

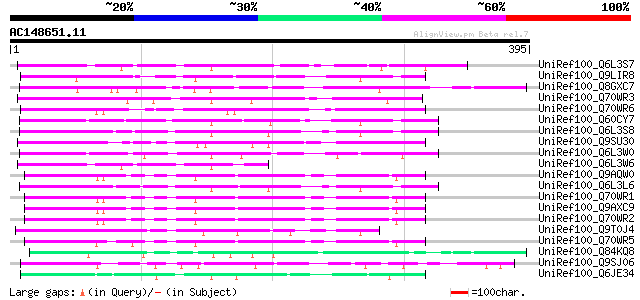
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148651.11 - phase: 0
(395 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q6L3S7 Putative F-Box protein [Solanum demissum] 117 7e-25
UniRef100_Q9LIR8 Gb|AAF30317.1 [Arabidopsis thaliana] 107 4e-22
UniRef100_Q8GXC7 Hypothetical protein At3g06240/F28L1_18 [Arabid... 106 1e-21
UniRef100_Q70WR3 S locus F-box (SLF)-S1E protein [Antirrhinum hi... 100 9e-20
UniRef100_Q70WR6 S locus F-box (SLF)-S4D protein [Antirrhinum hi... 94 9e-18
UniRef100_Q60CY7 Putative F-box protein [Solanum demissum] 91 7e-17
UniRef100_Q6L3S8 Putative F-Box protein [Solanum demissum] 90 1e-16
UniRef100_Q9SU30 Hypothetical protein AT4g12560 [Arabidopsis tha... 89 2e-16
UniRef100_Q6L3W0 Putative F-box protein [Solanum demissum] 89 2e-16
UniRef100_Q6L3W6 Putative S haplotype-specific F-box protein [So... 88 4e-16
UniRef100_Q9AQW0 SLF-S2 protein (S locus F-box (SLF)-S2 protein)... 87 6e-16
UniRef100_Q6L3L6 Putative F-box-like protein [Solanum demissum] 86 2e-15
UniRef100_Q70WR1 S locus F-box (SLF)-S5 protein [Antirrhinum his... 86 2e-15
UniRef100_Q9AXC9 S locus F-box (SLF)-S2-like protein [Antirrhinu... 85 4e-15
UniRef100_Q70WR2 S locus F-box (SLF)-S1 protein [Antirrhinum his... 85 4e-15
UniRef100_Q9T0J4 Hypothetical protein AT4g38870 [Arabidopsis tha... 83 2e-14
UniRef100_Q70WR5 S locus F-box (SLF)-S4 protein [Antirrhinum his... 81 6e-14
UniRef100_Q84KQ8 F-box [Prunus mume] 80 1e-13
UniRef100_Q9SJ06 Hypothetical protein At2g21930 [Arabidopsis tha... 75 2e-12
UniRef100_Q6JE34 S2-locus linked F-box protein [Petunia integrif... 75 4e-12
>UniRef100_Q6L3S7 Putative F-Box protein [Solanum demissum]
Length = 372
Score = 117 bits (292), Expect = 7e-25
Identities = 109/367 (29%), Positives = 165/367 (44%), Gaps = 55/367 (14%)
Query: 7 SVLPSELMTKILSLLPVKPLMRFRCVNKFYNTLISDPHFIQMHLKNSARNPNLMVIARQH 66
SVLP E++ +IL +P K L++F CV+K + LIS FI+ HL+ A + H
Sbjct: 7 SVLPHEIIIEILLKVPPKSLLKFMCVSKTWLELISSAKFIKTHLELIANDKEY-----SH 61
Query: 67 NFNSFDENVLNLPISLL---LENSLSTVPYDPYYRLKNENPHCPWLFAGSCNGLICLCLD 123
+ F E+ N + L L ST +D ENP GS NGLICL
Sbjct: 62 HRIIFQESACNFKVCCLPSMLNKERSTELFD--IGSPMENPTIYTWIVGSVNGLICLYSK 119
Query: 124 IDTSHGSRLCLWNPATRTKSEFDLASQEC-------FVFAFGYDNLNGNYKVIAFD-IKV 175
I+ + LWNPA + + + + FGYD +YKV+ I
Sbjct: 120 IEET-----VLWNPAVKKSKKLPTLGAKLRNGCSYYLKYGFGYDETRDDYKVVVIQCIYE 174
Query: 176 KSGNARSVVKVFSMRDNCWRNIQCFPVLPLYMFVSTQNGVYFSSTVNWLALQDYFGLDYF 235
SG+ SVV ++S++ + WR I F F+ G + + + W D +D F
Sbjct: 175 DSGSCDSVVNIYSLKADSWRTINKFQ----GNFLVNSPGKFVNGKIYWALSAD---VDTF 227
Query: 236 HLNYSSITPEKYVILSLDLSTETYTQLLLPRGFNKVSRHQPKLAVL---MDCLCFGHDYE 292
++ I+SLDL+ ET+ +L LP + K S + L V+ + LC + E
Sbjct: 228 NM---------CNIISLDLADETWRRLELPDSYGKGS-YPLALGVVGSHLSVLCL-NCIE 276
Query: 293 ETYFVIWQMKDFGVQSSWIQLFK----------ITYQNFFSYYCDFARESKWLDFL-PLC 341
T +W KD GV+ SW ++F I + + FS C + + + L L P+
Sbjct: 277 GTNSDVWIRKDCGVEVSWTKIFTVDHPKDLGEFIFFTSIFSVPCYQSNKDEILLLLPPVI 336
Query: 342 LSKNGDT 348
L+ NG T
Sbjct: 337 LTYNGST 343
>UniRef100_Q9LIR8 Gb|AAF30317.1 [Arabidopsis thaliana]
Length = 364
Score = 107 bits (268), Expect = 4e-22
Identities = 88/327 (26%), Positives = 150/327 (44%), Gaps = 58/327 (17%)
Query: 9 LPSELMTKILSLLPVKPLMRFRCVNKFYNTLISDPHFIQMH--------LKNSARNP-NL 59
LP E+M +IL LPVK L RF+CV + +LIS+ F H S ++P +
Sbjct: 14 LPLEMMEEILLRLPVKSLTRFKCVCSSWRSLISETLFALKHALILETSKATTSTKSPYGV 73
Query: 60 MVIARQHNFNSFDENVLNLPISLLLENSLSTVPYDPYYRLKNENPHCPWLFAGSCNGLIC 119
+ +R H + ++ N + E+ + D YY++ G+C+GL+C
Sbjct: 74 ITTSRYHLKSCCIHSLYNASTVYVSEHDGELLGRD-YYQV-----------VGTCHGLVC 121
Query: 120 LCLDIDTSHGSRLCLWNPATR-----TKSEFDLASQECFV-FAFGYDNLNGNYKVIAFDI 173
+D D S L LWNP + + S+ + + EC V + FGYD +YKV+A +
Sbjct: 122 FHVDYDKS----LYLWNPTIKLQQRLSSSDLETSDDECVVTYGFGYDESEDDYKVVA--L 175
Query: 174 KVKSGNARSVVKVFSMRDNCWRNIQCFPVLPLYMFVSTQNGVYFSSTVNWLALQDYFGLD 233
+ + K++S R WR+ FP + + +++G+Y + T+NW A
Sbjct: 176 LQQRHQVKIETKIYSTRQKLWRSNTSFP-SGVVVADKSRSGIYINGTLNWAA-------- 226
Query: 234 YFHLNYSSITPEKYVILSLDLSTETYTQLLLP----RGFNKVSRHQPKLAVLMDCLCFGH 289
+ + + I+S D+S + + +L P RG ++ + + M C C G
Sbjct: 227 -------TSSSSSWTIISYDMSRDEFKELPGPVCCGRGCFTMTLGDLRGCLSMVCYCKGA 279
Query: 290 DYEETYFVIWQMKDFGVQSSWIQLFKI 316
+ + +W MK+FG SW +L I
Sbjct: 280 NAD-----VWVMKEFGEVYSWSKLLSI 301
>UniRef100_Q8GXC7 Hypothetical protein At3g06240/F28L1_18 [Arabidopsis thaliana]
Length = 427
Score = 106 bits (264), Expect = 1e-21
Identities = 109/427 (25%), Positives = 183/427 (42%), Gaps = 83/427 (19%)
Query: 8 VLPSELMTKILSLLPVKPLMRFRCVNKFYNTLISDPHFIQMHL-----KNSARNPNLMVI 62
VLP E++T+IL LP K + RFRCV+K + TL SDP F ++HL S R+ + +I
Sbjct: 35 VLPPEIITEILLRLPAKSIGRFRCVSKLFCTLSSDPGFAKIHLDLILRNESVRSLHRKLI 94
Query: 63 ARQHNFNSFDENVL------------NLPI-------SLLLENSLSTVPYDP---YYRLK 100
HN S D N + N P+ S ++ N + YD +L
Sbjct: 95 VSSHNLYSLDFNSIGDGIRDLAAVEHNYPLKDDPSIFSEMIRNYVGDHLYDDRRVMLKLN 154
Query: 101 NENPHCPWL-FAGSCNGLICLCLDIDTSHGSRLCLWNPAT----RTKSEFDLASQE---- 151
++ W+ GS NGL+C I G+ + L+NP T R F S E
Sbjct: 155 AKSYRRNWVEIVGSSNGLVC----ISPGEGA-VFLYNPTTGDSKRLPENFRPKSVEYERD 209
Query: 152 -CFVFAFGYDNLNGNYKVIAFDIKVKSGNARSVVKVFSMRDNCWRNIQCFPVLPLYMFVS 210
+ FG+D L +YK++ V + V+S++ + WR I + + S
Sbjct: 210 NFQTYGFGFDGLTDDYKLVKL---VATSEDILDASVYSLKADSWRRI--CNLNYEHNDGS 264
Query: 211 TQNGVYFSSTVNWLALQDYFGLDYFHLNYSSITPEKYVILSLDLSTETYTQLLLPRGFNK 270
+GV+F+ ++W+ ++ + V+++ D+ TE + ++ +P
Sbjct: 265 YTSGVHFNGAIHWV--------------FTESRHNQRVVVAFDIQTEEFREMPVPDEAED 310
Query: 271 VSRHQPKLAV--LMDCLCFGHDYEETYFVIWQMKDFGVQSSWIQL-FKITYQNFFSYYCD 327
S V L LC + + + IW M ++G SW ++ + Y++
Sbjct: 311 CSHRFSNFVVGSLNGRLCVVNSCYDVHDDIWVMSEYGEAKSWSRIRINLLYRS------- 363
Query: 328 FARESKWLDFLPLCLSKNGDTLILANNQENEAFIYNRRDDRVEKIGITNKNML-WFEAWD 386
PLC +KN + ++L + + +YN + +GI + FEA
Sbjct: 364 ---------MKPLCSTKNDEEVLL--ELDGDLVLYNFETNASSNLGICGVKLSDGFEANT 412
Query: 387 YVESLVS 393
YVESL+S
Sbjct: 413 YVESLIS 419
>UniRef100_Q70WR3 S locus F-box (SLF)-S1E protein [Antirrhinum hispanicum]
Length = 384
Score = 100 bits (248), Expect = 9e-20
Identities = 89/330 (26%), Positives = 147/330 (43%), Gaps = 41/330 (12%)
Query: 7 SVLPSELMTKILSLLPVKPLMRFRCVNKFYNTLISDPHFIQMHLKNSARNPNLMVIARQH 66
S+LP +++ +I+ LPVK L+RFRCV+K + +I +FI H L++I R
Sbjct: 4 SLLPLDMVIEIMVQLPVKSLVRFRCVSKSFCVIIKSSNFINNHFLRRQTRDTLLLIRRYF 63
Query: 67 NFNSFDENV-LNLPISLLLENSL---STVPYDPYYRLKNENPHCPW--LFAGSCNGLICL 120
D+ + + P S LE + ++P+ RL+ + P+ P + G CNGLIC+
Sbjct: 64 PSPQEDDALSFHKPDSPGLEEEVWAKLSIPFLSDLRLRYDQPYFPQSVIILGPCNGLICI 123
Query: 121 CLDIDTSHGSRLCLWNPATRTKSEFDLASQEC------FVFAFGYDNLNGNYKVIAFDIK 174
D + NPA R + C + G+ N + N+ I
Sbjct: 124 FYD------DFIISCNPALREFKKLPPCPFCCPKRFYSNIIGQGFGNCDSNFFKIVLVRT 177
Query: 175 VKSGNARS------VVKVFSMRDNCWRNIQCFPVLPLYMFVSTQNGVYFSSTVNWLALQD 228
+KS + + +V +++ WR I+ VL Y+F S V+F+ +W A
Sbjct: 178 IKSVSDYNRDKPYIMVHLYNSNTQSWRLIEGEAVLVQYIFSSPCTDVFFNGACHWNA--G 235
Query: 229 YFGLDYFHLNYSSITPEKYVILSLDLSTETYTQLLLPRGFNKVSRHQPKLAVLMDCLC-- 286
FG+ Y IL+ D+STE +++ P GF ++ L L +CL
Sbjct: 236 VFGIPY-----------PGSILTFDISTEIFSEFEYPDGFRELYGGCLCLTALSECLSVI 284
Query: 287 -FGHDYEETYFV-IWQMKDFGVQSSWIQLF 314
+ ++ F+ IW MK +G SW + F
Sbjct: 285 RYNDSTKDPQFIEIWVMKVYGNSDSWTKDF 314
>UniRef100_Q70WR6 S locus F-box (SLF)-S4D protein [Antirrhinum hispanicum]
Length = 374
Score = 93.6 bits (231), Expect = 9e-18
Identities = 81/334 (24%), Positives = 141/334 (41%), Gaps = 57/334 (17%)
Query: 9 LPSELMTKILSLLPVKPLMRFRCVNKFYNTLISDPHFIQMHLKNSARNPNLMVIAR---- 64
+P +++ +IL LPVK L+R +C +K + LI FI H+ RN ++++ R
Sbjct: 7 IPEDILKEILVWLPVKSLIRLKCASKHLDMLIKSQAFITSHMIKQRRNDGMLLVRRILPP 66
Query: 65 --------QHNFNS--FDENVLNLPISLLLENSLSTVPYDPYYRLKNENPHCPWLFAGSC 114
H+ NS +E + LPI+LL ++ NP+ + G C
Sbjct: 67 STYNDVFSFHDVNSPELEEVLPKLPITLLSNPDEASF-----------NPNIVDVL-GPC 114
Query: 115 NGLICLCLDIDTSHGSRLCLWNPATRTKSEFDLASQECFVFAFGYDNLNG-------NYK 167
NG++C+ D + L NPA R + A C + G N+K
Sbjct: 115 NGIVCITGQED------IILCNPALREFRKLPSAPISCRPPCYSIRTGGGFGSTCTNNFK 168
Query: 168 VI----AFDIKVKSGNARSVVKVFSMRDNCWRNIQCFPVLPLYMFVSTQNGVYFSSTVNW 223
VI + +V +A+ + +++ ++ WR I F ++ +F + ++F +W
Sbjct: 169 VILMNTLYTARVDGRDAQHRIHLYNSNNDSWREINDFAIVMPVVFSYQCSELFFKGACHW 228
Query: 224 LALQDYFGLDYFHLNYSSITPEKYVILSLDLSTETYTQLLLPRGFNKVSRHQPKLAVLMD 283
TP+ VIL+ D+STE + Q P GF + Q +L +
Sbjct: 229 NGRTS-----------GETTPD--VILTFDVSTEVFGQFEHPSGFKLCTGLQHNFMILNE 275
Query: 284 CLC-FGHDYEETYFVIWQMKDFGVQSSWIQLFKI 316
C + +W MK++G++ SW + F I
Sbjct: 276 CFASVRSEVVRCLIEVWVMKEYGIKQSWTKKFVI 309
>UniRef100_Q60CY7 Putative F-box protein [Solanum demissum]
Length = 383
Score = 90.5 bits (223), Expect = 7e-17
Identities = 93/332 (28%), Positives = 143/332 (43%), Gaps = 44/332 (13%)
Query: 8 VLPSELMTKILSLLPVKPLMRFRCVNKFYNTLISDPHFIQMHLKNSARNPNLMVIARQHN 67
+LP EL+T+IL LP+K L +F CV+K + LIS P F++ H+K +A + I +
Sbjct: 10 LLPDELITEILLRLPIKSLSKFMCVSKSWLQLISSPAFVKKHIKLTANDKG--YIYHRLI 67
Query: 68 FNSFDENVLNLPISLLLENSLSTVPYDPYYRLKNENPHCPWLFAGSCNGLICLCLDIDTS 127
F + + + P+ L N + + E GS NGLIC +
Sbjct: 68 FRNTNNDFKFCPLPPLFTNQ-QLIEEILHIDSPIERTTLSTHIVGSVNGLIC----VAHV 122
Query: 128 HGSRLCLWNPA-TRTKSEFDLASQEC---FVFAFGYDNLNGNYKVIAFDIKVKSGNARSV 183
+WNPA T++K S C FGYD +YKV+ D ++ N R+V
Sbjct: 123 RQREAYIWNPAITKSKELPKSTSNLCSDGIKCGFGYDESRDDYKVVFIDYPIRH-NHRTV 181
Query: 184 VKVFSMRDNCWRNI--QCFPVLPLYMFVSTQNG-VYFSSTVNWLALQDYFGLDYFHLNYS 240
V ++S+R N W + Q + L + NG +Y++S+ + +Y
Sbjct: 182 VNIYSLRTNSWTTLHDQLQGIFLLNLHGRFVNGKLYWTSST---CINNY----------- 227
Query: 241 SITPEKYVILSLDLSTETYTQLLLP-----RGFNKVSRHQPKLAVLMDCLCFGHDYEETY 295
+ I S DL+ T+ L LP + V L++L C +
Sbjct: 228 ----KVCNITSFDLADGTWGSLELPSCGKDNSYINVGVVGSDLSLLYTCQLGAATSD--- 280
Query: 296 FVIWQMKDFGVQSSWIQLFKITY-QNFFSYYC 326
+W MK GV SW +LF I Y QN + C
Sbjct: 281 --VWIMKHSGVNVSWTKLFTIKYPQNIKIHRC 310
>UniRef100_Q6L3S8 Putative F-Box protein [Solanum demissum]
Length = 327
Score = 89.7 bits (221), Expect = 1e-16
Identities = 96/334 (28%), Positives = 139/334 (40%), Gaps = 47/334 (14%)
Query: 8 VLPSELMTKILSLLPVKPLMRFRCVNKFYNTLISDPHFIQMHLKNSARNPNLMVIARQHN 67
+LP EL+T+IL LP+K L++F CV+K + LIS P F++ H+K +A + +
Sbjct: 11 LLPDELITEILLKLPIKSLLKFMCVSKSWLQLISSPAFVKNHIKLTADDKGYIYHRLIFR 70
Query: 68 FNSFDENVLNLPISLLLENSLSTVPYDPYYRLKNENPHCPWLFAGSCNGLICLCLDIDTS 127
+ D LP L + L Y + E GS NGLIC
Sbjct: 71 NTNDDFKFCPLP-PLFTQQQLIKELY--HIDSPIERTTLSTHIVGSVNGLICAA----HV 123
Query: 128 HGSRLCLWNPA-TRTKSEFDLASQEC---FVFAFGYDNLNGNYKVIAFDIKVKSGNARSV 183
+WNP T++K S C FGYD +YKV+ D + N R+V
Sbjct: 124 RQREAYIWNPTITKSKELPKSRSNLCSDGIKCGFGYDESRDDYKVVFIDYPIHRHNHRTV 183
Query: 184 VKVFSMRDNCWR----NIQCFPVLPLY-MFVSTQNGVYFSSTVNWLALQDYFGLDYFHLN 238
V ++S+R W +Q F +L L+ FV+ + SS +N N
Sbjct: 184 VNIYSLRTKSWTTLHDQLQGFFLLNLHGRFVNGKLYWTSSSCIN---------------N 228
Query: 239 YSSITPEKYVILSLDLSTETYTQLLLP-----RGFNKVSRHQPKLAVLMDCLCFGHDYEE 293
Y I S DL+ T+ +L LP + V L++L C +
Sbjct: 229 YKVCN-----ITSFDLADGTWERLELPSCGKDNSYINVGVVGSDLSLLYTCQRGAATSD- 282
Query: 294 TYFVIWQMKDFGVQSSWIQLFKITY-QNFFSYYC 326
+W MK GV SW +LF I Y QN + C
Sbjct: 283 ----VWIMKHSGVNVSWTKLFTIKYPQNIKIHRC 312
>UniRef100_Q9SU30 Hypothetical protein AT4g12560 [Arabidopsis thaliana]
Length = 408
Score = 89.0 bits (219), Expect = 2e-16
Identities = 88/333 (26%), Positives = 154/333 (45%), Gaps = 54/333 (16%)
Query: 7 SVLPSELMTKILSLLPVKPLMRFRCVNKFYNTLISDPHFIQMHLKNSARNPNLMVIARQH 66
+ +P +++ I LP K L+R R ++K LI+DP FI+ HL + + ++I +
Sbjct: 2 ATIPMDIVNDIFLRLPAKTLVRCRALSKPCYHLINDPDFIESHLHRVLQTGDHLMILLRG 61
Query: 67 NFNSFDENVLNLPISLLLENSLSTVPYDPYYRLKNENPHCPWLFAGSCNGLICLCLDIDT 126
+ ++ +SL +V D + +K P + GS NGLI L +
Sbjct: 62 ALRLYSVDL----------DSLDSVS-DVEHPMKRGGPTEVF---GSSNGLIGL-----S 102
Query: 127 SHGSRLCLWNPATRT-----KSEFDL---ASQECFVF-AFGYDNLNGNYKVIAF-DIKVK 176
+ + L ++NP+TR S DL +S +VF GYD+++ +YKV+ K+
Sbjct: 103 NSPTDLAVFNPSTRQIHRLPPSSIDLPDGSSTRGYVFYGLGYDSVSDDYKVVRMVQFKID 162
Query: 177 SGNARSV-----VKVFSMRDNCWR-------NIQCFPVLPLYMFVSTQNGVYFSSTVNWL 224
S + VKVFS++ N W+ +IQ ++ GV ++++W+
Sbjct: 163 SEDELGCSFPYEVKVFSLKKNSWKRIESVASSIQLLFYFYYHLLYRRGYGVLAGNSLHWV 222
Query: 225 ALQDYFGLDYFHLNYSSITPEKYVILSLDLSTETYTQLLLPRGF-NKVSRHQPKLAVLMD 283
L GL F+L I+ DL+ E + + P N Q + VL
Sbjct: 223 -LPRRPGLIAFNL-----------IVRFDLALEEFEIVRFPEAVANGNVDIQMDIGVLDG 270
Query: 284 CLCFGHDYEETYFVIWQMKDFGVQSSWIQLFKI 316
CLC +Y+++Y +W MK++ V+ SW ++F +
Sbjct: 271 CLCLMCNYDQSYVDVWMMKEYNVRDSWTKVFTV 303
>UniRef100_Q6L3W0 Putative F-box protein [Solanum demissum]
Length = 427
Score = 89.0 bits (219), Expect = 2e-16
Identities = 94/335 (28%), Positives = 139/335 (41%), Gaps = 48/335 (14%)
Query: 8 VLPSELMTKILSLLPVKPLMRFRCVNKFYNTLISDPHFIQMHLKNSARNPNLMVIARQHN 67
+LP EL+T+IL LPVK L +F CV+K + LIS P F++ H+K +A + I +
Sbjct: 52 LLPDELITEILLKLPVKSLSKFMCVSKSWLQLISSPTFVKNHIKLTADDKG--YIHHRLI 109
Query: 68 FNSFDENVLNLPISLLLENSLSTVPYDPYYRLKN--ENPHCPWLFAGSCNGLICLCLDID 125
F + D N + L T + + + + E GS NGLIC+
Sbjct: 110 FRNIDGNFKFCSLPPLFTKQQHT---EELFHIDSPIERSTLSTHIVGSVNGLICVV---- 162
Query: 126 TSHGSR-LCLWNPA-TRTKSEFDLASQEC---FVFAFGYDNLNGNYKVIAFDIKV---KS 177
HG + +WNP T++K S C + FGYD +YKV+ S
Sbjct: 163 --HGQKEAYIWNPTITKSKELPKFTSNMCSSSIKYGFGYDESRDDYKVVFIHYPYNHSSS 220
Query: 178 GNARSVVKVFSMRDNCWRNIQCFPVLPLYMFVSTQNGVYFSSTVNWLALQDYFGLDYFHL 237
N +VV ++S+R+N W + L F+ G + + + W
Sbjct: 221 SNMTTVVHIYSLRNNSWTTFRD----QLQCFLVNHYGRFVNGKLYWT------------- 263
Query: 238 NYSSITPEKYV---ILSLDLSTETYTQLLLPRGFNKVSRHQPKLAVLMDCLCFGHDYEET 294
SS KY I S DL+ T+ L LP L V+ L + +
Sbjct: 264 --SSTCINKYKVCNITSFDLADGTWGSLDLP--ICGKDNFDINLGVVGSDLSLLYTCQRG 319
Query: 295 YFV--IWQMKDFGVQSSWIQLFKITY-QNFFSYYC 326
+W MK V SW +LF I Y QN ++ C
Sbjct: 320 AATSDVWIMKHSAVNVSWTKLFTIKYPQNIKTHRC 354
>UniRef100_Q6L3W6 Putative S haplotype-specific F-box protein [Solanum demissum]
Length = 240
Score = 88.2 bits (217), Expect = 4e-16
Identities = 65/201 (32%), Positives = 93/201 (45%), Gaps = 30/201 (14%)
Query: 7 SVLPSELMTKILSLLPVKPLMRFRCVNKFYNTLISDPHFIQMHLKNSARNPNLMVIARQH 66
SVLP E++ +IL LP K L++FRCV+K + LIS FI+ HLK +A + H
Sbjct: 7 SVLPHEIIKEILLNLPPKSLLKFRCVSKSWLELISSAKFIKNHLKQTANDKEY-----SH 61
Query: 67 NFNSFDENVLNLPISLL---LENSLSTVPYDPYYRLKNENPHCPWLFAGSCNGLICLCLD 123
+ F E+ N + L L ST +D ENP GS NGLICL
Sbjct: 62 HRIIFQESACNFKVCCLRSMLNKEQSTELFD--IGSPMENPSIYTWIVGSVNGLICLYSK 119
Query: 124 IDTSHGSRLCLWNPATRTKSEFDLASQEC-------FVFAFGYDNLNGNYKVIAFDIKVK 176
I+ + LWNPA + + + + FGYD +YKV++
Sbjct: 120 IEET-----VLWNPAVKKSKKLPTLGAKLRNGCSYYLKYGFGYDETRDDYKVMS------ 168
Query: 177 SGNARSVVKVFSMRDNCWRNI 197
N R VV ++S+R + W +
Sbjct: 169 --NMRIVVNIYSLRTDSWTTL 187
>UniRef100_Q9AQW0 SLF-S2 protein (S locus F-box (SLF)-S2 protein) [Antirrhinum
hispanicum]
Length = 376
Score = 87.4 bits (215), Expect = 6e-16
Identities = 89/330 (26%), Positives = 139/330 (41%), Gaps = 55/330 (16%)
Query: 12 ELMTKILSLLPVKPLMRFRCVNKFYNTLISDPHFIQMHLKNSARNPNLMVIARQ------ 65
+++++IL VK L+RFRCV+K + +LI FI HL N N+MV+ R
Sbjct: 10 DVISEILLFSSVKSLLRFRCVSKSWCSLIKSNDFIDNHLLRRQTNGNVMVVKRYVRTPER 69
Query: 66 -----HNFNS--FDENVLNLPISLLLENSLSTVPYDPYYRLKNENPHCPWLFAGSCNGLI 118
+N NS DE + +LP YD +Y + N G CNGLI
Sbjct: 70 DMFSFYNINSPELDELLPDLPNPYFKNIKFD---YDYFYLPQRVN------LMGPCNGLI 120
Query: 119 CLCLDIDTSHGSRLCLWNPATR-----TKSEFDLASQECF-VFAFGYDN-LNGNYKVIAF 171
CL ++G + L NPA R + F C + +G+ N N YKV+
Sbjct: 121 CL------AYGDCVLLSNPALREIKRLPPTPFANPEGHCTDIIGYGFGNTCNDCYKVVLI 174
Query: 172 DIKVKSGNARSVVKVFSMRDNCWRNIQCFPVLPLYMFVSTQNGVYFSSTVNWLALQDYFG 231
+ V + + V+ N W++I+ Y+ N ++F +W A
Sbjct: 175 E-SVGPEDHHINIYVYYSDTNSWKHIEDDSTPIKYICHFPCNELFFKGAFHWNA----NS 229
Query: 232 LDYFHLNYSSITPEKYVILSLDLSTETYTQLLLPRGFNKVSRHQPKLAVLMDCLCFGHDY 291
D F+ ++ IL+ D+ TE + ++ P + S L L +CL Y
Sbjct: 230 TDIFYADF---------ILTFDIITEVFKEMAYPHCLAQFSNSFLSLMSLNECLAMVR-Y 279
Query: 292 EE-----TYFVIWQMKDFGVQSSWIQLFKI 316
+E F IW M +GV+ SW + + I
Sbjct: 280 KEWMEDPELFDIWVMNQYGVRESWTKQYVI 309
>UniRef100_Q6L3L6 Putative F-box-like protein [Solanum demissum]
Length = 384
Score = 85.9 bits (211), Expect = 2e-15
Identities = 96/334 (28%), Positives = 138/334 (40%), Gaps = 48/334 (14%)
Query: 8 VLPSELMTKILSLLPVKPLMRFRCVNKFYNTLISDPHFIQMHLKNSARNPNLMVIARQHN 67
+LP EL+T+IL LP+K L +F CV+K + LIS P F++ H+K +A +
Sbjct: 11 LLPDELITEILLRLPIKSLSKFMCVSKSWLQLISSPAFVKNHIKLTANGKGYIYHRLIFR 70
Query: 68 FNSFDENVLNLPISLLLENSLSTVPYDPYYRLKNENPHCPWLFAGSCNGLICLCLDIDTS 127
+ D LP SL + L +D + E GS NGLIC
Sbjct: 71 NTNDDFKFCPLP-SLFTKQQLIEELFDIVSPI--ERTTLSTHIVGSVNGLICAA----HV 123
Query: 128 HGSRLCLWNPA-TRTKSEFDLASQEC---FVFAFGYDNLNGNYKVIAFDIKVKSGNARSV 183
+WNP T++K S C FGYD + +YKV+ + N RSV
Sbjct: 124 RQREAYIWNPTITKSKELPKSRSNLCSDGIKCGFGYDESHDDYKVVFINYP-SHHNHRSV 182
Query: 184 VKVFSMRDNCWR----NIQCFPVLPLYMFVSTQNGVYFSST-VNWLALQDYFGLDYFHLN 238
V ++S+R N W +Q +L L+ + + SST +N N
Sbjct: 183 VNIYSLRTNSWTTLHDQLQGIFLLNLHCRFVKEKLYWTSSTCIN---------------N 227
Query: 239 YSSITPEKYVILSLDLSTETYTQLLLP-----RGFNKVSRHQPKLAVLMDCLCFGHDYEE 293
Y I S DL+ T+ L LP + V L++L C + +
Sbjct: 228 YKVCN-----ITSFDLADGTWESLELPSCGKDNSYINVGVVGSDLSLLYTCQRGAANSD- 281
Query: 294 TYFVIWQMKDFGVQSSWIQLFKITY-QNFFSYYC 326
+W MK GV SW +LF I Y QN + C
Sbjct: 282 ----VWIMKHSGVNVSWTKLFTIKYPQNIKIHRC 311
>UniRef100_Q70WR1 S locus F-box (SLF)-S5 protein [Antirrhinum hispanicum]
Length = 376
Score = 85.9 bits (211), Expect = 2e-15
Identities = 88/330 (26%), Positives = 139/330 (41%), Gaps = 55/330 (16%)
Query: 12 ELMTKILSLLPVKPLMRFRCVNKFYNTLISDPHFIQMHLKNSARNPNLMVIARQ------ 65
+++++IL VK L+RFRCV+K + +LI FI HL N N+MV+ R
Sbjct: 10 DVISEILLFSSVKSLLRFRCVSKSWCSLIKSNDFIDNHLLRRQTNGNVMVVKRYVRTPER 69
Query: 66 -----HNFNS--FDENVLNLPISLLLENSLSTVPYDPYYRLKNENPHCPWLFAGSCNGLI 118
+N NS +E + +LP YD +Y + N G CNGLI
Sbjct: 70 DMFSFYNINSPKLEELLPDLPNPYFKNIKFD---YDYFYLPQRVN------LMGPCNGLI 120
Query: 119 CLCLDIDTSHGSRLCLWNPATR-----TKSEFDLASQECF-VFAFGYDN-LNGNYKVIAF 171
CL ++G + L NPA R + F C + +G+ N N YKV+
Sbjct: 121 CL------AYGDCVLLSNPALREIKRLPPTPFANPEGHCTDIIGYGFGNTCNDCYKVVLI 174
Query: 172 DIKVKSGNARSVVKVFSMRDNCWRNIQCFPVLPLYMFVSTQNGVYFSSTVNWLALQDYFG 231
+ V + + V+ N W++I+ Y+ N ++F +W A
Sbjct: 175 E-SVGPEDHHINIYVYYSDTNSWKHIEDDSTPIKYICHFPCNELFFKGAFHWNA----NS 229
Query: 232 LDYFHLNYSSITPEKYVILSLDLSTETYTQLLLPRGFNKVSRHQPKLAVLMDCLCFGHDY 291
D F+ ++ IL+ D+ TE + ++ P + S L L +CL Y
Sbjct: 230 TDIFYADF---------ILTFDIITEVFKEMAYPHCLAQFSNSFLSLMSLNECLAMVR-Y 279
Query: 292 EE-----TYFVIWQMKDFGVQSSWIQLFKI 316
+E F IW M +GV+ SW + + I
Sbjct: 280 KEWMEEPELFDIWVMNQYGVRESWTKQYVI 309
>UniRef100_Q9AXC9 S locus F-box (SLF)-S2-like protein [Antirrhinum hispanicum]
Length = 376
Score = 84.7 bits (208), Expect = 4e-15
Identities = 89/330 (26%), Positives = 138/330 (40%), Gaps = 55/330 (16%)
Query: 12 ELMTKILSLLPVKPLMRFRCVNKFYNTLISDPHFIQMHLKNSARNPNLMVIARQ------ 65
++ ++IL VK L+RFR V+K + +LI FI HL N N+MV+ R
Sbjct: 10 DVTSEILLFSSVKSLLRFRLVSKSWCSLIKSNDFIDNHLLRRQTNGNVMVVKRYVRTPER 69
Query: 66 -----HNFNS--FDENVLNLPISLLLENSLSTVPYDPYYRLKNENPHCPWLFAGSCNGLI 118
+N NS DE + +LP YD +Y + N G CNGLI
Sbjct: 70 DMFSFYNINSPELDELLPDLPNPYFKNIKFD---YDYFYLPQRVN------LMGPCNGLI 120
Query: 119 CLCLDIDTSHGSRLCLWNPATR-----TKSEFDLASQECF-VFAFGYDN-LNGNYKVIAF 171
CL ++G + L NPA R + F C + +G+ N N YKV+
Sbjct: 121 CL------AYGDCVLLSNPALREIKRLPPTPFANPEGHCTDIIGYGFGNTCNDCYKVVLI 174
Query: 172 DIKVKSGNARSVVKVFSMRDNCWRNIQCFPVLPLYMFVSTQNGVYFSSTVNWLALQDYFG 231
+ V + + V+ N W++I+ Y+ N ++F +W A
Sbjct: 175 E-SVGPEDHHINIYVYYSDTNSWKHIEDDSTPIKYICHFPCNELFFKGAFHWNA----NS 229
Query: 232 LDYFHLNYSSITPEKYVILSLDLSTETYTQLLLPRGFNKVSRHQPKLAVLMDCLCFGHDY 291
D F+ ++ IL+ D+ TE + ++ P + S L L +CL Y
Sbjct: 230 TDIFYADF---------ILTFDIITEVFKEMAYPHCLAQFSNSFLSLMSLNECLAMVR-Y 279
Query: 292 EE-----TYFVIWQMKDFGVQSSWIQLFKI 316
+E F IW MK +GV+ SW + + I
Sbjct: 280 KEWMEEPELFDIWVMKQYGVRESWTKQYVI 309
>UniRef100_Q70WR2 S locus F-box (SLF)-S1 protein [Antirrhinum hispanicum]
Length = 376
Score = 84.7 bits (208), Expect = 4e-15
Identities = 89/330 (26%), Positives = 138/330 (40%), Gaps = 55/330 (16%)
Query: 12 ELMTKILSLLPVKPLMRFRCVNKFYNTLISDPHFIQMHLKNSARNPNLMVIARQ------ 65
++ ++IL VK L+RFR V+K + +LI FI HL N N+MV+ R
Sbjct: 10 DVTSEILLFSSVKSLLRFRLVSKSWCSLIKSNDFIDNHLLRRQTNGNVMVVKRYVRTPER 69
Query: 66 -----HNFNS--FDENVLNLPISLLLENSLSTVPYDPYYRLKNENPHCPWLFAGSCNGLI 118
+N NS DE + +LP YD +Y + N G CNGLI
Sbjct: 70 DMFSFYNINSPELDELLPDLPNPYFKNIKFD---YDYFYLPQRVN------LMGPCNGLI 120
Query: 119 CLCLDIDTSHGSRLCLWNPATR-----TKSEFDLASQECF-VFAFGYDN-LNGNYKVIAF 171
CL ++G + L NPA R + F C + +G+ N N YKV+
Sbjct: 121 CL------AYGDCVLLSNPALREIKRLPPTPFANPEGHCTDIIGYGFGNTCNDCYKVVLI 174
Query: 172 DIKVKSGNARSVVKVFSMRDNCWRNIQCFPVLPLYMFVSTQNGVYFSSTVNWLALQDYFG 231
+ V + + V+ N W++I+ Y+ N ++F +W A
Sbjct: 175 E-SVGPEDHHINIYVYYSDTNSWKHIEDDSTPIKYICHFPCNELFFKGAFHWNA----NS 229
Query: 232 LDYFHLNYSSITPEKYVILSLDLSTETYTQLLLPRGFNKVSRHQPKLAVLMDCLCFGHDY 291
D F+ ++ IL+ D+ TE + ++ P + S L L +CL Y
Sbjct: 230 TDIFYADF---------ILTFDIITEVFKEMAYPHCLAQFSNSFLSLMSLNECLAMVR-Y 279
Query: 292 EE-----TYFVIWQMKDFGVQSSWIQLFKI 316
+E F IW MK +GV+ SW + + I
Sbjct: 280 KEWMEEPELFDIWVMKQYGVRESWTKQYVI 309
>UniRef100_Q9T0J4 Hypothetical protein AT4g38870 [Arabidopsis thaliana]
Length = 426
Score = 82.8 bits (203), Expect = 2e-14
Identities = 79/292 (27%), Positives = 135/292 (46%), Gaps = 42/292 (14%)
Query: 5 HQSVLPSELMTKILSLLPVKPLMRFRCVNKFYNTLISDPHFIQMHLKNSARNP-NLMVIA 63
+ +LP +L+ +IL L +KPL+RF CV+K + ++I DP+F+++ L S + P +L+ +
Sbjct: 49 NSELLPVDLIMEILKKLSLKPLIRFLCVSKLWASIIRDPYFMKLFLNESLKRPKSLVFVF 108
Query: 64 RQHNFNSFDENVLNLPISLLLENSLSTVPYDPYYRLKNENPHCPWLFAGSCNGLICLCLD 123
R + S +V + +S S+ Y + + S +GLIC
Sbjct: 109 RAQSLGSIFSSVHLKSTREISSSSSSSSASSITYHVTCYTQQ-RMTISPSVHGLICY--- 164
Query: 124 IDTSHGSRLCLWNPATRTKSEFD--LASQECFVFAFGYDNLNGNYKVIAFD-----IKVK 176
S L ++NP TR A + GYD L+GNYKV+ ++ +
Sbjct: 165 ---GPPSSLVIYNPCTRRSITLPKIKAGRRAINQYIGYDPLDGNYKVVCITRGMPMLRNR 221
Query: 177 SGNARSV-VKVFSMRDNCWRNIQCFPVLPLYMFVSTQ---NGVYFSSTVNWLALQDYFGL 232
G A + V RD+ WR I ++P + VS + NGV + Y
Sbjct: 222 RGLAEEIQVLTLGTRDSSWRMIH--DIIPPHSPVSEELCINGVLY-----------YRAF 268
Query: 233 DYFHLNYSSITPEKYVILSLDLSTETYTQLLLP---RGFNKVSRHQPKLAVL 281
LN S+ I+S D+ +E + + +P R F+K+++++ KLAV+
Sbjct: 269 IGTKLNESA-------IMSFDVRSEKFDLIKVPCNFRSFSKLAKYEGKLAVI 313
>UniRef100_Q70WR5 S locus F-box (SLF)-S4 protein [Antirrhinum hispanicum]
Length = 376
Score = 80.9 bits (198), Expect = 6e-14
Identities = 87/333 (26%), Positives = 139/333 (41%), Gaps = 61/333 (18%)
Query: 12 ELMTKILSLLPVKPLMRFRCVNKFYNTLISDPHFIQMHLKNSARNPNLMVIAR------Q 65
++ ++IL VK L+RFR V+K + +LI FI HL N N+MV+ R +
Sbjct: 10 DVTSEILLFSSVKSLLRFRLVSKSWCSLIKSHDFIDNHLLRRQTNGNVMVVKRYVRTPER 69
Query: 66 HNFNSFDENVLNLPISLLLENSLSTVP----------YDPYYRLKNENPHCPWLFAGSCN 115
F+ +D N S L+ L +P YD +Y + N G CN
Sbjct: 70 DMFSFYDIN------SPELDELLPDLPNPYFKNIKFDYDYFYLPQRVN------LMGPCN 117
Query: 116 GLICLCLDIDTSHGSRLCLWNPATR-----TKSEFDLASQECF-VFAFGYDN-LNGNYKV 168
GL+CL ++G + L NPA R + F C + +G+ N N YKV
Sbjct: 118 GLVCL------AYGDCVLLSNPALREIKRLPPTPFANPEGHCTDIIGYGFGNTCNDCYKV 171
Query: 169 IAFDIKVKSGNARSVVKVFSMRDNCWRNIQCFPVLPLYMFVSTQNGVYFSSTVNWLALQD 228
+ + V + + V+ N W++I+ Y+ N ++F +W A
Sbjct: 172 VLIE-SVGPEDHHINIYVYYSDTNSWKHIEDDSTPIKYICHFPCNELFFKGAFHWNA--- 227
Query: 229 YFGLDYFHLNYSSITPEKYVILSLDLSTETYTQLLLPRGFNKVSRHQPKLAVLMDCLCFG 288
D F+ ++ IL+ D+ TE + ++ P + S L L +CL
Sbjct: 228 -NSTDIFYADF---------ILTFDIITEVFKEMAYPHCLAQFSNSFLSLMSLNECLAMV 277
Query: 289 HDYEE-----TYFVIWQMKDFGVQSSWIQLFKI 316
Y+E F IW M +GV+ SW + + I
Sbjct: 278 R-YKEWMEEPELFDIWVMNQYGVRESWTKQYVI 309
>UniRef100_Q84KQ8 F-box [Prunus mume]
Length = 397
Score = 80.1 bits (196), Expect = 1e-13
Identities = 95/424 (22%), Positives = 169/424 (39%), Gaps = 78/424 (18%)
Query: 16 KILSLLPVKPLMRFRCVNKFYNTLISDPHFIQMHLKNSARNPN----------------- 58
+IL+ LP K LMRF+CV K ++ L+++PHF+ HL P+
Sbjct: 4 QILARLPPKSLMRFKCVCKSWHALLNNPHFVAKHLHLYNNQPSSTCVLFKRSVLSRTEHN 63
Query: 59 ------LMVIARQHNFNSFDENVLNLPI-SLLLENSLSTVPYDPYYRLK----NENPHCP 107
+I R N + D N++N + L S+ + L E+ H
Sbjct: 64 KEELVFTFLILRNDNEINADHNLINCKVEDLHFPRSMGLKSRGQFIELPGLELGESVH-- 121
Query: 108 WLFAGSCNGLICLCLDIDTSHGSRLCLWNPATRTKSEFDLASQECF------VFAFGYDN 161
G C+GL CL L + L +NPA + EF + Q C FGYD
Sbjct: 122 --IVGHCDGLFCLSL-----YTGELVFYNPAIK---EFRVLPQSCLEDACSCTLGFGYDP 171
Query: 162 LNGNY---KVIAFDIKVKSGNARSV----VKVFSMRDNCWRNIQC--FPVLPLYMFVSTQ 212
+Y ++++ ++ + +++++ N WR I+ Y + +
Sbjct: 172 KRKDYVLLSIVSYGEEIFDDERLVIHPPQAEIYTLSTNSWREIETHYLETETTYFWGNET 231
Query: 213 NGVYFSSTVNWLALQDYFGLDYFHLNYSSITPE-KYVILSLDLSTETYTQLLLPRGFNKV 271
YF+ WL ++ F Y + E K VI+ D E + + LP F +
Sbjct: 232 FSAYFNGVFYWLGYEEKKEFVSF---YDRLEEEKKQVIILFDTFDEVFHNMPLPDCFYEF 288
Query: 272 SRHQPKLAVLMDCLC-FG-HDYEETYFVIWQMKDFGVQSSWIQLFKITYQNFFSYYCDFA 329
H+ L V + + FG + E F +W M +F W + +
Sbjct: 289 PSHEMSLTVWNESIALFGFYRCEFEPFEVWVMDEF---DGWTKHLSVV-----------P 334
Query: 330 RESKWLDFLPLCLSKNGDTLILANNQENEAFIYNRRDDRVEKIGITNKNMLWFEAWDYVE 389
+ + +D +PL L + + L++ +++ F YN + ++ + + + F+A V
Sbjct: 335 KVDQEVD-IPLALWRRNEVLLV--DRDGRIFSYNFDTENLKYLPVHGASRGDFQAVVCVN 391
Query: 390 SLVS 393
S+VS
Sbjct: 392 SIVS 395
>UniRef100_Q9SJ06 Hypothetical protein At2g21930 [Arabidopsis thaliana]
Length = 396
Score = 75.5 bits (184), Expect = 2e-12
Identities = 102/419 (24%), Positives = 178/419 (42%), Gaps = 109/419 (26%)
Query: 9 LPSELMTKILSLLPVKPLMRFRCVNKFYNTLISDPHFIQMHLKNSARNPNLMVIARQ--- 65
+P +L+ +IL LPVK L RF V+K Y ++I + F++ +L NS+ P ++
Sbjct: 25 IPFDLIPEILKRLPVKTLARFLSVSKEYTSIIRNRDFMKSYLINSSTRPQSLIFTIAGGG 84
Query: 66 -HNFNSFDENVLNLPISLLLENSLSTVPYDPYYRLKNENPHCPWL----FAGSCNGLICL 120
H F S L++ ST P Y + +CP L FA S +GLIC
Sbjct: 85 IHCFFS------------LIDQGESTSSSKPTYLM-----NCPHLQLKTFAPSVHGLIC- 126
Query: 121 CLDIDTSHG---------SRLCLWNPATRTK---SEFDLASQECFVFAFGYDNLNGNYKV 168
HG RL + NP+TR + D A+ EC GYD ++G+YKV
Sbjct: 127 -------HGPPSTLIVSSPRLIVSNPSTRRSIILPKID-ANHECIYHHMGYDPIDGDYKV 178
Query: 169 IAFDIKVKSGNARSVVK---VFSMR-DNCWRNIQCFPVLPLYMFVSTQNGVYFSSTVNWL 224
+ + R + K VF++R N WR ++ FP P + + + + ++
Sbjct: 179 LCMMKGMHVYQRRYLAKELQVFTLRKGNSWRMVEDFP--PHCLCHEDTPDLCINGVLYYV 236
Query: 225 ALQDYFGLDYFHLNYSSITPEKYVILSLDLSTETYTQLLLPRGFN-----KVSRHQPKLA 279
A+ D T + ++S D+ +E + L+ G + K++R++ K A
Sbjct: 237 AMLD--------------TASNHAVMSFDVRSEKFD--LIKGGPDGDLNPKLTRYEGKPA 280
Query: 280 VLMDCLCFGHDYEETYFVI-------WQMKDFGVQSSWIQLFKITYQNFFSYYCDFARES 332
+L G DY +VI W + V S+ + +N + ++C F
Sbjct: 281 LLFP----GSDYRINLWVIEDAAKHEWSKMSYDVSSTSL------IRNPYFHHCVF---- 326
Query: 333 KWLDFLPLCLSKNGDTLILANNQENEAFI---YNRRDDRVEKI---GITNKNM-LWFEA 384
C + G+ ++ + + F+ Y+ + + + + GI ++ + LW EA
Sbjct: 327 --------CTNDAGEIVLAPDFVRTKTFVVLYYHPKKNTMRSVVIKGIRDRKIPLWDEA 377
>UniRef100_Q6JE34 S2-locus linked F-box protein [Petunia integrifolia subsp. inflata]
Length = 389
Score = 74.7 bits (182), Expect = 4e-12
Identities = 78/335 (23%), Positives = 128/335 (37%), Gaps = 54/335 (16%)
Query: 9 LPSELMTKILSLLPVKPLMRFRCVNKFYNTLISDPHFIQMHLKNSARNPNLMVIARQHNF 68
LP +L+ IL PVK LMRF+C++K ++ LI FI H+ + ++ ++
Sbjct: 9 LPEDLVFLILLTFPVKSLMRFKCISKAWSILIQSTTFINRHINRKTNTKDEFILFKR-AI 67
Query: 69 NSFDENVLNLPISLLLENSLSTVPYDPYYRLKNENPHCPWLF---AGSCNGLICLCLDID 125
+E +N+ +S + P P + C F G C+GLI L I
Sbjct: 68 KDDEEEFINI-LSFFSGHVDVLNPLFPDMDVSYMTSKCDCTFNPLIGPCDGLIALTDTII 126
Query: 126 TSHGSRLCLWNPATRTKSEFDLASQEC--------FVFAFGYDNLNGNYKVIAF------ 171
T + NPATR + C FG D ++ YKV+
Sbjct: 127 T------IVLNPATRNFRVLPASPFGCPKGYHRSVEGVGFGLDTISNYYKVVRISEVYCE 180
Query: 172 DIKVKSGNARSVVKVFSMRDNCWRNIQCFPVLPLYMFVSTQNGVYFSSTVNWLALQDYFG 231
+ G S + V + + WR + + +Y G+ + V+W A D
Sbjct: 181 EADGYPGPKDSKIDVCDLSTDSWRELDHVQLPSIYWVPCA--GMLYKEMVHWFATTDM-- 236
Query: 232 LDYFHLNYSSITPEKYVILSLDLSTETYTQLLLPRGFNKVSRHQPKLAVLMDCLCF---- 287
VIL D+STE + + +P ++++ H+ +++ C F
Sbjct: 237 --------------SMVILCFDMSTEMFHDMKMPDTCSRIT-HELYYGLVILCESFTLIG 281
Query: 288 ------GHDYEETYFVIWQMKDFGVQSSWIQLFKI 316
D IW M ++GV SWI + I
Sbjct: 282 YSNPISSTDPAHDKMHIWVMMEYGVSESWIMKYTI 316
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.325 0.139 0.445
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 698,039,607
Number of Sequences: 2790947
Number of extensions: 29398207
Number of successful extensions: 86254
Number of sequences better than 10.0: 310
Number of HSP's better than 10.0 without gapping: 189
Number of HSP's successfully gapped in prelim test: 121
Number of HSP's that attempted gapping in prelim test: 85805
Number of HSP's gapped (non-prelim): 386
length of query: 395
length of database: 848,049,833
effective HSP length: 129
effective length of query: 266
effective length of database: 488,017,670
effective search space: 129812700220
effective search space used: 129812700220
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 76 (33.9 bits)
Medicago: description of AC148651.11


