

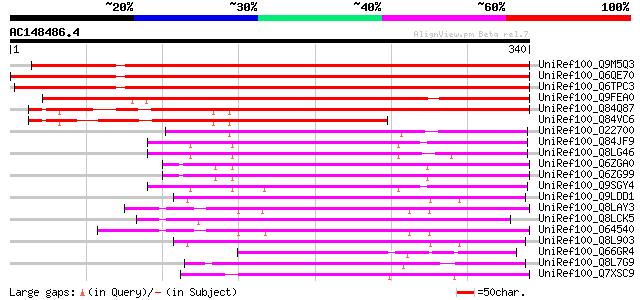
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148486.4 - phase: 0
(340 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9M5Q3 S-ribonuclease binding protein SBP1 [Petunia hy... 483 e-135
UniRef100_Q6QE70 S-RNase binding protein 1 [Solanum chacoense] 467 e-130
UniRef100_Q6TPC3 S-RNase-binding protein [Solanum chacoense] 457 e-127
UniRef100_Q9FEA0 S-ribonuclease binding protein SBP1, putative [... 433 e-120
UniRef100_Q84Q87 Hypothetical protein OJ1041F02.18 [Oryza sativa] 335 1e-90
UniRef100_Q84VC6 S-ribonuclease-binding protein SBP1-like protei... 198 2e-49
UniRef100_O22700 F8A5.13 protein [Arabidopsis thaliana] 161 3e-38
UniRef100_Q84JF9 Putative S-ribonuclease binding protein SBP1 [A... 154 3e-36
UniRef100_Q8LG46 S-ribonuclease binding protein SBP1, putative [... 153 7e-36
UniRef100_Q6ZGA0 Putative S-ribonuclease binding protein SBP1 [O... 140 5e-32
UniRef100_Q6ZG99 Putative S-ribonuclease binding protein SBP1 [O... 140 5e-32
UniRef100_Q9SGY4 F20B24.9 [Arabidopsis thaliana] 139 1e-31
UniRef100_Q9LDD1 Arabidopsis thaliana genomic DNA, chromosome 3,... 119 1e-25
UniRef100_Q8LAY3 Inhibitor of apoptosis-like protein [Arabidopsi... 117 5e-25
UniRef100_Q8LCK5 Hypothetical protein [Arabidopsis thaliana] 115 2e-24
UniRef100_O64540 YUP8H12R.27 protein [Arabidopsis thaliana] 115 2e-24
UniRef100_Q8L903 Hypothetical protein [Arabidopsis thaliana] 114 3e-24
UniRef100_Q66GR4 At4g35070 [Arabidopsis thaliana] 114 3e-24
UniRef100_Q8L7G9 Hypothetical protein At4g17680 [Arabidopsis tha... 110 6e-23
UniRef100_Q7XSC9 OSJNBb0118P14.4 protein [Oryza sativa] 109 1e-22
>UniRef100_Q9M5Q3 S-ribonuclease binding protein SBP1 [Petunia hybrida]
Length = 332
Score = 483 bits (1242), Expect = e-135
Identities = 232/328 (70%), Positives = 283/328 (85%), Gaps = 7/328 (2%)
Query: 15 QQQPQPQTKSFRNL-QTIEGQMSQQMAFYNPTDLQDQSQHPPYIPPFHVVGFAPGPVIPA 73
QQQPQ Q+KS+R++ ++GQ+S +A++N ++L +QSQHPPYIPPF VVG APG V
Sbjct: 10 QQQPQQQSKSYRDIYNNMDGQISTPVAYFNCSNLPEQSQHPPYIPPFQVVGLAPGLV--- 66
Query: 74 DGSDGGVDLHWNFGLEPERKRLKEQDFLENN-SQISSVDFLQPRSVSTGLGLSLDNTRLA 132
DGG+DL WN+GLEP+RKR KEQDFLENN SQISS+DFLQPRSVSTGLGLSLDN RLA
Sbjct: 67 --DDGGLDLQWNYGLEPKRKRPKEQDFLENNNSQISSIDFLQPRSVSTGLGLSLDNGRLA 124
Query: 133 STGDSALLSLIGDDIDRELQQQDLEMDRFLKLQGEQLRQTILEKVQATQLQSVSIIEDKV 192
S+GDSA L L+GDDI+RELQ+QD E+DR++K+QG++LRQ ILEKVQA QLQ+V+ +E+KV
Sbjct: 125 SSGDSAFLGLVGDDIERELQRQDAEIDRYIKVQGDRLRQAILEKVQANQLQTVTYVEEKV 184
Query: 193 LQKLREKETEVENINKRNMELEDQMEQLSVEAGAWQQRARYNENMIAALKFNLQQAYLQG 252
+QKLREKETEVE+INK+NMELE + EQL++EA AWQQRA+YNEN+I LK NL+ Y Q
Sbjct: 185 IQKLREKETEVEDINKKNMELELRTEQLALEANAWQQRAKYNENLINTLKVNLEHVYAQS 244
Query: 253 RDSKEGCGDSEVDDTASCCNGRSLDFHLLSNENSNMKDLMKCKACRVNEVTMVLLPCKHL 312
RDSKEGCGDSEVDDTASCCNGR+ D HLL +++ MK+LM CK CRVNEV+M+LLPCKHL
Sbjct: 245 RDSKEGCGDSEVDDTASCCNGRATDLHLLCRDSNEMKELMTCKVCRVNEVSMLLLPCKHL 304
Query: 313 CLCKDCESKLSFCPLCQSSKFIGMEVYM 340
CLCK+CESKLS CPLCQS+K+IGME+YM
Sbjct: 305 CLCKECESKLSLCPLCQSTKYIGMEIYM 332
>UniRef100_Q6QE70 S-RNase binding protein 1 [Solanum chacoense]
Length = 337
Score = 467 bits (1202), Expect = e-130
Identities = 225/342 (65%), Positives = 278/342 (80%), Gaps = 7/342 (2%)
Query: 1 MAFLQDQFQRHYQQQQQPQPQTKSFRNL-QTIEGQMSQQMAFYNPTDLQDQSQHPPYIPP 59
MA Q H QQQQ Q Q+KS+R+L ++GQ++ + ++N ++L +QSQHPPYIPP
Sbjct: 1 MALPHHHLQLHIQQQQPHQQQSKSYRDLYNNMDGQITTPVVYFNGSNLPEQSQHPPYIPP 60
Query: 60 FHVVGFAPGPVIPADGSDGGVDLHWNFGLEPERKRLKEQDFLENN-SQISSVDFLQPRSV 118
F VVG APG DGG+DL WN+GLEP++KR KEQDF+ENN SQISSVD Q RSV
Sbjct: 61 FQVVGLAPGTA-----DDGGLDLQWNYGLEPKKKRPKEQDFMENNNSQISSVDLFQRRSV 115
Query: 119 STGLGLSLDNTRLASTGDSALLSLIGDDIDRELQQQDLEMDRFLKLQGEQLRQTILEKVQ 178
STGLGLSLDN RLAS+ DSA L L+GDDI+RELQ+QD E+DR++K+QG++LRQ +LEKVQ
Sbjct: 116 STGLGLSLDNGRLASSCDSAFLGLVGDDIERELQRQDAEIDRYIKVQGDRLRQAVLEKVQ 175
Query: 179 ATQLQSVSIIEDKVLQKLREKETEVENINKRNMELEDQMEQLSVEAGAWQQRARYNENMI 238
A Q+Q+++ +E+KVLQKLRE++TEV++INK+NMELE +MEQL +EA AWQQRA+YNEN+I
Sbjct: 176 ANQIQAITYVEEKVLQKLRERDTEVDDINKKNMELELRMEQLDLEANAWQQRAKYNENLI 235
Query: 239 AALKFNLQQAYLQGRDSKEGCGDSEVDDTASCCNGRSLDFHLLSNENSNMKDLMKCKACR 298
LK NLQ Y Q RDSKEGCGDSEVDDTASCCNGR+ D HLL ++ MK+LM C+ CR
Sbjct: 236 NTLKVNLQHVYAQSRDSKEGCGDSEVDDTASCCNGRATDLHLLCRDSKEMKELMTCRVCR 295
Query: 299 VNEVTMVLLPCKHLCLCKDCESKLSFCPLCQSSKFIGMEVYM 340
NEV M+LLPCKHLCLCK+CESKLS CPLCQS+K+IGMEVYM
Sbjct: 296 TNEVCMLLLPCKHLCLCKECESKLSLCPLCQSTKYIGMEVYM 337
>UniRef100_Q6TPC3 S-RNase-binding protein [Solanum chacoense]
Length = 342
Score = 457 bits (1175), Expect = e-127
Identities = 222/339 (65%), Positives = 275/339 (80%), Gaps = 7/339 (2%)
Query: 4 LQDQFQRHYQQQQQPQPQTKSFRNL-QTIEGQMSQQMAFYNPTDLQDQSQHPPYIPPFHV 62
LQ Q+ QQQ Q Q+KS+R+L ++GQ++ + ++N ++L +QSQHPPYIPPF V
Sbjct: 9 LQLHIQQQQPHQQQQQQQSKSYRDLYNNMDGQITTPVVYFNGSNLPEQSQHPPYIPPFQV 68
Query: 63 VGFAPGPVIPADGSDGGVDLHWNFGLEPERKRLKEQDFLENN-SQISSVDFLQPRSVSTG 121
VG APG DGG+DL WN+GLEP++KR KEQDF+ENN SQISSVD LQ RSVSTG
Sbjct: 69 VGLAPGTA-----DDGGLDLQWNYGLEPKKKRPKEQDFMENNNSQISSVDLLQRRSVSTG 123
Query: 122 LGLSLDNTRLASTGDSALLSLIGDDIDRELQQQDLEMDRFLKLQGEQLRQTILEKVQATQ 181
LGLSLDN RLAS+ DSA L L+GDDI+RELQ+QD E+DR++K+QG++LRQ +LEKVQA Q
Sbjct: 124 LGLSLDNGRLASSCDSAFLGLVGDDIERELQRQDAEIDRYIKVQGDRLRQAVLEKVQANQ 183
Query: 182 LQSVSIIEDKVLQKLREKETEVENINKRNMELEDQMEQLSVEAGAWQQRARYNENMIAAL 241
+Q+++ +E+KVLQKLRE++TEV++INK+NMELE +MEQL +EA AWQQRA+YNEN+I L
Sbjct: 184 IQAITYVEEKVLQKLRERDTEVDDINKKNMELELRMEQLDLEANAWQQRAKYNENLINTL 243
Query: 242 KFNLQQAYLQGRDSKEGCGDSEVDDTASCCNGRSLDFHLLSNENSNMKDLMKCKACRVNE 301
K NLQ Y Q RDSKEGCGDSEVDDTASCCNGR+ D HLL ++ MK+LM C+ CR NE
Sbjct: 244 KVNLQHVYAQSRDSKEGCGDSEVDDTASCCNGRATDLHLLCRDSKEMKELMTCRVCRTNE 303
Query: 302 VTMVLLPCKHLCLCKDCESKLSFCPLCQSSKFIGMEVYM 340
V M+ LPCKHL LCK+CESKLS CPLCQS K+IGMEVYM
Sbjct: 304 VGMLWLPCKHLGLCKECESKLSLCPLCQSIKYIGMEVYM 342
>UniRef100_Q9FEA0 S-ribonuclease binding protein SBP1, putative [Arabidopsis
thaliana]
Length = 325
Score = 433 bits (1114), Expect = e-120
Identities = 212/323 (65%), Positives = 261/323 (80%), Gaps = 11/323 (3%)
Query: 22 TKSFRNLQTIEGQMSQQMAFYNPTDLQDQSQHPPYIPPFHVVGFAPGPVIPADGSDGG-- 79
+K+FR+ I+GQ+S ++ F +L DQSQHPPYIPPFHV GFAPGPV+ DGSDGG
Sbjct: 10 SKNFRDFCGIDGQISPELGFNRSENLHDQSQHPPYIPPFHVAGFAPGPVVQIDGSDGGNG 69
Query: 80 VDLHWNFGL--EPERKRLKEQDFLENNSQISSVDFLQPRSVSTGLGLSLDNTRLASTGDS 137
D WN+GL EP R+R+KEQDFLENNSQISS+DFLQ RSVSTGLGLSLDN R+AS+ S
Sbjct: 70 ADFEWNYGLGLEPRRERIKEQDFLENNSQISSIDFLQARSVSTGLGLSLDNARVASSDGS 129
Query: 138 ALLSLIGDDIDRELQQQDLEMDRFLKLQGEQLRQTILEKVQATQLQSVSIIEDKVLQKLR 197
ALLSL+GDDIDRELQ+QD ++DRFLK+QG+QLR IL+K++ Q ++VS++E+KV+QKLR
Sbjct: 130 ALLSLVGDDIDRELQRQDADIDRFLKIQGDQLRHAILDKIKRGQQKTVSLMEEKVVQKLR 189
Query: 198 EKETEVENINKRNMELEDQMEQLSVEAGAWQQRARYNENMIAALKFNLQQAYLQGRDSKE 257
EK+ E+E IN++N ELE +MEQL++EA AWQQRA+YNENMIAAL +NL +A + RDS E
Sbjct: 190 EKDEELERINRKNKELEVRMEQLTMEAEAWQQRAKYNENMIAALNYNLDRAQGRPRDSIE 249
Query: 258 GCGDSEVDDTASCCNGRSLDFHLLSNENSNMKDLMKCKACRVNEVTMVLLPCKHLCLCKD 317
GCGDSEVDDTASC NGR N N+N K +M C+ C V E+ M+LLPC H+CLCK+
Sbjct: 250 GCGDSEVDDTASCFNGR-------DNSNNNTKTMMMCRFCGVREMCMLLLPCNHMCLCKE 302
Query: 318 CESKLSFCPLCQSSKFIGMEVYM 340
CE KLS CPLCQSSKF+GMEVYM
Sbjct: 303 CERKLSSCPLCQSSKFLGMEVYM 325
>UniRef100_Q84Q87 Hypothetical protein OJ1041F02.18 [Oryza sativa]
Length = 342
Score = 335 bits (859), Expect = 1e-90
Identities = 183/344 (53%), Positives = 235/344 (68%), Gaps = 40/344 (11%)
Query: 13 QQQQQPQPQTKSFRNLQTI--EGQMSQQMAFYNPTD-LQDQSQHPPYIPPFHVVGFAPGP 69
QQQQQP P SFRN + +GQ+ + F+NP QDQ PP
Sbjct: 23 QQQQQPVPP--SFRNALPVPVDGQIPAPLPFFNPPPAFQDQPAQPP-------------- 66
Query: 70 VIPADGSDGGVDLHWNFGLEPERKRLKEQDFLENNSQISSVDFLQPRS-VSTGLGLSLDN 128
++ A G L W ++ +EQ+ L NSQ+SS+DFLQ S VSTGL LSL++
Sbjct: 67 LVDAMGLTAAAGLGW--------RQPREQELLGENSQMSSIDFLQTGSAVSTGLALSLED 118
Query: 129 TRLA---------STGDSALLSL--IGDDIDRELQQQDLEMDRFLKLQGEQLRQTILEKV 177
R S+GDS LL L + DDI RE+Q+ D +MDRF+K Q E+LRQ+ILEKV
Sbjct: 119 RRHGGGSGAGAGNSSGDSPLLLLPMLDDDISREVQRLDADMDRFIKAQSERLRQSILEKV 178
Query: 178 QATQLQSVSIIEDKVLQKLREKETEVENINKRNMELEDQMEQLSVEAGAWQQRARYNENM 237
QA Q ++++ +EDK+L+K+R+KE EVENINKRN ELEDQ++QL+VE GAWQQRA+YNE+M
Sbjct: 179 QAKQFEALASVEDKILRKIRDKEAEVENINKRNSELEDQIKQLAVEVGAWQQRAKYNESM 238
Query: 238 IAALKFNLQQAYL-QGRDSKEGCGDSEVDDTASCCNGRSLDFHLLSNENSNMKDLMKCKA 296
I ALK+NL+Q Q +D KEGCGDSEVDDTASCCNG + + L+ EN + KDL C+
Sbjct: 239 INALKYNLEQVCAHQSKDFKEGCGDSEVDDTASCCNGGAANLQLMPKENRHSKDLTACRV 298
Query: 297 CRVNEVTMVLLPCKHLCLCKDCESKLSFCPLCQSSKFIGMEVYM 340
C+ +E M+LLPC+HLCLCK+CESKLSFCPLCQSSK +GME+YM
Sbjct: 299 CKSSEACMLLLPCRHLCLCKECESKLSFCPLCQSSKILGMEIYM 342
>UniRef100_Q84VC6 S-ribonuclease-binding protein SBP1-like protein [Oryza sativa]
Length = 252
Score = 198 bits (504), Expect = 2e-49
Identities = 121/249 (48%), Positives = 159/249 (63%), Gaps = 37/249 (14%)
Query: 13 QQQQQPQPQTKSFRNLQTI--EGQMSQQMAFYNPTDLQDQSQHPPYIPPFHVVGFAPGPV 70
QQQQQP P SFRN + +GQ+ + F+NP PP A P+
Sbjct: 18 QQQQQPVPP--SFRNALPVPVDGQIPAPLPFFNP-------------PPAFQGQPAQPPL 62
Query: 71 IPADGSDGGVDLHWNFGLEPERKRLKEQDFLENNSQISSVDFLQPRS-VSTGLGLSLDNT 129
+ A G L W + +EQ+ L NSQ+SS+DFLQ S VSTGL LSL++
Sbjct: 63 VDAMGLTAAAGLGWG--------QPREQELLGENSQMSSIDFLQTGSAVSTGLALSLEDR 114
Query: 130 RLA---------STGDSALLSL--IGDDIDRELQQQDLEMDRFLKLQGEQLRQTILEKVQ 178
R S+GDS LL L + DDI RE+Q+ D +MDRF+K Q E+LRQ+ILEKVQ
Sbjct: 115 RHGGGSGAGAGNSSGDSPLLLLPMLDDDISREVQRLDADMDRFIKAQSERLRQSILEKVQ 174
Query: 179 ATQLQSVSIIEDKVLQKLREKETEVENINKRNMELEDQMEQLSVEAGAWQQRARYNENMI 238
A Q ++++ +EDK+L+K+R+KE EVENINKRN ELEDQ++QL+VE GAWQQRA+YNE+M
Sbjct: 175 AKQFEALASVEDKILRKIRDKEAEVENINKRNSELEDQIKQLAVEVGAWQQRAKYNESMT 234
Query: 239 AALKFNLQQ 247
ALK+NL+Q
Sbjct: 235 NALKYNLEQ 243
>UniRef100_O22700 F8A5.13 protein [Arabidopsis thaliana]
Length = 372
Score = 161 bits (407), Expect = 3e-38
Identities = 93/257 (36%), Positives = 147/257 (57%), Gaps = 28/257 (10%)
Query: 103 NNSQISSVDFLQPRSVSTGLGLSLDNTRLASTGDSALLSL-------IGDDIDRELQQQD 155
NN+ + + + VSTGL LS D+ S+ SA S+ +GD+I +L +Q+
Sbjct: 122 NNNTVVQDEVPKQNLVSTGLRLSYDDDERNSSVTSANGSITTPVYQSLGDNIRLDLNRQN 181
Query: 156 LEMDRFLKLQGEQLRQTILEKVQATQLQSVSIIEDKVLQKLREKETEVENINKRNMELED 215
E+D+F+K + +Q+ + + + Q V+ +E V +KL+EK+ E+E++NK+N EL D
Sbjct: 182 DELDQFIKFRADQMAKGVRDIKQRHVTSFVTALEKDVSKKLQEKDHEIESMNKKNRELVD 241
Query: 216 QMEQLSVEAGAWQQRARYNENMIAALKFNLQQAYLQGRDS-------------KEGCGDS 262
+++Q++VEA W +A+YNE+++ ALK NLQQ G D+ KEG GDS
Sbjct: 242 KIKQVAVEAQNWHYKAKYNESVVNALKVNLQQVMSHGNDNNAVGGGVADHHQMKEGFGDS 301
Query: 263 EVDDTASCCNGRSLDFHLLSNENSNMKDLMKCKACRVNEVTMVLLPCKHLCLCKDCESKL 322
E+DD A+ N N M+CK C V V+++L+PC+HL LCKDC+
Sbjct: 302 EIDDEAASYN--------YLNIPGMPSTGMRCKLCNVKNVSVLLVPCRHLSLCKDCDVFT 353
Query: 323 SFCPLCQSSKFIGMEVY 339
CP+CQS K ++V+
Sbjct: 354 GVCPVCQSLKTSSVQVF 370
>UniRef100_Q84JF9 Putative S-ribonuclease binding protein SBP1 [Arabidopsis thaliana]
Length = 339
Score = 154 bits (390), Expect = 3e-36
Identities = 93/267 (34%), Positives = 152/267 (56%), Gaps = 22/267 (8%)
Query: 91 ERKRLKEQDFLENNSQISSVDFLQPRS--VSTGLGLSLDNTRLASTGDSALLSLIG---- 144
+R++ K Q L N +SV P+ VSTGL LS D+ S+ SA S++
Sbjct: 75 QRQQQKLQMSLNYNYNNTSVREEVPKENLVSTGLRLSYDDDEHNSSVTSASGSILAASPI 134
Query: 145 -----DDIDRELQQQDLEMDRFLKLQGEQLRQTILEKVQATQLQSVSIIEDKVLQKLREK 199
D + +L +Q E D+F+K+Q Q+ + + + Q ++ +E V +KL+EK
Sbjct: 135 FQSLDDSLRIDLHRQKDEFDQFIKIQAAQMAKGVRDMKQRHIASFLTTLEKGVSKKLQEK 194
Query: 200 ETEVENINKRNMELEDQMEQLSVEAGAWQQRARYNENMIAALKFNLQQAYLQGR------ 253
+ E+ ++NK+N EL ++++Q+++EA W RA+YNE+++ LK NLQQA
Sbjct: 195 DHEINDMNKKNKELVERIKQVAMEAQNWHYRAKYNESVVNVLKANLQQAMSHNNSVIAAA 254
Query: 254 -DSKEGCGDSEVDDTASCCNGRSLDFHLLSNENSNMKDLMKCKACRVNEVTMVLLPCKHL 312
KEG GDSE+DD AS +D + +N N + M+CK C V EV+++++PC+HL
Sbjct: 255 DQGKEGFGDSEIDDAAS----SYIDPNNNNNNNMGIHQRMRCKMCNVKEVSVLIVPCRHL 310
Query: 313 CLCKDCESKLSFCPLCQSSKFIGMEVY 339
LCK+C+ CP+C+S K ++V+
Sbjct: 311 SLCKECDVFTKICPVCKSLKSSCVQVF 337
>UniRef100_Q8LG46 S-ribonuclease binding protein SBP1, putative [Arabidopsis
thaliana]
Length = 337
Score = 153 bits (386), Expect = 7e-36
Identities = 94/269 (34%), Positives = 152/269 (55%), Gaps = 28/269 (10%)
Query: 91 ERKRLKEQDFLENNSQISSVDFLQPRS--VSTGLGLSLDNTRLASTGDSALLSLIG---- 144
+R++ K Q L N +SV P+ VSTGL LS D+ S+ SA S++
Sbjct: 75 QRQQQKLQMSLNYNYNNTSVREEVPKENLVSTGLRLSYDDDEHNSSVTSASGSILAASPI 134
Query: 145 -----DDIDRELQQQDLEMDRFLKLQGEQLRQTILEKVQATQLQSVSIIEDKVLQKLREK 199
D + +L +Q E D+F+K+Q Q+ + + + Q ++ +E V +KL+EK
Sbjct: 135 FQSLDDSLRIDLHRQKDEFDQFIKIQAAQMAKGVRDMKQRHIASFLTTLEKGVSKKLQEK 194
Query: 200 ETEVENINKRNMELEDQMEQLSVEAGAWQQRARYNENMIAALKFNLQQAYLQGR------ 253
+ E+ ++NK+N EL ++++Q+++EA W RA+YNE+++ LK NLQQA
Sbjct: 195 DHEINDMNKKNKELVERIKQVAMEAQNWHYRAKYNESVVNVLKANLQQAMSHNNSVIAAA 254
Query: 254 -DSKEGCGDSEVDDTASCCNGRSLDFHLLSNENSNM--KDLMKCKACRVNEVTMVLLPCK 310
KEG GDSE+DD AS ++ N N+NM M+CK C V EV+++++PC+
Sbjct: 255 DQGKEGFGDSEIDDAASS--------YIDPNNNNNMGIHQRMRCKMCNVKEVSVLIVPCR 306
Query: 311 HLCLCKDCESKLSFCPLCQSSKFIGMEVY 339
HL LCK+C+ CP+C+S K ++V+
Sbjct: 307 HLSLCKECDVFTKICPVCKSLKSSCVQVF 335
>UniRef100_Q6ZGA0 Putative S-ribonuclease binding protein SBP1 [Oryza sativa]
Length = 343
Score = 140 bits (353), Expect = 5e-32
Identities = 83/253 (32%), Positives = 144/253 (56%), Gaps = 15/253 (5%)
Query: 101 LENNSQISSVDFLQPRSVSTGLGLSLDNTRLAS----TGDSALLSLIG---DDIDRELQQ 153
+EN +++ + P +VSTGL LS +N S +G+ + L ++ D++ EL +
Sbjct: 92 VENLDRLARIG--NPSAVSTGLRLSYENNEHTSITSGSGNMSSLPIMASFVDEVMAELDK 149
Query: 154 QDLEMDRFLKLQGEQLRQTILEKVQATQLQSVSIIEDKVLQKLREKETEVENINKRNMEL 213
++ E + + LQ EQL + + + Q ++ ++ +E V +KL+EKE EVE +N+++ EL
Sbjct: 150 ENKEFNCYFGLQVEQLVKCMKDVKQRQMVEFLASLERGVGKKLKEKELEVEAMNRKSKEL 209
Query: 214 EDQMEQLSVEAGAWQQRARYNENMIAALKFNLQQAYLQGRD-SKEGCGDSEVDDTASCCN 272
+Q+ Q+++E +WQ A +N+++ ++K L Q + ++EG GDSEVD+TAS N
Sbjct: 210 NEQIRQVALEVQSWQSVALHNQSVANSMKSKLMQMVAHSSNLTREGSGDSEVDNTASSQN 269
Query: 273 -----GRSLDFHLLSNENSNMKDLMKCKACRVNEVTMVLLPCKHLCLCKDCESKLSFCPL 327
G LL + L C+ CR+ E ++++PC+HLCLC DCE CP+
Sbjct: 270 VNAVPGVFFQSGLLGINSMADGGLGACRLCRMKEAAVLVMPCRHLCLCADCEKNADVCPV 329
Query: 328 CQSSKFIGMEVYM 340
C+ K +E+ M
Sbjct: 330 CRFPKSCSVEINM 342
>UniRef100_Q6ZG99 Putative S-ribonuclease binding protein SBP1 [Oryza sativa]
Length = 279
Score = 140 bits (353), Expect = 5e-32
Identities = 83/253 (32%), Positives = 144/253 (56%), Gaps = 15/253 (5%)
Query: 101 LENNSQISSVDFLQPRSVSTGLGLSLDNTRLAS----TGDSALLSLIG---DDIDRELQQ 153
+EN +++ + P +VSTGL LS +N S +G+ + L ++ D++ EL +
Sbjct: 28 VENLDRLARIG--NPSAVSTGLRLSYENNEHTSITSGSGNMSSLPIMASFVDEVMAELDK 85
Query: 154 QDLEMDRFLKLQGEQLRQTILEKVQATQLQSVSIIEDKVLQKLREKETEVENINKRNMEL 213
++ E + + LQ EQL + + + Q ++ ++ +E V +KL+EKE EVE +N+++ EL
Sbjct: 86 ENKEFNCYFGLQVEQLVKCMKDVKQRQMVEFLASLERGVGKKLKEKELEVEAMNRKSKEL 145
Query: 214 EDQMEQLSVEAGAWQQRARYNENMIAALKFNLQQAYLQGRD-SKEGCGDSEVDDTASCCN 272
+Q+ Q+++E +WQ A +N+++ ++K L Q + ++EG GDSEVD+TAS N
Sbjct: 146 NEQIRQVALEVQSWQSVALHNQSVANSMKSKLMQMVAHSSNLTREGSGDSEVDNTASSQN 205
Query: 273 -----GRSLDFHLLSNENSNMKDLMKCKACRVNEVTMVLLPCKHLCLCKDCESKLSFCPL 327
G LL + L C+ CR+ E ++++PC+HLCLC DCE CP+
Sbjct: 206 VNAVPGVFFQSGLLGINSMADGGLGACRLCRMKEAAVLVMPCRHLCLCADCEKNADVCPV 265
Query: 328 CQSSKFIGMEVYM 340
C+ K +E+ M
Sbjct: 266 CRFPKSCSVEINM 278
>UniRef100_Q9SGY4 F20B24.9 [Arabidopsis thaliana]
Length = 368
Score = 139 bits (350), Expect = 1e-31
Identities = 93/296 (31%), Positives = 152/296 (50%), Gaps = 51/296 (17%)
Query: 91 ERKRLKEQDFLENNSQISSVDFLQPRS--VSTGLGLSLDNTRLASTGDSALLSLIG---- 144
+R++ K Q L N +SV P+ VSTGL LS D+ S+ SA S++
Sbjct: 75 QRQQQKLQMSLNYNYNNTSVREEVPKENLVSTGLRLSYDDDEHNSSVTSASGSILAASPI 134
Query: 145 -----DDIDRELQQQDLEMDRFLKLQ-----------------------------GEQLR 170
D + +L +Q E D+F+K+Q Q+
Sbjct: 135 FQSLDDSLRIDLHRQKDEFDQFIKIQVLIVSACRLCFYVKRFFDSNVFVCFYVVQAAQMA 194
Query: 171 QTILEKVQATQLQSVSIIEDKVLQKLREKETEVENINKRNMELEDQMEQLSVEAGAWQQR 230
+ + + Q ++ +E V +KL+EK+ E+ ++NK+N EL ++++Q+++EA W R
Sbjct: 195 KGVRDMKQRHIASFLTTLEKGVSKKLQEKDHEINDMNKKNKELVERIKQVAMEAQNWHYR 254
Query: 231 ARYNENMIAALKFNLQQAYLQGR-------DSKEGCGDSEVDDTASCCNGRSLDFHLLSN 283
A+YNE+++ LK NLQQA KEG GDSE+DD AS +D + +N
Sbjct: 255 AKYNESVVNVLKANLQQAMSHNNSVIAAADQGKEGFGDSEIDDAAS----SYIDPNNNNN 310
Query: 284 ENSNMKDLMKCKACRVNEVTMVLLPCKHLCLCKDCESKLSFCPLCQSSKFIGMEVY 339
N + M+CK C V EV+++++PC+HL LCK+C+ CP+C+S K ++V+
Sbjct: 311 NNMGIHQRMRCKMCNVKEVSVLIVPCRHLSLCKECDVFTKICPVCKSLKSSCVQVF 366
>UniRef100_Q9LDD1 Arabidopsis thaliana genomic DNA, chromosome 3, P1 clone: MGH6
[Arabidopsis thaliana]
Length = 335
Score = 119 bits (298), Expect = 1e-25
Identities = 72/246 (29%), Positives = 123/246 (49%), Gaps = 15/246 (6%)
Query: 108 SSVDFLQP-----RSVSTGLGLSLDNTRLASTGDSALLSLIGDDIDRELQQQDLEMDRFL 162
++VDFL+P RS + L+ + L +G D+ +QQ ++DR +
Sbjct: 86 NNVDFLRPVSSRKRSREESVVLNPSAYMQIQKNPTDPLMFLGQDLSSNVQQHHFDIDRLI 145
Query: 163 KLQGEQLRQTILEKVQATQLQSVSIIEDKVLQKLREKETEVENINKRNMELEDQMEQLSV 222
E++R I EK + + V +E +++ LR K+ E+ +I K N+ LE++++ L V
Sbjct: 146 SNHVERMRMEIEEKRKTQGRRIVEAVEQGLMKTLRAKDDEINHIGKLNLFLEEKVKSLCV 205
Query: 223 EAGAWQQRARYNENMIAALKFNLQQAYLQGRDSKEGCGDSEVDDTASCCNGR-----SLD 277
E W+ A+ NE + AL+ NLQQ ++ + DD SCC +
Sbjct: 206 ENQIWRDVAQSNEATVNALRSNLQQVLAAVERNRWEEPPTVADDAQSCCGSNDEGDSEEE 265
Query: 278 FHLLSNENSNMKDLMK-----CKACRVNEVTMVLLPCKHLCLCKDCESKLSFCPLCQSSK 332
L+ E + K + + C++C E +++LLPC+H+CLC C S L+ CP+C+S K
Sbjct: 266 RWKLAGEAQDTKKMCRVGMSMCRSCGKGEASVLLLPCRHMCLCSVCGSSLNTCPICKSPK 325
Query: 333 FIGMEV 338
+ V
Sbjct: 326 TASLHV 331
>UniRef100_Q8LAY3 Inhibitor of apoptosis-like protein [Arabidopsis thaliana]
Length = 358
Score = 117 bits (293), Expect = 5e-25
Identities = 86/285 (30%), Positives = 130/285 (45%), Gaps = 31/285 (10%)
Query: 76 SDGGVDLHWNFGLEPERKRLKEQDFLENNSQISSVDFLQPRSVSTGLGLSLDNTRLASTG 135
+DG +L F P RKR ++ + S + PRS S +T
Sbjct: 83 ADGATNLDCEFFPLPTRKRSRDS----SRSNYHHLLLQNPRSSSCV-------NAATTTT 131
Query: 136 DSALLSLIGDDID--RELQQQDLEMDRFLKL---QGEQLRQTILEKVQATQLQSVSIIED 190
+ S +G DID + QQ E+DRF+ L Q E+++ I EK + + IE
Sbjct: 132 TTTPFSFLGQDIDISSHMNQQQHEIDRFVSLHLYQMERVKYEIEEKRKRQARTIMEAIEQ 191
Query: 191 KVLQKLREKETEVENINKRNMELEDQMEQLSVEAGAWQQRARYNENMIAALKFNLQQAYL 250
++++LR KE E E I K N LE++++ LS+E W+ A+ NE L+ NL+
Sbjct: 192 GLVKRLRVKEEERERIGKVNHALEERVKSLSIENQIWRDLAQTNEATANHLRTNLEHVLA 251
Query: 251 QGRDSKEGCG----DSEVDDTASCCNG-----------RSLDFHLLSNENSNMKDLMKCK 295
Q +D G G +E DD SCC R + + + + + C+
Sbjct: 252 QVKDVSRGAGLEKNMNEEDDAESCCGSSCGGGGEETVRRRVGLEREAQDKAERRRRRMCR 311
Query: 296 ACRVNEVTMVLLPCKHLCLCKDCESKLSFCPLCQSSKFIGMEVYM 340
C E ++LLPC+HLCLC C S + CP+C S K + V M
Sbjct: 312 NCGEEESCVLLLPCRHLCLCGVCGSSVHTCPICTSPKNASVHVNM 356
>UniRef100_Q8LCK5 Hypothetical protein [Arabidopsis thaliana]
Length = 312
Score = 115 bits (287), Expect = 2e-24
Identities = 82/258 (31%), Positives = 125/258 (47%), Gaps = 17/258 (6%)
Query: 84 WNFGLEPERKRLKEQDFLENNSQISSVDFLQ-PRSVSTGL---------GLSLDNTRLAS 133
+N G +RKR +E NN Q+ + Q P+ + L + RL S
Sbjct: 46 FNNGSSNQRKRRRET----NNHQLLPMQSHQFPQVIDLSLLHNYNHPPSNMVHTGLRLFS 101
Query: 134 TGDSAL-LSLIGDDI-DRELQQQDLEMDRFLKLQGEQLRQTILEKVQATQLQSVSIIEDK 191
D A +S + +D+ + +Q E+D FL Q E+LR+T+ EK + + +E+
Sbjct: 102 GEDQAQKISHLSEDVFAAHINRQSEELDEFLHAQAEELRRTLAEKRKMHYKALLGAVEES 161
Query: 192 VLQKLREKETEVENINKRNMELEDQMEQLSVEAGAWQQRARYNENMIAALKFNLQQAYLQ 251
+++KLREKE E+E +R+ EL + QL E WQ+RA+ +E+ A+L+ LQQA Q
Sbjct: 162 LVRKLREKEVEIERATRRHNELVARDSQLRAEVQVWQERAKAHEDAAASLQSQLQQAVNQ 221
Query: 252 GRDSKEGCGDSEVDDTASCCNGRS-LDFHLLSNENSNMKDLMKCKACRVNEVTMVLLPCK 310
DS + C S +D + CKACR E T+V+LPC+
Sbjct: 222 CAGGCVSAQDSRAAEEGLLCTTISGVDDAESVYVDPERVKRPNCKACREREATVVVLPCR 281
Query: 311 HLCLCKDCESKLSFCPLC 328
HL +C C+ CPLC
Sbjct: 282 HLSICPGCDRTALACPLC 299
>UniRef100_O64540 YUP8H12R.27 protein [Arabidopsis thaliana]
Length = 347
Score = 115 bits (287), Expect = 2e-24
Identities = 91/315 (28%), Positives = 136/315 (42%), Gaps = 43/315 (13%)
Query: 58 PPFHVVGFAPG-PVIPADGSDGGVDLHWNFGLEPERKRLKEQDFLENNSQISSVDFLQPR 116
P HV G + P +DG +L F P RKR ++ + S + PR
Sbjct: 42 PLLHVYGGSDTIPTTAGYYADGATNLDCEFFPLPTRKRSRDS----SRSNYHHLLLQNPR 97
Query: 117 SVSTGLGLSLDNTRLASTGDSALLSLIGDDID--RELQQQDLEMDRFLKLQG-------- 166
S S +T + L S +G DID + QQ E+DRF+ L
Sbjct: 98 SSSCV-------NAATTTTTTTLFSFLGQDIDISSHMNQQQHEIDRFVSLHVSFASTAEF 150
Query: 167 ------EQLRQTILEKVQATQLQSVSIIEDKVLQKLREKETEVENINKRNMELEDQMEQL 220
E+++ I EK + + IE ++++LR KE E E I K N LE++++ L
Sbjct: 151 VAMCIMERVKYEIEEKRKRQARTIMEAIEQGLVKRLRVKEEERERIGKVNHALEERVKSL 210
Query: 221 SVEAGAWQQRARYNENMIAALKFNLQQAYLQGRDSKEGCG----DSEVDDTASCCNG--- 273
S+E W+ A+ NE L+ NL+ Q +D G G +E DD SCC
Sbjct: 211 SIENQIWRDLAQTNEATANHLRTNLEHVLAQVKDVSRGAGLEKNMNEEDDAESCCGSSCG 270
Query: 274 --------RSLDFHLLSNENSNMKDLMKCKACRVNEVTMVLLPCKHLCLCKDCESKLSFC 325
R + + + + + C+ C E ++LLPC+HLCLC C S + C
Sbjct: 271 GGGEETVRRRVGLEREAQDKAERRRRRMCRNCGEEESCVLLLPCRHLCLCGVCGSSVHTC 330
Query: 326 PLCQSSKFIGMEVYM 340
P+C S K + V M
Sbjct: 331 PICTSPKNASVHVNM 345
>UniRef100_Q8L903 Hypothetical protein [Arabidopsis thaliana]
Length = 335
Score = 114 bits (286), Expect = 3e-24
Identities = 71/246 (28%), Positives = 121/246 (48%), Gaps = 15/246 (6%)
Query: 108 SSVDFLQP-----RSVSTGLGLSLDNTRLASTGDSALLSLIGDDIDRELQQQDLEMDRFL 162
++VDFL+P RS + L + L +G D+ +QQ ++DR +
Sbjct: 86 NNVDFLRPVSSRKRSREESVVLKPSAYMQIQKNPTDPLMFLGQDLSSNVQQHHFDIDRLI 145
Query: 163 KLQGEQLRQTILEKVQATQLQSVSIIEDKVLQKLREKETEVENINKRNMELEDQMEQLSV 222
E++R I EK + + V +E +++ LR K+ E+ +I K N+ LE++++ L V
Sbjct: 146 SNHVERMRMEIEEKRKTQGRRIVEAVEQGLMKTLRAKDDEINHIGKLNLFLEEKVKSLCV 205
Query: 223 EAGAWQQRARYNENMIAALKFNLQQAYLQGRDSKEGCGDSEVDDTASCCNGR-----SLD 277
E W+ A+ NE + AL+ NLQQ ++ + DD SC +
Sbjct: 206 ENQIWRDVAQSNEATVNALRSNLQQVLAAVERNRWEEPPTVADDAQSCYGSNDEGDSEEE 265
Query: 278 FHLLSNENSNMKDLMK-----CKACRVNEVTMVLLPCKHLCLCKDCESKLSFCPLCQSSK 332
L+ E + K + + C++C E +++LLPC+H+CLC C S L+ CP+C+S K
Sbjct: 266 RWKLAGEAQDTKKMCRVGMSMCRSCGKGEASVLLLPCRHMCLCSVCGSSLNTCPICKSPK 325
Query: 333 FIGMEV 338
+ V
Sbjct: 326 TASLHV 331
>UniRef100_Q66GR4 At4g35070 [Arabidopsis thaliana]
Length = 265
Score = 114 bits (286), Expect = 3e-24
Identities = 66/187 (35%), Positives = 101/187 (53%), Gaps = 11/187 (5%)
Query: 150 ELQQQDLEMDRFLKLQGEQLRQTILEKVQATQLQSVSIIEDKVLQKLREKETEVENINKR 209
++++Q E+D+F+K+Q E+LR + E+ + + +E K L + +KE E+ +
Sbjct: 77 QMEKQKQEIDQFIKIQNERLRYVLQEQRKREMEMILRKMESKALLLMSQKEEEMSKALNK 136
Query: 210 NMELEDQMEQLSVEAGAWQQRARYNENMIAALKFNLQQAYLQGRDSKEGC---GDSEVDD 266
NMELED + ++ +E WQ+ AR NE ++ L L+Q R+ C G++EV+D
Sbjct: 137 NMELEDLLRKMEMENQTWQRMARENEAIVQTLNTTLEQV----RERAATCYDAGEAEVED 192
Query: 267 TASCCNGRSLDFHLLSNENSNMKDLMKCKACRVNEVTMVL-LPCKHLCLCKDCESKLSFC 325
S C G D + L + M C C N VT VL LPC+HLC C DCE L C
Sbjct: 193 EGSFCGGEG-DGNSLPAKKMKMSSC--CCNCGSNGVTRVLFLPCRHLCCCMDCEEGLLLC 249
Query: 326 PLCQSSK 332
P+C + K
Sbjct: 250 PICNTPK 256
>UniRef100_Q8L7G9 Hypothetical protein At4g17680 [Arabidopsis thaliana]
Length = 314
Score = 110 bits (275), Expect = 6e-23
Identities = 73/232 (31%), Positives = 116/232 (49%), Gaps = 21/232 (9%)
Query: 115 PRSVSTGLGLSLDNTRLASTGDSALLSLIGDDIDRELQQQDLEMDRFLKLQGEQLRQTIL 174
P VSTGL L D S S + D+ ++++Q E+DRF++ QGE+LR+T+
Sbjct: 94 PNVVSTGLRLFHDQ----SQNQQQFFSSLPGDVTGKIKRQRDELDRFIQTQGEELRRTLA 149
Query: 175 EKVQATQLQSVSIIEDKVLQKLREKETEVENINKRNMELEDQMEQLSVEAGAWQQRARYN 234
+ + ++ + E+ V +KLR+KE E+E +R+ ELE ++ + EA WQ RA
Sbjct: 150 DNRERRYVELLCAAEEIVGRKLRKKEAELEKATRRHAELEARVAHIVEEARNWQLRAATR 209
Query: 235 ENMIAALKFNLQQAYLQGRDSK-------EGCGDSEVDDTASCCNGRSLDFHLLSNENSN 287
E +++L +LQQA D+ E GD+E + A L+
Sbjct: 210 EAEVSSLHAHLQQAIANRLDTAAKQSTFGEDGGDAEEAEDAESVYVDPERIELIG----- 264
Query: 288 MKDLMKCKACRVNEVTMVLLPCKHLCLCKDCE-SKLSFCPLCQSSKFIGMEV 338
C+ CR T++ LPC+HL LC C+ + CP+C + K G+EV
Sbjct: 265 ----PSCRICRRKSATVMALPCQHLILCNGCDVGAVRVCPICLAVKTSGVEV 312
>UniRef100_Q7XSC9 OSJNBb0118P14.4 protein [Oryza sativa]
Length = 347
Score = 109 bits (272), Expect = 1e-22
Identities = 69/237 (29%), Positives = 113/237 (47%), Gaps = 17/237 (7%)
Query: 113 LQPRSVSTGLGLSLDNTRLASTGDSALLSLIGDDIDRELQQQDLEMDRFLKLQGEQLRQT 172
+Q R+V G + A+ LLS +L Q +E+D ++L+ E++R
Sbjct: 118 VQSRAVGCGAASTSGRAGNAAGLSQGLLS--------QLYHQGVEIDALVRLESERMRAG 169
Query: 173 ILEKVQATQLQSVSIIEDKVLQKLREKETEVENINKRNMELEDQMEQLSVEAGAWQQRAR 232
+ E + VS +E +LR E E+E RNMELE+++ Q++ E AW A+
Sbjct: 170 LEEARRRHVRAVVSTVERAAAGRLRAAEAELERARCRNMELEERLRQMTAEGQAWLSVAK 229
Query: 233 YNENMIAALKFNLQQ-------AYLQGRDSKEGCGDSEVDDTASCCNGRSLDFHLLSNEN 285
+E + A L+ L Q A + G + + +D SCC ++
Sbjct: 230 SHEAVAAGLRATLDQLLQSPCAALAVAGAAGAGGAEGDAEDAQSCCYETPCGGDNAGADD 289
Query: 286 SNMKD--LMKCKACRVNEVTMVLLPCKHLCLCKDCESKLSFCPLCQSSKFIGMEVYM 340
+ K CKAC E +M+LLPC+HLCLC+ CE+ + CP+C ++K + V +
Sbjct: 290 AASKTPAAALCKACGAGEASMLLLPCRHLCLCRGCEAAVDACPVCAATKNASLHVLL 346
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.317 0.133 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 551,652,216
Number of Sequences: 2790947
Number of extensions: 23523220
Number of successful extensions: 114999
Number of sequences better than 10.0: 1628
Number of HSP's better than 10.0 without gapping: 588
Number of HSP's successfully gapped in prelim test: 1055
Number of HSP's that attempted gapping in prelim test: 111037
Number of HSP's gapped (non-prelim): 4205
length of query: 340
length of database: 848,049,833
effective HSP length: 128
effective length of query: 212
effective length of database: 490,808,617
effective search space: 104051426804
effective search space used: 104051426804
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 75 (33.5 bits)
Medicago: description of AC148486.4


