

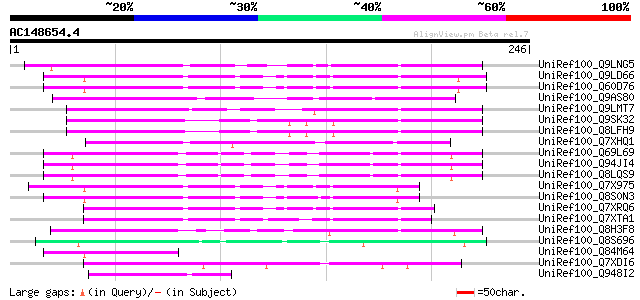
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148654.4 + phase: 0
(246 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9LNG5 F21D18.16 [Arabidopsis thaliana] 83 7e-15
UniRef100_Q9LD66 Similar to maize transposon MuDR mudrA protein ... 82 9e-15
UniRef100_Q60D76 Putative polyprotein [Oryza sativa] 82 9e-15
UniRef100_Q9AS80 P0028E10.12 protein [Oryza sativa] 79 8e-14
UniRef100_Q9LMT7 F2H15.15 protein [Arabidopsis thaliana] 77 3e-13
UniRef100_Q9SK32 Expressed protein [Arabidopsis thaliana] 76 9e-13
UniRef100_Q8LFH9 Hypothetical protein [Arabidopsis thaliana] 76 9e-13
UniRef100_Q7XHQ1 Calcineurin-like phosphoesterase-like [Oryza sa... 72 1e-11
UniRef100_Q69L69 Calcineurin-like phosphoesterase-like protein [... 72 1e-11
UniRef100_Q94JI4 P0638D12.5 protein [Oryza sativa] 70 5e-11
UniRef100_Q8LQS9 P0702H08.15 protein [Oryza sativa] 70 5e-11
UniRef100_Q7X975 Putative Mutator protein [Oryza sativa] 62 1e-08
UniRef100_Q8S0N3 Putative mutator-like transposase [Oryza sativa] 62 2e-08
UniRef100_Q7XRQ6 OSJNBb0096E05.18 protein [Oryza sativa] 62 2e-08
UniRef100_Q7XTA1 OSJNBa0008A08.9 protein [Oryza sativa] 61 3e-08
UniRef100_Q8H3F8 Serine/threonine phosphatase PP7-related-like p... 57 3e-07
UniRef100_Q8S696 Putative mutator-like transposase [Oryza sativa] 57 6e-07
UniRef100_Q84M64 Putative mutator-like transposase [Oryza sativa] 57 6e-07
UniRef100_Q7XDI6 Hypothetical protein [Oryza sativa] 55 1e-06
UniRef100_Q948I2 Putative transposable element [Oryza sativa] 55 1e-06
>UniRef100_Q9LNG5 F21D18.16 [Arabidopsis thaliana]
Length = 1340
Score = 82.8 bits (203), Expect = 7e-15
Identities = 67/222 (30%), Positives = 104/222 (46%), Gaps = 25/222 (11%)
Query: 8 GNENWFWDPLR-----ESGLHDLVYLGYATVPHALLMTLCEMWHPETSTFHMPLGEMTVT 62
G +W DPL E GL+ + + + + +AL+ L E W PET TFH+P GE+TVT
Sbjct: 50 GMRDWPLDPLVCQKLIEFGLYGVYKVAFIQLDYALITALVERWRPETHTFHLPAGEITVT 109
Query: 63 LDDVACLTHLPIEGRMLSHEKKMLKHKGATLLMRHLGVSQQEAEKICGQEYGGYISYPSL 122
L DV L L ++G ++ K+ A L LG + +G ++S L
Sbjct: 110 LQDVNILLGLRVDGPAVTGS---TKYNWADLCEDLLGHRPGPKDL-----HGSHVSLAWL 161
Query: 123 MDFYTTYLGRANLLAGTEDPEELVRVRPYCVRCYLLYLVECLWFGDRSNKRIELVYLTTM 182
R N DP+E V ++ + R ++L L+ +GD+S + L +L +
Sbjct: 162 ---------RENFRNLPADPDE-VTLKCH-TRAFVLALMSGFLYGDKSKHDVALTFLPLL 210
Query: 183 EEGYAGMRNYSWGGMTLAYLYGELAEACRPGDRALGGSVTLL 224
+ + + SWG TLA LY EL A + + G + LL
Sbjct: 211 RD-FDEVAKLSWGSATLALLYRELCRASKRTVSTICGPLVLL 251
>UniRef100_Q9LD66 Similar to maize transposon MuDR mudrA protein [Oryza sativa]
Length = 1230
Score = 82.4 bits (202), Expect = 9e-15
Identities = 70/215 (32%), Positives = 101/215 (46%), Gaps = 19/215 (8%)
Query: 17 LRESGLHDLVYLGYATVP---HALLMTLCEMWHPETSTFHMPLGEMTVTLDDVACLTHLP 73
LR + L V + VP A L L + W PET TFH+P GE+TVTL+DVA + LP
Sbjct: 552 LRRAKLLAFVTMAQRPVPLYNAAALTALVDRWRPETHTFHLPCGELTVTLEDVAMILGLP 611
Query: 74 IEGRMLSHEKKMLKHKGATLLMRHLGVSQQEAEKICGQEYGGYISYPSLMDFYTTYLGRA 133
I G+ ++ + + +LG+ A GQ P ++L RA
Sbjct: 612 IRGQAVTGD--TASGNWRERVEEYLGLEPPVAPD--GQRQTKTSGVP------LSWL-RA 660
Query: 134 NLLAGTEDPEELVRVRPYCVRCYLLYLVECLWFGDRSNKRIELVYLTTMEEGYAGMRNYS 193
N + +E V+ YC R Y+LY+ + F D ++L + + + +S
Sbjct: 661 NFGQCPAEADE-ATVQRYC-RAYVLYIFGSILFPDSGGDMASWMWLPLLAD-WDEAGTFS 717
Query: 194 WGGMTLAYLYGELAEACR--PGDRALGGSVTLLTV 226
WG LA+LY +L +ACR GD L G V LL V
Sbjct: 718 WGSAALAWLYRQLCDACRRQGGDANLAGCVWLLQV 752
>UniRef100_Q60D76 Putative polyprotein [Oryza sativa]
Length = 1754
Score = 82.4 bits (202), Expect = 9e-15
Identities = 70/215 (32%), Positives = 101/215 (46%), Gaps = 19/215 (8%)
Query: 17 LRESGLHDLVYLGYATVP---HALLMTLCEMWHPETSTFHMPLGEMTVTLDDVACLTHLP 73
LR + L V + VP A L L + W PET TFH+P GE+TVTL+DVA + LP
Sbjct: 934 LRRAKLLAFVTMAQRPVPLYNAAALTALVDRWRPETHTFHLPCGELTVTLEDVAMILGLP 993
Query: 74 IEGRMLSHEKKMLKHKGATLLMRHLGVSQQEAEKICGQEYGGYISYPSLMDFYTTYLGRA 133
I G+ ++ + + +LG+ A GQ P ++L RA
Sbjct: 994 IRGQAVTGD--TASGNWRERVEEYLGLEPPVAPD--GQRQTKTSGVP------LSWL-RA 1042
Query: 134 NLLAGTEDPEELVRVRPYCVRCYLLYLVECLWFGDRSNKRIELVYLTTMEEGYAGMRNYS 193
N + +E V+ YC R Y+LY+ + F D ++L + + + +S
Sbjct: 1043 NFGQCPAEADE-ATVQRYC-RAYVLYIFGSILFPDSGGDMASWMWLPLLAD-WDEAGTFS 1099
Query: 194 WGGMTLAYLYGELAEACR--PGDRALGGSVTLLTV 226
WG LA+LY +L +ACR GD L G V LL V
Sbjct: 1100 WGSAALAWLYRQLCDACRRQGGDANLAGCVWLLQV 1134
>UniRef100_Q9AS80 P0028E10.12 protein [Oryza sativa]
Length = 792
Score = 79.3 bits (194), Expect = 8e-14
Identities = 59/191 (30%), Positives = 85/191 (43%), Gaps = 21/191 (10%)
Query: 21 GLHDLVYLGYATVPHALLMTLCEMWHPETSTFHMPLGEMTVTLDDVACLTHLPIEGRMLS 80
GL ++ G ++ AL+ L + W PET TFHMP GE+T+TL DVA + LPI G ++
Sbjct: 245 GLANICRAGLPSIDRALVSALVDRWRPETHTFHMPCGEITITLQDVAMILGLPIAGHAVT 304
Query: 81 HEKKMLKHKGATLLMRHLGVSQQEAEKICGQEYGGYISYPSLMDFYTTYLGRANLLAGTE 140
+++ L+ R+LG + G RA E
Sbjct: 305 VNPTEPQNE---LVERYLGKAPPPDRPRPGLRVSWV---------------RAEFNNCPE 346
Query: 141 DPEELVRVRPYCVRCYLLYLVECLWFGDRSNKRIELVYLTTMEEGYAGMRNYSWGGMTLA 200
D +E R Y+L L+ L F D S + Y + + +YSWG TLA
Sbjct: 347 DADE--ETIKQHARAYILSLISGLLFPDASGD-LYTFYPFPLIADLENIGSYSWGSATLA 403
Query: 201 YLYGELAEACR 211
YLY + +ACR
Sbjct: 404 YLYRAMCDACR 414
>UniRef100_Q9LMT7 F2H15.15 protein [Arabidopsis thaliana]
Length = 478
Score = 77.4 bits (189), Expect = 3e-13
Identities = 54/198 (27%), Positives = 95/198 (47%), Gaps = 21/198 (10%)
Query: 28 LGYATVPHALLMTLCEMWHPETSTFHMPLGEMTVTLDDVACLTHLPIEGRMLSHEKKMLK 87
+G ++ ++L+ L E W ET+TFH P GEMT+TLD+V+ + L ++G+ + K+
Sbjct: 57 VGSISLNNSLISALVERWRRETNTFHFPCGEMTITLDEVSLILGLAVDGKPVVGVKEK-D 115
Query: 88 HKGATLLMRHLGVSQQEAEKICGQEYGGYISYPSLMDFYTTYLGRANLLAGTEDPE-ELV 146
+ + +R LG + G+ G ++ L + + E P+ +
Sbjct: 116 EDPSQVCLRLLGKLPK------GELSGNRVTAKWLKESF------------AECPKGATM 157
Query: 147 RVRPYCVRCYLLYLVECLWFGDRSNKRIELVYLTTMEEGYAGMRNYSWGGMTLAYLYGEL 206
+ Y R YL+Y+V F +I + YL E+ + Y+WG LA+LY ++
Sbjct: 158 KEIEYHTRAYLIYIVGSTIFATTDPSKISVDYLILFED-FEKAGEYAWGAAALAFLYRQI 216
Query: 207 AEACRPGDRALGGSVTLL 224
A + +GG +TLL
Sbjct: 217 GNASQRSQSIIGGCLTLL 234
>UniRef100_Q9SK32 Expressed protein [Arabidopsis thaliana]
Length = 509
Score = 75.9 bits (185), Expect = 9e-13
Identities = 62/201 (30%), Positives = 90/201 (43%), Gaps = 24/201 (11%)
Query: 28 LGYATVPHALLMTLCEMWHPETSTFHMPLGEMTVTLDDVACLTHLPIEGRMLSHEKKMLK 87
+G ++ ++L+ L E W ET+TFH+PLGEMT+TLD+VA + L I+G + K
Sbjct: 66 IGPMSLNNSLISALVERWRRETNTFHLPLGEMTITLDEVALVLGLEIDGDPIVGSK---- 121
Query: 88 HKGATLLMRHLGVSQQEAEKICGQEYGGYISYPSLMDFYTTYLG-RANLLAGT--EDPEE 144
V + A +CG+ G PS + + N L T E PE+
Sbjct: 122 ------------VGDEVAMDMCGRLLG---KLPSAANKEVNCSRVKLNWLKRTFSECPED 166
Query: 145 LVRVRPYC-VRCYLLYLVECLWFGDRSNKRIELVYLTTMEEGYAGMRNYSWGGMTLAYLY 203
C R YLLYL+ F ++ + YL E+ + Y+WG LA LY
Sbjct: 167 ASFDVVKCHTRAYLLYLIGSTIFATTDGDKVSVKYLPLFED-FDQAGRYAWGAAALACLY 225
Query: 204 GELAEACRPGDRALGGSVTLL 224
L A + G +TLL
Sbjct: 226 RALGNASLKSQSNICGCLTLL 246
>UniRef100_Q8LFH9 Hypothetical protein [Arabidopsis thaliana]
Length = 509
Score = 75.9 bits (185), Expect = 9e-13
Identities = 62/201 (30%), Positives = 90/201 (43%), Gaps = 24/201 (11%)
Query: 28 LGYATVPHALLMTLCEMWHPETSTFHMPLGEMTVTLDDVACLTHLPIEGRMLSHEKKMLK 87
+G ++ ++L+ L E W ET+TFH+PLGEMT+TLD+VA + L I+G + K
Sbjct: 66 IGPMSLNNSLISALVERWRRETNTFHLPLGEMTITLDEVALVLGLEIDGDPIVGSK---- 121
Query: 88 HKGATLLMRHLGVSQQEAEKICGQEYGGYISYPSLMDFYTTYLG-RANLLAGT--EDPEE 144
V + A +CG+ G PS + + N L T E PE+
Sbjct: 122 ------------VDDEVAMDMCGRLLG---KLPSAANKEVNCSRVKLNWLKRTFSECPED 166
Query: 145 LVRVRPYC-VRCYLLYLVECLWFGDRSNKRIELVYLTTMEEGYAGMRNYSWGGMTLAYLY 203
C R YLLYL+ F ++ + YL E+ + Y+WG LA LY
Sbjct: 167 ASFDVVKCHTRAYLLYLIGSTIFATTDGDKVSVKYLPLFED-FDQAGRYAWGAAALACLY 225
Query: 204 GELAEACRPGDRALGGSVTLL 224
L A + G +TLL
Sbjct: 226 RALGNASLKSQSNICGCLTLL 246
>UniRef100_Q7XHQ1 Calcineurin-like phosphoesterase-like [Oryza sativa]
Length = 327
Score = 72.0 bits (175), Expect = 1e-11
Identities = 50/177 (28%), Positives = 84/177 (47%), Gaps = 12/177 (6%)
Query: 37 LLMTLCEMWHPETSTFHMPLGEMTVTLDDVACLTHLPIEGRMLSHEKKMLKHKGATLLMR 96
L+ L E W P+T+TFH+P+GEMT+TL DV+CL LPI GR ++ + ++ R
Sbjct: 70 LISALVERWRPKTNTFHLPVGEMTITLQDVSCLWGLPIHGRPITGQ---ADGSWVDMIER 126
Query: 97 HLGVSQQE----AEKICGQEYGGYISYPSLMDFYTTYLGRANLLAGTEDPEELVRVRPYC 152
LG+ +E +K ++ +SY + R ++ E+ +
Sbjct: 127 LLGIPMEEQHMKQKKRKKEDDMTMVSYSRYSISLSKLRDRFRVMPKNATEREI----NWY 182
Query: 153 VRCYLLYLVECLWFGDRSNKRIELVYLTTMEEGYAGMRNYSWGGMTLAYLYGELAEA 209
R +L ++ + F D S + +YL M + Y+WG L+ LY +L+ A
Sbjct: 183 TRALVLDIIGSMVFTDTSGDGVPAMYLQFM-MNLSEQTEYNWGAAALSMLYRQLSIA 238
>UniRef100_Q69L69 Calcineurin-like phosphoesterase-like protein [Oryza sativa]
Length = 766
Score = 72.0 bits (175), Expect = 1e-11
Identities = 67/211 (31%), Positives = 101/211 (47%), Gaps = 23/211 (10%)
Query: 17 LRESGLHDLVYL--GYATVPHALLMTLCEMWHPETSTFHMPLGEMTVTLDDVACLTHLPI 74
LR++GL L + + AL+ L E W PET +FH+ GEMTVTL DV+ L LPI
Sbjct: 111 LRDAGLLGLSQICQKMPQLDKALITALVERWRPETHSFHLASGEMTVTLQDVSMLLALPI 170
Query: 75 EGRMLSHEKKMLKHKGATLLMRHLGVSQQEAEKICGQEYGGYISYPSLMDFYTTYLGRAN 134
+GR + H A +++ LG + G+ ++ Y L +
Sbjct: 171 DGRPVC---STTDHDYAQMVIDCLG-HDPRGPSMPGK---SFLHYKWLKKHF------YE 217
Query: 135 LLAGTEDPEELVRVRPYCVRCYLLYLVECLWFGDRSNKRIELVYLTTMEEGYAGMRNYSW 194
L GT+D + VR Y+L L+ + F D + R+ L+YL + + + + YSW
Sbjct: 218 LPEGTDD-----QTVERHVRAYILSLLCGVLFPDGTG-RMSLIYLPLIAD-LSLVGTYSW 270
Query: 195 GGMTLAYLYGELAE-ACRPGDRALGGSVTLL 224
G LA+LY L A + +GGS+ LL
Sbjct: 271 GSAVLAFLYRSLCSVASSHNIKNIGGSLLLL 301
>UniRef100_Q94JI4 P0638D12.5 protein [Oryza sativa]
Length = 761
Score = 70.1 bits (170), Expect = 5e-11
Identities = 66/211 (31%), Positives = 99/211 (46%), Gaps = 23/211 (10%)
Query: 17 LRESGLHDLVYL--GYATVPHALLMTLCEMWHPETSTFHMPLGEMTVTLDDVACLTHLPI 74
LR++GL L + + AL+ L E W PET +FH+ GEM VTL DVA L LPI
Sbjct: 128 LRDAGLLGLSQICQKMPQLDKALITALVERWRPETHSFHLASGEMAVTLQDVAMLLALPI 187
Query: 75 EGRMLSHEKKMLKHKGATLLMRHLGVSQQEAEKICGQEYGGYISYPSLMDFYTTYLGRAN 134
+GR + H A +++ LG + G+ ++ Y L +
Sbjct: 188 DGRPVC---STTDHDYAQMVIDCLG-HDPRGPSMPGK---SFLHYKWLKKHF------YE 234
Query: 135 LLAGTEDPEELVRVRPYCVRCYLLYLVECLWFGDRSNKRIELVYLTTMEEGYAGMRNYSW 194
L G +D + VR Y+L L+ + F D + R+ L+YL + + + + YSW
Sbjct: 235 LPEGADD-----QTVERHVRAYILSLLCGVLFPDGTG-RMSLIYLPLIAD-LSRVGTYSW 287
Query: 195 GGMTLAYLYGELAE-ACRPGDRALGGSVTLL 224
G LA+LY L A + +GGS+ LL
Sbjct: 288 GSAVLAFLYRSLCSVASSHNIKNIGGSLLLL 318
>UniRef100_Q8LQS9 P0702H08.15 protein [Oryza sativa]
Length = 718
Score = 70.1 bits (170), Expect = 5e-11
Identities = 66/211 (31%), Positives = 99/211 (46%), Gaps = 23/211 (10%)
Query: 17 LRESGLHDLVYL--GYATVPHALLMTLCEMWHPETSTFHMPLGEMTVTLDDVACLTHLPI 74
LR++GL L + + AL+ L E W PET +FH+ GEM VTL DVA L LPI
Sbjct: 142 LRDAGLLGLSQICQKMPQLDKALITALVERWRPETHSFHLASGEMAVTLQDVAMLLALPI 201
Query: 75 EGRMLSHEKKMLKHKGATLLMRHLGVSQQEAEKICGQEYGGYISYPSLMDFYTTYLGRAN 134
+GR + H A +++ LG + G+ ++ Y L +
Sbjct: 202 DGRPVC---STTDHDYAQMVIDCLG-HDPRGPSMPGK---SFLHYKWLKKHF------YE 248
Query: 135 LLAGTEDPEELVRVRPYCVRCYLLYLVECLWFGDRSNKRIELVYLTTMEEGYAGMRNYSW 194
L G +D + VR Y+L L+ + F D + R+ L+YL + + + + YSW
Sbjct: 249 LPEGADD-----QTVERHVRAYILSLLCGVLFPDGTG-RMSLIYLPLIAD-LSRVGTYSW 301
Query: 195 GGMTLAYLYGELAE-ACRPGDRALGGSVTLL 224
G LA+LY L A + +GGS+ LL
Sbjct: 302 GSAVLAFLYRSLCSVASSHNIKNIGGSLLLL 332
>UniRef100_Q7X975 Putative Mutator protein [Oryza sativa]
Length = 1456
Score = 62.4 bits (150), Expect = 1e-08
Identities = 55/191 (28%), Positives = 87/191 (44%), Gaps = 19/191 (9%)
Query: 10 ENWFWDPLRESGLHDLVYLGYATVP---HALLMTLCEMWHPETSTFHMPLGEMTVTLDDV 66
++W+ L+ + L V + VP A L L + W PET TFH+P GE+TVTL+DV
Sbjct: 866 DDWYVPYLQRAKLLAFVTMAQRPVPLYNAAALTALVDRWRPETHTFHLPCGELTVTLEDV 925
Query: 67 ACLTHLPIEGRMLSHEKKMLKHKGATLLMRHLGVSQQEAEKICGQEYGGYISYPSLMDFY 126
A + LPI G+ ++ + + +LG+ A GQ P
Sbjct: 926 AMILGLPIRGQAVTGD--TASGNWRERVEEYLGLEPPVAPD--GQRQTKTSGVP------ 975
Query: 127 TTYLGRANLLAGTEDPEELVRVRPYCVRCYLLYLVECLWFGDRSNKRIELVYLTTM---E 183
++L RAN + +E V+ YC R Y+LY+ + F D ++L + +
Sbjct: 976 LSWL-RANFGQCPAEADE-ATVQRYC-RAYVLYIFGSILFPDSGGDMASWMWLPLLADWD 1032
Query: 184 EGYAGMRNYSW 194
E G RN ++
Sbjct: 1033 EAVVGRRNLAY 1043
>UniRef100_Q8S0N3 Putative mutator-like transposase [Oryza sativa]
Length = 1556
Score = 61.6 bits (148), Expect = 2e-08
Identities = 55/184 (29%), Positives = 84/184 (44%), Gaps = 19/184 (10%)
Query: 17 LRESGLHDLVYLGYATVP---HALLMTLCEMWHPETSTFHMPLGEMTVTLDDVACLTHLP 73
LR + L V + VP A L L + W PET TFH+P GE+TVTL+DVA + LP
Sbjct: 942 LRRAKLLAFVTMAQRPVPLYNAAALTALVDRWRPETHTFHLPCGELTVTLEDVAMILGLP 1001
Query: 74 IEGRMLSHEKKMLKHKGATLLMRHLGVSQQEAEKICGQEYGGYISYPSLMDFYTTYLGRA 133
I G+ ++ + + +LG+ A GQ P ++L RA
Sbjct: 1002 IRGQAVTGD--TASGNWRERVEEYLGLEPPVAPD--GQRQTKTSGVP------LSWL-RA 1050
Query: 134 NLLAGTEDPEELVRVRPYCVRCYLLYLVECLWFGDRSNKRIELVYLTTM---EEGYAGMR 190
N + +E + V+ YC R Y+LY+ + F D ++L + +E G R
Sbjct: 1051 NFGQCPAEADEAI-VQRYC-RAYVLYIFGSILFPDSGGNMASWMWLPLLADWDEAVVGRR 1108
Query: 191 NYSW 194
N ++
Sbjct: 1109 NLAY 1112
>UniRef100_Q7XRQ6 OSJNBb0096E05.18 protein [Oryza sativa]
Length = 567
Score = 61.6 bits (148), Expect = 2e-08
Identities = 50/166 (30%), Positives = 77/166 (46%), Gaps = 14/166 (8%)
Query: 36 ALLMTLCEMWHPETSTFHMPLGEMTVTLDDVACLTHLPIEGRMLSHEKKMLKHKGATLLM 95
A L L + W PET TFH+P GE+TVTL+DVA + LPI G+ ++ + +
Sbjct: 12 AALTALVDRWRPETHTFHLPCGELTVTLEDVAMILGLPIRGQAVTGD--TASGNWRERVE 69
Query: 96 RHLGVSQQEAEKICGQEYGGYISYPSLMDFYTTYLGRANLLAGTEDPEELVRVRPYCVRC 155
+LG+ A GQ P ++L RAN + +E V+ YC R
Sbjct: 70 EYLGLEPPVAPD--GQRQTKTSGVP------LSWL-RANFGQCPAEADE-ATVQRYC-RA 118
Query: 156 YLLYLVECLWFGDRSNKRIELVYLTTMEEGYAGMRNYSWGGMTLAY 201
Y+LY+ + F D ++L + + + +SWG LA+
Sbjct: 119 YVLYIFGSILFPDSGGDMASWMWLPLLAD-WDEAGTFSWGSAALAW 163
>UniRef100_Q7XTA1 OSJNBa0008A08.9 protein [Oryza sativa]
Length = 1560
Score = 60.8 bits (146), Expect = 3e-08
Identities = 48/165 (29%), Positives = 73/165 (44%), Gaps = 14/165 (8%)
Query: 36 ALLMTLCEMWHPETSTFHMPLGEMTVTLDDVACLTHLPIEGRMLSHEKKMLKHKGATLLM 95
A L L + W PET TFH+P GE+TVTL+DVA + LPI G+ ++ + +
Sbjct: 875 AALTALVDRWRPETHTFHLPCGELTVTLEDVAMILGLPIRGQAVTSD--TASGNWRERVE 932
Query: 96 RHLGVSQQEAEKICGQEYGGYISYPSLMDFYTTYLGRANLLAGTEDPEELVRVRPYCVRC 155
+LGV A GQ P + + G+ A + V+ YC R
Sbjct: 933 EYLGVEPPVAPD--GQRQTKTSGVP--LSWLRANFGQCPAEA------DKATVQRYC-RA 981
Query: 156 YLLYLVECLWFGDRSNKRIELVYLTTMEEGYAGMRNYSWGGMTLA 200
Y+LY+ + F D ++L + + + +SWG LA
Sbjct: 982 YVLYIFGSILFPDSGGDMASWMWLPLLAD-WDEAGTFSWGSAALA 1025
>UniRef100_Q8H3F8 Serine/threonine phosphatase PP7-related-like protein [Oryza
sativa]
Length = 589
Score = 57.4 bits (137), Expect = 3e-07
Identities = 54/212 (25%), Positives = 89/212 (41%), Gaps = 31/212 (14%)
Query: 20 SGLHDLVYLGYATVPHALLMTLCEMWHPETSTFHMPLGEMTVTLDDVACLTHLPIEGRML 79
SGL +L++ + LM E+W ET+T + EM +L D A + +P+ GR++
Sbjct: 45 SGLGNLIHTAGLVIDRIALMAFFELWSKETNTAQLNGFEMAPSLRDAAYILGIPVTGRVV 104
Query: 80 SHEKKMLKHKGATLLMRHLGVSQQEAEKICGQEYGGYISYPSLMDFYTTYLGRANLLAGT 139
+ GA L + E +C Q G P D +++ L+
Sbjct: 105 T--------TGAVL--------NKSVEDLCFQYLG---QVPDCRDCRGSHV----KLSWL 141
Query: 140 EDPEELVRVRP------YCVRCYLLYLVECLWFGDRSNKRIELVYLTTMEEGYAGMRNYS 193
+ + RP Y R YLL+L+ +R + YL + + + ++ Y+
Sbjct: 142 QSKFSRIPERPTNDQTMYGTRAYLLFLIGSALLPERDRGYVSPKYLPLLSD-FDKVQEYA 200
Query: 194 WGGMTLAYLYGELAEA-CRPGDRALGGSVTLL 224
WG LA+LY L+ A + L GS LL
Sbjct: 201 WGAAALAHLYKALSIAVAHSARKRLFGSAALL 232
>UniRef100_Q8S696 Putative mutator-like transposase [Oryza sativa]
Length = 1530
Score = 56.6 bits (135), Expect = 6e-07
Identities = 62/231 (26%), Positives = 92/231 (38%), Gaps = 26/231 (11%)
Query: 13 FWDPLRESGLHDLVYLGYA-----------TVPHALLMTLCEMWHPETSTFHMPLGEMTV 61
F + LRE+GL + L A +V +LL TL + W PET TFH+P GE+
Sbjct: 934 FVERLREAGLLPMCRLVEAAAGDADPARRWSVDRSLLATLVDRWRPETHTFHLPCGEVAP 993
Query: 62 TLDDVACLTHLPIEGRMLSHEKKMLKHKGATLLMRHLGVSQQEAEKICGQEYGGYISYPS 121
TL DV+ L LP+ G + + + L R V Q A + + Y +
Sbjct: 994 TLQDVSYLLGLPLAGDAVGPVTTAVDWQD-DLTARFALV--QRAPHLPLEPLAHYRNTGP 1050
Query: 122 LMDFYTTYLGRANLLAGTEDPEELVRVRPYCVRCYLLYLVECLWF----GDRSNKRIELV 177
+ + L D R C+ YLL+L + F G +K +
Sbjct: 1051 TKRWLLQF--TVEQLQAEADEYSYSR----CLEAYLLWLFGWVMFCGGHGHAVDKGLVHY 1104
Query: 178 YLTTMEEGYAGMRNYSWGGMTLAYLYGELAEACRPGD--RALGGSVTLLTV 226
+ + + +SWG LA LY L E+C D GG L++
Sbjct: 1105 TRSIADAAVGEVPQWSWGSALLAALYHALCESCTKTDPSATFGGCPLFLSI 1155
>UniRef100_Q84M64 Putative mutator-like transposase [Oryza sativa]
Length = 1204
Score = 56.6 bits (135), Expect = 6e-07
Identities = 30/67 (44%), Positives = 40/67 (58%), Gaps = 3/67 (4%)
Query: 17 LRESGLHDLVYLGYATVP---HALLMTLCEMWHPETSTFHMPLGEMTVTLDDVACLTHLP 73
LR + L V + VP A L L + W PET TFH+P GE+TVTL+DVA + LP
Sbjct: 811 LRRAKLLAFVTMAQRPVPLYNAAALTALVDRWRPETHTFHLPCGELTVTLEDVAMILGLP 870
Query: 74 IEGRMLS 80
I G+ ++
Sbjct: 871 IRGQAVT 877
>UniRef100_Q7XDI6 Hypothetical protein [Oryza sativa]
Length = 1104
Score = 55.5 bits (132), Expect = 1e-06
Identities = 52/191 (27%), Positives = 77/191 (40%), Gaps = 16/191 (8%)
Query: 36 ALLMTLCEMWHPETSTFHMPLGEMTVTLDDVACLTHLPIEGRMLSHEKKMLKHKG---AT 92
+LL L + W PET TFH+P GEM TL DV+ L LP+ G + + K A
Sbjct: 428 SLLAALVDRWRPETHTFHLPCGEMAPTLQDVSYLLGLPLAGAPVGPVDGVFGWKEDITAR 487
Query: 93 LLMRHLGVSQQEAEKICGQEYGGYISYP-----SLMDFYTTYLGRANLLAGTEDPEELVR 147
L + Q+ + G G P + T +A+LL D + R
Sbjct: 488 FEQVMLAPTLQDVSYLLGLPLAGAPVGPVDGVFGWKEDITARFEQADLLHPDADDNSVRR 547
Query: 148 VRPYCVRCYLLYLVECLWFGDRSNKRIE---LVYLTTMEEGYA-GMRNYSWGGMTLAYLY 203
+ YLL+L + F ++ + Y ++ + + +SWG LA Y
Sbjct: 548 ----SLEAYLLWLFGWVMFTSTHGHVVDFRLVHYARSIADAQPQDVPQWSWGSAVLAATY 603
Query: 204 GELAEACRPGD 214
L EAC D
Sbjct: 604 RALCEACTKTD 614
>UniRef100_Q948I2 Putative transposable element [Oryza sativa]
Length = 1270
Score = 55.5 bits (132), Expect = 1e-06
Identities = 29/68 (42%), Positives = 39/68 (56%), Gaps = 2/68 (2%)
Query: 38 LMTLCEMWHPETSTFHMPLGEMTVTLDDVACLTHLPIEGRMLSHEKKMLKHKGATLLMRH 97
L L + W PET TFH+P GE+TVTL+DVA LPI+GR ++ + + +
Sbjct: 881 LTALVDRWRPETHTFHLPCGELTVTLEDVAMNLALPIQGRAVTGDTS--SGNWRLTVANY 938
Query: 98 LGVSQQEA 105
LGV EA
Sbjct: 939 LGVEPPEA 946
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.326 0.142 0.461
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 434,369,472
Number of Sequences: 2790947
Number of extensions: 17613063
Number of successful extensions: 42718
Number of sequences better than 10.0: 126
Number of HSP's better than 10.0 without gapping: 113
Number of HSP's successfully gapped in prelim test: 13
Number of HSP's that attempted gapping in prelim test: 42510
Number of HSP's gapped (non-prelim): 206
length of query: 246
length of database: 848,049,833
effective HSP length: 124
effective length of query: 122
effective length of database: 501,972,405
effective search space: 61240633410
effective search space used: 61240633410
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 73 (32.7 bits)
Medicago: description of AC148654.4


