

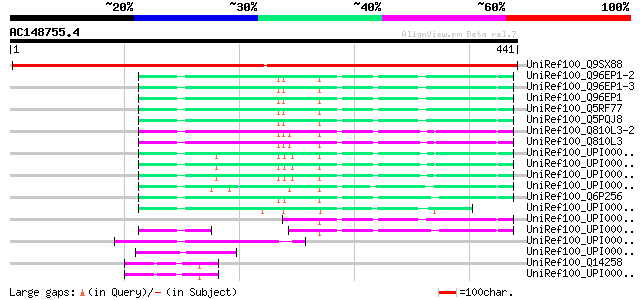
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148755.4 + phase: 1 /partial
(441 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9SX88 F16N3.15 [Arabidopsis thaliana] 544 e-153
UniRef100_Q96EP1-2 Splice isoform 2 of Q96EP1 [Homo sapiens] 129 2e-28
UniRef100_Q96EP1-3 Splice isoform 3 of Q96EP1 [Homo sapiens] 129 2e-28
UniRef100_Q96EP1 Ubiquitin ligase protein CHFR [Homo sapiens] 129 2e-28
UniRef100_Q5RF77 Hypothetical protein DKFZp469H0121 [Pongo pygma... 128 3e-28
UniRef100_Q5PQJ8 Hypothetical protein [Rattus norvegicus] 128 4e-28
UniRef100_Q810L3-2 Splice isoform 2 of Q810L3 [Mus musculus] 125 2e-27
UniRef100_Q810L3 Ubiquitin ligase protein CHFR [Mus musculus] 125 3e-27
UniRef100_UPI00003AAFA6 UPI00003AAFA6 UniRef100 entry 118 4e-25
UniRef100_UPI00003AAFA5 UPI00003AAFA5 UniRef100 entry 118 4e-25
UniRef100_UPI00003AAFA4 UPI00003AAFA4 UniRef100 entry 118 4e-25
UniRef100_UPI000036105F UPI000036105F UniRef100 entry 117 7e-25
UniRef100_Q6P256 Hypothetical protein MGC75959 [Xenopus tropicalis] 115 2e-24
UniRef100_UPI0000438729 UPI0000438729 UniRef100 entry 90 1e-16
UniRef100_UPI0000368C6B UPI0000368C6B UniRef100 entry 82 3e-14
UniRef100_UPI0000337E34 UPI0000337E34 UniRef100 entry 82 3e-14
UniRef100_UPI00001CFE64 UPI00001CFE64 UniRef100 entry 68 6e-10
UniRef100_UPI00002218AE UPI00002218AE UniRef100 entry 65 5e-09
UniRef100_Q14258 Tripartite motif protein 25 [Homo sapiens] 56 2e-06
UniRef100_UPI000036B09C UPI000036B09C UniRef100 entry 55 4e-06
>UniRef100_Q9SX88 F16N3.15 [Arabidopsis thaliana]
Length = 473
Score = 544 bits (1402), Expect = e-153
Identities = 252/439 (57%), Positives = 320/439 (72%), Gaps = 2/439 (0%)
Query: 3 DSRYPDVEIRSDKGQIRSEISTASCDKHRWCKIVRNSDLCSATLKNKSSNTILVDGAEVG 62
D+R+ D+EIR + I SEI +S +KH WC+I +N SAT+ NKSS+ ILVD A V
Sbjct: 35 DTRFSDIEIRCNDMVICSEIKPSSLEKHEWCRITKNLGQSSATIHNKSSDAILVDKAVVP 94
Query: 63 KGDTVVIKDRSEIIPGPAKEGFVSYKFQIVSSPEICQTLLKICVDVDHAKCSICLNIWHD 122
K V I SEI+PGP ++G++ Y+F I+ +PE LL+I +D +HAKCSICLNIWHD
Sbjct: 95 KDGAVDIISGSEIVPGPEEQGYLQYRFTIMPAPESRTQLLQISIDPEHAKCSICLNIWHD 154
Query: 123 VVTVAPCLHNFCNGCFSEWLRRSQEKRSTVLCPQCRAVVQFVGKNHFLRTIAEDMVKADS 182
VVT APCLHNFCNGCFSEW+RRS+EK VLCPQCR VQ+VGKNHFL+ I E+++K D+
Sbjct: 155 VVTAAPCLHNFCNGCFSEWMRRSEEKHKHVLCPQCRTTVQYVGKNHFLKNIQEEILKVDA 214
Query: 183 SLRRSHDEVALLDTYASVRSNLVIGSGKKSRKRERANTTMVNQSDGPDHHCSQCVTVVGG 242
+LRR +++A+LD+ AS++SNL+IGS +K R A T + D C QCV +GG
Sbjct: 215 ALRRPAEDIAVLDSSASIQSNLIIGSKRKRRLNMPAPTH--EERDSLRLQCPQCVANIGG 272
Query: 243 FQCDPSTIHLQCQECGGMMPSRTGLGVPQYCLGCDRPFCGAYWSALGVTGSGSYPVCSPD 302
++C+ HLQC C GMMP R L VP +C GCDRPFCGAYWS+ VT S PVC +
Sbjct: 273 YRCEHHGAHLQCHLCQGMMPFRANLQVPLHCKGCDRPFCGAYWSSENVTQGVSGPVCVRE 332
Query: 303 TLRPISEHTISGIPLVAHEKNLHEQNITESCIRQMGRTLQDVISEWIAKLNNREIDTTRL 362
T RPISE TI+ IP + HE N HEQ+IT+ CI M +T+ DV++EW+ NNREID +R+
Sbjct: 333 TFRPISERTITRIPFITHEMNRHEQDITQRCIAHMEKTVPDVVAEWLRLFNNREIDRSRM 392
Query: 363 MLNHAEMMTAGTLVCSDCYQKLVSFFLYWFRISTPKHLLPPEASTREDCWYGYACRTQHR 422
LNHAE +TA T VC+DCY KLV F LYWFRI+ P++ LP + + REDCWYGYACRTQH
Sbjct: 393 PLNHAETITASTHVCNDCYDKLVGFLLYWFRITLPRNHLPADVAAREDCWYGYACRTQHH 452
Query: 423 SEEHARKRNHVCRPTRGSH 441
+E+HARKRNHVCRPTRG+H
Sbjct: 453 NEDHARKRNHVCRPTRGNH 471
>UniRef100_Q96EP1-2 Splice isoform 2 of Q96EP1 [Homo sapiens]
Length = 652
Score = 129 bits (323), Expect = 2e-28
Identities = 94/378 (24%), Positives = 151/378 (39%), Gaps = 72/378 (19%)
Query: 113 CSICLNIWHDVVTVAPCLHNFCNGCFSEWLRRSQEKRSTVLCPQCRAVVQFVGKNHFLRT 172
C IC ++ HD V++ PC+H FC C+S W+ RS LCP CR V+ + KNH L
Sbjct: 292 CIICQDLLHDCVSLQPCMHTFCAACYSGWMERSS------LCPTCRCPVERICKNHILNN 345
Query: 173 IAEDMVKADSSLRRSHDEVALLDTYASVRSNLVIGSGKKSRKRERANTTMVNQSDGPDHH 232
+ E + RS ++V +D + +++ ++S E ++ + + D
Sbjct: 346 LVEAYLIQHPDKSRSEEDVQSMDARNKITQDMLQPKVRRSFSDEEGSSEDLLELSDVDSE 405
Query: 233 ----------CSQC------------------------------------VTVVGGFQCD 246
C QC T V + C
Sbjct: 406 SSDISQPYVVCRQCPEYRRQAAQPPHCPAPEGEPGAPQALGDAPSTSVSLTTAVQDYVCP 465
Query: 247 PSTIHLQCQECGGMMPSRTGLG------VPQYCLGCDRPFCGAYWSALGVTGSGSYPVCS 300
H C C MP R PQ C C +PFC YW G T +G Y +
Sbjct: 466 LQGSHALCTCCFQPMPDRRAEREQDPRVAPQQCAVCLQPFCHLYW---GCTRTGCYGCLA 522
Query: 301 PDTLRPISEHTISGIPLVAHEKNLHEQNITESCIRQMGRTLQDVISEWIAKLNNREIDTT 360
P + + + G+ N +E +I ++ + G T +++++E + L
Sbjct: 523 PFCELNLGDKCLDGVL----NNNSYESDILKNYLATRGLTWKNMLTESLVALQRG----- 573
Query: 361 RLMLNHAEMMTAGTLVCSDCYQKLVSFFLYWFRISTPKHLLPPEASTREDCWYGYACRTQ 420
+ L +T T++C C + Y +R + P LP ++R DC++G CRTQ
Sbjct: 574 -VFLLSDYRVTGDTVLCYCCGLRSFRELTYQYRQNIPASELPVAVTSRPDCYWGRNCRTQ 632
Query: 421 HRSEEHARKRNHVCRPTR 438
++ HA K NH+C TR
Sbjct: 633 VKA-HHAMKFNHICEQTR 649
>UniRef100_Q96EP1-3 Splice isoform 3 of Q96EP1 [Homo sapiens]
Length = 623
Score = 129 bits (323), Expect = 2e-28
Identities = 94/378 (24%), Positives = 151/378 (39%), Gaps = 72/378 (19%)
Query: 113 CSICLNIWHDVVTVAPCLHNFCNGCFSEWLRRSQEKRSTVLCPQCRAVVQFVGKNHFLRT 172
C IC ++ HD V++ PC+H FC C+S W+ RS LCP CR V+ + KNH L
Sbjct: 263 CIICQDLLHDCVSLQPCMHTFCAACYSGWMERSS------LCPTCRCPVERICKNHILNN 316
Query: 173 IAEDMVKADSSLRRSHDEVALLDTYASVRSNLVIGSGKKSRKRERANTTMVNQSDGPDHH 232
+ E + RS ++V +D + +++ ++S E ++ + + D
Sbjct: 317 LVEAYLIQHPDKSRSEEDVQSMDARNKITQDMLQPKVRRSFSDEEGSSEDLLELSDVDSE 376
Query: 233 ----------CSQC------------------------------------VTVVGGFQCD 246
C QC T V + C
Sbjct: 377 SSDISQPYVVCRQCPEYRRQAAQPPHCPAPEGEPGAPQALGDAPSTSVSLTTAVQDYVCP 436
Query: 247 PSTIHLQCQECGGMMPSRTGLG------VPQYCLGCDRPFCGAYWSALGVTGSGSYPVCS 300
H C C MP R PQ C C +PFC YW G T +G Y +
Sbjct: 437 LQGSHALCTCCFQPMPDRRAEREQDPRVAPQQCAVCLQPFCHLYW---GCTRTGCYGCLA 493
Query: 301 PDTLRPISEHTISGIPLVAHEKNLHEQNITESCIRQMGRTLQDVISEWIAKLNNREIDTT 360
P + + + G+ N +E +I ++ + G T +++++E + L
Sbjct: 494 PFCELNLGDKCLDGVL----NNNSYESDILKNYLATRGLTWKNMLTESLVALQRG----- 544
Query: 361 RLMLNHAEMMTAGTLVCSDCYQKLVSFFLYWFRISTPKHLLPPEASTREDCWYGYACRTQ 420
+ L +T T++C C + Y +R + P LP ++R DC++G CRTQ
Sbjct: 545 -VFLLSDYRVTGDTVLCYCCGLRSFRELTYQYRQNIPASELPVAVTSRPDCYWGRNCRTQ 603
Query: 421 HRSEEHARKRNHVCRPTR 438
++ HA K NH+C TR
Sbjct: 604 VKA-HHAMKFNHICEQTR 620
>UniRef100_Q96EP1 Ubiquitin ligase protein CHFR [Homo sapiens]
Length = 664
Score = 129 bits (323), Expect = 2e-28
Identities = 94/378 (24%), Positives = 151/378 (39%), Gaps = 72/378 (19%)
Query: 113 CSICLNIWHDVVTVAPCLHNFCNGCFSEWLRRSQEKRSTVLCPQCRAVVQFVGKNHFLRT 172
C IC ++ HD V++ PC+H FC C+S W+ RS LCP CR V+ + KNH L
Sbjct: 304 CIICQDLLHDCVSLQPCMHTFCAACYSGWMERSS------LCPTCRCPVERICKNHILNN 357
Query: 173 IAEDMVKADSSLRRSHDEVALLDTYASVRSNLVIGSGKKSRKRERANTTMVNQSDGPDHH 232
+ E + RS ++V +D + +++ ++S E ++ + + D
Sbjct: 358 LVEAYLIQHPDKSRSEEDVQSMDARNKITQDMLQPKVRRSFSDEEGSSEDLLELSDVDSE 417
Query: 233 ----------CSQC------------------------------------VTVVGGFQCD 246
C QC T V + C
Sbjct: 418 SSDISQPYVVCRQCPEYRRQAAQPPHCPAPEGEPGAPQALGDAPSTSVSLTTAVQDYVCP 477
Query: 247 PSTIHLQCQECGGMMPSRTGLG------VPQYCLGCDRPFCGAYWSALGVTGSGSYPVCS 300
H C C MP R PQ C C +PFC YW G T +G Y +
Sbjct: 478 LQGSHALCTCCFQPMPDRRAEREQDPRVAPQQCAVCLQPFCHLYW---GCTRTGCYGCLA 534
Query: 301 PDTLRPISEHTISGIPLVAHEKNLHEQNITESCIRQMGRTLQDVISEWIAKLNNREIDTT 360
P + + + G+ N +E +I ++ + G T +++++E + L
Sbjct: 535 PFCELNLGDKCLDGVL----NNNSYESDILKNYLATRGLTWKNMLTESLVALQRG----- 585
Query: 361 RLMLNHAEMMTAGTLVCSDCYQKLVSFFLYWFRISTPKHLLPPEASTREDCWYGYACRTQ 420
+ L +T T++C C + Y +R + P LP ++R DC++G CRTQ
Sbjct: 586 -VFLLSDYRVTGDTVLCYCCGLRSFRELTYQYRQNIPASELPVAVTSRPDCYWGRNCRTQ 644
Query: 421 HRSEEHARKRNHVCRPTR 438
++ HA K NH+C TR
Sbjct: 645 VKA-HHAMKFNHICEQTR 661
>UniRef100_Q5RF77 Hypothetical protein DKFZp469H0121 [Pongo pygmaeus]
Length = 571
Score = 128 bits (322), Expect = 3e-28
Identities = 93/377 (24%), Positives = 152/377 (39%), Gaps = 71/377 (18%)
Query: 113 CSICLNIWHDVVTVAPCLHNFCNGCFSEWLRRSQEKRSTVLCPQCRAVVQFVGKNHFLRT 172
C IC ++ HD V++ PC+H FC C+S W+ RS LCP CR V+ + KNH L
Sbjct: 212 CIICQDLLHDCVSLQPCMHTFCAACYSGWMERSS------LCPTCRCPVERICKNHILNN 265
Query: 173 IAEDMVKADSSLRRSHDEVALLDTYASVRSNLVIGSGKKSRKRERANTTMVNQSDGPDHH 232
+ E + RS ++V +D + +++ ++S E ++ + + D
Sbjct: 266 LVEAYLIQHPDKSRSEEDVQSMDARNKITQDMLQPKVRRSFSDEEGSSEDLLELSDVDSE 325
Query: 233 ----------CSQC-----------------------------------VTVVGGFQCDP 247
C QC +T V + C
Sbjct: 326 SSDISQPYVVCRQCPEYRRQAAQPPPCPAPEGEPGVPQALVDAPSTSVSLTTVQDYVCPL 385
Query: 248 STIHLQCQECGGMMPSRTGLG------VPQYCLGCDRPFCGAYWSALGVTGSGSYPVCSP 301
H C C MP R PQ C C +PFC YW G T +G + +P
Sbjct: 386 QGSHALCTCCFQPMPDRRAEREQNPRVAPQQCAVCLQPFCHLYW---GCTRTGCFGCLAP 442
Query: 302 DTLRPISEHTISGIPLVAHEKNLHEQNITESCIRQMGRTLQDVISEWIAKLNNREIDTTR 361
+ + + G+ N +E +I ++ + G T +++++E + L
Sbjct: 443 FCELNLGDKCLDGVL----NNNSYESDILKNYLATRGLTWKNMLTESLVALQRG------ 492
Query: 362 LMLNHAEMMTAGTLVCSDCYQKLVSFFLYWFRISTPKHLLPPEASTREDCWYGYACRTQH 421
+ L +T T++C C + Y +R + P LP ++R DC++G CRTQ
Sbjct: 493 VFLLSDYRVTGDTILCYCCGLRSFRELTYQYRQNIPASELPVAVTSRPDCYWGRNCRTQV 552
Query: 422 RSEEHARKRNHVCRPTR 438
++ HA K NH+C TR
Sbjct: 553 KA-HHAMKFNHICEQTR 568
>UniRef100_Q5PQJ8 Hypothetical protein [Rattus norvegicus]
Length = 663
Score = 128 bits (321), Expect = 4e-28
Identities = 92/378 (24%), Positives = 152/378 (39%), Gaps = 72/378 (19%)
Query: 113 CSICLNIWHDVVTVAPCLHNFCNGCFSEWLRRSQEKRSTVLCPQCRAVVQFVGKNHFLRT 172
C IC ++ HD V++ PC+H FC C+S W+ RS LCP CR V+ + KNH L
Sbjct: 303 CIICQDLLHDCVSLQPCMHTFCAACYSGWMERSS------LCPTCRCPVERICKNHILNN 356
Query: 173 IAEDMVKADSSLRRSHDEVALLDTYASVRSNLVIGSGKKSRKRERANTTMVNQSDGPDHH 232
+ E + RS ++V +D + +++ ++S E ++ + + D
Sbjct: 357 LVEAYLLQHPDKSRSEEDVRSMDARNKITQDMLQPKVRRSFSDEEGSSEDLLELSDVDSE 416
Query: 233 ----------CSQC------------------------------------VTVVGGFQCD 246
C QC +T G+ C
Sbjct: 417 SSDISQPYIVCRQCPEYRRQAVQSLPCPVPESELGATQALGGEAPSTSASLTTAPGYTCP 476
Query: 247 PSTIHLQCQECGGMMPSRTGLG------VPQYCLGCDRPFCGAYWSALGVTGSGSYPVCS 300
H C C MP R PQ C C +PFC YW G T +G + +
Sbjct: 477 LQGSHAICTCCFQPMPDRRAEHEQDSRIAPQQCAVCLQPFCHLYW---GCTRTGCFGCLA 533
Query: 301 PDTLRPISEHTISGIPLVAHEKNLHEQNITESCIRQMGRTLQDVISEWIAKLNNREIDTT 360
P + + + G+ N +E +I ++ + G T +++++E + L
Sbjct: 534 PFCELNLGDKCLDGVL----NNNNYESDILKNYLTTRGLTWKNMLTESLLALQRG----- 584
Query: 361 RLMLNHAEMMTAGTLVCSDCYQKLVSFFLYWFRISTPKHLLPPEASTREDCWYGYACRTQ 420
+ + +T T++C C + Y +R + P LP ++R DC++G CRTQ
Sbjct: 585 -VFILSDYRITGNTVLCYCCGLRSFRELTYQYRQNIPASELPVTVTSRPDCYWGRNCRTQ 643
Query: 421 HRSEEHARKRNHVCRPTR 438
++ HA K NH+C TR
Sbjct: 644 VKA-HHAMKFNHICEQTR 660
>UniRef100_Q810L3-2 Splice isoform 2 of Q810L3 [Mus musculus]
Length = 663
Score = 125 bits (314), Expect = 2e-27
Identities = 95/378 (25%), Positives = 152/378 (40%), Gaps = 72/378 (19%)
Query: 113 CSICLNIWHDVVTVAPCLHNFCNGCFSEWLRRSQEKRSTVLCPQCRAVVQFVGKNHFLRT 172
C IC ++ HD V++ PC+H FC C+S W+ RS LCP CR V+ + KNH L
Sbjct: 303 CIICQDLLHDCVSLQPCMHTFCAACYSGWMERSS------LCPTCRCPVERICKNHILNN 356
Query: 173 IAEDMVKADSSLRRSHDEVALLDTYASVRSNLVIGSGKKSRKRERANTTMVNQSDGPDHH 232
+ E + RS ++V +D + +++ ++S E ++ + + D
Sbjct: 357 LVEAYLIQHPDKSRSEEDVRSMDARNKITQDMLQPKVRRSFSDEEGSSEDLLELSDVDSE 416
Query: 233 ----------CSQC----------------------VTVVGG--------------FQCD 246
C QC +GG + C
Sbjct: 417 SSDISQPYIVCRQCPEYRRQAVQSLPCPVPESELGATLALGGEAPSTSASLPTAPDYMCP 476
Query: 247 PSTIHLQCQECGGMMPSRTGLG------VPQYCLGCDRPFCGAYWSALGVTGSGSYPVCS 300
H C C MP R PQ C C +PFC YW G T +G + +
Sbjct: 477 LQGSHAICTCCFQPMPDRRAEREQDPRVAPQQCAVCLQPFCHLYW---GCTRTGCFGCLA 533
Query: 301 PDTLRPISEHTISGIPLVAHEKNLHEQNITESCIRQMGRTLQDVISEWIAKLNNREIDTT 360
P + + + G+ N +E +I ++ + G T + V++E + L
Sbjct: 534 PFCELNLGDKCLDGVL----NNNNYESDILKNYLATRGLTWKSVLTESLLALQRGVF--- 586
Query: 361 RLMLNHAEMMTAGTLVCSDCYQKLVSFFLYWFRISTPKHLLPPEASTREDCWYGYACRTQ 420
ML+ + T T++C C + Y +R + P LP ++R DC++G CRTQ
Sbjct: 587 --MLSDYRI-TGNTVLCYCCGLRSFRELTYQYRQNIPASELPVTVTSRPDCYWGRNCRTQ 643
Query: 421 HRSEEHARKRNHVCRPTR 438
++ HA K NH+C TR
Sbjct: 644 VKA-HHAMKFNHICEQTR 660
>UniRef100_Q810L3 Ubiquitin ligase protein CHFR [Mus musculus]
Length = 664
Score = 125 bits (313), Expect = 3e-27
Identities = 95/379 (25%), Positives = 152/379 (40%), Gaps = 73/379 (19%)
Query: 113 CSICLNIWHDVVTVAPCLHNFCNGCFSEWLRRSQEKRSTVLCPQCRAVVQFVGKNHFLRT 172
C IC ++ HD V++ PC+H FC C+S W+ RS LCP CR V+ + KNH L
Sbjct: 303 CIICQDLLHDCVSLQPCMHTFCAACYSGWMERSS------LCPTCRCPVERICKNHILNN 356
Query: 173 IAEDMVKADSSLRRSHDEVALLDTYASVRSNLVIGSGKKSRKRERANTTMVNQSDGPDHH 232
+ E + RS ++V +D + +++ ++S E ++ + + D
Sbjct: 357 LVEAYLIQHPDKSRSEEDVRSMDARNKITQDMLQPKVRRSFSDEEGSSEDLLELSDVDSE 416
Query: 233 ----------CSQC----------------------VTVVGG---------------FQC 245
C QC +GG + C
Sbjct: 417 SSDISQPYIVCRQCPEYRRQAVQSLPCPVPESELGATLALGGEAPSTSASLPTAAPDYMC 476
Query: 246 DPSTIHLQCQECGGMMPSRTGLG------VPQYCLGCDRPFCGAYWSALGVTGSGSYPVC 299
H C C MP R PQ C C +PFC YW G T +G +
Sbjct: 477 PLQGSHAICTCCFQPMPDRRAEREQDPRVAPQQCAVCLQPFCHLYW---GCTRTGCFGCL 533
Query: 300 SPDTLRPISEHTISGIPLVAHEKNLHEQNITESCIRQMGRTLQDVISEWIAKLNNREIDT 359
+P + + + G+ N +E +I ++ + G T + V++E + L
Sbjct: 534 APFCELNLGDKCLDGVL----NNNNYESDILKNYLATRGLTWKSVLTESLLALQRGVF-- 587
Query: 360 TRLMLNHAEMMTAGTLVCSDCYQKLVSFFLYWFRISTPKHLLPPEASTREDCWYGYACRT 419
ML+ + T T++C C + Y +R + P LP ++R DC++G CRT
Sbjct: 588 ---MLSDYRI-TGNTVLCYCCGLRSFRELTYQYRQNIPASELPVTVTSRPDCYWGRNCRT 643
Query: 420 QHRSEEHARKRNHVCRPTR 438
Q ++ HA K NH+C TR
Sbjct: 644 QVKA-HHAMKFNHICEQTR 661
>UniRef100_UPI00003AAFA6 UPI00003AAFA6 UniRef100 entry
Length = 583
Score = 118 bits (295), Expect = 4e-25
Identities = 96/383 (25%), Positives = 151/383 (39%), Gaps = 77/383 (20%)
Query: 113 CSICLNIWHDVVTVAPCLHNFCNGCFSEWLRRSQEKRSTVLCPQCRAVVQFVGKNHFLRT 172
C IC + HD V++ PC+H FC C+S W+ RS LCP CR V+ + KNH L
Sbjct: 218 CIICQELLHDCVSLQPCMHTFCAACYSGWMERSS------LCPTCRCPVERICKNHILNN 271
Query: 173 IAEDMV-----KADSSLRRSHDEVALLDTYASVRSNLVIGSGKKSRKRERANTTMVNQSD 227
+ E + K R+ D+V +D + +++ ++S E ++ + +
Sbjct: 272 LVEAYLIQHPGKFQVYKCRNEDDVRSMDARNKITQDMLQPKVRRSFSDEEGSSEDLLELS 331
Query: 228 GPDHH----------CSQCV----------------TVVGGFQ----------------- 244
D C QC T GG Q
Sbjct: 332 DVDSESSDISQPYIVCRQCPGYRRHPVPALPGTGQETEAGGMQALGDAPSTSANFPAAVQ 391
Query: 245 ---CDPSTIHLQCQECGGMMPSRTGLG------VPQYCLGCDRPFCGAYWSALGVTGSGS 295
C H+ C C MP R PQ C C +PFC YW G T
Sbjct: 392 EYVCPAQGSHVICTCCFQPMPDRRAEREQNPHVAPQQCTVCLQPFCHLYW---GCTRMAC 448
Query: 296 YPVCSPDTLRPISEHTISGIPLVAHEKNLHEQNITESCIRQMGRTLQDVISEWIAKLNNR 355
+ +P + + + G+ N +E +I + + G T +++++E + L
Sbjct: 449 FGCLAPFCEINLGDKCLDGVL----NNNHYESDILKDYLASRGLTWKNMLNESLLALQRG 504
Query: 356 EIDTTRLMLNHAEMMTAGTLVCSDCYQKLVSFFLYWFRISTPKHLLPPEASTREDCWYGY 415
ML+ + T T++C C + Y +R + P LP ++R DC++G
Sbjct: 505 VF-----MLSDYRI-TGNTVLCYCCGLRSFRELAYQYRQNIPVAELPVTVTSRPDCYWGR 558
Query: 416 ACRTQHRSEEHARKRNHVCRPTR 438
CRTQ ++ HA K NH+C TR
Sbjct: 559 NCRTQVKA-HHAMKFNHICEQTR 580
>UniRef100_UPI00003AAFA5 UPI00003AAFA5 UniRef100 entry
Length = 614
Score = 118 bits (295), Expect = 4e-25
Identities = 96/383 (25%), Positives = 151/383 (39%), Gaps = 77/383 (20%)
Query: 113 CSICLNIWHDVVTVAPCLHNFCNGCFSEWLRRSQEKRSTVLCPQCRAVVQFVGKNHFLRT 172
C IC + HD V++ PC+H FC C+S W+ RS LCP CR V+ + KNH L
Sbjct: 249 CIICQELLHDCVSLQPCMHTFCAACYSGWMERSS------LCPTCRCPVERICKNHILNN 302
Query: 173 IAEDMV-----KADSSLRRSHDEVALLDTYASVRSNLVIGSGKKSRKRERANTTMVNQSD 227
+ E + K R+ D+V +D + +++ ++S E ++ + +
Sbjct: 303 LVEAYLIQHPGKFQVYKCRNEDDVRSMDARNKITQDMLQPKVRRSFSDEEGSSEDLLELS 362
Query: 228 GPDHH----------CSQCV----------------TVVGGFQ----------------- 244
D C QC T GG Q
Sbjct: 363 DVDSESSDISQPYIVCRQCPGYRRHPVPALPGTGQETEAGGMQALGDAPSTSANFPAAVQ 422
Query: 245 ---CDPSTIHLQCQECGGMMPSRTGLG------VPQYCLGCDRPFCGAYWSALGVTGSGS 295
C H+ C C MP R PQ C C +PFC YW G T
Sbjct: 423 EYVCPAQGSHVICTCCFQPMPDRRAEREQNPHVAPQQCTVCLQPFCHLYW---GCTRMAC 479
Query: 296 YPVCSPDTLRPISEHTISGIPLVAHEKNLHEQNITESCIRQMGRTLQDVISEWIAKLNNR 355
+ +P + + + G+ N +E +I + + G T +++++E + L
Sbjct: 480 FGCLAPFCEINLGDKCLDGVL----NNNHYESDILKDYLASRGLTWKNMLNESLLALQRG 535
Query: 356 EIDTTRLMLNHAEMMTAGTLVCSDCYQKLVSFFLYWFRISTPKHLLPPEASTREDCWYGY 415
ML+ + T T++C C + Y +R + P LP ++R DC++G
Sbjct: 536 VF-----MLSDYRI-TGNTVLCYCCGLRSFRELAYQYRQNIPVAELPVTVTSRPDCYWGR 589
Query: 416 ACRTQHRSEEHARKRNHVCRPTR 438
CRTQ ++ HA K NH+C TR
Sbjct: 590 NCRTQVKA-HHAMKFNHICEQTR 611
>UniRef100_UPI00003AAFA4 UPI00003AAFA4 UniRef100 entry
Length = 669
Score = 118 bits (295), Expect = 4e-25
Identities = 96/383 (25%), Positives = 151/383 (39%), Gaps = 77/383 (20%)
Query: 113 CSICLNIWHDVVTVAPCLHNFCNGCFSEWLRRSQEKRSTVLCPQCRAVVQFVGKNHFLRT 172
C IC + HD V++ PC+H FC C+S W+ RS LCP CR V+ + KNH L
Sbjct: 304 CIICQELLHDCVSLQPCMHTFCAACYSGWMERSS------LCPTCRCPVERICKNHILNN 357
Query: 173 IAEDMV-----KADSSLRRSHDEVALLDTYASVRSNLVIGSGKKSRKRERANTTMVNQSD 227
+ E + K R+ D+V +D + +++ ++S E ++ + +
Sbjct: 358 LVEAYLIQHPGKFQVYKCRNEDDVRSMDARNKITQDMLQPKVRRSFSDEEGSSEDLLELS 417
Query: 228 GPDHH----------CSQCV----------------TVVGGFQ----------------- 244
D C QC T GG Q
Sbjct: 418 DVDSESSDISQPYIVCRQCPGYRRHPVPALPGTGQETEAGGMQALGDAPSTSANFPAAVQ 477
Query: 245 ---CDPSTIHLQCQECGGMMPSRTGLG------VPQYCLGCDRPFCGAYWSALGVTGSGS 295
C H+ C C MP R PQ C C +PFC YW G T
Sbjct: 478 EYVCPAQGSHVICTCCFQPMPDRRAEREQNPHVAPQQCTVCLQPFCHLYW---GCTRMAC 534
Query: 296 YPVCSPDTLRPISEHTISGIPLVAHEKNLHEQNITESCIRQMGRTLQDVISEWIAKLNNR 355
+ +P + + + G+ N +E +I + + G T +++++E + L
Sbjct: 535 FGCLAPFCEINLGDKCLDGVL----NNNHYESDILKDYLASRGLTWKNMLNESLLALQRG 590
Query: 356 EIDTTRLMLNHAEMMTAGTLVCSDCYQKLVSFFLYWFRISTPKHLLPPEASTREDCWYGY 415
ML+ + T T++C C + Y +R + P LP ++R DC++G
Sbjct: 591 VF-----MLSDYRI-TGNTVLCYCCGLRSFRELAYQYRQNIPVAELPVTVTSRPDCYWGR 644
Query: 416 ACRTQHRSEEHARKRNHVCRPTR 438
CRTQ ++ HA K NH+C TR
Sbjct: 645 NCRTQVKA-HHAMKFNHICEQTR 666
>UniRef100_UPI000036105F UPI000036105F UniRef100 entry
Length = 606
Score = 117 bits (293), Expect = 7e-25
Identities = 99/395 (25%), Positives = 152/395 (38%), Gaps = 91/395 (23%)
Query: 113 CSICLNIWHDVVTVAPCLHNFCNGCFSEWLRRSQEKRSTVLCPQCRAVVQFVGKNHFLRT 172
C IC ++ +D V++ PC+H FC C+S W+ RS LCP CR V+ + KNH L
Sbjct: 231 CVICQDLLYDCVSLQPCMHVFCAACYSGWMERSP------LCPTCRCPVERIRKNHILNN 284
Query: 173 IA--------------EDMVKADSSLRRSHD------EVALLDTYASVRSNLVIGSGKKS 212
+ ED+ DS + + D E + D S L+ S S
Sbjct: 285 LVEAYLIQHPEKCRSEEDLKSMDSRNKITQDMLQPKVERSFSDEEGS-SDYLLEFSDNDS 343
Query: 213 RKRERANTTMVNQSDGPDHHCSQCVTVVGG------------------------------ 242
+ + M Q G SQ + G
Sbjct: 344 DSSDMSQPIMCRQCPGYKADVSQLLFATGSNYWFPGLPAPAPVPPPPKPAATEGCAKAAG 403
Query: 243 ---------------FQCDPSTIHLQCQECGGMMPSRTG-LG---VPQYCLGCDRPFCGA 283
++C P IHL C C MP R LG V Q C+ C R FC
Sbjct: 404 EQPSTSSDGPSAPQEYRCPPRGIHLICTCCLQPMPDRRAELGSQQVAQQCVLCQRSFCHM 463
Query: 284 YWSALGVTGSGSYPVCSPDTLRPISEHTISGIPLVAHEKNLHEQNITESCIRQMGRTLQD 343
YW G G +P +++ + G A N +E ++ ++ + G++ +D
Sbjct: 464 YW---GCQRIGCQGCLAPFGELNLTDKCLDG----ALNSNAYESDVLQNYLSSRGKSWKD 516
Query: 344 VISEWIAKLNNREIDTTRLMLNHAEMMTAGTLVCSDCYQKLVSFFLYWFRISTPKHLLPP 403
+ E + + + ++ T++CSDC + + Y +R P LP
Sbjct: 517 LRQEALQNVEQGKYPADC-------RISGSTIMCSDCGLRALRDLAYKYRQDIPSSELPA 569
Query: 404 EASTREDCWYGYACRTQHRSEEHARKRNHVCRPTR 438
+ R DC++G CRTQ ++ HA K NH+C TR
Sbjct: 570 AVTARPDCYWGRNCRTQVKA-HHAMKFNHICEQTR 603
>UniRef100_Q6P256 Hypothetical protein MGC75959 [Xenopus tropicalis]
Length = 626
Score = 115 bits (289), Expect = 2e-24
Identities = 88/377 (23%), Positives = 144/377 (37%), Gaps = 71/377 (18%)
Query: 113 CSICLNIWHDVVTVAPCLHNFCNGCFSEWLRRSQEKRSTVLCPQCRAVVQFVGKNHFLRT 172
C IC + HD V++ PC+H FC C+S W+ RS LCP CR V+ + KNH L
Sbjct: 267 CIICQELLHDCVSLQPCMHTFCAACYSGWMERSS------LCPTCRCPVERICKNHILNN 320
Query: 173 IAEDMVKADSSLRRSHDEVALLDTYASVRSNLVIGSGKKSRKRERANTTMVNQSDGPDHH 232
+ E + RS ++ +D + +++ ++S E ++ + + D
Sbjct: 321 LVEAYLIQHPEKCRSEEDRCSMDARNKITQDMLQPKVRRSFSDEEGSSEDLLELSDVDSE 380
Query: 233 ----------CSQCV-----------------------------------TVVGGFQCDP 247
C QC T + C
Sbjct: 381 SSDISQPYTVCRQCPGYVRHNIQPPPYPPPSDTEASRTQGDAPSTSTNFPTATQEYVCPS 440
Query: 248 STIHLQCQECGGMMPSRTGLG------VPQYCLGCDRPFCGAYWSALGVTGSGSYPVCSP 301
H+ C C MP R PQ C C PFC YW G G + +P
Sbjct: 441 HGSHVICTCCFQPMPDRRAEREHNSHVAPQQCTICLEPFCHMYW---GCNRMGCFGCLAP 497
Query: 302 DTLRPISEHTISGIPLVAHEKNLHEQNITESCIRQMGRTLQDVISEWIAKLNNREIDTTR 361
+ + + G+ N +E +I ++ + G T +D+++E ++
Sbjct: 498 FCELNLGDKCLDGVL----NNNNYESDILKNYLASRGLTWKDMLNESLSAAQRGVFMLPD 553
Query: 362 LMLNHAEMMTAGTLVCSDCYQKLVSFFLYWFRISTPKHLLPPEASTREDCWYGYACRTQH 421
+N T++C C + Y +R + P LP ++R +C++G CRTQ
Sbjct: 554 YRINGT------TVLCYFCGLRNFRILTYQYRQNIPASELPVTVTSRPNCYWGRNCRTQV 607
Query: 422 RSEEHARKRNHVCRPTR 438
++ HA K NH+C TR
Sbjct: 608 KA-HHAMKFNHICEQTR 623
>UniRef100_UPI0000438729 UPI0000438729 UniRef100 entry
Length = 420
Score = 89.7 bits (221), Expect = 1e-16
Identities = 74/344 (21%), Positives = 127/344 (36%), Gaps = 75/344 (21%)
Query: 113 CSICLNIWHDVVTVAPCLHNFCNGCFSEWLRRSQEKRSTVLCPQCRAVVQFVGKNHFLRT 172
C IC ++ +D ++V PC+H FC C+S W+ RS CP CR V+ + KNH L
Sbjct: 69 CIICQDLLYDCISVQPCMHTFCAACYSGWMERSS------FCPTCRCPVERIRKNHILNN 122
Query: 173 IAEDMVKADSSLRRSHDEVALLDTYASVRSNLVIGSGKKSRKRERA----------NTTM 222
+ E + R+ D++ +D + +++ ++S E A N +
Sbjct: 123 LVEAYLLQHPEKCRTEDDLRSMDARNKITQDMLQPKVERSFSDEEASSDYLFELSDNDSD 182
Query: 223 VNQSDGPDHHCSQC-------------------------------------VTVVGGFQC 245
++ P C QC F+C
Sbjct: 183 ISDMSQPYMMCRQCPGYRKELSSALWICESAQSESLAKTAGDGPSTSSDSTTAAPQEFRC 242
Query: 246 DPSTIHLQCQECGGMMPSRTGLGV-----PQYCLGCDRPFCGAYWSALGVTGSGSYPVCS 300
P HL C C MP R + PQ+CL C +PFC YW G G + +
Sbjct: 243 PPQASHLICTCCLQPMPDRRFEHLPPQVSPQHCLVCQKPFCHVYW---GCPRIGCHGCLA 299
Query: 301 PDTLRPISEHTISGIPLVAHEKNLHEQNITESCIRQMGRTLQDVISEWIAKLNNREIDTT 360
+ +++ + G+ N +E + ++ + G + + ++ + + L
Sbjct: 300 RFSELNLNDKCLDGV----FNGNQYESEVLQNYLSCRGMSWRHLLQDSLQALQQG----- 350
Query: 361 RLMLNHAE--MMTAGTLVCSDCYQKLVSFFLYWFRISTPKHLLP 402
L H +TA + +C C + Y +R P LP
Sbjct: 351 ---LYHLSDYRITANSFLCYCCGLRTFRELAYKYRERIPPSELP 391
>UniRef100_UPI0000368C6B UPI0000368C6B UniRef100 entry
Length = 295
Score = 82.0 bits (201), Expect = 3e-14
Identities = 57/207 (27%), Positives = 91/207 (43%), Gaps = 20/207 (9%)
Query: 238 TVVGGFQCDPSTIHLQCQECGGMMPSRTGLG------VPQYCLGCDRPFCGAYWSALGVT 291
T V + C H C C MP R PQ C+ C +PFC YW G T
Sbjct: 100 TAVQDYVCPLQGSHALCTCCFQPMPDRRAEREQDPRVAPQQCVVCLQPFCHLYW---GCT 156
Query: 292 GSGSYPVCSPDTLRPISEHTISGIPLVAHEKNLHEQNITESCIRQMGRTLQDVISEWIAK 351
+G Y +P + + + G+ N +E +I ++ + G T +++++E +
Sbjct: 157 RTGCYGCLAPFCELNLGDKCLDGVL----NNNSYESDILKNYLATRGLTWKNMLTESLVA 212
Query: 352 LNNREIDTTRLMLNHAEMMTAGTLVCSDCYQKLVSFFLYWFRISTPKHLLPPEASTREDC 411
L + L +T T++C C + Y +R + P LP ++R DC
Sbjct: 213 LQRG------VFLLSDYRVTGDTVLCYCCGLRSFRELTYQYRQNIPASELPVAVTSRPDC 266
Query: 412 WYGYACRTQHRSEEHARKRNHVCRPTR 438
++G CRTQ ++ HA K NH+C TR
Sbjct: 267 YWGRNCRTQVKA-HHAMKFNHICEQTR 292
>UniRef100_UPI0000337E34 UPI0000337E34 UniRef100 entry
Length = 709
Score = 82.0 bits (201), Expect = 3e-14
Identities = 55/200 (27%), Positives = 91/200 (45%), Gaps = 18/200 (9%)
Query: 243 FQCDPSTIHLQCQECGGMMPSRTG-LG---VPQYCLGCDRPFCGAYWSALGVTGSGSYPV 298
++C P H+ C C MP R LG V Q C C R FC YW G G
Sbjct: 521 YRCPPRGFHVFCTCCLQPMPDRRAELGSQQVAQQCGLCQRSFCHMYW---GCRRIGCRGC 577
Query: 299 CSPDTLRPISEHTISGIPLVAHEKNLHEQNITESCIRQMGRTLQDVISEWIAKLNNREID 358
+P + ++ + G+ N +E + ++ + G++ +D+ E + + + D
Sbjct: 578 LAPFSELNFTDKCLDGVL----NNNSYESEVLQNYLSSRGKSWKDLRQEALQSIEQGKYD 633
Query: 359 TTRLMLNHAEMMTAGTLVCSDCYQKLVSFFLYWFRISTPKHLLPPEASTREDCWYGYACR 418
T ++ + T++C C + ++ Y +R S P LP + R DC++G CR
Sbjct: 634 LTDRRISRS------TIMCIYCGLRALADLAYKYRQSIPPSELPAAVTARPDCYWGRNCR 687
Query: 419 TQHRSEEHARKRNHVCRPTR 438
TQ ++ HA K NH+C TR
Sbjct: 688 TQVKA-HHAMKFNHICEQTR 706
Score = 64.7 bits (156), Expect = 5e-09
Identities = 26/63 (41%), Positives = 37/63 (58%), Gaps = 6/63 (9%)
Query: 113 CSICLNIWHDVVTVAPCLHNFCNGCFSEWLRRSQEKRSTVLCPQCRAVVQFVGKNHFLRT 172
C IC ++ HD +++ PC+H FC C+S W+ RS LCP CR V+ + KNH L
Sbjct: 268 CVICQDLLHDCISLQPCMHVFCAACYSGWMERSS------LCPTCRCPVERIRKNHILNN 321
Query: 173 IAE 175
+ E
Sbjct: 322 LVE 324
>UniRef100_UPI00001CFE64 UPI00001CFE64 UniRef100 entry
Length = 398
Score = 67.8 bits (164), Expect = 6e-10
Identities = 42/166 (25%), Positives = 73/166 (43%), Gaps = 17/166 (10%)
Query: 92 VSSPEICQTLLKICVDVDHAKCSICLNIWHDVVTVAPCLHNFCNGCFSEWLRRSQEKRST 151
+SS ++ LK + C IC ++ HD +T+ PC+H FC C+S W+ RS
Sbjct: 202 ISSEDVKDASLKPNKMEETLTCIICQDLLHDSMTLQPCMHTFCVACYSGWMERSS----- 256
Query: 152 VLCPQCRAVVQFVGKNHFLRTIAEDMVKADSSLRRSHDEVALLDTYASVRSNLVIGSGKK 211
LCP C V+ + KNH L + + + RS +V +D + +++ ++
Sbjct: 257 -LCPTCXCPVERIYKNHILNNLMQAYLIQHPEKSRSEXDVRSMDAGNKITQDMLQPKVRR 315
Query: 212 SRKRERANTTMVNQSDGPDHHCSQCVTVVGGFQCDPSTIHLQCQEC 257
S E ++ + Q D S D S ++ C++C
Sbjct: 316 SFSDEEWSSEDLLQLSDVDIESS-----------DISQPYIVCRQC 350
>UniRef100_UPI00002218AE UPI00002218AE UniRef100 entry
Length = 458
Score = 64.7 bits (156), Expect = 5e-09
Identities = 27/88 (30%), Positives = 47/88 (52%), Gaps = 5/88 (5%)
Query: 110 HAKCSICLNIWHDVVTVAPCLHNFCNGCFSEWLRRSQEKRSTVLCPQCRAVVQFVGKNHF 169
H+ CSIC+ ++ V+ +PCLH FC GC +W ++ K CP CR V + N
Sbjct: 21 HSGCSICMATLYNAVSCSPCLHTFCAGCIVQWTDQNASK-----CPMCRIAVLDISPNCM 75
Query: 170 LRTIAEDMVKADSSLRRSHDEVALLDTY 197
+R + + + + +L R+ ++ LD +
Sbjct: 76 MRELVDSCLLVNPALERTEEDKIALDKF 103
>UniRef100_Q14258 Tripartite motif protein 25 [Homo sapiens]
Length = 630
Score = 56.2 bits (134), Expect = 2e-06
Identities = 33/85 (38%), Positives = 45/85 (52%), Gaps = 9/85 (10%)
Query: 101 LLKICVDVDHAKCSICLNIWHDVVTVAPCLHNFCNGCFSE-WLRRSQEKRSTVLCPQCRA 159
+ ++C + CSICL + + VT PC HNFC C +E W + S LCPQCRA
Sbjct: 1 MAELCPLAEELSCSICLEPFKEPVTT-PCGHNFCGSCLNETWAVQG----SPYLCPQCRA 55
Query: 160 VVQF---VGKNHFLRTIAEDMVKAD 181
V Q + KN L + E ++AD
Sbjct: 56 VYQARPQLHKNTVLCNVVEQFLQAD 80
>UniRef100_UPI000036B09C UPI000036B09C UniRef100 entry
Length = 630
Score = 55.1 bits (131), Expect = 4e-06
Identities = 32/85 (37%), Positives = 45/85 (52%), Gaps = 9/85 (10%)
Query: 101 LLKICVDVDHAKCSICLNIWHDVVTVAPCLHNFCNGCFSE-WLRRSQEKRSTVLCPQCRA 159
+ ++C + CSICL + + VT PC HNFC C +E W + + LCPQCRA
Sbjct: 1 MAELCPLAEELSCSICLEPFKEPVTT-PCGHNFCGSCLNETWAVQG----APYLCPQCRA 55
Query: 160 VVQF---VGKNHFLRTIAEDMVKAD 181
V Q + KN L + E ++AD
Sbjct: 56 VYQARPQLHKNTVLCNVVEQFLQAD 80
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.321 0.134 0.429
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 757,315,935
Number of Sequences: 2790947
Number of extensions: 31740365
Number of successful extensions: 73213
Number of sequences better than 10.0: 1442
Number of HSP's better than 10.0 without gapping: 231
Number of HSP's successfully gapped in prelim test: 1212
Number of HSP's that attempted gapping in prelim test: 72394
Number of HSP's gapped (non-prelim): 1612
length of query: 441
length of database: 848,049,833
effective HSP length: 130
effective length of query: 311
effective length of database: 485,226,723
effective search space: 150905510853
effective search space used: 150905510853
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 76 (33.9 bits)
Medicago: description of AC148755.4


