

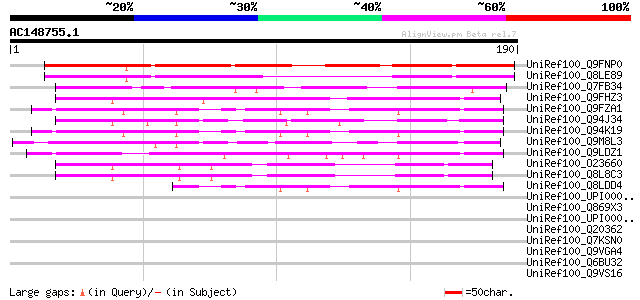
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148755.1 + phase: 0
(190 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9FNP0 Gb|AAF27124.1 [Arabidopsis thaliana] 174 1e-42
UniRef100_Q8LE89 Hypothetical protein [Arabidopsis thaliana] 134 1e-30
UniRef100_Q7FB34 OSJNBa0070O11.3 protein [Oryza sativa] 92 5e-18
UniRef100_Q9FHZ3 Gb|AAF34839.1 [Arabidopsis thaliana] 77 2e-13
UniRef100_Q9FZA1 F3H9.7 protein [Arabidopsis thaliana] 68 1e-10
UniRef100_Q94J34 P0481E12.14 protein [Oryza sativa] 66 4e-10
UniRef100_Q94K19 Hypothetical protein F3H9.7 [Arabidopsis thaliana] 65 9e-10
UniRef100_Q9M8L3 Hypothetical protein T21F11.22 [Arabidopsis tha... 61 1e-08
UniRef100_Q9LDZ1 Gb|AAF34839.1 [Arabidopsis thaliana] 59 9e-08
UniRef100_O23660 Hypothetical protein At2g33780 [Arabidopsis tha... 54 2e-06
UniRef100_Q8L8C3 Hypothetical protein At2g33780/T1B8.28 [Arabido... 54 2e-06
UniRef100_Q8LDD4 Hypothetical protein [Arabidopsis thaliana] 52 1e-05
UniRef100_UPI000042E22F UPI000042E22F UniRef100 entry 44 0.002
UniRef100_Q869X3 Similar to Dictyostelium discoideum (Slime mold... 40 0.032
UniRef100_UPI0000235C75 UPI0000235C75 UniRef100 entry 39 0.054
UniRef100_Q20362 Hypothetical protein F43D9.1 [Caenorhabditis el... 39 0.054
UniRef100_Q7KSN0 CG10042-PA, isoform A [Drosophila melanogaster] 39 0.054
UniRef100_Q9VGA4 CG10042-PB, isoform B [Drosophila melanogaster] 39 0.054
UniRef100_Q6BU32 Similar to CA5229|IPF10425 Candida albicans [De... 39 0.054
UniRef100_Q9VS16 CG32384-PA [Drosophila melanogaster] 39 0.070
>UniRef100_Q9FNP0 Gb|AAF27124.1 [Arabidopsis thaliana]
Length = 173
Score = 174 bits (441), Expect = 1e-42
Identities = 102/179 (56%), Positives = 120/179 (66%), Gaps = 22/179 (12%)
Query: 14 CKPLTTFVHTNTGAFREVVQRLTGPSEGN---ATKEEQTAKVAPITKRTPPKLHERRKCM 70
CKP+TTFV T+T FRE+VQRLTGP+E N AT E K A I KR KLHERR+CM
Sbjct: 13 CKPVTTFVQTDTNTFREIVQRLTGPTENNAAAATPEATVIKTA-IQKRPTSKLHERRQCM 71
Query: 71 KPKLEIVKPNLQYHKQPGASPSSKSRNSSFPSSPVSGSGSSSLLPSPTTPSTLFSRLTIM 130
+PKLEIVKP L + K G +PSSKS N++ +SPV TPS+LFS L+++
Sbjct: 72 RPKLEIVKPPLSF-KPTGTTPSSKSGNTNLLTSPVG------------TPSSLFSNLSLI 118
Query: 131 EDEKKQGSVINEINIEEEEKAIKERRFYLHPSPRSKASGFNEPELLNLFPLASPKACEK 189
E E + NIEEEEKAIKERRFYLHPSPRSK G+ EPELL LFPL SP + K
Sbjct: 119 EGEPDSCTT----NIEEEEKAIKERRFYLHPSPRSK-PGYTEPELLTLFPLTSPNSSGK 172
>UniRef100_Q8LE89 Hypothetical protein [Arabidopsis thaliana]
Length = 142
Score = 134 bits (337), Expect = 1e-30
Identities = 84/179 (46%), Positives = 94/179 (51%), Gaps = 53/179 (29%)
Query: 14 CKPLTTFVHTNTGAFREVVQRLTGPSEGN---ATKEEQTAKVAPITKRTPPKLHERRKCM 70
CKP+TTFV T+T FRE+VQRLTGP+E N AT E K A I KR KLHERR+CM
Sbjct: 13 CKPVTTFVQTDTNTFREIVQRLTGPTENNAAAATPEATVIKTA-IQKRPTSKLHERRQCM 71
Query: 71 KPKLEIVKPNLQYHKQPGASPSSKSRNSSFPSSPVSGSGSSSLLPSPTTPSTLFSRLTIM 130
+PKLEIVKP L + G S +
Sbjct: 72 RPKLEIVKPPLSFKPTEGEPDSCTT----------------------------------- 96
Query: 131 EDEKKQGSVINEINIEEEEKAIKERRFYLHPSPRSKASGFNEPELLNLFPLASPKACEK 189
NIEEEEKAIKERRFYLHPSPRSK G+ EPELL LFPL SP + K
Sbjct: 97 -------------NIEEEEKAIKERRFYLHPSPRSK-PGYTEPELLTLFPLTSPNSSGK 141
>UniRef100_Q7FB34 OSJNBa0070O11.3 protein [Oryza sativa]
Length = 202
Score = 92.4 bits (228), Expect = 5e-18
Identities = 71/186 (38%), Positives = 94/186 (50%), Gaps = 39/186 (20%)
Query: 18 TTFVHTNTGAFREVVQRLTGPSEGNATKEEQTAKVAPITKRTPPKLHERRKCMKPKLEIV 77
TTFV + FR +VQ+LTG +K A AP+ +R PKL ERR+ +LE+
Sbjct: 33 TTFVQADPATFRALVQKLTGAPGSGGSKP---APAAPVMRR--PKLQERRRAAPARLELA 87
Query: 78 KPNLQY----HKQPGASP------------SSKSRNSSFPSSPVSGSGSSSLLPSPTTPS 121
+P Y H + SP SS S +SS PSS SSSL PSP S
Sbjct: 88 RPQPLYYSHHHHRLMHSPVSPMDYAYVMASSSSSSSSSLPSS------SSSLSPSPPASS 141
Query: 122 TLFSRLTIMEDEKKQGSVINEINIEEEEKAIKERRFYLHPSPRSKASGFNE-PELLNLFP 180
+ + I ++E+ E EEKAI + FYLH SPRS +G E P+LL LFP
Sbjct: 142 SSCGVVVITKEEE-----------EREEKAIASKGFYLHSSPRSGGAGDGERPKLLPLFP 190
Query: 181 LASPKA 186
+ SP++
Sbjct: 191 VHSPRS 196
>UniRef100_Q9FHZ3 Gb|AAF34839.1 [Arabidopsis thaliana]
Length = 243
Score = 77.0 bits (188), Expect = 2e-13
Identities = 63/198 (31%), Positives = 87/198 (43%), Gaps = 38/198 (19%)
Query: 18 TTFVHTNTGAFREVVQRLTG-------------PSEGNATKEEQTAKVAPITKRTPPKLH 64
TTFV +T F++VVQ LTG PS N + + + PI K KL+
Sbjct: 49 TTFVQADTSTFKQVVQMLTGSSTDTTTGKHHEAPSPVNNNNKGSSFSIPPIKKTNSFKLY 108
Query: 65 ERRKCMK------------------PKLEIVKPNLQYHKQPGASPSSKSRNSSFPSSPVS 106
ERR+ +L N +H+ P SP + S + + SP
Sbjct: 109 ERRQNNNNMFAKNDLMINTLRLQNSQRLMFTGGNSSHHQSPRFSPRNSSSSENILLSPSM 168
Query: 107 GSGSSSLLPSPTTPSTLFSRLTIMEDEKKQGSVINEINIEEEEKAIKERRFYLHPSPRSK 166
L SP TP L +D + S ++ N EE+KAI ++ FYLHPSP S
Sbjct: 169 LDFPKLGLNSPVTP------LRSNDDPFNKSSPLSLGNSSEEDKAIADKGFYLHPSPVST 222
Query: 167 ASGFNEPELLNLFPLASP 184
++P LL LFP+ASP
Sbjct: 223 PRD-SQPLLLPLFPVASP 239
>UniRef100_Q9FZA1 F3H9.7 protein [Arabidopsis thaliana]
Length = 259
Score = 68.2 bits (165), Expect = 1e-10
Identities = 70/209 (33%), Positives = 94/209 (44%), Gaps = 52/209 (24%)
Query: 9 STTGDCKPLTTFVHTNTGAFREVVQRLTGPSEG---------NATKEEQTAKVAPITKRT 59
S +G+ P TTFV +T +F++VVQ LTG +E N T + + P +
Sbjct: 61 SESGNPYP-TTFVQADTSSFKQVVQMLTGSAERPKHGSSLKPNPTHHQPDPRSTPSSFSI 119
Query: 60 PP-----------------KLHERRKCMKPKLEIVKPNLQYHKQPGASPSSKSRNSSF-P 101
PP +L+ERR MK NL+ + +P NS+F P
Sbjct: 120 PPIKAVPNKKQSSSSASGFRLYERRNSMK--------NLKINP---LNPVFNPVNSAFSP 168
Query: 102 SSPVSGSGS----SSLLPSPTTPSTLFSRLTIMEDEKKQGSVINEIN-IEEEEKAIKERR 156
P S S SL+ SP TP I + + GS N + EEKA+KER
Sbjct: 169 RKPEILSPSILDFPSLVLSPVTP-------LIPDPFDRSGSSNQSPNELAAEEKAMKERG 221
Query: 157 FYLHPSPRSKASGFNEPELLNLFPLASPK 185
FYLHPSP + EP LL LFP+ SP+
Sbjct: 222 FYLHPSPATTPMD-PEPRLLPLFPVTSPR 249
>UniRef100_Q94J34 P0481E12.14 protein [Oryza sativa]
Length = 231
Score = 66.2 bits (160), Expect = 4e-10
Identities = 67/189 (35%), Positives = 83/189 (43%), Gaps = 44/189 (23%)
Query: 18 TTFVHTNTGAFREVVQRLTG---PSEGNATKEEQTA-------------KVAPITKRTPP 61
TTFV +T +F++VVQ LTG PS+ AT A P + P
Sbjct: 60 TTFVQADTASFKQVVQMLTGAEQPSKNAATAATAAAGNSSAAGIGGGQGANGPCRPKKPA 119
Query: 62 -KLHERRKCMKPKLEIVKPNLQYHKQPGASPSSKSRNSSFPS--SPVSGSGSSSLLPSPT 118
KL+ERR +K L+++ P GA PS R P SP S SL SP
Sbjct: 120 FKLYERRSSLK-NLKMIAPLAM-----GALPSPTGRKVGTPEILSP-SVLDFPSLKLSPV 172
Query: 119 TPST--LFSRLTIMEDEKKQGSVINEINIEEEEKAIKERRFYLHPSPRSKASGFNEPELL 176
TP T F+R E + E AI ER F+LHPSPR G P LL
Sbjct: 173 TPLTGEPFNRSPASSSE------------DAERAAISERGFFLHPSPR----GAEPPRLL 216
Query: 177 NLFPLASPK 185
LFP+ SP+
Sbjct: 217 PLFPVTSPR 225
>UniRef100_Q94K19 Hypothetical protein F3H9.7 [Arabidopsis thaliana]
Length = 247
Score = 65.1 bits (157), Expect = 9e-10
Identities = 69/209 (33%), Positives = 92/209 (44%), Gaps = 52/209 (24%)
Query: 9 STTGDCKPLTTFVHTNTGAFREVVQRLTGPSEG---------NATKEEQTAKVAPITKRT 59
S +G+ P TTF +T +F++VVQ LTG E N T + + P +
Sbjct: 49 SESGNPYP-TTFDQADTSSFKQVVQMLTGSGERPKHGSSIKPNPTHHQPDPRSTPSSFSI 107
Query: 60 PP-----------------KLHERRKCMKPKLEIVKPNLQYHKQPGASPSSKSRNSSF-P 101
PP +L+ERR MK NL+ + +P NS+F P
Sbjct: 108 PPIKAVPNKKQSSSSASGFRLYERRNSMK--------NLKINP---LNPVFNPVNSAFSP 156
Query: 102 SSPVSGSGS----SSLLPSPTTPSTLFSRLTIMEDEKKQGSVINEIN-IEEEEKAIKERR 156
P S S SL+ SP TP I + + GS N + EEKA+KER
Sbjct: 157 RKPEILSPSILDFPSLVLSPVTP-------LIPDPFDRSGSSNQSPNELAAEEKAMKERG 209
Query: 157 FYLHPSPRSKASGFNEPELLNLFPLASPK 185
FYLHPSP + EP LL LFP+ SP+
Sbjct: 210 FYLHPSPATTPMD-PEPRLLPLFPVTSPR 237
>UniRef100_Q9M8L3 Hypothetical protein T21F11.22 [Arabidopsis thaliana]
Length = 177
Score = 61.2 bits (147), Expect = 1e-08
Identities = 62/188 (32%), Positives = 85/188 (44%), Gaps = 34/188 (18%)
Query: 2 EKPPVHGSTTGDCKPLTTFVHTNTGAFREVVQRLTGPSEGNATKEEQTAKVA----PITK 57
++PP + + +P T FV + FR +VQ+LTG ++ A P+T
Sbjct: 4 QQPPSYAT-----EPNTMFVQADPSNFRNIVQKLTGAPPDISSSSFSAVSAAHQKLPLTP 58
Query: 58 RTPP-KLHERRKCMKPKLEIVKPNLQYHKQPGASPSSKSRNSSFPSSPVSGSGSSSLLPS 116
+ P KLHERR+ K K+E+ N+ P + S R F SPVS S
Sbjct: 59 KKPAFKLHERRQSSK-KMELKVNNIT---NPNDAFSHFHRG--FLVSPVSHLDPFWARVS 112
Query: 117 PTTPSTLFSRLTIMEDEKKQGSVINEINIEEEEKAIKERRFYLHPSPRSKASGFNEPELL 176
P + E+ Q +EE+KAI E+ FY PSPRS + PELL
Sbjct: 113 PHSAR---------EEHHAQPD-------KEEQKAIAEKGFYFLPSPRSGSE--PAPELL 154
Query: 177 NLFPLASP 184
LFPL SP
Sbjct: 155 PLFPLRSP 162
>UniRef100_Q9LDZ1 Gb|AAF34839.1 [Arabidopsis thaliana]
Length = 219
Score = 58.5 bits (140), Expect = 9e-08
Identities = 59/200 (29%), Positives = 94/200 (46%), Gaps = 33/200 (16%)
Query: 7 HGSTTGDCKPLTTFVHTNTGAFREVVQRLTGPSEGNATKEEQTAKVAPITKRTPPKLHER 66
H T D P TTFV ++ +F++VVQ LTG S +P + R P +
Sbjct: 27 HIITRSDHYP-TTFVQADSSSFKQVVQMLTGSSSPR----------SPDSPRPPTTPSGK 75
Query: 67 RKCMKPKLEIVKP-----NLQYHKQPGASPSSKSRNSSFPSS-PVSGSGSSSLLPSP--- 117
+ P ++ +P N Y ++ ++ +NS ++ + G G+ S SP
Sbjct: 76 GNFVIPPIKTAQPKKHSGNKLYERRSHGGFNNNLKNSLMINTLMIGGGGAGSPRFSPRNQ 135
Query: 118 --TTPSTL-FSRLTIME--DEKKQGSVINEIN-------IEEEEKAIKERRFYLHPSPRS 165
+PS L F +L + KQG+ NE + + EEE+ I ++ +YLH SP S
Sbjct: 136 EILSPSCLDFPKLALNSPVTPLKQGTNGNEGDPFDKMSPLSEEERGIADKGYYLHRSPIS 195
Query: 166 KASGFNEPELLNLFPLASPK 185
+EP+LL LFP+ SP+
Sbjct: 196 TPRD-SEPQLLPLFPVTSPR 214
>UniRef100_O23660 Hypothetical protein At2g33780 [Arabidopsis thaliana]
Length = 204
Score = 53.9 bits (128), Expect = 2e-06
Identities = 55/184 (29%), Positives = 76/184 (40%), Gaps = 48/184 (26%)
Query: 18 TTFVHTNTGAFREVVQRLTG----PSEGNATKEEQTAKVAPITKRTPPK------LHERR 67
TTF+ T+ +F++VVQ LTG P+ + + PI T K L ERR
Sbjct: 36 TTFIRTDPSSFKQVVQLLTGIPKNPTHQPDPRFPPFHSIPPIKAVTNKKQSSSFRLSERR 95
Query: 68 KCMKPKL----------EIVKPNLQYHKQPGASPSSKSRNSSFPSSPVSGSGSSSLLPSP 117
MK L EI+ P + SP ++ S P GS S PS
Sbjct: 96 NSMKHYLNINPTHSGPPEILTPTILNFPALDLSP-----DTPLMSDPFYRPGSFSQSPSD 150
Query: 118 TTPSTLFSRLTIMEDEKKQGSVINEINIEEEEKAIKERRFYLHPSPRSKASGFNEPELLN 177
+ PS +++E++IKE+ FYL PSP S EP LL+
Sbjct: 151 SKPSF----------------------DDDQERSIKEKGFYLRPSP-STTPRDTEPRLLS 187
Query: 178 LFPL 181
LFP+
Sbjct: 188 LFPM 191
>UniRef100_Q8L8C3 Hypothetical protein At2g33780/T1B8.28 [Arabidopsis thaliana]
Length = 204
Score = 53.9 bits (128), Expect = 2e-06
Identities = 55/184 (29%), Positives = 76/184 (40%), Gaps = 48/184 (26%)
Query: 18 TTFVHTNTGAFREVVQRLTG----PSEGNATKEEQTAKVAPITKRTPPK------LHERR 67
TTF+ T+ +F++VVQ LTG P+ + + PI T K L ERR
Sbjct: 36 TTFIRTDPSSFKQVVQLLTGIPKNPTHQPDPRFPPFHSIPPIKAVTNKKQSSSFRLSERR 95
Query: 68 KCMKPKL----------EIVKPNLQYHKQPGASPSSKSRNSSFPSSPVSGSGSSSLLPSP 117
MK L EI+ P + SP ++ S P GS S PS
Sbjct: 96 NSMKHYLNINPTHSGPPEILTPTILNFPALDLSP-----DTPLMSDPFYXPGSFSQSPSX 150
Query: 118 TTPSTLFSRLTIMEDEKKQGSVINEINIEEEEKAIKERRFYLHPSPRSKASGFNEPELLN 177
+ PS +++E++IKE+ FYL PSP S EP LL+
Sbjct: 151 SKPSF----------------------DDDQERSIKEKGFYLRPSP-STTPRDTEPRLLS 187
Query: 178 LFPL 181
LFP+
Sbjct: 188 LFPM 191
>UniRef100_Q8LDD4 Hypothetical protein [Arabidopsis thaliana]
Length = 141
Score = 51.6 bits (122), Expect = 1e-05
Identities = 49/130 (37%), Positives = 63/130 (47%), Gaps = 25/130 (19%)
Query: 62 KLHERRKCMKPKLEIVKPNLQYHKQPGASPSSKSRNSSF-PSSPVSGSGS----SSLLPS 116
+L+ERR MK NL+ + +P NS+F P P S S SL+ S
Sbjct: 21 RLYERRNSMK--------NLKINP---LNPVFNPVNSAFSPRKPEILSPSILDFPSLVLS 69
Query: 117 PTTPSTLFSRLTIMEDEKKQGSVINEIN-IEEEEKAIKERRFYLHPSPRSKASGFNEPEL 175
P TP I + + GS N + EEKA+KER FYLHPSP + EP L
Sbjct: 70 PVTP-------LIPDPFDRSGSSNQSPNELAAEEKAMKERGFYLHPSPATTPMD-PEPRL 121
Query: 176 LNLFPLASPK 185
L LFP+ SP+
Sbjct: 122 LPLFPVTSPR 131
>UniRef100_UPI000042E22F UPI000042E22F UniRef100 entry
Length = 572
Score = 44.3 bits (103), Expect = 0.002
Identities = 32/119 (26%), Positives = 55/119 (45%), Gaps = 2/119 (1%)
Query: 29 REVVQRLTGPSEGNATKEE-QTAKVAPITKRTPPKLHERRKCMKPKLEIVKPNLQYHKQP 87
+EV+Q + P EG T+ + +A P ++++PPK + P+ + +
Sbjct: 109 KEVIQVPSVPKEGGNTQSQTSSAPSMPSSQQSPPKQSSSQPAPPPQQSSSQQSSSQQSSS 168
Query: 88 GASPSSKSRNSSFPSSPVSGSGSSSLLPSPTTPSTLFSRLTIMEDEKKQGSVINEINIE 146
SP +S PS P +SS PS +TPS+ S+ T M+ +K I+ I+
Sbjct: 169 QQSPPKQSPPQQSPSQPAPPPRNSSSQPSSSTPSSKPSQ-TQMDSQKSNSQKISPQKIQ 226
>UniRef100_Q869X3 Similar to Dictyostelium discoideum (Slime mold). protein kinase
[Dictyostelium discoideum]
Length = 921
Score = 40.0 bits (92), Expect = 0.032
Identities = 28/114 (24%), Positives = 49/114 (42%), Gaps = 4/114 (3%)
Query: 8 GSTTGDCKPLTTFVHTNTGAFREVVQRLTGPSEGNATKEEQTAKVAPITKRTPPKLHERR 67
G T + + + +T A + + LT S + Q + + TK+ P +
Sbjct: 802 GDTVSHRRSRSLDIKNSTKAIIKKFEELTRFSSQQSNLVHQPSSSSSSTKQPPTSQFLQN 861
Query: 68 KCMKPKLEIVKPNLQYHKQPGASPSSKSRNSSFPSSPVSGSGSSSLLPSPTTPS 121
M+ KL + L + K P SP S P+S S S+S +P+P++P+
Sbjct: 862 PRMEAKLSFAQETLVFEKSPSTSPKS----LDIPTSSSLTSNSNSSIPAPSSPT 911
>UniRef100_UPI0000235C75 UPI0000235C75 UniRef100 entry
Length = 690
Score = 39.3 bits (90), Expect = 0.054
Identities = 30/84 (35%), Positives = 42/84 (49%), Gaps = 8/84 (9%)
Query: 79 PNLQYHKQPGASPSSKSRNSSFPSSPVS-GSG-----SSSLLPSPTTPSTLFSRLTIMED 132
P + +QP S SK R+SSF S VS GSG S++L SP+ P + E
Sbjct: 145 PRAGHTRQPSLSIQSKMRSSSFRKSSVSQGSGVPSTSPSAMLKSPSLPPLSADGEAVQEV 204
Query: 133 EKKQGSVINEINIEEE--EKAIKE 154
+KQ I E+ E + EK ++E
Sbjct: 205 YRKQSGRIEELEKENKRLEKEVEE 228
>UniRef100_Q20362 Hypothetical protein F43D9.1 [Caenorhabditis elegans]
Length = 1226
Score = 39.3 bits (90), Expect = 0.054
Identities = 35/128 (27%), Positives = 52/128 (40%), Gaps = 15/128 (11%)
Query: 54 PITKRTPPKLHERRKCMKPKLEIVKPNLQYHKQPGASPSSKSRNSSFP-SSPVSGSGSSS 112
PI++++P + +RK V P P P S + P +S + S+S
Sbjct: 13 PISRQSPAAVRFQRKVSTGSAPTVPP-------PAVPPPESSADEEEPRASSNNNYSSTS 65
Query: 113 LLPSPTTPSTLFSRLTIMEDEKKQGSVINEINIEEEEKAI-------KERRFYLHPSPRS 165
+ +PT T S L E EKKQ + E EEK + +E +HPS
Sbjct: 66 TVAAPTREPTRVSTLKREEKEKKQKERMERERRELEEKRVQEDEMKKEEHNHTVHPSLGE 125
Query: 166 KASGFNEP 173
+ S EP
Sbjct: 126 RTSYVEEP 133
>UniRef100_Q7KSN0 CG10042-PA, isoform A [Drosophila melanogaster]
Length = 1081
Score = 39.3 bits (90), Expect = 0.054
Identities = 42/148 (28%), Positives = 62/148 (41%), Gaps = 26/148 (17%)
Query: 31 VVQRLTGPSEGNATKEEQTAKVAPITKRTPPKLHERRKCMKPKLEIVKPNLQYHKQPGAS 90
V Q L+ +E AT+E +T K A K TP + K K+P +
Sbjct: 45 VAQILSESAERKATEEGKTGKAADDVKNTPESGEDANKAAA-------------KEPSGT 91
Query: 91 PSSKSRNSSFPSSPVSGSGSSSLLPSPTTPS--------TLFSRLTIMEDEKK-QGSVIN 141
P + S ++ P+ GS SSS P P TP T+ +RL + E + I
Sbjct: 92 PVAASSEATVPA---PGSASSSNSPLPGTPPKYSNPHNLTIGARLEALSVEGNWLPARIV 148
Query: 142 EINIEEEEKAIK-ERRFYLHPSPRSKAS 168
E+N E+ ++ ER L SP + S
Sbjct: 149 EVNETEQTLLVRFERNHKLKVSPSTSGS 176
>UniRef100_Q9VGA4 CG10042-PB, isoform B [Drosophila melanogaster]
Length = 1169
Score = 39.3 bits (90), Expect = 0.054
Identities = 42/148 (28%), Positives = 62/148 (41%), Gaps = 26/148 (17%)
Query: 31 VVQRLTGPSEGNATKEEQTAKVAPITKRTPPKLHERRKCMKPKLEIVKPNLQYHKQPGAS 90
V Q L+ +E AT+E +T K A K TP + K K+P +
Sbjct: 133 VAQILSESAERKATEEGKTGKAADDVKNTPESGEDANKAAA-------------KEPSGT 179
Query: 91 PSSKSRNSSFPSSPVSGSGSSSLLPSPTTPS--------TLFSRLTIMEDEKK-QGSVIN 141
P + S ++ P+ GS SSS P P TP T+ +RL + E + I
Sbjct: 180 PVAASSEATVPA---PGSASSSNSPLPGTPPKYSNPHNLTIGARLEALSVEGNWLPARIV 236
Query: 142 EINIEEEEKAIK-ERRFYLHPSPRSKAS 168
E+N E+ ++ ER L SP + S
Sbjct: 237 EVNETEQTLLVRFERNHKLKVSPSTSGS 264
>UniRef100_Q6BU32 Similar to CA5229|IPF10425 Candida albicans [Debaryomyces hansenii]
Length = 554
Score = 39.3 bits (90), Expect = 0.054
Identities = 32/115 (27%), Positives = 54/115 (46%), Gaps = 6/115 (5%)
Query: 43 ATKEEQTAKVAPITKRTPPKLHERRKCMKPKLEIVKPNLQYHKQPGASPSSKSRNSSFPS 102
A+ E K A +K+T PK++E +K +K K K +++ P A PS + + +
Sbjct: 249 ASTESPKKKKAVKSKQTDPKINETKKSVKSK----KKSVEAVTPPSADPSKSVVHPNMTT 304
Query: 103 SPVSGSGSSSLLPSPTTPSTLFSRLTIMEDEK--KQGSVINEINIEEEEKAIKER 155
+ LLPSP S ++ ED K K + +E + E E++ KE+
Sbjct: 305 ERNVIKDNDILLPSPQIIDLTMSPPSLKEDVKIEKDNNKTSENDKEREKEKEKEK 359
>UniRef100_Q9VS16 CG32384-PA [Drosophila melanogaster]
Length = 1627
Score = 38.9 bits (89), Expect = 0.070
Identities = 22/49 (44%), Positives = 30/49 (60%), Gaps = 1/49 (2%)
Query: 80 NLQYHKQPGASPSSKSRNSSFPSSPVSGSGSSSLLPSPTTPSTLFSRLT 128
N++ P PS S ++S +SPVSGS S+S+ PSP +P L RLT
Sbjct: 47 NVESSASPSPPPSLSSESASSATSPVSGSASTSVTPSP-SPLHLQRRLT 94
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.309 0.127 0.360
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 347,229,636
Number of Sequences: 2790947
Number of extensions: 14966932
Number of successful extensions: 61509
Number of sequences better than 10.0: 379
Number of HSP's better than 10.0 without gapping: 45
Number of HSP's successfully gapped in prelim test: 336
Number of HSP's that attempted gapping in prelim test: 60981
Number of HSP's gapped (non-prelim): 690
length of query: 190
length of database: 848,049,833
effective HSP length: 120
effective length of query: 70
effective length of database: 513,136,193
effective search space: 35919533510
effective search space used: 35919533510
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 71 (32.0 bits)
Medicago: description of AC148755.1


