

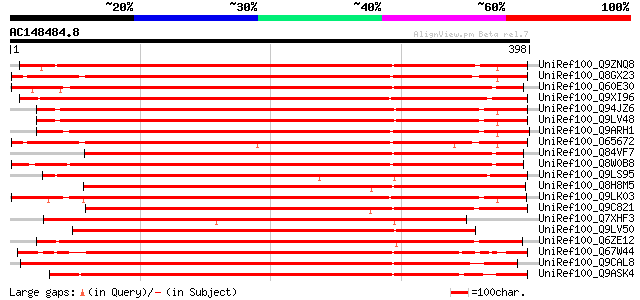
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148484.8 + phase: 0
(398 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9ZNQ8 Hypothetical protein At2g18470 [Arabidopsis tha... 521 e-146
UniRef100_Q8GX23 Putative serine/threonine protein kinase [Arabi... 512 e-144
UniRef100_Q60E30 Hypothetical protein OSJNBb0012L23.7 [Oryza sat... 507 e-142
UniRef100_Q9XI96 Similar to somatic embryogenesis receptor-like ... 505 e-141
UniRef100_Q94JZ6 Protein kinase-like protein [Arabidopsis thaliana] 501 e-140
UniRef100_Q9LV48 Protein kinase-like protein [Arabidopsis thaliana] 501 e-140
UniRef100_Q9ARH1 Receptor protein kinase PERK1 [Brassica napus] 500 e-140
UniRef100_O65672 Putative serine/threonine protein kinase [Arabi... 495 e-139
UniRef100_Q84VF7 Putative receptor protein kinase [Oryza sativa] 494 e-138
UniRef100_Q8W0B8 Putative receptor protein kinase PERK1 [Oryza s... 492 e-138
UniRef100_Q9LS95 Somatic embryogenesis receptor kinase-like prot... 491 e-137
UniRef100_Q8H8M5 Putative kinase [Oryza sativa] 478 e-133
UniRef100_Q9LK03 Somatic embryogenesis receptor kinase-like prot... 459 e-128
UniRef100_Q9C821 Protein kinase, putative; 60711-62822 [Arabidop... 456 e-127
UniRef100_Q7XHF3 Hypothetical protein [Oryza sativa] 456 e-127
UniRef100_Q9LV50 Protein kinase-like protein [Arabidopsis thaliana] 424 e-117
UniRef100_Q6ZE12 Hypothetical protein P0495H05.37 [Oryza sativa] 424 e-117
UniRef100_Q67W44 Hypothetical protein OJ1568_D07.15 [Oryza sativa] 420 e-116
UniRef100_Q9CAL8 Hypothetical protein F24J13.3 [Arabidopsis thal... 415 e-114
UniRef100_Q9ASK4 Putative LRR receptor-like protein kinase [Oryz... 408 e-112
>UniRef100_Q9ZNQ8 Hypothetical protein At2g18470 [Arabidopsis thaliana]
Length = 633
Score = 521 bits (1343), Expect = e-146
Identities = 269/395 (68%), Positives = 318/395 (80%), Gaps = 10/395 (2%)
Query: 8 QQNPNGASGVWGAPHP---LMNSGEMSSNYSYGMGPPGSMQSSPGLSLTLKGGTFTYEEL 64
QQ+ G G +P P + SGE SS YS G P SP L+L TFTY+EL
Sbjct: 219 QQSTGGWGGGGPSPPPPPRMPTSGEDSSMYS-GPSRPVLPPPSPALALGFNKSTFTYQEL 277
Query: 65 ASATKGFANENIIGQGGFGYVHKGILPTGKEIAVKSLKAGSGQGEREFQAEIDIISRVHH 124
A+AT GF + N++GQGGFGYVHKG+LP+GKE+AVKSLKAGSGQGEREFQAE+DIISRVHH
Sbjct: 278 AAATGGFTDANLLGQGGFGYVHKGVLPSGKEVAVKSLKAGSGQGEREFQAEVDIISRVHH 337
Query: 125 RHLVSLVGYCVSGGQRMLVYEFVPNKTLEYHLHGKGVPTMDWPTRMRIALGSARGLAYLH 184
R+LVSLVGYC++ GQRMLVYEFVPNKTLEYHLHGK +P M++ TR+RIALG+A+GLAYLH
Sbjct: 338 RYLVSLVGYCIADGQRMLVYEFVPNKTLEYHLHGKNLPVMEFSTRLRIALGAAKGLAYLH 397
Query: 185 EDCSPRIIHRDIKAANVLIDDSFEAKVADFGLAKLTTDTNTHVSTRVMGTFGYMAPEYAS 244
EDC PRIIHRDIK+AN+L+D +F+A VADFGLAKLT+D NTHVSTRVMGTFGY+APEYAS
Sbjct: 398 EDCHPRIIHRDIKSANILLDFNFDAMVADFGLAKLTSDNNTHVSTRVMGTFGYLAPEYAS 457
Query: 245 SGKLTEKSDVFSFGVMLLELLTGKRPLDLTNAMDESLVDWARPLLSRALEEDGNFAELVD 304
SGKLTEKSDVFS+GVMLLEL+TGKRP+D + MD++LVDWARPL++RAL EDGNF EL D
Sbjct: 458 SGKLTEKSDVFSYGVMLLELITGKRPVDNSITMDDTLVDWARPLMARAL-EDGNFNELAD 516
Query: 305 PFLEGNYDHQEMIRLAACAASSIRHSAKKRSKMSQIVRALEGDVSLEDLKESMIKSPAPQ 364
LEGNY+ QEM R+ CAA+SIRHS +KR KMSQIVRALEG+VSL+ L E +
Sbjct: 517 ARLEGNYNPQEMARMVTCAAASIRHSGRKRPKMSQIVRALEGEVSLDALNEGV---KPGH 573
Query: 365 TGVYTTTG--SEYDTMQYNADMAKFRKQIMSSQEF 397
+ VY + G S+Y YNADM KFR+ +SSQEF
Sbjct: 574 SNVYGSLGASSDYSQTSYNADMKKFRQIALSSQEF 608
>UniRef100_Q8GX23 Putative serine/threonine protein kinase [Arabidopsis thaliana]
Length = 670
Score = 512 bits (1318), Expect = e-144
Identities = 256/399 (64%), Positives = 312/399 (78%), Gaps = 15/399 (3%)
Query: 2 DHVVRVQQNPNGASGVWGAPHPLMNSGEMSSNYSYGMGPPGSMQSSPGLSLTLKGGTFTY 61
DHVV + G WG P+ +SN + P ++ G + + TFTY
Sbjct: 250 DHVVNMAGQ---GGGNWGPQQPVSGPHSDASNLTGRTAIPSPQAATLGHNQS----TFTY 302
Query: 62 EELASATKGFANENIIGQGGFGYVHKGILPTGKEIAVKSLKAGSGQGEREFQAEIDIISR 121
+EL+ AT+GFA N++GQGGFGYVHKG+LP+GKE+AVKSLK GSGQGEREFQAE+DIISR
Sbjct: 303 DELSIATEGFAQSNLLGQGGFGYVHKGVLPSGKEVAVKSLKLGSGQGEREFQAEVDIISR 362
Query: 122 VHHRHLVSLVGYCVSGGQRMLVYEFVPNKTLEYHLHGKGVPTMDWPTRMRIALGSARGLA 181
VHHRHLVSLVGYC+SGGQR+LVYEF+PN TLE+HLHGKG P +DWPTR++IALGSARGLA
Sbjct: 363 VHHRHLVSLVGYCISGGQRLLVYEFIPNNTLEFHLHGKGRPVLDWPTRVKIALGSARGLA 422
Query: 182 YLHEDCSPRIIHRDIKAANVLIDDSFEAKVADFGLAKLTTDTNTHVSTRVMGTFGYMAPE 241
YLHEDC PRIIHRDIKAAN+L+D SFE KVADFGLAKL+ D THVSTRVMGTFGY+APE
Sbjct: 423 YLHEDCHPRIIHRDIKAANILLDFSFETKVADFGLAKLSQDNYTHVSTRVMGTFGYLAPE 482
Query: 242 YASSGKLTEKSDVFSFGVMLLELLTGKRPLDLTNAMDESLVDWARPLLSRALEEDGNFAE 301
YASSGKL++KSDVFSFGVMLLEL+TG+ PLDLT M++SLVDWARPL +A +DG++ +
Sbjct: 483 YASSGKLSDKSDVFSFGVMLLELITGRPPLDLTGEMEDSLVDWARPLCLKA-AQDGDYNQ 541
Query: 302 LVDPFLEGNYDHQEMIRLAACAASSIRHSAKKRSKMSQIVRALEGDVSLEDLKESMIKSP 361
L DP LE NY HQEM+++A+CAA++IRHSA++R KMSQIVRALEGD+S++DL E
Sbjct: 542 LADPRLELNYSHQEMVQMASCAAAAIRHSARRRPKMSQIVRALEGDMSMDDLSE----GT 597
Query: 362 APQTGVYTTTG---SEYDTMQYNADMAKFRKQIMSSQEF 397
P Y + G SEYD Y ADM KF+K + ++E+
Sbjct: 598 RPGQSTYLSPGSVSSEYDASSYTADMKKFKKLALENKEY 636
>UniRef100_Q60E30 Hypothetical protein OSJNBb0012L23.7 [Oryza sativa]
Length = 471
Score = 507 bits (1306), Expect = e-142
Identities = 257/402 (63%), Positives = 318/402 (78%), Gaps = 17/402 (4%)
Query: 2 DHVVRVQQNPNGASG------VWGAPHPLMNSGEMSSNYSYG--MGPPGSMQSSPGLSLT 53
+HVV++ +P A P ++NS S +YS G + PP SPG +L
Sbjct: 25 EHVVKMHPSPPPAYANRPPQPPSTPPAAMINSSGGSGSYSGGEILPPP-----SPGAALG 79
Query: 54 LKGGTFTYEELASATKGFANENIIGQGGFGYVHKGILPTGKEIAVKSLKAGSGQGEREFQ 113
TFTYEEL AT GF++ N++GQGGFGYVH+G+LPTGKEIAVK LK GSGQGEREFQ
Sbjct: 80 FSKSTFTYEELLRATDGFSDANLLGQGGFGYVHRGVLPTGKEIAVKQLKVGSGQGEREFQ 139
Query: 114 AEIDIISRVHHRHLVSLVGYCVSGGQRMLVYEFVPNKTLEYHLHGKGVPTMDWPTRMRIA 173
AE++IISRVHH+HLVSLVGYC+SGG+R+LVYEFVPN TLE+HLHGKG PTM+WPTR++IA
Sbjct: 140 AEVEIISRVHHKHLVSLVGYCISGGKRLLVYEFVPNNTLEFHLHGKGRPTMEWPTRLKIA 199
Query: 174 LGSARGLAYLHEDCSPRIIHRDIKAANVLIDDSFEAKVADFGLAKLTTDTNTHVSTRVMG 233
LG+A+GLAYLHEDC P+IIHRDIKA+N+L+D FE+KVADFGLAK T+D NTHVSTRVMG
Sbjct: 200 LGAAKGLAYLHEDCHPKIIHRDIKASNILLDFKFESKVADFGLAKFTSDNNTHVSTRVMG 259
Query: 234 TFGYMAPEYASSGKLTEKSDVFSFGVMLLELLTGKRPLDLTNA-MDESLVDWARPLLSRA 292
TFGY+APEYASSGKLTEKSDVFS+GVMLLEL+TG+RP+D + MD+SLVDWARPLL +A
Sbjct: 260 TFGYLAPEYASSGKLTEKSDVFSYGVMLLELITGRRPVDTSQTYMDDSLVDWARPLLMQA 319
Query: 293 LEEDGNFAELVDPFLEGNYDHQEMIRLAACAASSIRHSAKKRSKMSQIVRALEGDVSLED 352
L E+GN+ ELVDP L +++ EM R+ ACAA+ +RHSA++R +MSQ+VRALEGDVSLED
Sbjct: 320 L-ENGNYEELVDPRLGKDFNPNEMARMIACAAACVRHSARRRPRMSQVVRALEGDVSLED 378
Query: 353 LKESMIKSPAPQTGVYTTTGSEYDTMQYNADMAKFRKQIMSS 394
L E + + G Y++ S+YD+ QYN DM KFRK ++
Sbjct: 379 LNEGVRPGHSRYFGSYSS--SDYDSGQYNEDMKKFRKMAFTN 418
>UniRef100_Q9XI96 Similar to somatic embryogenesis receptor-like kinase [Arabidopsis
thaliana]
Length = 699
Score = 505 bits (1300), Expect = e-141
Identities = 253/391 (64%), Positives = 309/391 (78%), Gaps = 6/391 (1%)
Query: 8 QQNPNGASGVWGAPHPLMNSGEMSSNYSYGMGPPGSMQSSPGLSLTLKGGTFTYEELASA 67
Q P+ + V G + ++ SGEMSSN+S G P P ++L TFTYEELASA
Sbjct: 274 QPMPSPPAPVSGGAN-VIQSGEMSSNFSSGPYAPSLPPPHPSVALGFNNSTFTYEELASA 332
Query: 68 TKGFANENIIGQGGFGYVHKGILPTGKEIAVKSLKAGSGQGEREFQAEIDIISRVHHRHL 127
T+GF+ + ++GQGGFGYVHKGILP GKEIAVKSLKAGSGQGEREFQAE++IISRVHHRHL
Sbjct: 333 TQGFSKDRLLGQGGFGYVHKGILPNGKEIAVKSLKAGSGQGEREFQAEVEIISRVHHRHL 392
Query: 128 VSLVGYCVS-GGQRMLVYEFVPNKTLEYHLHGKGVPTMDWPTRMRIALGSARGLAYLHED 186
VSLVGYC + GGQR+LVYEF+PN TLE+HLHGK MDWPTR++IALGSA+GLAYLHED
Sbjct: 393 VSLVGYCSNAGGQRLLVYEFLPNDTLEFHLHGKSGTVMDWPTRLKIALGSAKGLAYLHED 452
Query: 187 CSPRIIHRDIKAANVLIDDSFEAKVADFGLAKLTTDTNTHVSTRVMGTFGYMAPEYASSG 246
C P+IIHRDIKA+N+L+D +FEAKVADFGLAKL+ D NTHVSTRVMGTFGY+APEYASSG
Sbjct: 453 CHPKIIHRDIKASNILLDHNFEAKVADFGLAKLSQDNNTHVSTRVMGTFGYLAPEYASSG 512
Query: 247 KLTEKSDVFSFGVMLLELLTGKRPLDLTNAMDESLVDWARPLLSRALEEDGNFAELVDPF 306
KLTEKSDVFSFGVMLLEL+TG+ P+DL+ M++SLVDWARPL R + +DG + ELVDPF
Sbjct: 513 KLTEKSDVFSFGVMLLELITGRGPVDLSGDMEDSLVDWARPLCMR-VAQDGEYGELVDPF 571
Query: 307 LEGNYDHQEMIRLAACAASSIRHSAKKRSKMSQIVRALEGDVSLEDLKESMIKSPAPQTG 366
LE Y+ EM R+ ACAA+++RHS ++R KMSQIVR LEGD SL+DL + + + G
Sbjct: 572 LEHQYEPYEMARMVACAAAAVRHSGRRRPKMSQIVRTLEGDASLDDLDDGVKPKQSSSGG 631
Query: 367 VYTTTGSEYDTMQYNADMAKFRKQIMSSQEF 397
S+Y+ Y A+M KFRK + S+++
Sbjct: 632 ---EGSSDYEMGTYGAEMRKFRKVTLESRDY 659
>UniRef100_Q94JZ6 Protein kinase-like protein [Arabidopsis thaliana]
Length = 652
Score = 501 bits (1291), Expect = e-140
Identities = 252/380 (66%), Positives = 304/380 (79%), Gaps = 10/380 (2%)
Query: 21 PHPLMNSGEMSSNYSYGMGPPGSMQSSPGLSLTLKGGTFTYEELASATKGFANENIIGQG 80
P P S S+YS P SPGL L TFTYEEL+ AT GF+ N++GQG
Sbjct: 233 PPPAFMSSSGGSDYS---DLPVLPPPSPGLVLGFSKSTFTYEELSRATNGFSEANLLGQG 289
Query: 81 GFGYVHKGILPTGKEIAVKSLKAGSGQGEREFQAEIDIISRVHHRHLVSLVGYCVSGGQR 140
GFGYVHKGILP+GKE+AVK LKAGSGQGEREFQAE++IISRVHHRHLVSL+GYC++G QR
Sbjct: 290 GFGYVHKGILPSGKEVAVKQLKAGSGQGEREFQAEVEIISRVHHRHLVSLIGYCMAGVQR 349
Query: 141 MLVYEFVPNKTLEYHLHGKGVPTMDWPTRMRIALGSARGLAYLHEDCSPRIIHRDIKAAN 200
+LVYEFVPN LE+HLHGKG PTM+W TR++IALGSA+GL+YLHEDC+P+IIHRDIKA+N
Sbjct: 350 LLVYEFVPNNNLEFHLHGKGRPTMEWSTRLKIALGSAKGLSYLHEDCNPKIIHRDIKASN 409
Query: 201 VLIDDSFEAKVADFGLAKLTTDTNTHVSTRVMGTFGYMAPEYASSGKLTEKSDVFSFGVM 260
+LID FEAKVADFGLAK+ +DTNTHVSTRVMGTFGY+APEYA+SGKLTEKSDVFSFGV+
Sbjct: 410 ILIDFKFEAKVADFGLAKIASDTNTHVSTRVMGTFGYLAPEYAASGKLTEKSDVFSFGVV 469
Query: 261 LLELLTGKRPLDLTNA-MDESLVDWARPLLSRALEEDGNFAELVDPFLEGNYDHQEMIRL 319
LLEL+TG+RP+D N +D+SLVDWARPLL+RA EE G+F L D + YD +EM R+
Sbjct: 470 LLELITGRRPVDANNVYVDDSLVDWARPLLNRASEE-GDFEGLADSKMGNEYDREEMARM 528
Query: 320 AACAASSIRHSAKKRSKMSQIVRALEGDVSLEDLKESMIKSPAPQTGVYTTTG--SEYDT 377
ACAA+ +RHSA++R +MSQIVRALEG+VSL DL E M + VY++ G ++YDT
Sbjct: 529 VACAAACVRHSARRRPRMSQIVRALEGNVSLSDLNEGMRPG---HSNVYSSYGGSTDYDT 585
Query: 378 MQYNADMAKFRKQIMSSQEF 397
QYN DM KFRK + +QE+
Sbjct: 586 SQYNDDMIKFRKMALGTQEY 605
>UniRef100_Q9LV48 Protein kinase-like protein [Arabidopsis thaliana]
Length = 652
Score = 501 bits (1291), Expect = e-140
Identities = 252/380 (66%), Positives = 304/380 (79%), Gaps = 10/380 (2%)
Query: 21 PHPLMNSGEMSSNYSYGMGPPGSMQSSPGLSLTLKGGTFTYEELASATKGFANENIIGQG 80
P P S S+YS P SPGL L TFTYEEL+ AT GF+ N++GQG
Sbjct: 233 PPPAFMSSSGGSDYS---DLPVLPPPSPGLVLGFSKSTFTYEELSRATNGFSEANLLGQG 289
Query: 81 GFGYVHKGILPTGKEIAVKSLKAGSGQGEREFQAEIDIISRVHHRHLVSLVGYCVSGGQR 140
GFGYVHKGILP+GKE+AVK LKAGSGQGEREFQAE++IISRVHHRHLVSL+GYC++G QR
Sbjct: 290 GFGYVHKGILPSGKEVAVKQLKAGSGQGEREFQAEVEIISRVHHRHLVSLIGYCMAGVQR 349
Query: 141 MLVYEFVPNKTLEYHLHGKGVPTMDWPTRMRIALGSARGLAYLHEDCSPRIIHRDIKAAN 200
+LVYEFVPN LE+HLHGKG PTM+W TR++IALGSA+GL+YLHEDC+P+IIHRDIKA+N
Sbjct: 350 LLVYEFVPNNNLEFHLHGKGRPTMEWSTRLKIALGSAKGLSYLHEDCNPKIIHRDIKASN 409
Query: 201 VLIDDSFEAKVADFGLAKLTTDTNTHVSTRVMGTFGYMAPEYASSGKLTEKSDVFSFGVM 260
+LID FEAKVADFGLAK+ +DTNTHVSTRVMGTFGY+APEYA+SGKLTEKSDVFSFGV+
Sbjct: 410 ILIDFKFEAKVADFGLAKIASDTNTHVSTRVMGTFGYLAPEYAASGKLTEKSDVFSFGVV 469
Query: 261 LLELLTGKRPLDLTNA-MDESLVDWARPLLSRALEEDGNFAELVDPFLEGNYDHQEMIRL 319
LLEL+TG+RP+D N +D+SLVDWARPLL+RA EE G+F L D + YD +EM R+
Sbjct: 470 LLELITGRRPVDANNVYVDDSLVDWARPLLNRASEE-GDFEGLADSKMGNEYDREEMARM 528
Query: 320 AACAASSIRHSAKKRSKMSQIVRALEGDVSLEDLKESMIKSPAPQTGVYTTTG--SEYDT 377
ACAA+ +RHSA++R +MSQIVRALEG+VSL DL E M + VY++ G ++YDT
Sbjct: 529 VACAAACVRHSARRRPRMSQIVRALEGNVSLSDLNEGMRPG---HSNVYSSYGGSTDYDT 585
Query: 378 MQYNADMAKFRKQIMSSQEF 397
QYN DM KFRK + +QE+
Sbjct: 586 SQYNDDMIKFRKMALGTQEY 605
>UniRef100_Q9ARH1 Receptor protein kinase PERK1 [Brassica napus]
Length = 647
Score = 500 bits (1287), Expect = e-140
Identities = 249/381 (65%), Positives = 304/381 (79%), Gaps = 11/381 (2%)
Query: 21 PHPLMNSGEMSSNYSYGMGPPGSMQSSPGLSLTLKGGTFTYEELASATKGFANENIIGQG 80
P P M+S S + PP SPGL L TFTYEELA AT GF+ N++GQG
Sbjct: 229 PPPFMSSSGGSDYSDRPVLPP----PSPGLVLGFSKSTFTYEELARATNGFSEANLLGQG 284
Query: 81 GFGYVHKGILPTGKEIAVKSLKAGSGQGEREFQAEIDIISRVHHRHLVSLVGYCVSGGQR 140
GFGYVHKG+LP+GKE+AVK LK GSGQGEREFQAE++IISRVHHRHLVSLVGYC++G +R
Sbjct: 285 GFGYVHKGVLPSGKEVAVKQLKVGSGQGEREFQAEVEIISRVHHRHLVSLVGYCIAGAKR 344
Query: 141 MLVYEFVPNKTLEYHLHGKGVPTMDWPTRMRIALGSARGLAYLHEDCSPRIIHRDIKAAN 200
+LVYEFVPN LE HLHG+G PTM+W TR++IALGSA+GL+YLHEDC+P+IIHRDIKA+N
Sbjct: 345 LLVYEFVPNNNLELHLHGEGRPTMEWSTRLKIALGSAKGLSYLHEDCNPKIIHRDIKASN 404
Query: 201 VLIDDSFEAKVADFGLAKLTTDTNTHVSTRVMGTFGYMAPEYASSGKLTEKSDVFSFGVM 260
+LID FEAKVADFGLAK+ +DTNTHVSTRVMGTFGY+APEYA+SGKLTEKSDVFSFGV+
Sbjct: 405 ILIDFKFEAKVADFGLAKIASDTNTHVSTRVMGTFGYLAPEYAASGKLTEKSDVFSFGVV 464
Query: 261 LLELLTGKRPLDLTNA-MDESLVDWARPLLSRALEEDGNFAELVDPFLEGNYDHQEMIRL 319
LLEL+TG+RP+D N +D+SLVDWARPLL+RA E G+F L D + YD +EM R+
Sbjct: 465 LLELITGRRPVDANNVYVDDSLVDWARPLLNRA-SEQGDFEGLADAKMNNGYDREEMARM 523
Query: 320 AACAASSIRHSAKKRSKMSQIVRALEGDVSLEDLKESMIKSPAPQTGVYTTTG--SEYDT 377
ACAA+ +RHSA++R +MSQIVRALEG+VSL DL E M Q+ VY++ G ++YD+
Sbjct: 524 VACAAACVRHSARRRPRMSQIVRALEGNVSLSDLNEGMRPG---QSNVYSSYGGSTDYDS 580
Query: 378 MQYNADMAKFRKQIMSSQEFD 398
QYN DM KFRK + +QE++
Sbjct: 581 SQYNEDMKKFRKMALGTQEYN 601
>UniRef100_O65672 Putative serine/threonine protein kinase [Arabidopsis thaliana]
Length = 674
Score = 495 bits (1275), Expect = e-139
Identities = 256/421 (60%), Positives = 312/421 (73%), Gaps = 37/421 (8%)
Query: 2 DHVVRVQQNPNGASGVWGAPHPLMNSGEMSSNYSYGMGPPGSMQSSPGLSLTLKGGTFTY 61
DHVV + G WG P+ +SN + P ++ G + + TFTY
Sbjct: 232 DHVVNMAGQ---GGGNWGPQQPVSGPHSDASNLTGRTAIPSPQAATLGHNQS----TFTY 284
Query: 62 EELASATKGFANENIIGQGGFGYVHKGILPTGKEIAVKSLKAGSGQGEREFQAEIDIISR 121
+EL+ AT+GFA N++GQGGFGYVHKG+LP+GKE+AVKSLK GSGQGEREFQAE+DIISR
Sbjct: 285 DELSIATEGFAQSNLLGQGGFGYVHKGVLPSGKEVAVKSLKLGSGQGEREFQAEVDIISR 344
Query: 122 VHHRHLVSLVGYCVSGGQRMLVYEFVPNKTLEYHLHGKGVPTMDWPTRMRIALGSARGLA 181
VHHRHLVSLVGYC+SGGQR+LVYEF+PN TLE+HLHGKG P +DWPTR++IALGSARGLA
Sbjct: 345 VHHRHLVSLVGYCISGGQRLLVYEFIPNNTLEFHLHGKGRPVLDWPTRVKIALGSARGLA 404
Query: 182 YLHEDCS-----------PRIIHRDIKAANVLIDDSFEAKVADFGLAKLTTDTNTHVSTR 230
YLHEDC PRIIHRDIKAAN+L+D SFE KVADFGLAKL+ D THVSTR
Sbjct: 405 YLHEDCKKIFISHICISHPRIIHRDIKAANILLDFSFETKVADFGLAKLSQDNYTHVSTR 464
Query: 231 VMGTFGYMAPEYASSGKLTEKSDVFSFGVMLLELLTGKRPLDLTNAMDESLVDWARPLLS 290
VMGTFGY+APEYASSGKL++KSDVFSFGVMLLEL+TG+ PLDLT M++SLVDWARPL
Sbjct: 465 VMGTFGYLAPEYASSGKLSDKSDVFSFGVMLLELITGRPPLDLTGEMEDSLVDWARPLCL 524
Query: 291 RALEEDGNFAELVDPFLEGNYDHQEMIRLAACAASSIRHSAKKRSKMSQ----------- 339
+A +DG++ +L DP LE NY HQEM+++A+CAA++IRHSA++R KMSQ
Sbjct: 525 KA-AQDGDYNQLADPRLELNYSHQEMVQMASCAAAAIRHSARRRPKMSQVQKLIPLVGSI 583
Query: 340 IVRALEGDVSLEDLKESMIKSPAPQTGVYTTTG---SEYDTMQYNADMAKFRKQIMSSQE 396
IVRALEGD+S++DL E P Y + G SEYD Y ADM KF+K + ++E
Sbjct: 584 IVRALEGDMSMDDLSE----GTRPGQSTYLSPGSVSSEYDASSYTADMKKFKKLALENKE 639
Query: 397 F 397
+
Sbjct: 640 Y 640
>UniRef100_Q84VF7 Putative receptor protein kinase [Oryza sativa]
Length = 394
Score = 494 bits (1272), Expect = e-138
Identities = 239/338 (70%), Positives = 291/338 (85%), Gaps = 4/338 (1%)
Query: 58 TFTYEELASATKGFANENIIGQGGFGYVHKGILPTGKEIAVKSLKAGSGQGEREFQAEID 117
TFTYEEL AT GF++ N++GQGGFGYVH+G+LPTGKEIAVK LK GSGQGEREFQAE++
Sbjct: 7 TFTYEELLRATDGFSDANLLGQGGFGYVHRGVLPTGKEIAVKQLKVGSGQGEREFQAEVE 66
Query: 118 IISRVHHRHLVSLVGYCVSGGQRMLVYEFVPNKTLEYHLHGKGVPTMDWPTRMRIALGSA 177
IISRVHH+HLVSLVGYC+SGG+R+LVYEFVPN TLE+HLHGKG PTM+WPTR++IALG+A
Sbjct: 67 IISRVHHKHLVSLVGYCISGGKRLLVYEFVPNNTLEFHLHGKGRPTMEWPTRLKIALGAA 126
Query: 178 RGLAYLHEDCSPRIIHRDIKAANVLIDDSFEAKVADFGLAKLTTDTNTHVSTRVMGTFGY 237
+GLAYLHEDC P+IIHRDIKA+N+L+D FE+KVADFGLAK T+D NTHVSTRVMGTFGY
Sbjct: 127 KGLAYLHEDCHPKIIHRDIKASNILLDFKFESKVADFGLAKFTSDNNTHVSTRVMGTFGY 186
Query: 238 MAPEYASSGKLTEKSDVFSFGVMLLELLTGKRPLDLTNA-MDESLVDWARPLLSRALEED 296
+APEYASSGKLTEKSDVFS+GVMLLEL+TG+RP+D + MD+SLVDWARPLL +AL E+
Sbjct: 187 LAPEYASSGKLTEKSDVFSYGVMLLELITGRRPVDTSQTYMDDSLVDWARPLLMQAL-EN 245
Query: 297 GNFAELVDPFLEGNYDHQEMIRLAACAASSIRHSAKKRSKMSQIVRALEGDVSLEDLKES 356
GN+ ELVDP L +++ EM R+ ACAA+ +RHSA++R +MSQ+VRALEGDVSLEDL E
Sbjct: 246 GNYEELVDPRLGKDFNPNEMARMIACAAACVRHSARRRPRMSQVVRALEGDVSLEDLNEG 305
Query: 357 MIKSPAPQTGVYTTTGSEYDTMQYNADMAKFRKQIMSS 394
+ + G Y++ S+YD+ QYN DM KFRK ++
Sbjct: 306 VRPGHSRYFGSYSS--SDYDSGQYNEDMKKFRKMAFTN 341
>UniRef100_Q8W0B8 Putative receptor protein kinase PERK1 [Oryza sativa]
Length = 597
Score = 492 bits (1266), Expect = e-138
Identities = 251/396 (63%), Positives = 311/396 (78%), Gaps = 12/396 (3%)
Query: 2 DHVVR-VQQNPNGASGVWGAPHPL-MNSGEMSSNYSYGMGPPGSMQSSPGLSLTLKGGTF 59
DHV++ V +P+ P PL ++SG SNYS G + SPG +L TF
Sbjct: 158 DHVLKKVPSHPSPPP----PPAPLNVHSGGSGSNYSGGDNSQPLV--SPGAALGFSRCTF 211
Query: 60 TYEELASATKGFANENIIGQGGFGYVHKGILPTGKEIAVKSLKAGSGQGEREFQAEIDII 119
TYE+L++AT GF++ N++GQGGFGYVHKG+LP G E+AVK L+ GSGQGEREFQAE++II
Sbjct: 212 TYEDLSAATDGFSDANLLGQGGFGYVHKGVLPNGTEVAVKQLRDGSGQGEREFQAEVEII 271
Query: 120 SRVHHRHLVSLVGYCVSGGQRMLVYEFVPNKTLEYHLHGKGVPTMDWPTRMRIALGSARG 179
SRVHH+HLV+LVGYC+SGG+R+LVYE+VPN TLE HLHG+G PTM+WPTR+RIALG+A+G
Sbjct: 272 SRVHHKHLVTLVGYCISGGKRLLVYEYVPNNTLELHLHGRGRPTMEWPTRLRIALGAAKG 331
Query: 180 LAYLHEDCSPRIIHRDIKAANVLIDDSFEAKVADFGLAKLTTDTNTHVSTRVMGTFGYMA 239
LAYLHEDC P+IIHRDIK+AN+L+D FEAKVADFGLAKLT+D NTHVSTRVMGTFGY+A
Sbjct: 332 LAYLHEDCHPKIIHRDIKSANILLDARFEAKVADFGLAKLTSDNNTHVSTRVMGTFGYLA 391
Query: 240 PEYASSGKLTEKSDVFSFGVMLLELLTGKRPLDLTNA-MDESLVDWARPLLSRALEEDGN 298
PEYASSG+LTEKSDVFSFGVMLLEL+TG+RP+ + MD+SLVDWARPL+ RA +DGN
Sbjct: 392 PEYASSGQLTEKSDVFSFGVMLLELITGRRPVRSNQSQMDDSLVDWARPLMMRA-SDDGN 450
Query: 299 FAELVDPFLEGNYDHQEMIRLAACAASSIRHSAKKRSKMSQIVRALEGDVSLEDLKESMI 358
+ LVDP L Y+ EM R+ ACAA+ +RHSA++R +MSQ+VRALEGDVSL+DL E +
Sbjct: 451 YDALVDPRLGQEYNGNEMARMIACAAACVRHSARRRPRMSQVVRALEGDVSLDDLNEGVR 510
Query: 359 KSPAPQTGVYTTTGSEYDTMQYNADMAKFRKQIMSS 394
+ G Y + +EYDT YN D+ KFRK S
Sbjct: 511 PGHSRFLGSYNS--NEYDTGHYNEDLKKFRKMAFGS 544
>UniRef100_Q9LS95 Somatic embryogenesis receptor kinase-like protein [Arabidopsis
thaliana]
Length = 714
Score = 491 bits (1263), Expect = e-137
Identities = 246/385 (63%), Positives = 302/385 (77%), Gaps = 16/385 (4%)
Query: 26 NSGEMSSNYSYGMGPPGSMQSSPGLSLTLKGGTFTYEELASATKGFANENIIGQGGFGYV 85
NS + SSNYS G P P ++L TFTY+ELA+AT+GF+ ++GQGGFGYV
Sbjct: 293 NSSDFSSNYS-GPHGPSVPPPHPSVALGFNKSTFTYDELAAATQGFSQSRLLGQGGFGYV 351
Query: 86 HKGILPTGKEIAVKSLKAGSGQGEREFQAEIDIISRVHHRHLVSLVGYCVSGGQRMLVYE 145
HKGILP GKEIAVKSLKAGSGQGEREFQAE+DIISRVHHR LVSLVGYC++GGQRMLVYE
Sbjct: 352 HKGILPNGKEIAVKSLKAGSGQGEREFQAEVDIISRVHHRFLVSLVGYCIAGGQRMLVYE 411
Query: 146 FVPNKTLEYHLHGKGVPTMDWPTRMRIALGSARGLAYLHEDCSPRIIHRDIKAANVLIDD 205
F+PN TLE+HLHGK +DWPTR++IALGSA+GLAYLHEDC PRIIHRDIKA+N+L+D+
Sbjct: 412 FLPNDTLEFHLHGKSGKVLDWPTRLKIALGSAKGLAYLHEDCHPRIIHRDIKASNILLDE 471
Query: 206 SFEAKVADFGLAKLTTDTNTHVSTRVMGTFG---------YMAPEYASSGKLTEKSDVFS 256
SFEAKVADFGLAKL+ D THVSTR+MGTFG Y+APEYASSGKLT++SDVFS
Sbjct: 472 SFEAKVADFGLAKLSQDNVTHVSTRIMGTFGISNCESNDRYLAPEYASSGKLTDRSDVFS 531
Query: 257 FGVMLLELLTGKRPLDLTNAMDESLVDWARPLLSRAL----EEDGNFAELVDPFLEGNYD 312
FGVMLLEL+TG+RP+DLT M++SLVDW R ++R + +DG+++ELVDP LE Y+
Sbjct: 532 FGVMLLELVTGRRPVDLTGEMEDSLVDWVRNHMARPICLNAAQDGDYSELVDPRLENQYE 591
Query: 313 HQEMIRLAACAASSIRHSAKKRSKMSQIVRALEGDVSLEDLKESMIKSPAPQTGVYTTTG 372
EM ++ ACAA+++RHSA++R KMSQIVRALEGD +L+DL E + G +
Sbjct: 592 PHEMAQMVACAAAAVRHSARRRPKMSQIVRALEGDATLDDLSEGGKAGQSSFLG--RGSS 649
Query: 373 SEYDTMQYNADMAKFRKQIMSSQEF 397
S+YD+ Y+ADM KFRK + S E+
Sbjct: 650 SDYDSSTYSADMKKFRKVALDSHEY 674
>UniRef100_Q8H8M5 Putative kinase [Oryza sativa]
Length = 466
Score = 478 bits (1230), Expect = e-133
Identities = 239/349 (68%), Positives = 286/349 (81%), Gaps = 11/349 (3%)
Query: 57 GTFTYEELASATKGFANENIIGQGGFGYVHKGILPTGKEIAVKSLKAGSGQGEREFQAEI 116
GTFTYE+LA+AT GFA EN++GQGGFGYVHKG+L GK +AVK LK+GSGQGEREFQAE+
Sbjct: 91 GTFTYEQLAAATGGFAEENLVGQGGFGYVHKGVLAGGKAVAVKQLKSGSGQGEREFQAEV 150
Query: 117 DIISRVHHRHLVSLVGYCVSGGQRMLVYEFVPNKTLEYHLHGKGVPTMDWPTRMRIALGS 176
DIISRVHHRHLVSLVGYC++G +R+LVYEFVPNKTLE+HLHGKG+P M WPTR+RIALGS
Sbjct: 151 DIISRVHHRHLVSLVGYCIAGARRVLVYEFVPNKTLEFHLHGKGLPVMPWPTRLRIALGS 210
Query: 177 ARGLAYLHEDCSPRIIHRDIKAANVLIDDSFEAKVADFGLAKLTTDTNTHVSTRVMGTFG 236
A+GLAYLHEDC PRIIHRDIK+AN+L+D++FEAKVADFGLAKLT+D NTHVSTRVMGTFG
Sbjct: 211 AKGLAYLHEDCHPRIIHRDIKSANILLDNNFEAKVADFGLAKLTSDNNTHVSTRVMGTFG 270
Query: 237 YMAPEYASSGKLTEKSDVFSFGVMLLELLTGKRPLDLTNA---------MDESLVDWARP 287
Y+APEYASSGKLTEKSDVFS+GVMLLEL+TG+RP+D A D+SLV+WARP
Sbjct: 271 YLAPEYASSGKLTEKSDVFSYGVMLLELVTGRRPIDAGAADHPWPASFMEDDSLVEWARP 330
Query: 288 LLSRALEEDGNFAELVDPFLEGNYDHQEMIRLAACAASSIRHSAKKRSKMSQIVRALEGD 347
++RAL DG++ + DP LEG+YD EM R+ A AA+S+RHSAKKR KMSQIVRALEGD
Sbjct: 331 AMARAL-ADGDYGGVADPRLEGSYDAVEMARVVASAAASVRHSAKKRPKMSQIVRALEGD 389
Query: 348 VSLEDLKESMIKSPAPQTGVYTTTGSEYDTM-QYNADMAKFRKQIMSSQ 395
+SLEDL E M + G T GS + Y DM + ++ +++
Sbjct: 390 MSLEDLNEGMRPGQSMVFGTAETGGSISEASGSYTFDMDRIIQEATAAR 438
>UniRef100_Q9LK03 Somatic embryogenesis receptor kinase-like protein [Arabidopsis
thaliana]
Length = 458
Score = 459 bits (1181), Expect = e-128
Identities = 240/404 (59%), Positives = 301/404 (74%), Gaps = 17/404 (4%)
Query: 2 DHVVRVQQNPNGASGVWGAPHPLMNSG---EMSSNYS-YGMGPPGSMQSSPGLSLTLK-- 55
DHVV P S P M+SG + SNYS + PP SPGL+L L
Sbjct: 24 DHVVMSVPPPKSPSSAPPRPPHFMSSGSSGDYDSNYSDQSVLPP----PSPGLALGLGIY 79
Query: 56 GGTFTYEELASATKGFANENIIGQGGFGYVHKGILPTGKEIAVKSLKAGSGQGEREFQAE 115
GTF YEEL+ AT GF+ N++GQGGFGYV KG+L GKE+AVK LK GS QGEREFQAE
Sbjct: 80 QGTFNYEELSRATNGFSEANLLGQGGFGYVFKGMLRNGKEVAVKQLKEGSSQGEREFQAE 139
Query: 116 IDIISRVHHRHLVSLVGYCVSGGQRMLVYEFVPNKTLEYHLHGKGVPTMDWPTRMRIALG 175
+ IISRVHHRHLV+LVGYC++ QR+LVYEFVPN TLE+HLHGKG PTM+W +R++IA+G
Sbjct: 140 VGIISRVHHRHLVALVGYCIADAQRLLVYEFVPNNTLEFHLHGKGRPTMEWSSRLKIAVG 199
Query: 176 SARGLAYLHEDCSPRIIHRDIKAANVLIDDSFEAKVADFGLAKLTTDTNTHVSTRVMGTF 235
SA+GL+YLHE+C+P+IIHRDIKA+N+LID FEAKVADFGLAK+ +DTNTHVSTRVMGTF
Sbjct: 200 SAKGLSYLHENCNPKIIHRDIKASNILIDFKFEAKVADFGLAKIASDTNTHVSTRVMGTF 259
Query: 236 GYMAPEYASSGKLTEKSDVFSFGVMLLELLTGKRPLDLTNA-MDESLVDWARPLLSRALE 294
GY+APEYASSGKLTEKSDVFSFGV+LLEL+TG+RP+D+ N D SLVDWARPLL++ +
Sbjct: 260 GYLAPEYASSGKLTEKSDVFSFGVVLLELITGRRPIDVNNVHADNSLVDWARPLLNQ-VS 318
Query: 295 EDGNFAELVDPFLEGNYDHQEMIRLAACAASSIRHSAKKRSKMSQIVRALEGDVSLEDLK 354
E GNF +VD L YD +EM R+ ACAA+ +R +A +R +M Q+ R LEG++S DL
Sbjct: 319 ELGNFEVVVDKKLNNEYDKEEMARMVACAAACVRSTAPRRPRMDQVARVLEGNISPSDLN 378
Query: 355 ESMIKSPAPQTGVYTTTG--SEYDTMQYNADMAKFRKQIMSSQE 396
+ + + VY ++G ++YD+ Q N M KFRK + +Q+
Sbjct: 379 QGITPG---HSNVYGSSGGSTDYDSSQDNEGMNKFRKVGLETQD 419
>UniRef100_Q9C821 Protein kinase, putative; 60711-62822 [Arabidopsis thaliana]
Length = 509
Score = 456 bits (1173), Expect = e-127
Identities = 221/342 (64%), Positives = 280/342 (81%), Gaps = 7/342 (2%)
Query: 59 FTYEELASATKGFANENIIGQGGFGYVHKGILPTGKEIAVKSLKAGSGQGEREFQAEIDI 118
FTYE+L+ AT F+N N++GQGGFGYVH+G+L G +A+K LK+GSGQGEREFQAEI
Sbjct: 131 FTYEDLSKATSNFSNTNLLGQGGFGYVHRGVLVDGTLVAIKQLKSGSGQGEREFQAEIQT 190
Query: 119 ISRVHHRHLVSLVGYCVSGGQRMLVYEFVPNKTLEYHLHGKGVPTMDWPTRMRIALGSAR 178
ISRVHHRHLVSL+GYC++G QR+LVYEFVPNKTLE+HLH K P M+W RM+IALG+A+
Sbjct: 191 ISRVHHRHLVSLLGYCITGAQRLLVYEFVPNKTLEFHLHEKERPVMEWSKRMKIALGAAK 250
Query: 179 GLAYLHEDCSPRIIHRDIKAANVLIDDSFEAKVADFGLAKLTTDTNTHVSTRVMGTFGYM 238
GLAYLHEDC+P+ IHRD+KAAN+LIDDS+EAK+ADFGLA+ + DT+THVSTR+MGTFGY+
Sbjct: 251 GLAYLHEDCNPKTIHRDVKAANILIDDSYEAKLADFGLARSSLDTDTHVSTRIMGTFGYL 310
Query: 239 APEYASSGKLTEKSDVFSFGVMLLELLTGKRPLDLTN--AMDESLVDWARPLLSRALEED 296
APEYASSGKLTEKSDVFS GV+LLEL+TG+RP+D + A D+S+VDWA+PL+ +AL D
Sbjct: 311 APEYASSGKLTEKSDVFSIGVVLLELITGRRPVDKSQPFADDDSIVDWAKPLMIQAL-ND 369
Query: 297 GNFAELVDPFLEGNYDHQEMIRLAACAASSIRHSAKKRSKMSQIVRALEGDVSLEDLKES 356
GNF LVDP LE ++D EM R+ ACAA+S+RHSAK+R KMSQIVRA EG++S++DL E
Sbjct: 370 GNFDGLVDPRLENDFDINEMTRMVACAAASVRHSAKRRPKMSQIVRAFEGNISIDDLTEG 429
Query: 357 MIKSPAPQTGVYTTTG-SEYDTMQYNADMAKFRKQIMSSQEF 397
+ Q+ +Y+ G S+Y + QY D+ KF+K S+ F
Sbjct: 430 ---AAPGQSTIYSLDGSSDYSSTQYKEDLKKFKKMAFESKTF 468
>UniRef100_Q7XHF3 Hypothetical protein [Oryza sativa]
Length = 568
Score = 456 bits (1172), Expect = e-127
Identities = 233/331 (70%), Positives = 270/331 (81%), Gaps = 7/331 (2%)
Query: 27 SGEMSSNYSYGMGPPGSMQSSPGLSLTLKGGTFTYEELASATKGFANENIIGQGGFGYVH 86
SGEM YS G P SP ++L +F+YEELA+AT GF+ N++GQGGFGYV+
Sbjct: 189 SGEMGMGYSSGPYGPALPPPSPNVALGFSKSSFSYEELAAATSGFSAANLLGQGGFGYVY 248
Query: 87 KGILP-TGKEIAVKSLKAGSGQGEREFQAEIDIISRVHHRHLVSLVGYCVSGGQRMLVYE 145
KG+L GKE+AVK LK+GSGQGEREFQAE+DIISRVHHRHLVSLVGYC++ QRMLVYE
Sbjct: 249 KGVLAGNGKEVAVKQLKSGSGQGEREFQAEVDIISRVHHRHLVSLVGYCIAANQRMLVYE 308
Query: 146 FVPNKTLEYHLH--GKGVPTMDWPTRMRIALGSARGLAYLHEDCSPRIIHRDIKAANVLI 203
FVPN TLE+HL+ G G +DW R RIALGSA+GLAYLHEDC PRIIHRDIKAAN+L+
Sbjct: 309 FVPNGTLEHHLYRGGNGDRVLDWSARHRIALGSAKGLAYLHEDCHPRIIHRDIKAANILL 368
Query: 204 DDSFEAKVADFGLAKLTTDTNTHVSTRVMGTFGYMAPEYASSGKLTEKSDVFSFGVMLLE 263
D ++EA VADFGLAKLTTDTNTHVSTRVMGTFGY+APEYAS+GKLTEKSDVFSFGVMLLE
Sbjct: 369 DANYEAMVADFGLAKLTTDTNTHVSTRVMGTFGYLAPEYASTGKLTEKSDVFSFGVMLLE 428
Query: 264 LLTGKRPLDLTNAMDESLVDWARPLLSRAL----EEDGNFAELVDPFLEGNYDHQEMIRL 319
LLTG+RP+D +N M++SLVDWARP+L+R L EE G ELVD L G Y E+ R+
Sbjct: 429 LLTGRRPVDTSNYMEDSLVDWARPVLARLLVAGGEEGGLIRELVDSRLGGEYSAVEVERM 488
Query: 320 AACAASSIRHSAKKRSKMSQIVRALEGDVSL 350
AACAA+SIRHSA++R KMSQIVRALEGD SL
Sbjct: 489 AACAAASIRHSARQRPKMSQIVRALEGDASL 519
>UniRef100_Q9LV50 Protein kinase-like protein [Arabidopsis thaliana]
Length = 567
Score = 424 bits (1091), Expect = e-117
Identities = 203/310 (65%), Positives = 256/310 (82%), Gaps = 2/310 (0%)
Query: 49 GLSLTLKGGTFTYEELASATKGFANENIIGQGGFGYVHKGILPTGKEIAVKSLKAGSGQG 108
GL L + TFTY ELA AT F+ N++G+GGFG+V+KGIL G E+AVK LK GS QG
Sbjct: 249 GLVLGIHQSTFTYGELARATNKFSEANLLGEGGFGFVYKGILNNGNEVAVKQLKVGSAQG 308
Query: 109 EREFQAEIDIISRVHHRHLVSLVGYCVSGGQRMLVYEFVPNKTLEYHLHGKGVPTMDWPT 168
E+EFQAE++IIS++HHR+LVSLVGYC++G QR+LVYEFVPN TLE+HLHGKG PTM+W
Sbjct: 309 EKEFQAEVNIISQIHHRNLVSLVGYCIAGAQRLLVYEFVPNNTLEFHLHGKGRPTMEWSL 368
Query: 169 RMRIALGSARGLAYLHEDCSPRIIHRDIKAANVLIDDSFEAKVADFGLAKLTTDTNTHVS 228
R++IA+ S++GL+YLHE+C+P+IIHRDIKAAN+LID FEAKVADFGLAK+ DTNTHVS
Sbjct: 369 RLKIAVSSSKGLSYLHENCNPKIIHRDIKAANILIDFKFEAKVADFGLAKIALDTNTHVS 428
Query: 229 TRVMGTFGYMAPEYASSGKLTEKSDVFSFGVMLLELLTGKRPLDLTNA-MDESLVDWARP 287
TRVMGTFGY+APEYA+SGKLTEKSDV+SFGV+LLEL+TG+RP+D N D+SLVDWARP
Sbjct: 429 TRVMGTFGYLAPEYAASGKLTEKSDVYSFGVVLLELITGRRPVDANNVYADDSLVDWARP 488
Query: 288 LLSRALEEDGNFAELVDPFLEGNYDHQEMIRLAACAASSIRHSAKKRSKMSQIVRALEGD 347
LL +ALEE NF L D L YD +EM R+ ACAA+ +R++A++R +M Q+VR LEG+
Sbjct: 489 LLVQALEE-SNFEGLADIKLNNEYDREEMARMVACAAACVRYTARRRPRMDQVVRVLEGN 547
Query: 348 VSLEDLKESM 357
+S DL + +
Sbjct: 548 ISPSDLNQGI 557
>UniRef100_Q6ZE12 Hypothetical protein P0495H05.37 [Oryza sativa]
Length = 517
Score = 424 bits (1090), Expect = e-117
Identities = 213/376 (56%), Positives = 281/376 (74%), Gaps = 7/376 (1%)
Query: 21 PHPLMNSGEMSSNYSYGMGPPGSMQSSPGLSLTLKGGTFTYEELASATKGFANENIIGQG 80
P+ +M+ M+ + + PP P + L F Y+ELA+AT GF+ N++GQG
Sbjct: 105 PNMMMHPTNMTGPHVV-VRPPLVPPPPPPVPAGLDENAFGYDELAAATGGFSEGNMLGQG 163
Query: 81 GFGYVHKGILPTGKEIAVKSLKAGSGQGEREFQAEIDIISRVHHRHLVSLVGYCVSGGQR 140
GFGYV++G+L GKE+AVK L AG GQGEREFQAE+D+ISRVHHRHLV LVGYC++G QR
Sbjct: 164 GFGYVYRGVLGDGKEVAVKQLSAGGGQGEREFQAEVDMISRVHHRHLVPLVGYCIAGAQR 223
Query: 141 MLVYEFVPNKTLEYHLHGKGVPTMDWPTRMRIALGSARGLAYLHEDCSPRIIHRDIKAAN 200
+LVY+FVPN+TLE+HLH KG+P M W TR+RIA+GSA+GLAYLHE+C+PRIIHRDIK+AN
Sbjct: 224 LLVYDFVPNRTLEHHLHEKGLPVMKWTTRLRIAVGSAKGLAYLHEECNPRIIHRDIKSAN 283
Query: 201 VLIDDSFEAKVADFGLAKLTTDTNTHVSTRVMGTFGYMAPEYASSGKLTEKSDVFSFGVM 260
+L+D++FE VADFG+AKLT++ THVSTRVMGTFGY+APEYASSGKLT+KSDVFS+GVM
Sbjct: 284 ILLDNNFEPLVADFGMAKLTSENVTHVSTRVMGTFGYLAPEYASSGKLTDKSDVFSYGVM 343
Query: 261 LLELLTGKRPLDLTNAMDESLVDWARPLLSRALEE--DGNFAELVDPFLEGNYDHQEMIR 318
LLELLTG+RP D ++ + LVDWAR L RA+ G + ++VDP L G YD E R
Sbjct: 344 LLELLTGRRPADRSSYGADCLVDWARQALPRAMAAGGGGGYDDIVDPRLRGEYDRAEAAR 403
Query: 319 LAACAASSIRHSAKKRSKMSQIVRALEGDVSLEDLKESMIKSPAPQTGVYTTTG-SEYDT 377
+AACA + +RH+ ++R KMSQ+V+ LEGDVS E+L + + Q+ + +++G S +
Sbjct: 404 VAACAVACVRHAGRRRPKMSQVVKVLEGDVSPEELGDG---ARPGQSAMSSSSGDSSSGS 460
Query: 378 MQYNADMAKFRKQIMS 393
Y A M + R+ S
Sbjct: 461 GSYTAQMERVRRTAAS 476
>UniRef100_Q67W44 Hypothetical protein OJ1568_D07.15 [Oryza sativa]
Length = 748
Score = 420 bits (1079), Expect = e-116
Identities = 223/392 (56%), Positives = 286/392 (72%), Gaps = 30/392 (7%)
Query: 7 VQQNPNGASGVWGAPHPLMNSGEMSSNYSYGMGPPGSMQSSPGLSLTLKGGTFTYEELAS 66
+ N GAS G + NSG S G GP GS F+YEEL
Sbjct: 363 IMLNSGGASADGGGYY---NSGTFSGGE--GTGPAGSKSR------------FSYEELTG 405
Query: 67 ATKGFANENIIGQGGFGYVHKGILPTGKEIAVKSLKAGSGQGEREFQAEIDIISRVHHRH 126
T F+ +N+IG+GGFG V+KG L GK +AVK LKAGSGQGEREFQAE++IISRVHHRH
Sbjct: 406 ITSNFSRDNVIGEGGFGCVYKGWLSDGKCVAVKQLKAGSGQGEREFQAEVEIISRVHHRH 465
Query: 127 LVSLVGYCVSGGQRMLVYEFVPNKTLEYHLHGKGVPTMDWPTRMRIALGSARGLAYLHED 186
LVSLVGYC++ RML+YEFVPN TLE+HLHG+G+P MDWPTR+RIA+G+A+GLAYLHED
Sbjct: 466 LVSLVGYCIAAHHRMLIYEFVPNGTLEHHLHGRGMPVMDWPTRLRIAIGAAKGLAYLHED 525
Query: 187 CSPRIIHRDIKAANVLIDDSFEAKVADFGLAKLTTDTNTHVSTRVMGTFGYMAPEYASSG 246
C PRIIHRDIK AN+L+D S+EA+VADFGLAKL DT+THVSTR+MGTFGY+APEYASSG
Sbjct: 526 CHPRIIHRDIKTANILLDYSWEAQVADFGLAKLANDTHTHVSTRIMGTFGYLAPEYASSG 585
Query: 247 KLTEKSDVFSFGVMLLELLTGKRPLDLTNAM-DESLVDWARPLLSRALEEDGNFAELVDP 305
KLT++SDVFSFGV+LLEL+TG++P+D T + +ESLV+WARP+L+ A+ E G+ +ELVDP
Sbjct: 586 KLTDRSDVFSFGVVLLELITGRKPVDQTQPLGEESLVEWARPVLADAV-ETGDLSELVDP 644
Query: 306 FLEGNYDHQEMIRLAACAASSIRHSAKKRSKMSQIVRALEGDVSLEDLKESMIKSPAPQT 365
LEG Y+ EM+ + AA+ +RHSA KR +M Q++R L+ + S+ DL + Q+
Sbjct: 645 RLEGAYNRNEMMTMVEAAAACVRHSAPKRPRMVQVMRVLD-EGSMTDLSNGI---KVGQS 700
Query: 366 GVYTTTGSEYDTMQYNADMAKFRKQIMSSQEF 397
V+ T GS+ AD+ + R+ +S+EF
Sbjct: 701 QVF-TGGSD------AADIQQLRRIAFASEEF 725
>UniRef100_Q9CAL8 Hypothetical protein F24J13.3 [Arabidopsis thaliana]
Length = 710
Score = 415 bits (1067), Expect = e-114
Identities = 216/384 (56%), Positives = 276/384 (71%), Gaps = 14/384 (3%)
Query: 9 QNPN-GASGVWGAPHPLMNSGEMSSNYSYGMGPPGSMQSSPGLSLTLKGGT-FTYEELAS 66
QNP G SG G ++ S G G S+P ++ G T FTYEEL
Sbjct: 289 QNPTKGYSGPGGYNSQQQSNSGNSFGSQRGGGGYTRSGSAPDSAVMGSGQTHFTYEELTD 348
Query: 67 ATKGFANENIIGQGGFGYVHKGILPTGKEIAVKSLKAGSGQGEREFQAEIDIISRVHHRH 126
T+GF+ NI+G+GGFG V+KG L GK +AVK LK GSGQG+REF+AE++IISRVHHRH
Sbjct: 349 ITEGFSKHNILGEGGFGCVYKGKLNDGKLVAVKQLKVGSGQGDREFKAEVEIISRVHHRH 408
Query: 127 LVSLVGYCVSGGQRMLVYEFVPNKTLEYHLHGKGVPTMDWPTRMRIALGSARGLAYLHED 186
LVSLVGYC++ +R+L+YE+VPN+TLE+HLHGKG P ++W R+RIA+GSA+GLAYLHED
Sbjct: 409 LVSLVGYCIADSERLLIYEYVPNQTLEHHLHGKGRPVLEWARRVRIAIGSAKGLAYLHED 468
Query: 187 CSPRIIHRDIKAANVLIDDSFEAKVADFGLAKLTTDTNTHVSTRVMGTFGYMAPEYASSG 246
C P+IIHRDIK+AN+L+DD FEA+VADFGLAKL T THVSTRVMGTFGY+APEYA SG
Sbjct: 469 CHPKIIHRDIKSANILLDDEFEAQVADFGLAKLNDSTQTHVSTRVMGTFGYLAPEYAQSG 528
Query: 247 KLTEKSDVFSFGVMLLELLTGKRPLDLTNAM-DESLVDWARPLLSRALEEDGNFAELVDP 305
KLT++SDVFSFGV+LLEL+TG++P+D + +ESLV+WARPLL +A+ E G+F+ELVD
Sbjct: 529 KLTDRSDVFSFGVVLLELITGRKPVDQYQPLGEESLVEWARPLLHKAI-ETGDFSELVDR 587
Query: 306 FLEGNYDHQEMIRLAACAASSIRHSAKKRSKMSQIVRALEGDVSLEDLKESMIKSPAPQT 365
LE +Y E+ R+ AA+ +RHS KR +M Q+VRAL+ + + D+
Sbjct: 588 RLEKHYVENEVFRMIETAAACVRHSGPKRPRMVQVVRALDSEGDMGDI----------SN 637
Query: 366 GVYTTTGSEYDTMQYNADMAKFRK 389
G S YD+ QYN D KFRK
Sbjct: 638 GNKVGQSSAYDSGQYNNDTMKFRK 661
>UniRef100_Q9ASK4 Putative LRR receptor-like protein kinase [Oryza sativa]
Length = 698
Score = 408 bits (1049), Expect = e-112
Identities = 208/368 (56%), Positives = 270/368 (72%), Gaps = 15/368 (4%)
Query: 31 SSNYSYGMGPPGSMQSSPGLSLTLKGGTFTYEELASATKGFANENIIGQGGFGYVHKGIL 90
S +Y G P ++ S+ FTYEEL T GFA +N++G+GGFG V+KG L
Sbjct: 321 SGKTNYSAGSPDYKETMSEFSMG-NCRFFTYEELHQITNGFAAKNLLGEGGFGSVYKGCL 379
Query: 91 PTGKEIAVKSLKAGSGQGEREFQAEIDIISRVHHRHLVSLVGYCVSGGQRMLVYEFVPNK 150
G+E+AVK LK G GQGEREFQAE++IISRVHHRHLVSLVGYC+SG QR+LVY+FVPN
Sbjct: 380 ADGREVAVKKLKGGGGQGEREFQAEVEIISRVHHRHLVSLVGYCISGDQRLLVYDFVPND 439
Query: 151 TLEYHLHGKGVPTMDWPTRMRIALGSARGLAYLHEDCSPRIIHRDIKAANVLIDDSFEAK 210
TL +HLHG+G+P ++W R++IA GSARG+AYLHEDC PRIIHRDIK++N+L+D++FEA+
Sbjct: 440 TLHHHLHGRGMPVLEWSARVKIAAGSARGIAYLHEDCHPRIIHRDIKSSNILLDNNFEAQ 499
Query: 211 VADFGLAKLTTDTNTHVSTRVMGTFGYMAPEYASSGKLTEKSDVFSFGVMLLELLTGKRP 270
VADFGLA+L D THV+TRVMGTFGY+APEYASSGKLTE+SDVFSFGV+LLEL+TG++P
Sbjct: 500 VADFGLARLAMDAVTHVTTRVMGTFGYLAPEYASSGKLTERSDVFSFGVVLLELITGRKP 559
Query: 271 LDLTNAM-DESLVDWARPLLSRALEEDGNFAELVDPFLEGNYDHQEMIRLAACAASSIRH 329
+D + + DESLV+WARPLL+ A+ E GN EL+D L+ N++ EM R+ AA+ IRH
Sbjct: 560 VDASKPLGDESLVEWARPLLTEAI-ETGNVGELIDSRLDKNFNEAEMFRMIEAAAACIRH 618
Query: 330 SAKKRSKMSQIVRALEGDVSLEDLKESMIKSPAPQTGVYTTTGSEYDTMQYNADMAKFRK 389
SA +R +MSQ+VR L+ SL D+ S P SE + A++ F++
Sbjct: 619 SASRRPRMSQVVRVLD---SLADVDLSNGIQPGK---------SEMFNVANTAEIRLFQR 666
Query: 390 QIMSSQEF 397
SQ+F
Sbjct: 667 MAFGSQDF 674
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.316 0.133 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 666,762,112
Number of Sequences: 2790947
Number of extensions: 29158363
Number of successful extensions: 113103
Number of sequences better than 10.0: 18296
Number of HSP's better than 10.0 without gapping: 10320
Number of HSP's successfully gapped in prelim test: 7978
Number of HSP's that attempted gapping in prelim test: 78518
Number of HSP's gapped (non-prelim): 20311
length of query: 398
length of database: 848,049,833
effective HSP length: 129
effective length of query: 269
effective length of database: 488,017,670
effective search space: 131276753230
effective search space used: 131276753230
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 76 (33.9 bits)
Medicago: description of AC148484.8


