

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148755.7 - phase: 0
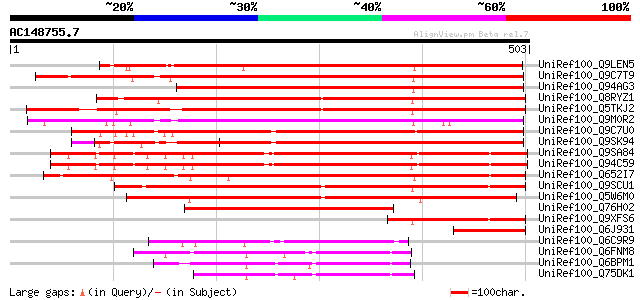
(503 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9LEN5 Hypothetical protein [Cicer arietinum] 536 e-151
UniRef100_Q9C7T9 Cytosolic factor, putative; 19554-17768 [Arabid... 475 e-132
UniRef100_Q94AG3 At1g72160/T9N14_8 [Arabidopsis thaliana] 470 e-131
UniRef100_Q8RYZ1 P0648C09.9 protein [Oryza sativa] 469 e-131
UniRef100_Q5TKJ2 Putative cellular retinaldehyde-binding/triple ... 453 e-126
UniRef100_Q9M0R2 Hypothetical protein AT4g09160 [Arabidopsis tha... 445 e-123
UniRef100_Q9C7U0 Cytosolic factor, putative; 12503-14597 [Arabid... 429 e-119
UniRef100_Q9SK94 F12K8.13 protein [Arabidopsis thaliana] 415 e-114
UniRef100_Q9SA84 T5I8.14 protein [Arabidopsis thaliana] 368 e-100
UniRef100_Q94C59 Hypothetical protein At1g30690 [Arabidopsis tha... 366 e-100
UniRef100_Q652I7 SEC14 cytosolic factor-like [Oryza sativa] 364 4e-99
UniRef100_Q9SCU1 Hypothetical protein T18N14.50 [Arabidopsis tha... 340 6e-92
UniRef100_Q5W6M0 Hypothetical protein P0015F11.17 [Oryza sativa] 326 8e-88
UniRef100_Q76H02 Hypothetical protein 5E08 [Solanum melongena] 267 5e-70
UniRef100_Q9XFS6 Hypothetical protein [Arabidopsis thaliana] 108 4e-22
UniRef100_Q6J931 Hypothetical protein [Xerophyta humilis] 100 2e-19
UniRef100_Q6C9R9 Similar to sp|P47008 Saccharomyces cerevisiae Y... 99 4e-19
UniRef100_Q6FNM8 Candida glabrata strain CBS138 chromosome J com... 95 4e-18
UniRef100_Q6BPM1 Similar to sp|P46250 Candida albicans SEC14 cyt... 95 4e-18
UniRef100_Q75DK1 SEC14 cytosolic factor [Ashbya gossypii] 94 7e-18
>UniRef100_Q9LEN5 Hypothetical protein [Cicer arietinum]
Length = 482
Score = 536 bits (1380), Expect = e-151
Identities = 277/430 (64%), Positives = 333/430 (77%), Gaps = 25/430 (5%)
Query: 88 KEQKQSTTTTVAAEPAQENKYQLE--DKK---------ENVVSSVEDDGAKTVEAIEESI 136
KE+KQ +++ EP E K + E +KK E + +S E+DGAKTVEAI+ESI
Sbjct: 53 KEEKQGSSSE---EPKTEAKPETEAVEKKVDVTVVELVEKIATSAEEDGAKTVEAIQESI 109
Query: 137 VAVSASVPPEQKPVVEKVEASLPLPPEQVSIYGIPLLADETSDVILLKFLRARDFKVKEA 196
V+V+ + ++P + VE P PE+V I+GIPLLADE SDVILLKFLRARDFKVKEA
Sbjct: 110 VSVTVTNGDGEQPAAD-VELP-PSTPEEVEIWGIPLLADERSDVILLKFLRARDFKVKEA 167
Query: 197 FTMIKNTILWRKEFGIEELMDEKLGDELE-------KVVYMHGFDKEGHPVCYNIYGEFQ 249
FTMIK T++WRKEFGIE L+ E LG + E KVV+ G+DKEGHPVCYN++GEF+
Sbjct: 168 FTMIKQTVIWRKEFGIEGLLQEDLGTDWEDLGTDWDKVVFTDGYDKEGHPVCYNVFGEFE 227
Query: 250 NKELYNKTFSDEEKRHNFLKWRIQFLEKSIRNLDFNHGGVCTIVHVNDLKDSPGPGKWEL 309
NK+LY KTFSDEEKR+ F++WRIQFLEKS+R L+F + T V VNDLK+SPG GK EL
Sbjct: 228 NKDLYQKTFSDEEKRNKFIRWRIQFLEKSVRKLNFAPSAISTFVQVNDLKNSPGLGKREL 287
Query: 310 RQATKQALQLFQDNYPEFVAKQVFINVPWWYLAVNRMISPFLTQRTKSKFVFAGPSKSTE 369
RQAT QALQL QDNYPEFVAKQ+FINVPWWYLA +RMIS FLT RTKSKF FAGPSKS +
Sbjct: 288 RQATNQALQLLQDNYPEFVAKQIFINVPWWYLAFSRMISAFLTPRTKSKFFFAGPSKSAD 347
Query: 370 TLLSYIAPEQLPVKYGGLSKDG--EFGNSDSVTEITIRPASKHTVEFPVTEKCLLSWEVR 427
TL YIAPEQ+PV+YGGLS++G EF +D TE+TI+PA+KH VEFP+ EK L WEVR
Sbjct: 348 TLFKYIAPEQVPVQYGGLSREGDQEFTTADPATEVTIKPATKHAVEFPIPEKSTLVWEVR 407
Query: 428 VIGWEVRYGAEFVPSNEGSYTVIVQKARKVASSEEAVLCNSFKINEPGKVVLTIDNTSSR 487
V+GW+V YGAEFVPS E YTVIVQK RK+A ++E V+ N+FKI EPGKVVLTIDN +S+
Sbjct: 408 VVGWDVSYGAEFVPSAEDGYTVIVQKNRKIAPADETVINNTFKIGEPGKVVLTIDNQTSK 467
Query: 488 KKKLLYRLKT 497
KKKLLYR KT
Sbjct: 468 KKKLLYRSKT 477
>UniRef100_Q9C7T9 Cytosolic factor, putative; 19554-17768 [Arabidopsis thaliana]
Length = 490
Score = 475 bits (1223), Expect = e-132
Identities = 247/483 (51%), Positives = 325/483 (67%), Gaps = 17/483 (3%)
Query: 26 PQKKNNPVTLQSHLSKPNTEQEQTFNKPSDNIPNNENNSLQELQNLIQQAFNNHAFSAPP 85
P+K +P S +S+ + T + + + E N + + A PP
Sbjct: 13 PEKLPSPSLTPSEVSESTQDALPTETETLEKV--TETNPPETADTTTKPEEETAAEHHPP 70
Query: 86 LIKEQKQSTTTTVAAEPAQENKYQLEDKKENV---VSSVEDDGAKTVEAIEESIVAVSAS 142
+ E + ++T + K E+KK + + S +++ +K + ++
Sbjct: 71 TVTETETASTEKQEVKDEASQKEVAEEKKSMIPQNLGSFKEESSKLSDLSNSEKKSLD-- 128
Query: 143 VPPEQKPVVEKV--EASLPLPPEQVSIYGIPLLADETSDVILLKFLRARDFKVKEAFTMI 200
E K +V + PE+V I+GIPLL D+ SDV+LLKFLRAR+FKVK++F M+
Sbjct: 129 ---ELKHLVREALDNHQFTNTPEEVKIWGIPLLEDDRSDVVLLKFLRAREFKVKDSFAML 185
Query: 201 KNTILWRKEFGIEELMDEKLGDELEKVVYMHGFDKEGHPVCYNIYGEFQNKELYNKTFSD 260
KNTI WRKEF I+EL++E L D+L+KVV+MHG D+EGHPVCYN+YGEFQNKELYNKTFSD
Sbjct: 186 KNTIKWRKEFKIDELVEEDLVDDLDKVVFMHGHDREGHPVCYNVYGEFQNKELYNKTFSD 245
Query: 261 EEKRHNFLKWRIQFLEKSIRNLDFNHGGVCTIVHVNDLKDSPGPGKWELRQATKQALQLF 320
EEKR +FL+ RIQFLE+SIR LDF+ GGV TI VND+K+SPG GK ELR ATKQA++L
Sbjct: 246 EEKRKHFLRTRIQFLERSIRKLDFSSGGVSTIFQVNDMKNSPGLGKKELRSATKQAVELL 305
Query: 321 QDNYPEFVAKQVFINVPWWYLAVNRMISPFLTQRTKSKFVFAGPSKSTETLLSYIAPEQL 380
QDNYPEFV KQ FINVPWWYL +I PF+T R+KSK VFAGPS+S ETL YI+PEQ+
Sbjct: 306 QDNYPEFVFKQAFINVPWWYLVFYTVIGPFMTPRSKSKLVFAGPSRSAETLFKYISPEQV 365
Query: 381 PVKYGGLSKD-----GEFGNSDSVTEITIRPASKHTVEFPVTEKCLLSWEVRVIGWEVRY 435
PV+YGGLS D +F DS +EIT++P +K TVE + EKC L WE+RV GWEV Y
Sbjct: 366 PVQYGGLSVDPCDCNPDFSLEDSASEITVKPGTKQTVEIIIYEKCELVWEIRVTGWEVSY 425
Query: 436 GAEFVPSNEGSYTVIVQKARKVASSEEAVLCNSFKINEPGKVVLTIDNTSSRKKKLLYRL 495
AEFVP + +YTV++QK RK+ S+E VL +SFK+NE GKV+LT+DN +S+KKKL+YR
Sbjct: 426 KAEFVPEEKDAYTVVIQKPRKMRPSDEPVLTHSFKVNELGKVLLTVDNPTSKKKKLVYRF 485
Query: 496 KTK 498
K
Sbjct: 486 NVK 488
>UniRef100_Q94AG3 At1g72160/T9N14_8 [Arabidopsis thaliana]
Length = 390
Score = 470 bits (1209), Expect = e-131
Identities = 225/342 (65%), Positives = 276/342 (79%), Gaps = 5/342 (1%)
Query: 162 PEQVSIYGIPLLADETSDVILLKFLRARDFKVKEAFTMIKNTILWRKEFGIEELMDEKLG 221
PE+V I+GIPLL D+ SDV+LLKFLRAR+FKVK++F M+KNTI WRKEF I+EL++E L
Sbjct: 47 PEEVKIWGIPLLEDDRSDVVLLKFLRAREFKVKDSFAMLKNTIKWRKEFKIDELVEEDLV 106
Query: 222 DELEKVVYMHGFDKEGHPVCYNIYGEFQNKELYNKTFSDEEKRHNFLKWRIQFLEKSIRN 281
D+L+KVV+MHG D+EGHPVCYN+YGEFQNKELYNKTFSDEEKR +FL+ RIQFLE+SIR
Sbjct: 107 DDLDKVVFMHGHDREGHPVCYNVYGEFQNKELYNKTFSDEEKRKHFLRTRIQFLERSIRK 166
Query: 282 LDFNHGGVCTIVHVNDLKDSPGPGKWELRQATKQALQLFQDNYPEFVAKQVFINVPWWYL 341
LDF+ GGV TI VND+K+SPG GK ELR ATKQA++L QDNYPEFV KQ FINVPWWYL
Sbjct: 167 LDFSSGGVSTIFQVNDMKNSPGLGKKELRSATKQAVELLQDNYPEFVFKQAFINVPWWYL 226
Query: 342 AVNRMISPFLTQRTKSKFVFAGPSKSTETLLSYIAPEQLPVKYGGLSKD-----GEFGNS 396
+I PF+T R+KSK VFAGPS+S ETL YI+PEQ+PV+YGGLS D +F
Sbjct: 227 VFYTVIGPFMTPRSKSKLVFAGPSRSAETLFKYISPEQVPVQYGGLSVDPCDCNPDFSLE 286
Query: 397 DSVTEITIRPASKHTVEFPVTEKCLLSWEVRVIGWEVRYGAEFVPSNEGSYTVIVQKARK 456
DS +EIT++P +K TVE + EKC L WE+RV GWEV Y AEFVP + +YTV++QK RK
Sbjct: 287 DSASEITVKPGTKQTVEIIIYEKCELVWEIRVTGWEVSYKAEFVPEEKDAYTVVIQKPRK 346
Query: 457 VASSEEAVLCNSFKINEPGKVVLTIDNTSSRKKKLLYRLKTK 498
+ S+E VL +SFK+NE GKV+LT+DN +S+KKKL+YR K
Sbjct: 347 MRPSDEPVLTHSFKVNELGKVLLTVDNPTSKKKKLVYRFNVK 388
>UniRef100_Q8RYZ1 P0648C09.9 protein [Oryza sativa]
Length = 613
Score = 469 bits (1207), Expect = e-131
Identities = 238/423 (56%), Positives = 307/423 (72%), Gaps = 13/423 (3%)
Query: 85 PLIKEQKQSTTTTVAAEPAQENKYQLEDKKENVVSSVEDDGAKTVEAIEESIVAVSAS-- 142
P +E K A+PA+ E +++ VV + E+ KTVEAIEE++V +A+
Sbjct: 193 PAKEESKAEAAPAEEAKPAEP-----EPEEKTVVVTEEEAATKTVEAIEETVVPAAAAPA 247
Query: 143 VPPEQKPVVEKVEASLPLPPEQVSIYGIPLLAD-ETSDVILLKFLRARDFKVKEAFTMIK 201
++ + E PE V I+G+PL+ D E +D +LLKFLRAR+FKVKEA M++
Sbjct: 248 AAATEEAAAPEPEVQAAAAPEPVLIWGVPLVGDDERTDTVLLKFLRAREFKVKEAMAMLR 307
Query: 202 NTILWRKEFGIEELMDEKLG-DELEKVVYMHGFDKEGHPVCYNIYGEFQNKELYNKTFSD 260
+ +LWRK FGIE L+D L EL+ VV+ G D+EGHPVCYN+YGEFQ+K+LY K F D
Sbjct: 308 SAVLWRKRFGIESLLDADLALPELDSVVFYRGADREGHPVCYNVYGEFQDKDLYEKAFGD 367
Query: 261 EEKRHNFLKWRIQFLEKSIRN-LDFNHGGVCTIVHVNDLKDSPGPGKWELRQATKQALQL 319
EEKR FLKWRIQ LE+ I + LDF+ G+C++V V DLK+SP P + R T+QA+ L
Sbjct: 368 EEKRERFLKWRIQLLERGILSQLDFSPSGICSMVQVTDLKNSP-PMLGKHRAVTRQAVAL 426
Query: 320 FQDNYPEFVAKQVFINVPWWYLAVNRMISPFLTQRTKSKFVFAGPSKSTETLLSYIAPEQ 379
QDNYPEF+AK+VFINVPWWYLA N+M+SPFLTQRTKSKF+FA P+KS ETL YIAPEQ
Sbjct: 427 LQDNYPEFIAKKVFINVPWWYLAANKMMSPFLTQRTKSKFIFASPAKSAETLFRYIAPEQ 486
Query: 380 LPVKYGGLSK--DGEFGNSDSVTEITIRPASKHTVEFPVTEKCLLSWEVRVIGWEVRYGA 437
+PV++GGL K D EF SD+VTE+TI+P+SK TVE PVTE + WE+RV+GWEV YGA
Sbjct: 487 VPVQFGGLFKEDDPEFTTSDAVTELTIKPSSKETVEIPVTENSTIGWELRVLGWEVSYGA 546
Query: 438 EFVPSNEGSYTVIVQKARKVASSEEAVLCNSFKINEPGKVVLTIDNTSSRKKKLLYRLKT 497
EF P EG YTVIVQK RKV ++EE ++ SFK+ EPGK+VLTI+N +S+KKKLLYR K
Sbjct: 547 EFTPDAEGGYTVIVQKTRKVPANEEPIMKGSFKVGEPGKIVLTINNPASKKKKLLYRSKV 606
Query: 498 KTS 500
K++
Sbjct: 607 KST 609
Score = 38.5 bits (88), Expect = 0.46
Identities = 32/102 (31%), Positives = 43/102 (41%), Gaps = 10/102 (9%)
Query: 56 NIPNNENNSLQELQNLIQQAFNNHAFSAPPLIKEQKQSTTTTVAAEPAQENKYQLEDKKE 115
++P+ E +L E + LI A F+ PP K V A+E K + E K E
Sbjct: 84 DLPDPEKKALDEFKQLIAAALAACEFNLPPPPPPPKAKVEAAVEETKAEETKAEEEPKAE 143
Query: 116 NVVSSVEDDGAKTVEAIEESIVAVSASVPPEQKPVVEKVEAS 157
+ AK E E VA +A+ PPE K EAS
Sbjct: 144 --------EPAKEEEPKAE--VAAAAAAPPEAGTEEPKAEAS 175
>UniRef100_Q5TKJ2 Putative cellular retinaldehyde-binding/triple function [Oryza
sativa]
Length = 585
Score = 453 bits (1166), Expect = e-126
Identities = 245/497 (49%), Positives = 319/497 (63%), Gaps = 41/497 (8%)
Query: 17 PQPLYSHPQPQKKNNPVTLQSHLSK-PNTEQEQTFNKPSDNIPNNENNSLQELQNLIQQA 75
P P +P K+ P ++ ++ P E E + P E +E
Sbjct: 113 PPPPAKAEEPAKEEEPKAAEAPAAEEPKAEAEAEAEAAATEEPKTEEPKTEE-------- 164
Query: 76 FNNHAFSAPPLIKEQKQSTTTTVAAEP-----AQENKYQLEDKKENVVSSVEDDG-AKTV 129
P +E+ ++ A EP A+E K + +E V ED+G +KTV
Sbjct: 165 ---------PAKEEEPKAAAAAAAEEPKAEAAAEEAKPAEPETEEKTVVVTEDEGTSKTV 215
Query: 130 EAIEESIVAVSASVPPEQKPVVEKVEASLPLPPEQVSIYGIPLLAD-ETSDVILLKFLRA 188
EAIEE++V + + E + K E I+G+PL D E +D +LLKFLRA
Sbjct: 216 EAIEETVVVAAPAAAAEAEAAAPKEEL----------IWGVPLTGDDERTDTVLLKFLRA 265
Query: 189 RDFKVKEAFTMIKNTILWRKEFGIEELMDEKLG-DELEKVVYMHGFDKEGHPVCYNIYGE 247
R+FKVKEA M+K +LWRK FGI+ ++ LG ELE VV+ G D+EGHPVCYN+YGE
Sbjct: 266 REFKVKEAMAMLKAAVLWRKRFGIDAVLAADLGLPELENVVFYRGADREGHPVCYNVYGE 325
Query: 248 FQNKELYNKTFSDEEKRHNFLKWRIQFLEKSIRN-LDFNHGGVCTIVHVNDLKDSPGPGK 306
FQ+K+LY K F DEEKR FLKWRIQ LE+ I + LDF+ G+C++V V DLK+SP P
Sbjct: 326 FQDKDLYEKAFGDEEKRERFLKWRIQLLERGILDQLDFSPSGICSMVQVTDLKNSP-PML 384
Query: 307 WELRQATKQALQLFQDNYPEFVAKQVFINVPWWYLAVNRMISPFLTQRTKSKFVFAGPSK 366
+ R T+QAL L QDNYPEF+AK++FINVPWWY+A N+M+SPFLTQRTKSK +F +K
Sbjct: 385 GKHRTVTRQALALLQDNYPEFIAKKIFINVPWWYIAANKMVSPFLTQRTKSKIIFCTAAK 444
Query: 367 STETLLSYIAPEQLPVKYGGLSK--DGEFGNSDSVTEITIRPASKHTVEFPVTEKCLLSW 424
S ETL YIAPEQ+PV++GGL K D EF SD+VTE+ I+P+SK TVE P TE + W
Sbjct: 445 SAETLFRYIAPEQVPVQFGGLYKEDDTEFSTSDAVTELPIKPSSKETVEIPATENSTVVW 504
Query: 425 EVRVIGWEVRYGAEFVPSNEGSYTVIVQKARKVASSEEAVLCNSFKINEPGKVVLTIDNT 484
E+RV+GWEV YGAEF P EG YTVIVQK RKV ++EE ++ SFK+ EPGK+VLT+DN
Sbjct: 505 ELRVLGWEVSYGAEFTPDAEGGYTVIVQKTRKVPANEEPIMKGSFKVGEPGKIVLTVDNA 564
Query: 485 SSRKKK-LLYRLKTKTS 500
+S+KKK LLYR K K+S
Sbjct: 565 ASKKKKQLLYRFKVKSS 581
>UniRef100_Q9M0R2 Hypothetical protein AT4g09160 [Arabidopsis thaliana]
Length = 723
Score = 445 bits (1144), Expect = e-123
Identities = 252/566 (44%), Positives = 341/566 (59%), Gaps = 95/566 (16%)
Query: 18 QPLYSHPQPQKKNNP----VTLQSHLSKPN-TEQEQTFNKPSDNIPNNENNSLQELQNLI 72
QP P P+ + + + + S +PN ++ + +PS + + E+++
Sbjct: 163 QPRGVTPTPETETSEADTSLLVTSETEEPNHAAEDYSETEPSQKLMLEQRRKYMEVEDWT 222
Query: 73 QQAFNNHA-FSAPPLIKEQKQ-----------STTTTVA----AEPAQENKYQLEDKKE- 115
+ + A A + E KQ +TT+TVA AE + ++E+K++
Sbjct: 223 EPELPDEAVLEAAASVPEPKQPEPQTPPPPPSTTTSTVASRSLAEMMNREEAEVEEKQKI 282
Query: 116 ---NVVSSVEDDGAKTVEAIEESIVAVSASVPPEQKPVVEKVEASLPLPPEQVSIYGIPL 172
+ S +++ K + E + A+ E + +++ + S + SI+G+PL
Sbjct: 283 QIPRSLGSFKEETNKISDLSETELNALQ-----ELRHLLQVSQDS-----SKTSIWGVPL 332
Query: 173 LADETSDVILLKFLRARDFKVKEAFTMIKNTILWRKEFGIEELMDEKLGDELEKVVYMHG 232
L D+ +DV+LLKFLRARDFK +EA++M+ T+ WR +F IEEL+DE LGD+L+KVV+M G
Sbjct: 333 LKDDRTDVVLLKFLRARDFKPQEAYSMLNKTLQWRIDFNIEELLDENLGDDLDKVVFMQG 392
Query: 233 FDKEGHPVCYNIYGEFQNKELYNKTFSDEEKRHNFLKWRIQFLEKSIRNLDFNHGGVCTI 292
DKE HPVCYN+YGEFQNK+LY KTFSDEEKR FL+WRIQFLEKSIRNLDF GGV TI
Sbjct: 393 QDKENHPVCYNVYGEFQNKDLYQKTFSDEEKRERFLRWRIQFLEKSIRNLDFVAGGVSTI 452
Query: 293 VHVNDLKDSPGPGKWELRQATKQALQLFQDNYPEFVAKQVFINVPWWYLAVNRMISPFLT 352
VNDLK+SPGPGK ELR ATKQAL L QDNYPEFV+KQ+FINVPWWYLA R+ISPF++
Sbjct: 453 CQVNDLKNSPGPGKTELRLATKQALHLLQDNYPEFVSKQIFINVPWWYLAFYRIISPFMS 512
Query: 353 QRTKSKFVFAGPSKSTETLLSYIAPEQLPVKYGGLSKD-----GEFGNSDSVTEITIRPA 407
QR+KSK VFAGPS+S ETLL YI+PE +PV+YGGLS D +F + D TEIT++P
Sbjct: 513 QRSKSKLVFAGPSRSAETLLKYISPEHVPVQYGGLSVDNCECNSDFTHDDIATEITVKPT 572
Query: 408 SKHTVEFPVTEK-----------------CLLSW-------------------------- 424
+K TVE V E C+ W
Sbjct: 573 TKQTVEIIVYEVRPFCIKTFYTTFSNAHFCVCVWKITNCGFNTKYLRFSYVLKSSYICAS 632
Query: 425 ------------EVRVIGWEVRYGAEFVPSNEGSYTVIVQKARKVASSEEAVLCNSFKIN 472
E+RV+GWEV YGAEFVP N+ YTVI+QK RK+ + E V+ +SFK+
Sbjct: 633 KHFFLQKCTIVWEIRVVGWEVSYGAEFVPENKEGYTVIIQKPRKMTAKNELVVSHSFKVG 692
Query: 473 EPGKVVLTIDNTSSRKKKLLYRLKTK 498
E G+++LT+DN +S KK L+YR K K
Sbjct: 693 EVGRILLTVDNPTSTKKMLIYRFKVK 718
>UniRef100_Q9C7U0 Cytosolic factor, putative; 12503-14597 [Arabidopsis thaliana]
Length = 573
Score = 429 bits (1104), Expect = e-119
Identities = 239/489 (48%), Positives = 319/489 (64%), Gaps = 59/489 (12%)
Query: 61 ENNSLQELQNLIQQAFNNHAFSAPPL---------------------IKEQKQSTTTTVA 99
+ +L+E + L+++A N F+AP +E+K+ TTT V
Sbjct: 93 QKKALEEFKELVREALNKREFTAPVTPVKEEKTEEKKTEEETKEEEKTEEKKEETTTEVK 152
Query: 100 AE------PAQENKYQLE------------DKKENVV----SSVEDDGAKTVEAIEESIV 137
E PA E + E ++K V SS E+DG KTVEAIEESIV
Sbjct: 153 VEEEKPAVPAAEEEKSSEAAPVETKSEEKPEEKAEVTTEKASSAEEDGTKTVEAIEESIV 212
Query: 138 AVSASVPPEQK--PVVEKVEA---SLPLPPEQVSIYGIPLLADETSDVILLKFLRARDFK 192
+VS PPE PVV + A + P+ PE+VSI+G+PLL DE SDVIL KFLRARDFK
Sbjct: 213 SVS---PPESAVAPVVVETVAVAEAEPVEPEEVSIWGVPLLQDERSDVILTKFLRARDFK 269
Query: 193 VKEAFTMIKNTILWRKEFGIEELMDE-KLGDELEKVVYMHGFDKEGHPVCYNIYGEFQNK 251
VKEA TM+KNT+ WRKE I+EL++ + E EK+V+ HG DKEGH V Y+ YGEFQNK
Sbjct: 270 VKEALTMLKNTVQWRKENKIDELVESGEEVSEFEKMVFAHGVDKEGHVVIYSSYGEFQNK 329
Query: 252 ELYNKTFSDEEKRHNFLKWRIQFLEKSIRNLDF-NHGGVCTIVHVNDLKDSPGPGKWELR 310
EL FSD+EK + FL WRIQ EK +R +DF N + V V+D +++PG GK L
Sbjct: 330 EL----FSDKEKLNKFLSWRIQLQEKCVRAIDFSNPEAKSSFVFVSDFRNAPGLGKRALW 385
Query: 311 QATKQALQLFQDNYPEFVAKQVFINVPWWYLAVNRMISPFLTQ-RTKSKFVFAGPSKSTE 369
Q ++A++ F+DNYPEF AK++FINVPWWY+ + +T RT+SK V AGPSKS +
Sbjct: 386 QFIRRAVKQFEDNYPEFAAKELFINVPWWYIPYYKTFGSIITSPRTRSKMVLAGPSKSAD 445
Query: 370 TLLSYIAPEQLPVKYGGLSKDGEFGNSDSVTEITIRPASKHTVEFPVTEKCLLSWEVRVI 429
T+ YIAPEQ+PVKYGGLSKD +++TE ++PA+ +T+E P +E C LSWE+RV+
Sbjct: 446 TIFKYIAPEQVPVKYGGLSKDTPL-TEETITEAIVKPAANYTIELPASEACTLSWELRVL 504
Query: 430 GWEVRYGAEFVPSNEGSYTVIVQKARKVASSEEAVLCNSFKINEPGKVVLTIDNTSSRKK 489
G +V YGA+F P+ EGSY VIV K RK+ S++E V+ +SFK+ EPGK+V+TIDN +S+KK
Sbjct: 505 GADVSYGAQFEPTTEGSYAVIVSKTRKIGSTDEPVITDSFKVGEPGKIVITIDNQTSKKK 564
Query: 490 KLLYRLKTK 498
K+LYR KT+
Sbjct: 565 KVLYRFKTQ 573
>UniRef100_Q9SK94 F12K8.13 protein [Arabidopsis thaliana]
Length = 683
Score = 415 bits (1067), Expect = e-114
Identities = 222/418 (53%), Positives = 290/418 (69%), Gaps = 21/418 (5%)
Query: 83 APPLIKEQKQSTTTTVAAEPAQENKYQLEDKKENVVSSVEDDGAKTVEAIEESIVAVSAS 142
A IK+ S TT+ E +E + E ++ +++ KTVEA+EESIV+++
Sbjct: 284 ASKFIKDIFVSVTTS---EKKKEEEKPAVVTIEKAFAADQEEETKTVEAVEESIVSITL- 339
Query: 143 VPPEQKPVVEKVEASLPLPPEQVSIYGIPLLADETSDVILLKFLRARDFKVKEAFTMIKN 202
PE VE PE+VSI+GIPLL DE SDVILLKFLRARDFKVKEAFTM+KN
Sbjct: 340 --PETAAYVE---------PEEVSIWGIPLLEDERSDVILLKFLRARDFKVKEAFTMLKN 388
Query: 203 TILWRKEFGIEELMDEKL-GDELEKVVYMHGFDKEGHPVCYNIYGEFQNKELYNKTFSDE 261
T+ WRKE I++L+ E L G E EK+V+ HG DK+GH V Y+ YGEFQNKE+ FSD+
Sbjct: 389 TVQWRKENKIDDLVSEDLEGSEFEKLVFTHGVDKQGHVVIYSSYGEFQNKEI----FSDK 444
Query: 262 EKRHNFLKWRIQFLEKSIRNLDFNHGGVCTIVHVNDLKDSPGPGKWELRQATKQALQLFQ 321
EK FLKWRIQF EK +R+LDF+ + V V+D +++PG G+ L Q K+A++ F+
Sbjct: 445 EKLSKFLKWRIQFQEKCVRSLDFSPEAKSSFVFVSDFRNAPGLGQRALWQFIKRAVKQFE 504
Query: 322 DNYPEFVAKQVFINVPWWYLAVNRMISPFLTQ-RTKSKFVFAGPSKSTETLLSYIAPEQL 380
DNYPEFVAK++FINVPWWY+ + +T RT+SK V +GPSKS ET+ Y+APE +
Sbjct: 505 DNYPEFVAKELFINVPWWYIPYYKTFGSIITSPRTRSKMVLSGPSKSAETIFKYVAPEVV 564
Query: 381 PVKYGGLSKDGEFGNSDSVTEITIRPASKHTVEFPVTEKCLLSWEVRVIGWEVRYGAEFV 440
PVKYGGLSKD F D VTE ++ SK+T++ P TE LSWE+RV+G +V YGA+F
Sbjct: 565 PVKYGGLSKDSPFTVEDGVTEAVVKSTSKYTIDLPATEGSTLSWELRVLGADVSYGAQFE 624
Query: 441 PSNEGSYTVIVQKARKVASSEEAVLCNSFKINEPGKVVLTIDNTSSRKKKLLYRLKTK 498
PSNE SYTVIV K RKV ++E V+ +SFK +E GKVV+TIDN + +KKK+LYR KT+
Sbjct: 625 PSNEASYTVIVSKNRKVGLTDEPVITDSFKASEAGKVVITIDNQTFKKKKVLYRSKTQ 682
Score = 48.5 bits (114), Expect = 4e-04
Identities = 42/150 (28%), Positives = 69/150 (46%), Gaps = 14/150 (9%)
Query: 61 ENNSLQELQNLIQQAFNNHAFSAPP----LIKEQKQSTTTTVAAEPAQENKYQLEDKKEN 116
E N+L EL+ L+++A N F+APP +KE+K T E +E + + E+K
Sbjct: 94 EKNALAELKELVREALNKREFTAPPPPPAPVKEEKVEEKKTEETEEKKE-EVKTEEKSLE 152
Query: 117 VVSSVEDDGA--KTVEAIEESIVAVSASVPPEQKPVVEKVEASLPLPPEQVSIY-GIPLL 173
+ E+ A TVE +E I+A A + V E + P+ P V P++
Sbjct: 153 AETKEEEKSAAPATVETKKEEILAAPAPI------VAETKKEETPVAPAPVETKPAAPVV 206
Query: 174 ADETSDVILLKFLRARDFKVKEAFTMIKNT 203
A+ + IL + KV+E ++ T
Sbjct: 207 AETKKEEILPAAPVTTETKVEEKVVPVETT 236
>UniRef100_Q9SA84 T5I8.14 protein [Arabidopsis thaliana]
Length = 540
Score = 368 bits (944), Expect = e-100
Identities = 213/507 (42%), Positives = 316/507 (62%), Gaps = 54/507 (10%)
Query: 40 SKPN-TEQEQTFNKPSD---NIPNNENNSLQELQNLIQQAFNNHAF-------SAPPLIK 88
SKP E+ +F + SD ++ +E +L +L++ +++A ++ S+P +K
Sbjct: 43 SKPEGVEKSASFKEESDFFADLKESEKKALSDLKSKLEEAIVDNTLLKTKKKESSP--MK 100
Query: 89 EQKQSTTTTVAA-----EPAQENKYQLEDKKENVVSSVEDDGAKTVEAI-------EESI 136
E+K+ A E A E K + E K E VV+ E A+TVEA+ +E +
Sbjct: 101 EKKEEVVKPEAEVEKKKEEAAEEKVEEEKKSEAVVTE-EAPKAETVEAVVTEEIIPKEEV 159
Query: 137 VAVSASVPPEQKP--------VVEKVEA-SLPLPPEQVSI------YGIPLLAD---ETS 178
V V E K V E+V+A ++ + E S+ +G+PLL E++
Sbjct: 160 TTVVEKVEEETKEEEKKTEDVVTEEVKAETIEVEDEDESVDKDIELWGVPLLPSKGAEST 219
Query: 179 DVILLKFLRARDFKVKEAFTMIKNTILWRKEFGIEELMDEKLGDELEKVVYMHGFDKEGH 238
DVILLKFLRARDFKV EAF M+K T+ WRK+ I+ ++ E+ G++L YM+G D+E H
Sbjct: 220 DVILLKFLRARDFKVNEAFEMLKKTLKWRKQNKIDSILGEEFGEDLATAAYMNGVDRESH 279
Query: 239 PVCYNIYGEFQNKELYNKTFSDEEKRHNFLKWRIQFLEKSIRNLDFNHGGVCTIVHVNDL 298
PVCYN++ E ELY +T E+ R FL+WR Q +EK I+ L+ GGV +++ ++DL
Sbjct: 280 PVCYNVHSE----ELY-QTIGSEKNREKFLRWRFQLMEKGIQKLNLKPGGVTSLLQIHDL 334
Query: 299 KDSPGPGKWELRQATKQALQLFQDNYPEFVAKQVFINVPWWYLAVNRMISPFLTQRTKSK 358
K++PG + E+ K+ ++ QDNYPEFV++ +FINVP+W+ A+ ++SPFLTQRTKSK
Sbjct: 335 KNAPGVSRTEIWVGIKKVIETLQDNYPEFVSRNIFINVPFWFYAMRAVLSPFLTQRTKSK 394
Query: 359 FVFAGPSKSTETLLSYIAPEQLPVKYGGLS--KDGEFGNSDSVTEITIRPASKHTVEFPV 416
FV A P+K ETLL YI ++LPV+YGG D EF N ++V+E+ ++P S T+E P
Sbjct: 395 FVVARPAKVRETLLKYIPADELPVQYGGFKTVDDTEFSN-ETVSEVVVKPGSSETIEIPA 453
Query: 417 TE-KCLLSWEVRVIGWEVRYGAEFVPSNEGSYTVIVQKARKVASSEEAVLCNSFKINEPG 475
E + L W++ V+GWEV Y EFVP+ EG+YTVIVQK +K+ ++E + NSFK ++ G
Sbjct: 454 PETEGTLVWDIAVLGWEVNYKEEFVPTEEGAYTVIVQKVKKMGANEGPIR-NSFKNSQAG 512
Query: 476 KVVLTIDNTSSRKKKLLYRLKTKTSLS 502
K+VLT+DN S +KKK+LYR +TKT S
Sbjct: 513 KIVLTVDNVSGKKKKVLYRYRTKTESS 539
>UniRef100_Q94C59 Hypothetical protein At1g30690 [Arabidopsis thaliana]
Length = 540
Score = 366 bits (940), Expect = e-100
Identities = 213/507 (42%), Positives = 315/507 (62%), Gaps = 54/507 (10%)
Query: 40 SKPN-TEQEQTFNKPSD---NIPNNENNSLQELQNLIQQAFNNHAF-------SAPPLIK 88
SKP E+ +F + SD ++ +E +L +L++ +++A ++ S+P +K
Sbjct: 43 SKPEGVEKSASFKEESDFFADLKESEKKALSDLKSKLEEAIVDNTLLKTKKKESSP--MK 100
Query: 89 EQKQSTTTTVAA-----EPAQENKYQLEDKKENVVSSVEDDGAKTVEAI-------EESI 136
E+K+ A E A E K + E K E VV+ E A+TVEA+ +E +
Sbjct: 101 EKKEEVVKPEAEVEKKKEEAAEEKVEEEKKSEAVVTE-EAPKAETVEAVVTEEIIPKEEV 159
Query: 137 VAVSASVPPEQKP--------VVEKVEA-SLPLPPEQVSI------YGIPLLAD---ETS 178
V V E K V E+V+A ++ + E S+ +G+PLL E++
Sbjct: 160 TTVVEKVEEETKEEEKKTEDVVTEEVKAETIEVEDEDESVDKDIELWGVPLLPSKGAEST 219
Query: 179 DVILLKFLRARDFKVKEAFTMIKNTILWRKEFGIEELMDEKLGDELEKVVYMHGFDKEGH 238
DVILLKFLRARDFKV EAF M+K T+ WRK+ I+ ++ E+ G++L YM+G D+E H
Sbjct: 220 DVILLKFLRARDFKVNEAFEMLKKTLKWRKQNKIDSILGEEFGEDLATAAYMNGVDRESH 279
Query: 239 PVCYNIYGEFQNKELYNKTFSDEEKRHNFLKWRIQFLEKSIRNLDFNHGGVCTIVHVNDL 298
PVCYN+ E ELY +T E+ R FL+WR Q +EK I+ L+ GGV +++ ++DL
Sbjct: 280 PVCYNVNSE----ELY-QTIGSEKNREKFLRWRFQLMEKGIQKLNLKPGGVTSLLQIHDL 334
Query: 299 KDSPGPGKWELRQATKQALQLFQDNYPEFVAKQVFINVPWWYLAVNRMISPFLTQRTKSK 358
K++PG + E+ K+ ++ QDNYPEFV++ +FINVP+W+ A+ ++SPFLTQRTKSK
Sbjct: 335 KNAPGVSRTEIWVGIKKVIETLQDNYPEFVSRNIFINVPFWFYAMRAVLSPFLTQRTKSK 394
Query: 359 FVFAGPSKSTETLLSYIAPEQLPVKYGGLS--KDGEFGNSDSVTEITIRPASKHTVEFPV 416
FV A P+K ETLL YI ++LPV+YGG D EF N ++V+E+ ++P S T+E P
Sbjct: 395 FVVARPAKVRETLLKYIPADELPVQYGGFKTVDDTEFSN-ETVSEVVVKPGSSETIEIPA 453
Query: 417 TE-KCLLSWEVRVIGWEVRYGAEFVPSNEGSYTVIVQKARKVASSEEAVLCNSFKINEPG 475
E + L W++ V+GWEV Y EFVP+ EG+YTVIVQK +K+ ++E + NSFK ++ G
Sbjct: 454 PETEGTLVWDIAVLGWEVNYKEEFVPTEEGAYTVIVQKVKKMGANEGPIR-NSFKNSQAG 512
Query: 476 KVVLTIDNTSSRKKKLLYRLKTKTSLS 502
K+VLT+DN S +KKK+LYR +TKT S
Sbjct: 513 KIVLTVDNVSGKKKKVLYRYRTKTESS 539
>UniRef100_Q652I7 SEC14 cytosolic factor-like [Oryza sativa]
Length = 517
Score = 364 bits (934), Expect = 4e-99
Identities = 210/484 (43%), Positives = 296/484 (60%), Gaps = 22/484 (4%)
Query: 33 VTLQSHLSKPNTEQEQTFNKPSDNIPNNENNSLQELQNLIQQAFNNHAFSAPPLIKEQKQ 92
VTL + +SK + +E++ D++ + E +L EL+ +++A + ++ +K+
Sbjct: 35 VTLAAVVSKNASFREES--NFLDDLKDGERKALAELRAKVEEAIVDGKLFDDGKVEAKKK 92
Query: 93 STTTT---VAAEPAQENKYQLEDKKENVVSSVEDDGAKTVEAIEESIVAVSASVPPEQKP 149
+ E A E K E K+E + E + E EE + E+KP
Sbjct: 93 AAAAEEEKAVEEAAGEKKDGEEKKEEEEPVTEEKKEEEQGEEEEEPKKEEADEGEKEEKP 152
Query: 150 VVEKVEASLPLPPEQVSIYGIPLL---ADETSDVILLKFLRARDFKVKEAFTMIKNTILW 206
E+ A + + ++++G+PLL D+ +DV+LLKFLRARDFK AF M++ T+ W
Sbjct: 153 AEEEAAAVVD---KDIALWGVPLLPSKGDDATDVVLLKFLRARDFKAGAAFDMLRKTLHW 209
Query: 207 RKEF-----GIEELMD-EKLGDELEKVVYMHGFDKEGHPVCYNIYGEFQNKELYNKTFSD 260
R+E+ G ++ D E L EL Y+ G D+EGHPVCYN G F + +Y K
Sbjct: 210 RREWKGFAAGTDDDDDGEALPAELADACYLDGADREGHPVCYNALGVFADDAVYKKALGT 269
Query: 261 EEKRHNFLKWRIQFLEKSIRNLDFNHGGVCTIVHVNDLKDSPGPGKWELRQATKQALQLF 320
EE + FL+WR++ +E + LD GGV +++ V DLK+SPGP K +LR A KQ L LF
Sbjct: 270 EEGKARFLRWRVRAMESHVAKLDLRPGGVASLLQVTDLKNSPGPAKKDLRVAMKQVLDLF 329
Query: 321 QDNYPEFVAKQVFINVPWWYLAVNRMISPFLTQRTKSKFVFAGPSKSTETLLSYIAPEQL 380
QDNYPE VA+ + INVP+WY A + + PF+TQRTKSKFV A PSK TETLL YI E +
Sbjct: 330 QDNYPELVARNILINVPFWYYAFSTLFYPFMTQRTKSKFVIARPSKVTETLLKYIPIEAI 389
Query: 381 PVKYGGLSKDG--EFGNSDS-VTEITIRPASKHTVEFPVTE-KCLLSWEVRVIGWEVRYG 436
PVKYGGL +D EF DS VTE+ ++ +S T+E TE L+W++ V+GWEV Y
Sbjct: 390 PVKYGGLKRDDDTEFSAEDSEVTELVVKASSTETIEIEATEGDTTLTWDLTVLGWEVNYK 449
Query: 437 AEFVPSNEGSYTVIVQKARKVASSEEAVLCNSFKINEPGKVVLTIDNTSSRKKKLLYRLK 496
EFVPS EGSYTVIV+K +K+ SSE AV NSF+ EPGKVVLT++N + RKKK+L+R K
Sbjct: 450 EEFVPSEEGSYTVIVKKGKKMGSSEAAVR-NSFRAGEPGKVVLTVENLTHRKKKVLFRHK 508
Query: 497 TKTS 500
K++
Sbjct: 509 AKSA 512
>UniRef100_Q9SCU1 Hypothetical protein T18N14.50 [Arabidopsis thaliana]
Length = 409
Score = 340 bits (872), Expect = 6e-92
Identities = 186/406 (45%), Positives = 261/406 (63%), Gaps = 14/406 (3%)
Query: 102 PAQENKYQLEDKKENVVSSVEDDGAKTVEAIEESIVAVSASVPPEQKPVVEKVEASLPLP 161
P K Q + K++ ++S+ + I+E VS P EQK + E E
Sbjct: 7 PFDHQKTQNTEPKKSFITSLITLRSNN---IKEDTYFVSELKPTEQKSLQELKEKLSASS 63
Query: 162 PEQVSIYGIPLLA-DETSDVILLKFLRARDFKVKEAFTMIKNTILWRKEFGIEELMDEKL 220
+ S++G+ LL D+ +DVILLKFLRARDFKV ++ M++ + WR+EF E+L +E L
Sbjct: 64 SKASSMWGVSLLGGDDKADVILLKFLRARDFKVADSLRMLEKCLEWREEFKAEKLTEEDL 123
Query: 221 G-DELE-KVVYMHGFDKEGHPVCYNIYGEFQNKELYNKTFSDEEKRHNFLKWRIQFLEKS 278
G +LE KV YM G+DKEGHPVCYN YG F+ KE+Y + F DEEK + FL+WR+Q LE+
Sbjct: 124 GFKDLEGKVAYMRGYDKEGHPVCYNAYGVFKEKEMYERVFGDEEKLNKFLRWRVQVLERG 183
Query: 279 IRNLDFNHGGVCTIVHVNDLKDSPGPGKWELRQATKQALQLFQDNYPEFVAKQVFINVPW 338
++ L F GGV +I+ V DLKD P K ELR A+ Q L LFQDNYPE VA ++FINVPW
Sbjct: 184 VKMLHFKPGGVNSIIQVTDLKDMP---KRELRVASNQILSLFQDNYPELVATKIFINVPW 240
Query: 339 WYLAVNRMISPFLTQRTKSKFVFAGPSKSTETLLSYIAPEQLPVKYGGLSK--DGEFGNS 396
++ + M SPFLTQRTKSKFV + + ETL +I PE +PV+YGGLS+ D + G
Sbjct: 241 YFSVIYSMFSPFLTQRTKSKFVMSKEGNAAETLYKFIRPEDIPVQYGGLSRPTDSQNGPP 300
Query: 397 DSVTEITIRPASKHTVEFPVTE-KCLLSWEVRVIGWEVRYGAEFVPSNEGSYTVIVQKAR 455
+E +I+ K ++ E ++W++ V GW++ Y AEFVP+ E SY ++V+K +
Sbjct: 301 KPASEFSIKGGEKVNIQIEGIEGGATITWDIVVGGWDLEYSAEFVPNAEESYAIVVEKPK 360
Query: 456 KVASSEEAVLCNSFKINEPGKVVLTIDNTSSRKKKL-LYRLKTKTS 500
K+ +++EAV CNSF E GK++L++DNT SRKKK+ YR + S
Sbjct: 361 KMKATDEAV-CNSFTTVEAGKLILSVDNTLSRKKKVAAYRYTVRKS 405
>UniRef100_Q5W6M0 Hypothetical protein P0015F11.17 [Oryza sativa]
Length = 435
Score = 326 bits (836), Expect = 8e-88
Identities = 175/399 (43%), Positives = 253/399 (62%), Gaps = 24/399 (6%)
Query: 114 KENVVSSVEDDGAKTVEAIEESIVAVSASVPPEQKPVVEKVEASLPLPPEQVSIYGIPL- 172
K +++SS+ + A + + V++++P + + A L P+ +SI+G+PL
Sbjct: 28 KRSLMSSLMEATALLRSSSFKEDSYVASALPASDLRALADLRALLSTHPDPISIWGVPLN 87
Query: 173 ---------------LADETSDVILLKFLRARDFKVKEAFTMIKNTILWRKEFGIEELMD 217
ADE +DV+LLKFLRARDF+V++A M+ WR EF + ++D
Sbjct: 88 PAPPQGGEGAPAPAAAADERADVVLLKFLRARDFRVRDAHAMLLRCAAWRAEFRADAVLD 147
Query: 218 EKLG-DELEKVV-YMHGFDKEGHPVCYNIYGEFQNKELYNKTFSDEEKRHNFLKWRIQFL 275
E LG +LE VV YMHG+D+EGHPVCYN YG F+++++Y++ F D E+ FL+WR+Q +
Sbjct: 148 EDLGFKDLEGVVAYMHGWDREGHPVCYNAYGVFKDRDMYDRVFGDGERLARFLRWRVQVM 207
Query: 276 EKSIRNLDFNHGGVCTIVHVNDLKDSPGPGKWELRQATKQALQLFQDNYPEFVAKQVFIN 335
E+ +R L GGV I+ V DLKD P K ELR A+ Q L LFQDNYPE VA++VFIN
Sbjct: 208 ERGVRALHLRPGGVNAIIQVTDLKDMP---KRELRAASNQILSLFQDNYPEMVARKVFIN 264
Query: 336 VPWWYLAVNRMISPFLTQRTKSKFVFAGPSKSTETLLSYIAPEQLPVKYGGLSKDGEFGN 395
VPW++ + MISPFLT+RTKSKFV A ETL +I PE +PV+YGGLS+ G+ N
Sbjct: 265 VPWYFSVLFSMISPFLTERTKSKFVIAREGNVAETLFKFIRPELVPVQYGGLSRAGDLEN 324
Query: 396 S--DSVTEITIRPASKHTVEFPVTEK-CLLSWEVRVIGWEVRYGAEFVPSNEGSYTVIVQ 452
+E TI+ K +E E ++W++ V GWE+ YGAE+VP+ E SYT+ V+
Sbjct: 325 GPPKPASEFTIKGGEKVFLEIDGIEAGATITWDLVVGGWELEYGAEYVPAAEDSYTLCVE 384
Query: 453 KARKVASSEEAVLCNSFKINEPGKVVLTIDNTSSRKKKL 491
+ RKV ++ + + N+F E GK+VL+IDN+ SRK+K+
Sbjct: 385 RTRKVPAAADEPVHNAFTAREAGKMVLSIDNSGSRKRKV 423
>UniRef100_Q76H02 Hypothetical protein 5E08 [Solanum melongena]
Length = 206
Score = 267 bits (683), Expect = 5e-70
Identities = 121/203 (59%), Positives = 158/203 (77%)
Query: 170 IPLLADETSDVILLKFLRARDFKVKEAFTMIKNTILWRKEFGIEELMDEKLGDELEKVVY 229
+P E ++V+LLKFLRARD+KV E+F M+K T+ WRK+F I+ +++E LG +L Y
Sbjct: 3 LPSKGGEKTNVVLLKFLRARDYKVNESFEMLKKTLQWRKDFKIQSILEEDLGSDLAPAAY 62
Query: 230 MHGFDKEGHPVCYNIYGEFQNKELYNKTFSDEEKRHNFLKWRIQFLEKSIRNLDFNHGGV 289
M G D +GHP+CYNI+G +++LYNKTF EEKR FL+WR+Q +EK I+ LDF GGV
Sbjct: 63 MSGIDNQGHPICYNIFGVLDDEKLYNKTFGTEEKRKQFLRWRVQLMEKGIQQLDFKAGGV 122
Query: 290 CTIVHVNDLKDSPGPGKWELRQATKQALQLFQDNYPEFVAKQVFINVPWWYLAVNRMISP 349
+++ +NDLK+SPGP K E+R ATKQA+ L QDNYPEFVAK +FINVP+WY AV+ ++SP
Sbjct: 123 SSLLQINDLKNSPGPSKKEVRVATKQAVDLLQDNYPEFVAKSIFINVPFWYYAVHSLLSP 182
Query: 350 FLTQRTKSKFVFAGPSKSTETLL 372
FLTQRTKSKFVFA P+K TET L
Sbjct: 183 FLTQRTKSKFVFARPAKVTETSL 205
>UniRef100_Q9XFS6 Hypothetical protein [Arabidopsis thaliana]
Length = 147
Score = 108 bits (270), Expect = 4e-22
Identities = 57/138 (41%), Positives = 89/138 (64%), Gaps = 5/138 (3%)
Query: 367 STETLLSYIAPEQLPVKYGGLSK--DGEFGNSDSVTEITIRPASKHTVEFPVTEK-CLLS 423
+ ETL +I PE +PV+YGGLS+ D + G +E +I+ K ++ E ++
Sbjct: 7 AAETLYKFIRPEDIPVQYGGLSRPTDSQNGPPKPASEFSIKGGEKVNIQIEGIEGGATIT 66
Query: 424 WEVRVIGWEVRYGAEFVPSNEGSYTVIVQKARKVASSEEAVLCNSFKINEPGKVVLTIDN 483
W++ V GW++ Y AEFVP+ E SY ++V+K +K+ +++EAV CNSF E GK++L++DN
Sbjct: 67 WDIVVGGWDLEYSAEFVPNAEESYAIVVEKPKKMKATDEAV-CNSFTTVEAGKLILSVDN 125
Query: 484 TSSRKKKL-LYRLKTKTS 500
T SRKKK+ YR + S
Sbjct: 126 TLSRKKKVAAYRYTVRKS 143
>UniRef100_Q6J931 Hypothetical protein [Xerophyta humilis]
Length = 74
Score = 99.8 bits (247), Expect = 2e-19
Identities = 47/70 (67%), Positives = 58/70 (82%)
Query: 431 WEVRYGAEFVPSNEGSYTVIVQKARKVASSEEAVLCNSFKINEPGKVVLTIDNTSSRKKK 490
WEV YGAEFVPS E YTVIVQKARK S++E V+ +SFKI EPGK+VLT++N +S+KKK
Sbjct: 1 WEVTYGAEFVPSAENGYTVIVQKARKFTSTDEPVVKSSFKIGEPGKIVLTVENNTSKKKK 60
Query: 491 LLYRLKTKTS 500
LLYR K K++
Sbjct: 61 LLYRSKAKST 70
>UniRef100_Q6C9R9 Similar to sp|P47008 Saccharomyces cerevisiae YJL145w [Yarrowia
lipolytica]
Length = 362
Score = 98.6 bits (244), Expect = 4e-19
Identities = 77/269 (28%), Positives = 131/269 (48%), Gaps = 27/269 (10%)
Query: 135 SIVAVSASVPPEQKPVVEKVEASLPLPPEQVS---IYGIPLLADETS----------DVI 181
SI A + P + ++K++ LP E+ +YG L E D+I
Sbjct: 2 SIPAHLQILQPSELEALKKLQDELPKILEKTDYNEMYGHKLSEAEDGPGKAYSEVKRDII 61
Query: 182 LLKFLRARDFKVKEAFTMIKNTILWRKEFGIEELMDEKLGDELEK--VVYMHGFDKEGHP 239
LLKFL+ARD+ + + M+ + + WRKEF + K + +K V+ G E
Sbjct: 62 LLKFLKARDYDIAQTKDMLTDALKWRKEFDPLDCASAKHDSKFDKLGVITDKGAGGEPQV 121
Query: 240 VCYNIYGEFQNKELYNKTFSDEEKRHNFLKWRIQFLEKSIRNLDFNHGGVCT-IVHVNDL 298
+N+YG N++ + F D + FL+WR+ +E+S+ LDF G + ++ ++D
Sbjct: 122 TNWNLYGAVSNRK---EIFGDLK---GFLRWRVGIMERSLALLDFTKPGAGSMLLQIHDY 175
Query: 299 KD-SPGPGKWELRQATKQALQLFQDNYPEFVAKQVFINVPWWYLAVNRMISPFLTQRTKS 357
K+ S E + A+K+ +++FQ YPE + ++ F+NVP V ++ FL++ T +
Sbjct: 176 KNVSFLRLDAETKAASKETIRVFQSYYPETLERKFFVNVPTLMQFVFGFVNKFLSRETVA 235
Query: 358 KFVFAGPSKSTETLLSYIAPEQLPVKYGG 386
KFV K L +P +YGG
Sbjct: 236 KFVVYSNGKDLHKSLG----SWVPAEYGG 260
>UniRef100_Q6FNM8 Candida glabrata strain CBS138 chromosome J complete sequence
[Candida glabrata]
Length = 306
Score = 95.1 bits (235), Expect = 4e-18
Identities = 84/282 (29%), Positives = 125/282 (43%), Gaps = 23/282 (8%)
Query: 121 VEDDGAKTVEAIEESIVAVSASVPPEQKP--VVEKVEASLPLPPEQVSIYGIPLLADETS 178
+ ++ K + I +S +V P P + + + +L EQ+ G L D
Sbjct: 1 MSEEAIKKEKEILDSFPSVCEPGSPSGTPGNLNDSQKKALAQLKEQLQKDGYKLRLD--- 57
Query: 179 DVILLKFLRARDFKVKEAFTMIKNTILWRKEFGIEELMDEKLGDELEKVV-----YMHGF 233
D LL+FLRAR F V A M WRK G++ ++++ +E V Y H
Sbjct: 58 DATLLRFLRARKFNVAMAKEMYVACEKWRKSAGVDTILEDFHYEEKPLVAKYYPQYYHKI 117
Query: 234 DKEGHPVCYNIYGEFQNKELYNKTFSDEEKRH------NFLKWRIQFLEKSIRNLDFNHG 287
DK+G PV + G E+Y T + ++ +F+K+R+ +S L
Sbjct: 118 DKDGRPVYFEELGTVNLNEMYKITTHERMIKNLVWEYESFVKYRLPACSRSRGYLIETS- 176
Query: 288 GVCTIVHVNDLKDSPGPGKWELRQATKQALQLFQDNYPEFVAKQVFINVPWWYLAVNRMI 347
CTI+ DLK + + K+A + Q+ YPE + K IN P+ + R+
Sbjct: 177 --CTIM---DLKGISISSAYHVLSYVKEASHIGQNYYPERMGKFYLINAPFGFSTAFRLF 231
Query: 348 SPFLTQRTKSKFVFAGPSKSTETLLSYIAPEQLPVKYGGLSK 389
PFL T SK G S E LL I E LPVKYGG S+
Sbjct: 232 KPFLDPVTVSKIFILGSSYKKE-LLKQIPAENLPVKYGGKSE 272
>UniRef100_Q6BPM1 Similar to sp|P46250 Candida albicans SEC14 cytosolic factor
[Debaryomyces hansenii]
Length = 301
Score = 95.1 bits (235), Expect = 4e-18
Identities = 78/259 (30%), Positives = 114/259 (43%), Gaps = 26/259 (10%)
Query: 140 SASVPPEQKPVVEKVEASLPLPPEQVSIYGIPLLADETSDVILLKFLRARDFKVKEAFTM 199
++++ +QK +E++ A L + + D LL+FLRAR F + +A M
Sbjct: 25 TSNLTDDQKKTLEQLRAELTADGYK----------ERLDDATLLRFLRARKFDIVKAKQM 74
Query: 200 IKNTILWRKEFGIEELMDEKLGDELEKVV-----YMHGFDKEGHPVCYNIYGEFQNKELY 254
WRK+FG ++ + DE V Y H DK+G PV + G+ E+
Sbjct: 75 YVKCETWRKDFGTNTILTDFHYDEKPLVAKLYPQYYHKIDKDGRPVYFEELGKVNLNEML 134
Query: 255 NKTFSDEEKRHNFLKWRIQ-FLEKSIRNLDFNHGGV----CTIVHVNDLKDSPGPGKWEL 309
T +E+ L W + F + G + CTI+ DLK +++
Sbjct: 135 KIT--TQERMLKNLVWEYESFALYRLPACSRQQGSLVETSCTIM---DLKGISLSAAYQV 189
Query: 310 RQATKQALQLFQDNYPEFVAKQVFINVPWWYLAVNRMISPFLTQRTKSKFVFAGPSKSTE 369
K+A + QD YPE + K IN P+ + R+ PFL T SK G S E
Sbjct: 190 VNYVKEASAIGQDYYPERMGKFYLINSPFGFSTAFRVFKPFLDPVTVSKIFILGSSYQKE 249
Query: 370 TLLSYIAPEQLPVKYGGLS 388
LL I PE LP KYGG S
Sbjct: 250 -LLKQIPPENLPAKYGGKS 267
>UniRef100_Q75DK1 SEC14 cytosolic factor [Ashbya gossypii]
Length = 311
Score = 94.4 bits (233), Expect = 7e-18
Identities = 76/224 (33%), Positives = 101/224 (44%), Gaps = 16/224 (7%)
Query: 179 DVILLKFLRARDFKVKEAFTMIKNTILWRKEFGIEELMDEKLGDELEKVV-----YMHGF 233
D LL+FLRAR F V A M +N WRKE G++ + ++ +E V Y H
Sbjct: 56 DSTLLRFLRARKFDVAAARAMFENCEKWRKENGVDTIFEDFHYEEKPLVAKFYPQYYHKT 115
Query: 234 DKEGHPVCYNIYGEFQNKELYNKTFSDEEKRHNFLKWRIQF-----LEKSIRNLDFNHGG 288
DK+G PV G E+Y T +E+ L W + L S R D
Sbjct: 116 DKDGRPVYIEELGAVNLTEMYKIT--TQERMLKNLIWEYESFSRYRLPASSRQADCLVET 173
Query: 289 VCTIVHVNDLKDSPGPGKWELRQATKQALQLFQDNYPEFVAKQVFINVPWWYLAVNRMIS 348
CTI+ DLK ++ ++A + Q+ YPE + K IN P+ + A R+
Sbjct: 174 SCTIL---DLKGISISAAAQVLSYVREASNIGQNYYPERMGKFYMINAPFGFSAAFRLFK 230
Query: 349 PFLTQRTKSKFVFAGPSKSTETLLSYIAPEQLPVKYGGLSKDGE 392
PFL T SK G S E LL I E LPVK+GG S E
Sbjct: 231 PFLDPVTVSKIFILGSSYQKE-LLKQIPAENLPVKFGGQSDVSE 273
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.313 0.131 0.377
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 882,592,028
Number of Sequences: 2790947
Number of extensions: 39567520
Number of successful extensions: 116791
Number of sequences better than 10.0: 607
Number of HSP's better than 10.0 without gapping: 220
Number of HSP's successfully gapped in prelim test: 396
Number of HSP's that attempted gapping in prelim test: 115380
Number of HSP's gapped (non-prelim): 1279
length of query: 503
length of database: 848,049,833
effective HSP length: 132
effective length of query: 371
effective length of database: 479,644,829
effective search space: 177948231559
effective search space used: 177948231559
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 77 (34.3 bits)
Medicago: description of AC148755.7


