLYPQEK----------- 346
Query: 472 RRELPPPILVWEWGWTKSAETWNGRAAMIAVLLLLFLEVTTGEGFLHQWGIL 523
W WGW S+E WNGR AM+ L +E+ +G G LH G+L
Sbjct: 347 ----------WSWGWNNSSEVWNGRLAMLGFSAFL-MELLSGHGPLHALGLL 387
>UniRef100_UPI00002FD15D UPI00002FD15D UniRef100 entry
Length = 391
Score = 414 bits (1065), Expect = e-114
Identities = 214/414 (51%), Positives = 278/414 (66%), Gaps = 27/414 (6%)
Query: 113 DKIGVLLLNLGGPETLDDVQPFLFNLFADPDIIRLPRLFSFLQKPLAQFVSVLRAPKSKE 172
DKIGVLL+NLGGPE + DV PFL+NLF+DP+IIR P F QKPLA +S LR+ S++
Sbjct: 2 DKIGVLLMNLGGPERITDVGPFLYNLFSDPEIIRTP--FPVFQKPLAWLISTLRSTTSQQ 59
Query: 173 GYASIGGGSPLRRMTDAQAEELRKSLFEKNVPANVYVGMRYWHPFTEEAIELIKRDGITK 232
Y SIGGGSP+RR+T+ QA EL+ L +K A Y+ MRYWHPFTE AI +K DGI +
Sbjct: 60 AYLSIGGGSPIRRITEQQARELQSKLRDKGFNATTYIAMRYWHPFTESAIADMKADGIDQ 119
Query: 233 LVVLPLYPQFSISTSGSSLRLLESIFREDEYLVNMQHTVIPSWYQREGYIKAMANLIEKE 292
+VV+PLYP FSISTSGSS R L+ + D+ + + SW+ + GY+K+M LI ++
Sbjct: 120 VVVIPLYPHFSISTSGSSFRELKKLRDADDEFKKVPMRCVRSWFSQSGYLKSMVELISEQ 179
Query: 293 LKGFDLPEKVMIFFSAHGVPVAYVEEAGDPYKAEMEECVDLIMEELEK-RKITNAYTLAY 351
+ +LP K IFF+AHGVP +YVEEAGDPYK ++E+C LI+ ELEK +N +TL+Y
Sbjct: 180 ISLCELPSKAHIFFTAHGVPKSYVEEAGDPYKQQIEDCSLLIINELEKCLGHSNPHTLSY 239
Query: 352 QSRVGPVEWLKPYTDETIIELGKKGVKSLLAVPISFVSEHIETLEEIDVEYKELALESGI 411
QSRVGPVEWLKPYT+E + +LG+ V L+ VPISFV EHIETL+EID+EYKE+A ++GI
Sbjct: 240 QSRVGPVEWLKPYTEEVLADLGRSNVNDLIVVPISFVGEHIETLQEIDIEYKEIAEKAGI 299
Query: 412 ENWGRVPALGCEPTFISDLADAVIESL--PYVGAMAVSNLEARQSLVPLGSVEELLAAYD 469
+N+ RV AL PTFI L+D VI L P + S L + L P
Sbjct: 300 KNFRRVKALNTHPTFIEGLSDLVISCLEGPLINIEEASQLPEKVKLYPQEK--------- 350
Query: 470 SQRRELPPPILVWEWGWTKSAETWNGRAAMIAVLLLLFLEVTTGEGFLHQWGIL 523
W+WGW S+E WNGR AMI + L+LF+E+ +G G LH+ GIL
Sbjct: 351 ------------WQWGWNNSSEVWNGRVAMI-IFLVLFIELISGSGPLHKLGIL 391
>UniRef100_UPI000031F6DF UPI000031F6DF UniRef100 entry
Length = 391
Score = 412 bits (1060), Expect = e-114
Identities = 214/414 (51%), Positives = 277/414 (66%), Gaps = 27/414 (6%)
Query: 113 DKIGVLLLNLGGPETLDDVQPFLFNLFADPDIIRLPRLFSFLQKPLAQFVSVLRAPKSKE 172
DKIGVLL+NLGGPE + DV PFL+NLF+DP+IIR P F QKPLA +S LR+ S++
Sbjct: 2 DKIGVLLMNLGGPERITDVGPFLYNLFSDPEIIRTP--FPVFQKPLAWLISTLRSTTSQQ 59
Query: 173 GYASIGGGSPLRRMTDAQAEELRKSLFEKNVPANVYVGMRYWHPFTEEAIELIKRDGITK 232
Y SIGGGSP+RR+T+ QA EL+ L +K A Y+ MRYWHPFTE AI +K DGI +
Sbjct: 60 AYLSIGGGSPIRRITEQQARELQSKLRDKGFNATTYIAMRYWHPFTESAIADMKADGIDQ 119
Query: 233 LVVLPLYPQFSISTSGSSLRLLESIFREDEYLVNMQHTVIPSWYQREGYIKAMANLIEKE 292
+VV+PLYP FSISTSGSS R L+ + D+ + + SW+ + GY+K+M LI ++
Sbjct: 120 VVVIPLYPHFSISTSGSSFRELKKLRDSDDEFKKVPMRCVRSWFSQSGYLKSMVELISEQ 179
Query: 293 LKGFDLPEKVMIFFSAHGVPVAYVEEAGDPYKAEMEECVDLIMEELEK-RKITNAYTLAY 351
+ + P K IFF+AHGVP +YVEEAGDPYK ++E+C LI+ ELEK +N +TL+Y
Sbjct: 180 ISLCESPSKAHIFFTAHGVPKSYVEEAGDPYKQQIEDCSLLIINELEKCLGHSNPHTLSY 239
Query: 352 QSRVGPVEWLKPYTDETIIELGKKGVKSLLAVPISFVSEHIETLEEIDVEYKELALESGI 411
QSRVGPVEWLKPYT+E + +LG+ V L+ VPISFV EHIETL+EID+EYKE+A ++GI
Sbjct: 240 QSRVGPVEWLKPYTEEVLADLGRSNVNDLVVVPISFVGEHIETLQEIDIEYKEIAEKAGI 299
Query: 412 ENWGRVPALGCEPTFISDLADAVIESL--PYVGAMAVSNLEARQSLVPLGSVEELLAAYD 469
+N+ RV AL PTFI L+D VI L P V S L + L P
Sbjct: 300 KNFRRVKALNTHPTFIEGLSDLVISCLEGPLVNIEEASQLPEKVKLYPQEK--------- 350
Query: 470 SQRRELPPPILVWEWGWTKSAETWNGRAAMIAVLLLLFLEVTTGEGFLHQWGIL 523
W+WGW S+E WNGR AMI + L+LF+E+ +G G LH+ GIL
Sbjct: 351 ------------WQWGWNNSSEVWNGRVAMI-IFLVLFIELISGSGPLHKLGIL 391
>UniRef100_Q7U5G0 Ferrochelatase [Synechococcus sp.]
Length = 391
Score = 411 bits (1057), Expect = e-113
Identities = 221/413 (53%), Positives = 274/413 (65%), Gaps = 27/413 (6%)
Query: 114 KIGVLLLNLGGPETLDDVQPFLFNLFADPDIIRLPRLFSFLQKPLAQFVSVLRAPKSKEG 173
++GV+LLNLGGPE + DV PFL+NLFADP+IIRLP LQKPLA +S LR+ KS+E
Sbjct: 3 RVGVVLLNLGGPERIQDVGPFLYNLFADPEIIRLPS--PALQKPLAWLISTLRSGKSQEA 60
Query: 174 YASIGGGSPLRRMTDAQAEELRKSLFEKNVPANVYVGMRYWHPFTEEAIELIKRDGITKL 233
Y SIGGGSPLRR+T+ QA EL+ L ++ + A YV MRYWHPFTE A+ +K DG+ ++
Sbjct: 61 YRSIGGGSPLRRITEQQARELQSLLRQRGLDATTYVAMRYWHPFTESAVADMKADGMDEV 120
Query: 234 VVLPLYPQFSISTSGSSLRLLESIFREDEYLVNMQHTVIPSWYQREGYIKAMANLIEKEL 293
VVLPLYP FSISTSGSS R L+ + + D + I SW+ GYIKAMA LI +E+
Sbjct: 121 VVLPLYPHFSISTSGSSFRELQRLRQGDAAFEQLPIRCIRSWFDHPGYIKAMAELIAEEV 180
Query: 294 KGFDLPEKVMIFFSAHGVPVAYVEEAGDPYKAEMEECVDLIMEEL-EKRKITNAYTLAYQ 352
+ D PEK +FFSAHGVP +YVEEAGDPY+ ++E C DLIM+ L E +N +TLAYQ
Sbjct: 181 RNSDDPEKAHVFFSAHGVPKSYVEEAGDPYQQQIEACTDLIMKSLAEHMGHSNPHTLAYQ 240
Query: 353 SRVGPVEWLKPYTDETIIELGKKGVKSLLAVPISFVSEHIETLEEIDVEYKELALESGIE 412
SRVGPVEWLKPYT+E + +LG+ L+ VPISFVSEHIETLEEID+EY+ELA E+G+
Sbjct: 241 SRVGPVEWLKPYTEEALEQLGEAKTNDLVVVPISFVSEHIETLEEIDIEYRELATEAGVV 300
Query: 413 NWGRVPALGCEPTFISDLADAVIESL--PYVGAMAVSNLEARQSLVPLGSVEELLAAYDS 470
N+ RV AL P FI LAD V SL P V A + L + L P
Sbjct: 301 NFRRVRALDTYPPFIEGLADLVTTSLEGPEVSLDAAAELPTKVKLYPQEK---------- 350
Query: 471 QRRELPPPILVWEWGWTKSAETWNGRAAMIAVLLLLFLEVTTGEGFLHQWGIL 523
WEWGW S+E WNGR AM+ L LE+ +G G LH G+L
Sbjct: 351 -----------WEWGWNNSSEVWNGRLAMLGFSAFL-LELISGHGPLHALGLL 391
>UniRef100_UPI00002B771B UPI00002B771B UniRef100 entry
Length = 391
Score = 410 bits (1055), Expect = e-113
Identities = 215/414 (51%), Positives = 276/414 (65%), Gaps = 27/414 (6%)
Query: 113 DKIGVLLLNLGGPETLDDVQPFLFNLFADPDIIRLPRLFSFLQKPLAQFVSVLRAPKSKE 172
DK+GVLL+NLGGPE ++DV PFL+NLF+DP+IIR P F QKPLA +S LR+ S++
Sbjct: 2 DKVGVLLMNLGGPERINDVGPFLYNLFSDPEIIRTP--FPAFQKPLAWLISTLRSTTSQQ 59
Query: 173 GYASIGGGSPLRRMTDAQAEELRKSLFEKNVPANVYVGMRYWHPFTEEAIELIKRDGITK 232
Y SIGGGSP+RR+T+ QA EL+ L +K + A Y+ MRYWHPFTE AI +K DGI +
Sbjct: 60 AYLSIGGGSPIRRITEQQARELQSKLRDKGLNATTYIAMRYWHPFTESAIADMKADGIDQ 119
Query: 233 LVVLPLYPQFSISTSGSSLRLLESIFREDEYLVNMQHTVIPSWYQREGYIKAMANLIEKE 292
+VVLPLYP FSISTSGSS R L+ + D + I SW+ + GY+K+M LI ++
Sbjct: 120 IVVLPLYPHFSISTSGSSFRELKKLRDSDNDFKKIPMRCIRSWFSQSGYLKSMVELISEQ 179
Query: 293 LKGFDLPEKVMIFFSAHGVPVAYVEEAGDPYKAEMEECVDLIMEELEK-RKITNAYTLAY 351
+ + P IFF+AHGVP +YVEEAGDPYK ++E+C LI+ ELEK N YTL+Y
Sbjct: 180 ILLCESPSNAHIFFTAHGVPKSYVEEAGDPYKQQIEDCSLLIINELEKYLGHGNPYTLSY 239
Query: 352 QSRVGPVEWLKPYTDETIIELGKKGVKSLLAVPISFVSEHIETLEEIDVEYKELALESGI 411
QSRVGPVEWLKPYT+E + +LG+ V L+ VPISFV EHIETL+EID+EYKE+A ++GI
Sbjct: 240 QSRVGPVEWLKPYTEEVLEDLGRSKVNDLIVVPISFVGEHIETLQEIDIEYKEIAEKAGI 299
Query: 412 ENWGRVPALGCEPTFISDLADAVIESL--PYVGAMAVSNLEARQSLVPLGSVEELLAAYD 469
+N+ RV AL PTFI L+D V+ L P V S L + L P
Sbjct: 300 KNFRRVKALNTHPTFIEGLSDLVVSCLEGPLVNIEDASKLPEKVKLYPQEK--------- 350
Query: 470 SQRRELPPPILVWEWGWTKSAETWNGRAAMIAVLLLLFLEVTTGEGFLHQWGIL 523
W+WGW S+E WNGR AMI V L+LF+E+ +G G LH+ GIL
Sbjct: 351 ------------WQWGWNNSSEVWNGRVAMI-VFLILFIELISGSGPLHRLGIL 391
>UniRef100_Q7V2F5 Ferrochelatase [Prochlorococcus marinus subsp. pastoris]
Length = 391
Score = 409 bits (1052), Expect = e-113
Identities = 211/414 (50%), Positives = 276/414 (65%), Gaps = 27/414 (6%)
Query: 113 DKIGVLLLNLGGPETLDDVQPFLFNLFADPDIIRLPRLFSFLQKPLAQFVSVLRAPKSKE 172
+K+GVLL+NLGGPE + DV PFL+NLF+DP+IIRLP QKPLA +S LR+ S++
Sbjct: 2 EKVGVLLMNLGGPERITDVGPFLYNLFSDPEIIRLP--VPAFQKPLAWLISTLRSTTSQQ 59
Query: 173 GYASIGGGSPLRRMTDAQAEELRKSLFEKNVPANVYVGMRYWHPFTEEAIELIKRDGITK 232
Y SIGGGSP+RR+T+ QA EL+ L +K + Y+ MRYWHPFTE AI +K DG+ +
Sbjct: 60 AYLSIGGGSPIRRITEQQARELQSKLRDKGLNVTTYIAMRYWHPFTESAIADMKADGVDQ 119
Query: 233 LVVLPLYPQFSISTSGSSLRLLESIFREDEYLVNMQHTVIPSWYQREGYIKAMANLIEKE 292
+VVLPLYP FSISTSGSS R L+ + D + + SW+ + GY+K+M LI ++
Sbjct: 120 IVVLPLYPHFSISTSGSSFRELKKLRDSDSEFQKIPMRCVRSWFSQSGYLKSMVELISEQ 179
Query: 293 LKGFDLPEKVMIFFSAHGVPVAYVEEAGDPYKAEMEECVDLIMEELEK-RKITNAYTLAY 351
+ + P+ IFF+AHGVP +YVEEAGDPYK ++E+C LI++ELEK TN YTL+Y
Sbjct: 180 ISLCESPDSAHIFFTAHGVPKSYVEEAGDPYKEQIEDCSLLIIDELEKYLGHTNPYTLSY 239
Query: 352 QSRVGPVEWLKPYTDETIIELGKKGVKSLLAVPISFVSEHIETLEEIDVEYKELALESGI 411
QSRVGPVEWLKPYT+E + +LGK V L+ VPISFV EHIETL+EID+EYKE+A ++GI
Sbjct: 240 QSRVGPVEWLKPYTEEVLTDLGKAKVNDLIVVPISFVGEHIETLQEIDIEYKEIAEKAGI 299
Query: 412 ENWGRVPALGCEPTFISDLADAVIESL--PYVGAMAVSNLEARQSLVPLGSVEELLAAYD 469
N+ RV AL PTFI L++ V+ L P + S L + L P
Sbjct: 300 VNFRRVKALNTHPTFIDGLSELVVSCLEGPIINIEKASELPEKVKLYPQEK--------- 350
Query: 470 SQRRELPPPILVWEWGWTKSAETWNGRAAMIAVLLLLFLEVTTGEGFLHQWGIL 523
W+WGW S+E WNGR AMI V L+LF+E+ +G G LH+ GIL
Sbjct: 351 ------------WQWGWNNSSEVWNGRVAMI-VFLILFIELISGSGPLHKLGIL 391
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.318 0.137 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 889,300,377
Number of Sequences: 2790947
Number of extensions: 37849362
Number of successful extensions: 100003
Number of sequences better than 10.0: 891
Number of HSP's better than 10.0 without gapping: 805
Number of HSP's successfully gapped in prelim test: 86
Number of HSP's that attempted gapping in prelim test: 97448
Number of HSP's gapped (non-prelim): 965
length of query: 527
length of database: 848,049,833
effective HSP length: 132
effective length of query: 395
effective length of database: 479,644,829
effective search space: 189459707455
effective search space used: 189459707455
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 77 (34.3 bits)
Medicago: description of AC148755.9


