

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148484.5 - phase: 0
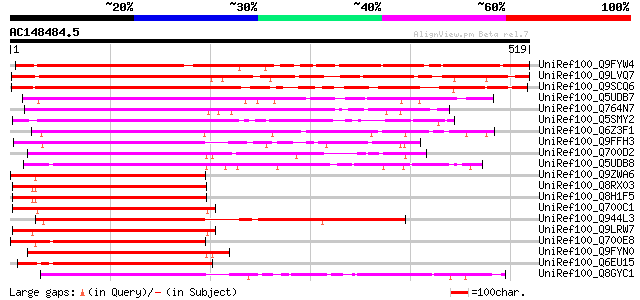
(519 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9FYW4 BAC19.12 [Lycopersicon esculentum] 575 e-162
UniRef100_Q9LVQ7 Zinc finger protein [Arabidopsis thaliana] 541 e-152
UniRef100_Q9SCQ6 Zinc finger protein [Arabidopsis thaliana] 533 e-150
UniRef100_Q5UDB7 INDETERMINATE-related protein 7 [Zea mays] 343 5e-93
UniRef100_Q764N7 Zinc finger protein [Malus domestica] 340 8e-92
UniRef100_Q5SMY2 Putative zinc finger protein ID1 [Oryza sativa] 338 2e-91
UniRef100_Q6Z3F1 Putative zinc finger protein [Oryza sativa] 337 5e-91
UniRef100_Q9FFH3 Similarity to zinc finger protein ID1 [Arabidop... 333 5e-90
UniRef100_Q700D2 Hypothetical protein [Arabidopsis thaliana] 333 7e-90
UniRef100_Q5UDB8 INDETERMINATE-related protein 10 [Zea mays] 331 4e-89
UniRef100_Q9ZWA6 F11M21.23 protein [Arabidopsis thaliana] 326 1e-87
UniRef100_Q8RX03 Putative zinc finger protein [Arabidopsis thali... 321 4e-86
UniRef100_Q8H1F5 Putative zinc finger protein [Arabidopsis thali... 321 4e-86
UniRef100_Q700C1 Hypothetical protein [Arabidopsis thaliana] 320 6e-86
UniRef100_Q944L3 AT3g45260/F18N11_20 [Arabidopsis thaliana] 319 1e-85
UniRef100_Q9LRW7 Similarity to zinc finger protein [Arabidopsis ... 318 2e-85
UniRef100_Q700E8 Hypothetical protein [Arabidopsis thaliana] 317 5e-85
UniRef100_Q9FYN0 Similarity to C2H2-type zinc finger protein [Ar... 313 6e-84
UniRef100_Q6EU15 Finger protein pcp1-like [Oryza sativa] 313 6e-84
UniRef100_Q8GYC1 Putative C2H2-type zinc finger protein [Arabido... 313 1e-83
>UniRef100_Q9FYW4 BAC19.12 [Lycopersicon esculentum]
Length = 519
Score = 575 bits (1481), Expect = e-162
Identities = 324/541 (59%), Positives = 380/541 (69%), Gaps = 60/541 (11%)
Query: 6 NVSTASGEASISSSGNNNIQSPIPKPTKKKRNLPGMPDPEAEVIALSPTTLLATNRFVCE 65
N ST SGEAS+SSSGN + P+ + KKKRNLPGMPDP+AEVIALSPTTLLATNRFVCE
Sbjct: 12 NDSTGSGEASVSSSGNQVV--PLKESAKKKRNLPGMPDPDAEVIALSPTTLLATNRFVCE 69
Query: 66 ICNKGFQRDQNLQLHRRGHNLPWKLRQRSSKEIRKRVYVCPEPTCVHHDPSRALGDLTGI 125
IC+KGFQRDQNLQLHRRGHNLPWKLRQRSS E++KRVYVCPE +CVHHDPSRALGDLTGI
Sbjct: 70 ICSKGFQRDQNLQLHRRGHNLPWKLRQRSSNEVKKRVYVCPESSCVHHDPSRALGDLTGI 129
Query: 126 KKHFCRKHGEKKWKCEKCSKKYAVQSDWKAHSKVCGSREYKCDCGTVFSRRDSFITHRAF 185
KKHFCRKHGEKKWKC+KCSKKYAVQSD KAHSK+CG+REYKCDCGT+FS RAF
Sbjct: 130 KKHFCRKHGEKKWKCDKCSKKYAVQSDLKAHSKICGTREYKCDCGTLFS--------RAF 181
Query: 186 CDALAEENAKSQNQAVGKANSESDSKVLTGDSLPAVITTTAAA----------------- 228
CDALA+E+AK+ + N E ++ + S P A
Sbjct: 182 CDALAQESAKTLPEKPPSTNEEPKTQAVASSSPPPSPPAPPPATESQPPPPPPAAPKSLM 241
Query: 229 AATTPQSNSGVSSALETQKLDLPENP---------PQIIEEPQVVVTTTASALNASCSSS 279
T P S + +S A Q +L ENP Q++EE VV ++L +CSSS
Sbjct: 242 TPTVPPSTAVMSFASSVQNRELRENPVTNSAGAATKQVVEEAAVV-----ASLTGNCSSS 296
Query: 280 TSSKSNGCAATSTGVFASLFASSTASASASLQPQAPAFSDLIRSMGCTDPRPTDFSAPPS 339
S+ S +S+ VF SLFASST AS SL QAP FSD+ R+M P T APPS
Sbjct: 297 CSNGS-----SSSNVFGSLFASST--ASGSLPSQAPVFSDIFRAMA---PEHTLEMAPPS 346
Query: 340 S-EAISLCLSTSHGSSIFGTGGQECRQYVPTHQPPAMSATALLQKAAQMGAAATNASLLR 398
S E ISL L+ SH SSIF GQE RQY P Q PAMSATALLQKAAQMGAAAT++S LR
Sbjct: 347 STEPISLGLAMSHSSSIFRPAGQERRQYAPAPQ-PAMSATALLQKAAQMGAAATSSSFLR 405
Query: 399 GLGIVSSSASTSSGQQDSLHWGLGPMEPEGSSLVSAGLGLGLPCDADSGLKELMLGTPSM 458
G+G++SS++S++ Q+ W P + G+SL +AGLGLGLPCDA SGLKELMLGTPS+
Sbjct: 406 GIGVMSSTSSSNGHQE----WSGRPSDANGASL-AAGLGLGLPCDAGSGLKELMLGTPSV 460
Query: 459 FGPKQTTLDFLGLGMAAGGSAGGGLSALITSIGGGSGLDVTAAAASFGNGEFSGKDIGRS 518
FGPK TLD LGLGMAA GLSAL+TS+ GS LD+ +A SFG+ +FSGKD+GR+
Sbjct: 461 FGPKHPTLDLLGLGMAASVGPSPGLSALLTSM--GSNLDMVTSAGSFGSADFSGKDLGRN 518
Query: 519 S 519
S
Sbjct: 519 S 519
>UniRef100_Q9LVQ7 Zinc finger protein [Arabidopsis thaliana]
Length = 500
Score = 541 bits (1395), Expect = e-152
Identities = 313/545 (57%), Positives = 371/545 (67%), Gaps = 74/545 (13%)
Query: 2 VDLDNVSTASGEASISSSGNNNIQSPIPKPT-KKKRNLPGMPDPEAEVIALSPTTLLATN 60
VDLDN ST SG+AS+SS+GN N+ PK KKKRNLPGMPDP+AEVIALSP TL+ATN
Sbjct: 3 VDLDNSSTVSGDASVSSTGNQNLT---PKSVGKKKRNLPGMPDPDAEVIALSPKTLMATN 59
Query: 61 RFVCEICNKGFQRDQNLQLHRRGHNLPWKLRQRSSKEIRKRVYVCPEPTCVHHDPSRALG 120
RFVCEICNKGFQRDQNLQLHRRGHNLPWKLRQRS+KE+RK+VYVCP CVHHDPSRALG
Sbjct: 60 RFVCEICNKGFQRDQNLQLHRRGHNLPWKLRQRSTKEVRKKVYVCPVSGCVHHDPSRALG 119
Query: 121 DLTGIKKHFCRKHGEKKWKCEKCSKKYAVQSDWKAHSKVCGSREYKCDCGTVFSRRDSFI 180
DLTGIKKHFCRKHGEKKWKCEKCSKKYAVQSDWKAHSK+CG++EYKCDCGT+FSRRDSFI
Sbjct: 120 DLTGIKKHFCRKHGEKKWKCEKCSKKYAVQSDWKAHSKICGTKEYKCDCGTLFSRRDSFI 179
Query: 181 THRAFCDALAEENAKSQNQA-------VGKANSESDSK---------VLTGDSLPAVITT 224
THRAFCDALAEE+AK+ Q+ V + N E + K L S P+V
Sbjct: 180 THRAFCDALAEESAKNHTQSKKLYPETVTRKNPEIEQKSPAAVESSPSLPPSSPPSVAIA 239
Query: 225 TAAAAATTPQSNSGVSSALETQKLDLP-ENPPQIIE---EPQVVVTTTASALNASCSSST 280
A A + +S +SS++ LP +N P+ E P+V++ + + + SSS
Sbjct: 240 PAPAISVETESVKIISSSV------LPIQNSPESQENNNHPEVIIEEASRTIGFNVSSSD 293
Query: 281 SSKSNGCAATSTGVFASLFASSTASASASLQPQAPAFSDLIRSMGCTDPRPTDFSAPPSS 340
S + + + G +A LF SSTAS S A P+ F+ S
Sbjct: 294 LSNDH---SNNNGGYAGLFVSSTASPSLYASSTA---------------SPSLFAPSSSM 335
Query: 341 EAISLCLSTSHGSSIFGTGGQECRQYV-PTHQPPAMSATALLQKAAQMGAAATNASLLRG 399
E ISLCLST+ S+FG ++ ++ P PAMSATALLQKAAQMG+ + SLLRG
Sbjct: 336 EPISLCLSTN--PSLFGPTIRDPPHFLTPLPPQPAMSATALLQKAAQMGSTGSGGSLLRG 393
Query: 400 LGIVSSSASTSSGQQDSLHWGLGPMEPEGSSLVSAGLGLGLPCD---ADSGLKELMLGTP 456
LGIVS +TSS + S H L ++ GLGLGLPC + SGLKELM+G
Sbjct: 394 LGIVS---TTSSSMELSNHDALS---------LAPGLGLGLPCSSGGSGSGLKELMMGNS 441
Query: 457 SMFGPKQTTLDFLGLGMAA--GGSAGGGLSALITSIGGGSGLDVTAAAASFGNGEFSGKD 514
S+FGPKQTTLDFLGLG A GG+ GGGLSAL+TSIGGG G+D+ FG+GEFSGKD
Sbjct: 442 SVFGPKQTTLDFLGLGRAVGNGGNTGGGLSALLTSIGGGGGIDL------FGSGEFSGKD 495
Query: 515 IGRSS 519
IGRSS
Sbjct: 496 IGRSS 500
>UniRef100_Q9SCQ6 Zinc finger protein [Arabidopsis thaliana]
Length = 452
Score = 533 bits (1373), Expect = e-150
Identities = 299/520 (57%), Positives = 356/520 (67%), Gaps = 77/520 (14%)
Query: 2 VDLDNVSTASGEASISSSGNNNIQSPIPKPT-KKKRNLPGMPDPEAEVIALSPTTLLATN 60
VDLDN ST SGEAS+S S N Q+P+P T KKKRNLPGMPDPE+EVIALSP TLLATN
Sbjct: 3 VDLDNSSTVSGEASVSISSTGN-QNPLPNSTGKKKRNLPGMPDPESEVIALSPKTLLATN 61
Query: 61 RFVCEICNKGFQRDQNLQLHRRGHNLPWKLRQRSSKEIRKRVYVCPEPTCVHHDPSRALG 120
RFVCEICNKGFQRDQNLQLHRRGHNLPWKLRQ+S+KE++K+VYVCPE +CVHHDPSRALG
Sbjct: 62 RFVCEICNKGFQRDQNLQLHRRGHNLPWKLRQKSNKEVKKKVYVCPEVSCVHHDPSRALG 121
Query: 121 DLTGIKKHFCRKHGEKKWKCEKCSKKYAVQSDWKAHSKVCGSREYKCDCGTVFSRRDSFI 180
DLTGIKKHFCRKHGEKKWKC+KCSKKYAVQSDWKAHSK+CG++EYKCDCGT+FSRRDSFI
Sbjct: 122 DLTGIKKHFCRKHGEKKWKCDKCSKKYAVQSDWKAHSKICGTKEYKCDCGTLFSRRDSFI 181
Query: 181 THRAFCDALAEENAKSQNQAVGKANSE-SDSKVLTGDSLPAVITTTAAAAATTPQSNSGV 239
THRAFCDALAEENA+S + K N E K + +PA + T +A +
Sbjct: 182 THRAFCDALAEENARSHHSQSKKQNPEILTRKNPVPNPVPAPVDTESAKIKS-------- 233
Query: 240 SSALETQKLDLPENPPQIIEEPQVVVTTTASALNASCSSSTSSKSNGCAATSTGVFASLF 299
SS L ++ + P+ PP+I++E ++LN TS GVFA LF
Sbjct: 234 SSTLTIKQSESPKTPPEIVQE-----APKPTSLN--------------VVTSNGVFAGLF 274
Query: 300 ASSTASASASLQPQAPAFSDLIRSMGCTDPRPTDFSAPPSSEAISLCLSTSHGSSIFGTG 359
SS+AS P S +S+ F++ S E ISL LSTSHGSS G+
Sbjct: 275 ESSSAS------PSIYTTSSSSKSL---------FASSSSIEPISLGLSTSHGSSFLGSN 319
Query: 360 GQECRQYVPTHQPPAMSATALLQKAAQMGAAATNASLLRGLGIVSSSASTSSGQQDSLHW 419
H PAMSATALLQKAAQMGAA++ SLL GLGIVSS++++
Sbjct: 320 --------RFHAQPAMSATALLQKAAQMGAASSGGSLLHGLGIVSSTSTSI--------- 362
Query: 420 GLGPMEPEGSSLVSAGLGLGLPC--DADSGLKELMLGTPSMFGPKQTTLDFLGLGMAAGG 477
++V GLGLGLPC ++ SGLKELM+G S+FGPKQTTLDFLGLG A G
Sbjct: 363 ---------DAIVPHGLGLGLPCGGESSSGLKELMMGNSSVFGPKQTTLDFLGLGRAV-G 412
Query: 478 SAGGGLSALITSIGGGSGLDVTAAAASFGNGEFSGKDIGR 517
+ G + L T +GGG+G+D+ A +FG+GEFSGKDI R
Sbjct: 413 NGNGPSNGLSTLVGGGTGIDM---ATTFGSGEFSGKDISR 449
>UniRef100_Q5UDB7 INDETERMINATE-related protein 7 [Zea mays]
Length = 518
Score = 343 bits (881), Expect = 5e-93
Identities = 221/523 (42%), Positives = 279/523 (53%), Gaps = 73/523 (13%)
Query: 13 EASISSSGNNNIQSP---IPKPTKKKRNLPGMPDPEAEVIALSPTTLLATNRFVCEICNK 69
+ S SS N+ +P + P KKKRN PG P+P+AEVIALSP TLLATNRFVCE+CNK
Sbjct: 5 QGSSSSLQQNSTSAPGAAVVPPPKKKRNQPGNPNPDAEVIALSPRTLLATNRFVCEVCNK 64
Query: 70 GFQRDQNLQLHRRGHNLPWKLRQRSSKEIRKRVYVCPEPTCVHHDPSRALGDLTGIKKHF 129
GFQR+QNLQLHRRGHNLPWKL+Q++ +E R+RVY+CPEPTCVHHDPSRALGDLTGIKKH+
Sbjct: 65 GFQREQNLQLHRRGHNLPWKLKQKNPREARRRVYLCPEPTCVHHDPSRALGDLTGIKKHY 124
Query: 130 CRKHGEKKWKCEKCSKKYAVQSDWKAHSKVCGSREYKCDCGTVFSRRDSFITHRAFCDAL 189
CRKHGEKKWKC+KCSK+YAVQSDWKAHSK CG+REY+CDCGT+FSRRDSFITHRAFCDAL
Sbjct: 125 CRKHGEKKWKCDKCSKRYAVQSDWKAHSKTCGTREYRCDCGTLFSRRDSFITHRAFCDAL 184
Query: 190 AEENAKSQNQAVGKANSESDSKVLTGDSLPAVITTTAAAAATTPQ------SNSGVSSAL 243
A E+A+ G + +++AA PQ S+SG SS
Sbjct: 185 ARESAQMPPLGAGLYVGPGSMSLGLSQIHGFADQAQSSSAAAAPQFDHIMPSSSGSSSMF 244
Query: 244 ETQ------------------KLDLPENPPQIIEEPQV--------VVTTTASALNASCS 277
+Q D E+ Q + P + ++ + S
Sbjct: 245 RSQASASSPSYFLGGSAPPAAAQDFSEDGSQGSQGPLLHGKAHFHGLMMQLPEQQHQPGS 304
Query: 278 SSTSSKSNGCAATSTGVFASLFASSTASASASLQPQAPAFSDLIRSMGCTDPRPTDFSAP 337
S+ + +NG + G F+ A ++ S SLQ A D G S
Sbjct: 305 SNAAVGANGSNILNLGFFS---AGNSGGTSGSLQDARIAIQDQFNLSG---------SGS 352
Query: 338 PSSEAISLCLSTSHGSSIFGTGGQECRQYVPTHQPPAMSATALLQKAAQMGAA------- 390
S+E + + S GS + G G P+ SATALL KAAQMG+
Sbjct: 353 GSAEHGNNVMVASIGSHL-GRGFPSLYSSSPSAGMAQNSATALLMKAAQMGSTTSSTTHN 411
Query: 391 -----ATNASLLRGLGIVSSSAS----TSSGQQDSLHWGLGPMEPEGSSLVSAGL-GLGL 440
T ++LLR G ++ S T+ G+ + G E L+ L G G
Sbjct: 412 NHNGPTTTSTLLRATGFSGATGSGQLGTTGGRAAAGEEGAASHEAHFHELIMNSLAGGGF 471
Query: 441 PCDADSGLKELMLGTPSMFGPKQTTLDFLGLGMAAGGSAGGGL 483
A G + K +T DFLG+G A A GL
Sbjct: 472 SGTAGFG--------GGVDDGKLSTRDFLGVGRGAATMAPPGL 506
>UniRef100_Q764N7 Zinc finger protein [Malus domestica]
Length = 522
Score = 340 bits (871), Expect = 8e-92
Identities = 208/448 (46%), Positives = 257/448 (56%), Gaps = 40/448 (8%)
Query: 15 SISSSGNNNIQ-SPIPKPTKKKRNLPGMPDPEAEVIALSPTTLLATNRFVCEICNKGFQR 73
+I+S+G + Q P P KKKRNLPG PDP AEVIALSP TL+ATNRFVCEIC KGFQR
Sbjct: 49 NINSNGISTTQVQKQPPPAKKKRNLPGTPDPTAEVIALSPKTLMATNRFVCEICKKGFQR 108
Query: 74 DQNLQLHRRGHNLPWKLRQRSSKEIRKRVYVCPEPTCVHHDPSRALGDLTGIKKHFCRKH 133
DQNLQLHRRGHNLPWKL+QR+S EI KRVY+CPE +CVHHDPSRALGDLTGIKKHF RKH
Sbjct: 109 DQNLQLHRRGHNLPWKLKQRTSTEIIKRVYICPESSCVHHDPSRALGDLTGIKKHFFRKH 168
Query: 134 GEKKWKCEKCSKKYAVQSDWKAHSKVCGSREYKCDCGTVFSRRDSFITHRAFCDALAEEN 193
GEK WKC+KCSKKYAVQSDWKAH K CG+REYKCDCGT+FSRRDSFITHRAFCDA+AEEN
Sbjct: 169 GEKTWKCDKCSKKYAVQSDWKAHLKTCGTREYKCDCGTIFSRRDSFITHRAFCDAIAEEN 228
Query: 194 AKSQ------NQAVGKANSE--SDSKVLTGDSLPA--VITTTAAAAATTPQSNSGVSSAL 243
++Q N +G N S+S+++ S P IT ++A + ++
Sbjct: 229 NRNQGVVPMSNNIMGAPNQGQLSNSELIIPASPPMNNKITDNPSSARSDTTTSDQFHHNF 288
Query: 244 ETQKLDLPENPPQIIEEPQVVVTTTASALNASCSSSTSSKSNGCAATSTGVFASLFASST 303
+ + L P Q P V L+ + SSSTS S G SL +S+
Sbjct: 289 DAKNAPLTLEPQQPFPMPTKPVNMLPRTLSNNTSSSTSPSSLLFGLNHGGHDQSLIPNSS 348
Query: 304 ASASASLQPQAPAFSDLIRSMGCTDPRPTDFSAPPSSEAISLCLSTSHGSSIFGTGGQEC 363
SA+ Q A S G P P PS I+ +++G++ TGG
Sbjct: 349 GHLSATQLLQKAAQMGATMSGG---PNPN-----PSGTTITSMAPSTYGTA---TGGYNM 397
Query: 364 RQYVPTHQPPAM-----SATALLQKAAQMGA-------AATNASLLRGLGIVSSSASTSS 411
++ P + S AQMG A N LL+ + ++ S
Sbjct: 398 NPFMQQRDPNNILQLETSPQFFGANYAQMGMFSGMLFDHAQNNGLLKNMEHENNGTSNKI 457
Query: 412 GQQDSLHWGLGPMEPEGSSLVSAGLGLG 439
G + + + G G +L LGLG
Sbjct: 458 GFRGASNVG------NGDTLTVDFLGLG 479
>UniRef100_Q5SMY2 Putative zinc finger protein ID1 [Oryza sativa]
Length = 487
Score = 338 bits (867), Expect = 2e-91
Identities = 207/447 (46%), Positives = 256/447 (56%), Gaps = 64/447 (14%)
Query: 3 DLDNVSTASGE-ASISSSGNNNIQSPIPKPTKKKRNLPGMPDPEAEVIALSPTTLLATNR 61
++ N+++ASG+ AS+SS P P P KKKR+LPG PDPEAEVIALSP TL+ATNR
Sbjct: 21 NMSNLTSASGDQASVSSH-------PAPPPAKKKRSLPGNPDPEAEVIALSPRTLMATNR 73
Query: 62 FVCEICNKGFQRDQNLQLHRRGHNLPWKLRQRSSKEI-RKRVYVCPEPTCVHHDPSRALG 120
+VCEIC KGFQRDQNLQLHRRGHNLPWKL+QR+ KE+ RK+VYVCPE CVHHDP+RALG
Sbjct: 74 YVCEICGKGFQRDQNLQLHRRGHNLPWKLKQRNPKEVVRKKVYVCPEAGCVHHDPARALG 133
Query: 121 DLTGIKKHFCRKHGEKKWKCEKCSKKYAVQSDWKAHSKVCGSREYKCDCGTVFSRRDSFI 180
DLTGIKKHF RKHGEKKWKC+KCSK+YAV SDWKAHSKVCG+REY+CDCGT+FSRRDSFI
Sbjct: 134 DLTGIKKHFSRKHGEKKWKCDKCSKRYAVHSDWKAHSKVCGTREYRCDCGTLFSRRDSFI 193
Query: 181 THRAFCDALAEENAKSQNQAVGKANSESDSKVLTGDSLPAVITTTAAAAATTPQSNSGVS 240
THRAFCDALAEE+A++ A A + + + V+ AA A G+
Sbjct: 194 THRAFCDALAEESARAVTAAAAVAGQQQHGGGMLFSQVADVLDHQAAMA----MGGHGL- 248
Query: 241 SALETQKLDLPENPPQIIEEPQVVVTTTASALNASCSSSTSSKSNGCAATSTGVFASLFA 300
Q+L L Q ++ Q + + + + N A + F
Sbjct: 249 ----MQELCLKREQQQ--QQQQFAPSWLTAQQQQQQLEAMAGAGNPAAMYGSARLDQEFI 302
Query: 301 SSTASASASLQPQAPAFSDLIRSMGCTDPRPTDFSAPPSSEAISLCLSTSHGSSIFGTGG 360
S+ S Q +F S SAPP A S++H
Sbjct: 303 GSSTPESGGAQQAGLSFGFSSTS-----------SAPPHPAA-----SSAH--------- 337
Query: 361 QECRQYVPTHQPPAMSATALLQKAAQMGAAATNASLLRGLGIVSSSASTSSGQQDSLHWG 420
MSATALLQKAAQMGA + S + +++AST + +
Sbjct: 338 --------------MSATALLQKAAQMGATLSRPSSHAHMA-AAAAASTHNSSSSAATTN 382
Query: 421 LGPMEPE---GSSLVSAGLGLGLPCDA 444
P P S+ V AG G GL +A
Sbjct: 383 APPPPPTSNVSSTCVGAG-GYGLAFEA 408
>UniRef100_Q6Z3F1 Putative zinc finger protein [Oryza sativa]
Length = 548
Score = 337 bits (864), Expect = 5e-91
Identities = 220/527 (41%), Positives = 272/527 (50%), Gaps = 78/527 (14%)
Query: 22 NNIQSPIPK--PTKKKRNLPGMPDPEAEVIALSPTTLLATNRFVCEICNKGFQRDQNLQL 79
NN +P+ P KKKRN PG P+P+AEV+ALSP TLLATNRFVCE+CNKGFQR+QNLQL
Sbjct: 28 NNAAAPVAATPPPKKKRNQPGNPNPDAEVVALSPHTLLATNRFVCEVCNKGFQREQNLQL 87
Query: 80 HRRGHNLPWKLRQRSSKEIRKRVYVCPEPTCVHHDPSRALGDLTGIKKHFCRKHGEKKWK 139
HRRGHNLPWKL+Q++ KE R+RVY+CPEP+CVHHDPSRALGDLTGIKKH+ RKHGEKKWK
Sbjct: 88 HRRGHNLPWKLKQKNPKETRRRVYLCPEPSCVHHDPSRALGDLTGIKKHYSRKHGEKKWK 147
Query: 140 CEKCSKKYAVQSDWKAHSKVCGSREYKCDCGTVFSRRDSFITHRAFCDALAEEN------ 193
C+KC+K+YAVQSDWKAHSK CG+REY+CDCGT+FSRRDSFITHRAFCDALA+E+
Sbjct: 148 CDKCNKRYAVQSDWKAHSKTCGTREYRCDCGTLFSRRDSFITHRAFCDALAQESGRIMPP 207
Query: 194 -------AKSQNQAVGKANSESDSKVLTGDSLPAVITTTAAAAATTPQSNSGVSSALETQ 246
A A+G + S L + TTAA AA SSA
Sbjct: 208 MGAALYAAAGAGMAIGGLTGMAASHQLQPFQDHSSAITTAANAAAQFDHLMATSSAAAGS 267
Query: 247 KLDLPENPPQIIEEP------------QVVVTTTASALNASCSSSTSSKSNGCAATSTGV 294
P P + A + SNG +
Sbjct: 268 PAFRAAQPTSSSSSPFYLGGGGDDGQAHTSLLHGKPAFHGLMQLPEQQGSNGGGLLN--- 324
Query: 295 FASLFASSTASASASLQPQAPAFSDLIRSMGCTDPRPTDFSAPPSSEAISLCLSTSHGSS 354
S F+ Q F D + + +S + S G+
Sbjct: 325 -LSYFSGGNGGHHHHHQEGRLVFPDQFNGVAAGNGARAGSGEHGNSGNNADSGSIFSGNM 383
Query: 355 IFGTGG-----QECRQYVPTHQPPAMSATALLQKAAQMGAAATNA------SLLRGLGIV 403
+ G GG Q VP PP MSATALLQKAAQMGA ++ SLLRGLG
Sbjct: 384 MGGGGGFSSLYSSSDQTVP---PPQMSATALLQKAAQMGATTSSGGAGSVNSLLRGLGSG 440
Query: 404 SSSASTSSGQQDSLHWGLGPMEPEGSSLVSAGLGLGLPCDADSGLKELMLGT-------- 455
+ + + + M E SS +A + +S L+ELM+ T
Sbjct: 441 GGGGALNGKPAGAAGF---IMSGESSSRSTA----SQTAENESQLRELMMNTLSATGGGT 493
Query: 456 -----------PSMFGPKQTTLDFLGLGMAA-------GGSAGGGLS 484
P + K +T DFLG+ A GG+AG G++
Sbjct: 494 GAGTVFVGGGFPGVDDGKLSTRDFLGVSGGAPGLQLRHGGAAGMGMA 540
>UniRef100_Q9FFH3 Similarity to zinc finger protein ID1 [Arabidopsis thaliana]
Length = 466
Score = 333 bits (855), Expect = 5e-90
Identities = 207/422 (49%), Positives = 246/422 (58%), Gaps = 64/422 (15%)
Query: 4 LDNVSTASGEASISSSGNNNIQSPIPKP--TKKKRNLPGMPDPEAEVIALSPTTLLATNR 61
L +S+ SG A SS + + P KKKRNLPG PDPEAEVIALSPTTL+ATNR
Sbjct: 6 LQTISSGSGFAQPQSSSTLDHDESLINPPLVKKKRNLPGNPDPEAEVIALSPTTLMATNR 65
Query: 62 FVCEICNKGFQRDQNLQLHRRGHNLPWKLRQRSSKEIRKRVYVCPEPTCVHHDPSRALGD 121
F+CE+C KGFQRDQNLQLHRRGHNLPWKL+QR+SKE+RKRVYVCPE TCVHH SRALGD
Sbjct: 66 FLCEVCGKGFQRDQNLQLHRRGHNLPWKLKQRTSKEVRKRVYVCPEKTCVHHHSSRALGD 125
Query: 122 LTGIKKHFCRKHGEKKWKCEKCSKKYAVQSDWKAHSKVCGSREYKCDCGTVFSRRDSFIT 181
LTGIKKHFCRKHGEKKW CEKC+K+YAVQSDWKAHSK CG+REY+CDCGT+FSRRDSFIT
Sbjct: 126 LTGIKKHFCRKHGEKKWTCEKCAKRYAVQSDWKAHSKTCGTREYRCDCGTIFSRRDSFIT 185
Query: 182 HRAFCDALAEENAKSQNQAVGKANSESDSKVLTGDSLPAVITTTAAAAATTPQSNSGVSS 241
HRAFCDALAEE AK + AV AAA P S +
Sbjct: 186 HRAFCDALAEETAK----------------------INAVSHLNGLAAAGAPGSVN---- 219
Query: 242 ALETQKLDLPENPP--QIIEEPQVVVTTTASALNASCSSSTSSKSNGCAATSTGVFASLF 299
L Q L PP + +PQ T + + TSS SL+
Sbjct: 220 -LNYQYLMGTFIPPLQPFVPQPQ----TNPNHHHQHFQPPTSSS------------LSLW 262
Query: 300 ASSTASASASLQPQAP---AFSDLIRSMGCTDPRPT-DFSAPPSSEAISLCLSTSHGSSI 355
A QPQ F + + C D T D ++ A +T S+
Sbjct: 263 MGQDI---APPQPQPDYDWVFGNAKAASACIDNNNTHDEQITQNANASLTTTTTLSAPSL 319
Query: 356 FGTGGQECRQYVPTHQPPAMSATALLQKAAQMGA-----AATN--ASLLRGLGIVSSSAS 408
F + + Q + MSATALLQKAA++GA AATN ++ L+ + S+ +
Sbjct: 320 FSS---DQPQNANANSNVNMSATALLQKAAEIGATSTTTAATNDPSTFLQSFPLKSTDQT 376
Query: 409 TS 410
TS
Sbjct: 377 TS 378
>UniRef100_Q700D2 Hypothetical protein [Arabidopsis thaliana]
Length = 503
Score = 333 bits (854), Expect = 7e-90
Identities = 207/416 (49%), Positives = 243/416 (57%), Gaps = 56/416 (13%)
Query: 18 SSGNNNIQSPIPKPTKKKRNLPGMPDPEAEVIALSPTTLLATNRFVCEICNKGFQRDQNL 77
+S N P KKKRN PG PDP+A+VIALSPTTL+ATNRFVCEICNKGFQRDQNL
Sbjct: 38 NSNPNPNAKPNSSSAKKKRNQPGTPDPDADVIALSPTTLMATNRFVCEICNKGFQRDQNL 97
Query: 78 QLHRRGHNLPWKLRQRSSKE-IRKRVYVCPEPTCVHHDPSRALGDLTGIKKHFCRKHGEK 136
QLHRRGHNLPWKL+QRS +E I+K+VY+CP TCVHHD SRALGDLTGIKKH+ RKHGEK
Sbjct: 98 QLHRRGHNLPWKLKQRSKQEVIKKKVYICPIKTCVHHDASRALGDLTGIKKHYSRKHGEK 157
Query: 137 KWKCEKCSKKYAVQSDWKAHSKVCGSREYKCDCGTVFSRRDSFITHRAFCDALAEENAK- 195
KWKCEKCSKKYAVQSDWKAH+K CG+REYKCDCGT+FSR+DSFITHRAFCDAL EE A+
Sbjct: 158 KWKCEKCSKKYAVQSDWKAHAKTCGTREYKCDCGTLFSRKDSFITHRAFCDALTEEGARM 217
Query: 196 ---SQNQAV---GKANSESDSKVLTGDSLPAVITTTAAAAATTPQSNSGVSSALETQKLD 249
S N V N ++S V+ +LP P N+ +S D
Sbjct: 218 SSLSNNNPVISTTNLNFGNESNVMNNPNLPHGF---VHRGVHHPDINAAISQFGLGFGHD 274
Query: 250 LPENPPQIIEEPQVVVTTTASALNASCSSSTSSKSN----GCAATSTGVFASLFASSTA- 304
L Q + E + +T L S SSS S TST +L +SST+
Sbjct: 275 LSAMHAQGLSEMVQMASTGNHHLFPSSSSSLPDFSGHHQFQIPMTSTNPSLTLSSSSTSQ 334
Query: 305 SASASLQPQAPAFSDLIRSMGCTDPRPTDFSAPPSSEAISLCLSTSHGSSIFGTGGQECR 364
SASLQ Q L S S +F + E +
Sbjct: 335 QTSASLQHQT--------------------------------LKDSSFSPLF-SSSSENK 361
Query: 365 QYVPTHQPPAMSATALLQKAAQMGAAATNA----SLLRGLGIVSSSASTSSGQQDS 416
Q P MSATALLQKAAQMG+ +N+ S G + SSSA+ S + S
Sbjct: 362 QNKPL---SPMSATALLQKAAQMGSTRSNSSTAPSFFAGPTMTSSSATASPPPRSS 414
>UniRef100_Q5UDB8 INDETERMINATE-related protein 10 [Zea mays]
Length = 583
Score = 331 bits (848), Expect = 4e-89
Identities = 216/512 (42%), Positives = 289/512 (56%), Gaps = 72/512 (14%)
Query: 14 ASISSSGNNNIQSPIPKPTKKKRNLPGMPDPEAEVIALSPTTLLATNRFVCEICNKGFQR 73
A+ SS+G P KKKRNLP DP+AEVIALSP TLLATNRFVCE+CNKGFQR
Sbjct: 49 AASSSAGPGQAAGATPPAVKKKRNLP---DPDAEVIALSPKTLLATNRFVCEVCNKGFQR 105
Query: 74 DQNLQLHRRGHNLPWKLRQRS-SKEIRKRVYVCPEPTCVHHDPSRALGDLTGIKKHFCRK 132
+QNLQLHRRGHNLPWKL+Q+ S+ R+RVY+CPEPTC HHDPSRALGDLTGIKKHFCRK
Sbjct: 106 EQNLQLHRRGHNLPWKLKQKDPSQAQRRRVYLCPEPTCAHHDPSRALGDLTGIKKHFCRK 165
Query: 133 HGEKKWKCEKCSKKYAVQSDWKAHSKVCGSREYKCDCGTVFSRRDSFITHRAFCDALAEE 192
HGEKKWKC+KCSK+YAVQSDWKAHSKVCG+REY+CDCGT+FSRRDSFITHRAFCDALA+E
Sbjct: 166 HGEKKWKCDKCSKRYAVQSDWKAHSKVCGTREYRCDCGTLFSRRDSFITHRAFCDALAQE 225
Query: 193 NAK------SQNQAVGKANSESDSKVLT--GDSLPAVITTTA------------AAAATT 232
+A+ + + G N+ + L+ G L + + A + AA+
Sbjct: 226 SARLPPPGLTASHLYGATNAANMGLSLSQVGSHLASTLGADAHGHHQDLLRLGGSNAASR 285
Query: 233 PQSNSGVSSALETQKLDLPENPPQIIEEPQVVVT-------------TTASALNASCSSS 279
+ G S+A + L P + ++ V+ +AS S
Sbjct: 286 FEHLLGPSNASAFRPLPPPPSSAFLMGRSAGVLAQGDGSGSHGFLQGKPVPQSHASAGSP 345
Query: 280 TSSKSNGCAATSTGVFASLFASSTASASASLQPQAPAFSDLIRSMGCTDPRPTDFSAPPS 339
+ A+++TG+F + +++A++S + + L T + ++
Sbjct: 346 RQRRRGASASSATGLFNLGYIANSANSSGTSSHGHASQGHL------TSDQFSEGGGGGG 399
Query: 340 SEAISLCLSTSHGSSIFGTGGQECRQYVPTHQP----PAMSATALLQKAAQMGAAAT--- 392
SE+ S + S G + G Q + + P P MSATALLQKA+QMG++A+
Sbjct: 400 SES-SAAMLFSGGGNFAGGDHQVAPGGMYNNDPAVMLPQMSATALLQKASQMGSSASAHG 458
Query: 393 NASLLRGLGIVSSSASTSSGQQDSLHWGLGPMEPEGSSLVSAGLGLGLPCDADSGLKELM 452
S+ GL + SS+ S + L ++ SSL + G+ G A+SG
Sbjct: 459 GVSVFGGL-VGSSAPSARHARAPMLDQSQMHLQSLMSSLAAGGMFGG----ANSG----S 509
Query: 453 LGTPSMF------------GPKQTTLDFLGLG 472
+ P M+ G + T DFLG+G
Sbjct: 510 MIDPRMYDMNQDVKFSQGRGGAEMTRDFLGVG 541
>UniRef100_Q9ZWA6 F11M21.23 protein [Arabidopsis thaliana]
Length = 506
Score = 326 bits (835), Expect = 1e-87
Identities = 152/203 (74%), Positives = 171/203 (83%), Gaps = 8/203 (3%)
Query: 1 MVDLDNVSTASGEASISSSGNNNI--------QSPIPKPTKKKRNLPGMPDPEAEVIALS 52
M D ++SG SSS +++ +S P KKKRNLPG PDPEAEVIALS
Sbjct: 1 MTTEDQTISSSGGYVQSSSTTDHVDHHHHDQHESLNPPLVKKKRNLPGNPDPEAEVIALS 60
Query: 53 PTTLLATNRFVCEICNKGFQRDQNLQLHRRGHNLPWKLRQRSSKEIRKRVYVCPEPTCVH 112
P TL+ATNRF+CEIC KGFQRDQNLQLHRRGHNLPWKL+QR+SKE+RKRVYVCPE +CVH
Sbjct: 61 PKTLMATNRFLCEICGKGFQRDQNLQLHRRGHNLPWKLKQRTSKEVRKRVYVCPEKSCVH 120
Query: 113 HDPSRALGDLTGIKKHFCRKHGEKKWKCEKCSKKYAVQSDWKAHSKVCGSREYKCDCGTV 172
H P+RALGDLTGIKKHFCRKHGEKKWKCEKC+K+YAVQSDWKAHSK CG+REY+CDCGT+
Sbjct: 121 HHPTRALGDLTGIKKHFCRKHGEKKWKCEKCAKRYAVQSDWKAHSKTCGTREYRCDCGTI 180
Query: 173 FSRRDSFITHRAFCDALAEENAK 195
FSRRDSFITHRAFCDALAEE A+
Sbjct: 181 FSRRDSFITHRAFCDALAEETAR 203
>UniRef100_Q8RX03 Putative zinc finger protein [Arabidopsis thaliana]
Length = 455
Score = 321 bits (822), Expect = 4e-86
Identities = 151/208 (72%), Positives = 180/208 (85%), Gaps = 14/208 (6%)
Query: 3 DLDNVSTASGEASISSSGN------NNI------QSPIPKPT-KKKRNLPGMPDPEAEVI 49
++ N+++ASG+ + SSGN +NI Q +P+ + K+KRN PG PDPEAEV+
Sbjct: 20 NMSNLTSASGDQASVSSGNRTETSGSNINQHHQEQCFVPQSSLKRKRNQPGNPDPEAEVM 79
Query: 50 ALSPTTLLATNRFVCEICNKGFQRDQNLQLHRRGHNLPWKLRQRSSKEI-RKRVYVCPEP 108
ALSP TL+ATNRF+CE+CNKGFQRDQNLQLH+RGHNLPWKL+QRS+K++ RK+VYVCPEP
Sbjct: 80 ALSPKTLMATNRFICEVCNKGFQRDQNLQLHKRGHNLPWKLKQRSNKDVVRKKVYVCPEP 139
Query: 109 TCVHHDPSRALGDLTGIKKHFCRKHGEKKWKCEKCSKKYAVQSDWKAHSKVCGSREYKCD 168
CVHH PSRALGDLTGIKKHF RKHGEKKWKCEKCSKKYAVQSDWKAH+K CG++EYKCD
Sbjct: 140 GCVHHHPSRALGDLTGIKKHFFRKHGEKKWKCEKCSKKYAVQSDWKAHAKTCGTKEYKCD 199
Query: 169 CGTVFSRRDSFITHRAFCDALAEENAKS 196
CGT+FSRRDSFITHRAFCDALAEE+A++
Sbjct: 200 CGTLFSRRDSFITHRAFCDALAEESARA 227
>UniRef100_Q8H1F5 Putative zinc finger protein [Arabidopsis thaliana]
Length = 455
Score = 321 bits (822), Expect = 4e-86
Identities = 151/208 (72%), Positives = 180/208 (85%), Gaps = 14/208 (6%)
Query: 3 DLDNVSTASGEASISSSGN------NNI------QSPIPKPT-KKKRNLPGMPDPEAEVI 49
++ N+++ASG+ + SSGN +NI Q +P+ + K+KRN PG PDPEAEV+
Sbjct: 20 NMSNLTSASGDQASVSSGNRTETSGSNINQHHQEQCFVPQSSLKRKRNQPGNPDPEAEVM 79
Query: 50 ALSPTTLLATNRFVCEICNKGFQRDQNLQLHRRGHNLPWKLRQRSSKEI-RKRVYVCPEP 108
ALSP TL+ATNRF+CE+CNKGFQRDQNLQLH+RGHNLPWKL+QRS+K++ RK+VYVCPEP
Sbjct: 80 ALSPKTLMATNRFICEVCNKGFQRDQNLQLHKRGHNLPWKLKQRSNKDVVRKKVYVCPEP 139
Query: 109 TCVHHDPSRALGDLTGIKKHFCRKHGEKKWKCEKCSKKYAVQSDWKAHSKVCGSREYKCD 168
CVHH PSRALGDLTGIKKHF RKHGEKKWKCEKCSKKYAVQSDWKAH+K CG++EYKCD
Sbjct: 140 GCVHHHPSRALGDLTGIKKHFFRKHGEKKWKCEKCSKKYAVQSDWKAHAKTCGTKEYKCD 199
Query: 169 CGTVFSRRDSFITHRAFCDALAEENAKS 196
CGT+FSRRDSFITHRAFCDALAEE+A++
Sbjct: 200 CGTLFSRRDSFITHRAFCDALAEESARA 227
>UniRef100_Q700C1 Hypothetical protein [Arabidopsis thaliana]
Length = 516
Score = 320 bits (820), Expect = 6e-86
Identities = 157/232 (67%), Positives = 180/232 (76%), Gaps = 29/232 (12%)
Query: 3 DLDNVSTASGEASISSSGNNNIQS------------------------PIPKPTKKKRNL 38
++ N+++ASG+ + SSGN S P +P KK+RN
Sbjct: 19 NMSNLTSASGDQASVSSGNITEASGSNYFPHHQQQQEQQQQQRQQLFVPDSQPQKKRRNQ 78
Query: 39 PGMPDPEAEVIALSPTTLLATNRFVCEICNKGFQRDQNLQLHRRGHNLPWKLRQRSSKE- 97
PG PDPE+EVIALSP TL+ATNRFVCEICNKGFQRDQNLQLHRRGHNLPWKL+QRS+KE
Sbjct: 79 PGNPDPESEVIALSPKTLMATNRFVCEICNKGFQRDQNLQLHRRGHNLPWKLKQRSNKEV 138
Query: 98 IRKRVYVCPEPTCVHHDPSRALGDLTGIKKHFCRKHGEKKWKCEKCSKKYAVQSDWKAHS 157
IRK+VYVCPE +CVHHDPSRALGDLTGIKKHFCRKHGEKKWKC+KCSKKYAVQSD KAHS
Sbjct: 139 IRKKVYVCPEASCVHHDPSRALGDLTGIKKHFCRKHGEKKWKCDKCSKKYAVQSDCKAHS 198
Query: 158 KVCGSREYKCDCGTVFSRRDSFITHRAFCDALAEENAKS----QNQAVGKAN 205
K CG++EY+CDCGT+FSRRDSFITHRAFC+ALAEE A+ QNQ + N
Sbjct: 199 KTCGTKEYRCDCGTLFSRRDSFITHRAFCEALAEETAREVVIPQNQNNNQPN 250
>UniRef100_Q944L3 AT3g45260/F18N11_20 [Arabidopsis thaliana]
Length = 446
Score = 319 bits (818), Expect = 1e-85
Identities = 183/384 (47%), Positives = 234/384 (60%), Gaps = 52/384 (13%)
Query: 26 SPIPKPT-----KKKRNLPGMPDPEAEVIALSPTTLLATNRFVCEICNKGFQRDQNLQLH 80
+P P PT K+KRNLPG PDP+AEVIALSP +L+ TNRF+CE+CNKGF+RDQNLQLH
Sbjct: 27 NPNPNPTSSNSAKRKRNLPGNPDPDAEVIALSPNSLMTTNRFICEVCNKGFKRDQNLQLH 86
Query: 81 RRGHNLPWKLRQRSSKE-IRKRVYVCPEPTCVHHDPSRALGDLTGIKKHFCRKHGEKKWK 139
RRGHNLPWKL+QR++KE ++K+VY+CPE TCVHHDP+RALGDLTGIKKHF RKHGEKKWK
Sbjct: 87 RRGHNLPWKLKQRTNKEQVKKKVYICPEKTCVHHDPARALGDLTGIKKHFSRKHGEKKWK 146
Query: 140 CEKCSKKYAVQSDWKAHSKVCGSREYKCDCGTVFSRRDSFITHRAFCDALAEENAKSQNQ 199
C+KCSKKYAV SDWKAHSK+CG++EY+CDCGT+FSR+DSFITHRAFCDALAEE+A+
Sbjct: 147 CDKCSKKYAVMSDWKAHSKICGTKEYRCDCGTLFSRKDSFITHRAFCDALAEESAR---- 202
Query: 200 AVGKANSESDSKVLTGDSLPAVITTTAAAAATTPQSNSGVSSALETQKLDLPENPPQIIE 259
+ P + + +++A LD+ N I +
Sbjct: 203 -----------------------------FVSVPPAPAYLNNA-----LDVEVNHGNINQ 228
Query: 260 -EPQVVVTTTASALNASCSSSTSSKSNGCAAT-STGVFASLFASSTASASASLQ-----P 312
Q + TT+S L+ ++ + T T VFAS + S SAS SLQ
Sbjct: 229 NHQQRQLNTTSSQLDQPGFNTNRNNIAFLGQTLPTNVFASSSSPSPRSASDSLQNLWHLQ 288
Query: 313 QAPAFSDLIRSMGCTDPRPTDFSAPPSSEAISLCLSTSHGSSIFGTGGQECRQYVPT-HQ 371
+ L+ + + E + S+GS Y Q
Sbjct: 289 GQSSHQWLLNENNNNNNNILQRGISKNQEEHEMKNVISNGSLFSSEARNNTNNYNQNGGQ 348
Query: 372 PPAMSATALLQKAAQMGAAATNAS 395
+MSATALLQKAAQMG+ +++S
Sbjct: 349 IASMSATALLQKAAQMGSKRSSSS 372
>UniRef100_Q9LRW7 Similarity to zinc finger protein [Arabidopsis thaliana]
Length = 513
Score = 318 bits (816), Expect = 2e-85
Identities = 155/229 (67%), Positives = 179/229 (77%), Gaps = 26/229 (11%)
Query: 3 DLDNVSTASGEASISSSGN---------------------NNIQSPIPKPTKKKRNLPGM 41
++ N+++ASG+ + SSGN + P + KK+RN PG
Sbjct: 19 NMSNLTSASGDQASVSSGNITEASGSNYFPHHQQQQEQQQQQLVVPDSQTQKKRRNQPGN 78
Query: 42 PDPEAEVIALSPTTLLATNRFVCEICNKGFQRDQNLQLHRRGHNLPWKLRQRSSKE-IRK 100
PDPE+EVIALSP TL+ATNRFVCEICNKGFQRDQNLQLHRRGHNLPWKL+QRS+KE IRK
Sbjct: 79 PDPESEVIALSPKTLMATNRFVCEICNKGFQRDQNLQLHRRGHNLPWKLKQRSNKEVIRK 138
Query: 101 RVYVCPEPTCVHHDPSRALGDLTGIKKHFCRKHGEKKWKCEKCSKKYAVQSDWKAHSKVC 160
+VYVCPE +CVHHDPSRALGDLTGIKKHFCRKHGEKKWKC+KCSKKYAVQSD KAHSK C
Sbjct: 139 KVYVCPEASCVHHDPSRALGDLTGIKKHFCRKHGEKKWKCDKCSKKYAVQSDCKAHSKTC 198
Query: 161 GSREYKCDCGTVFSRRDSFITHRAFCDALAEENAKS----QNQAVGKAN 205
G++EY+CDCGT+FSRRDSFITHRAFC+ALAEE A+ QNQ + N
Sbjct: 199 GTKEYRCDCGTLFSRRDSFITHRAFCEALAEETAREVVIPQNQNNNQPN 247
>UniRef100_Q700E8 Hypothetical protein [Arabidopsis thaliana]
Length = 504
Score = 317 bits (812), Expect = 5e-85
Identities = 150/203 (73%), Positives = 170/203 (82%), Gaps = 10/203 (4%)
Query: 1 MVDLDNVSTASGEASISSSGNNNI--------QSPIPKPTKKKRNLPGMPDPEAEVIALS 52
M D ++SG SSS +++ +S P KKKRNLPG +PEAEVIALS
Sbjct: 1 MTTEDQTISSSGGYVQSSSTTDHVDHHHHDQHESLNPPLVKKKRNLPG--NPEAEVIALS 58
Query: 53 PTTLLATNRFVCEICNKGFQRDQNLQLHRRGHNLPWKLRQRSSKEIRKRVYVCPEPTCVH 112
P TL+ATNRF+CEIC KGFQRDQNLQLHRRGHNLPWKL+QR+SKE+RKRVYVCPE +CVH
Sbjct: 59 PKTLMATNRFLCEICGKGFQRDQNLQLHRRGHNLPWKLKQRTSKEVRKRVYVCPEKSCVH 118
Query: 113 HDPSRALGDLTGIKKHFCRKHGEKKWKCEKCSKKYAVQSDWKAHSKVCGSREYKCDCGTV 172
H P+RALGDLTGIKKHFCRKHGEKKWKCEKC+K+YAVQSDWKAHSK CG+REY+CDCGT+
Sbjct: 119 HHPTRALGDLTGIKKHFCRKHGEKKWKCEKCAKRYAVQSDWKAHSKTCGTREYRCDCGTI 178
Query: 173 FSRRDSFITHRAFCDALAEENAK 195
FSRRDSFITHRAFCDALAEE A+
Sbjct: 179 FSRRDSFITHRAFCDALAEETAR 201
>UniRef100_Q9FYN0 Similarity to C2H2-type zinc finger protein [Arabidopsis thaliana]
Length = 412
Score = 313 bits (803), Expect = 6e-84
Identities = 150/210 (71%), Positives = 169/210 (80%), Gaps = 8/210 (3%)
Query: 18 SSGNNNIQSPIPKPTKKKRNLPGMPDPEAEVIALSPTTLLATNRFVCEICNKGFQRDQNL 77
+S N P KKKRN PG PDP+A+VIALSPTTL+ATNRFVCEICNKGFQRDQNL
Sbjct: 38 NSNPNPNAKPNSSSAKKKRNQPGTPDPDADVIALSPTTLMATNRFVCEICNKGFQRDQNL 97
Query: 78 QLHRRGHNLPWKLRQRSSKE-IRKRVYVCPEPTCVHHDPSRALGDLTGIKKHFCRKHGEK 136
QLHRRGHNLPWKL+QRS +E I+K+VY+CP TCVHHD SRALGDLTGIKKH+ RKHGEK
Sbjct: 98 QLHRRGHNLPWKLKQRSKQEVIKKKVYICPIKTCVHHDASRALGDLTGIKKHYSRKHGEK 157
Query: 137 KWKCEKCSKKYAVQSDWKAHSKVCGSREYKCDCGTVFSRRDSFITHRAFCDALAEENAK- 195
KWKCEKCSKKYAVQSDWKAH+K CG+REYKCDCGT+FSR+DSFITHRAFCDAL EE A+
Sbjct: 158 KWKCEKCSKKYAVQSDWKAHAKTCGTREYKCDCGTLFSRKDSFITHRAFCDALTEEGARM 217
Query: 196 ---SQNQAV---GKANSESDSKVLTGDSLP 219
S N V N ++S V+ +LP
Sbjct: 218 SSLSNNNPVISTTNLNFGNESNVMNNPNLP 247
Score = 35.8 bits (81), Expect = 3.1
Identities = 23/51 (45%), Positives = 29/51 (56%), Gaps = 4/51 (7%)
Query: 370 HQPPAMSATALLQKAAQMGAAATNA----SLLRGLGIVSSSASTSSGQQDS 416
H AM A ALLQKAAQMG+ +N+ S G + SSSA+ S + S
Sbjct: 273 HDLSAMHAQALLQKAAQMGSTRSNSSTAPSFFAGPTMTSSSATASPPPRSS 323
>UniRef100_Q6EU15 Finger protein pcp1-like [Oryza sativa]
Length = 615
Score = 313 bits (803), Expect = 6e-84
Identities = 145/196 (73%), Positives = 170/196 (85%), Gaps = 7/196 (3%)
Query: 8 STASGEASISSSGNNNIQSPIPKPTKKKRNLPGMPDPEAEVIALSPTTLLATNRFVCEIC 67
+ A+ A+ S++G + +P P KKKR LP DP+AEVIALSP TLLATNRFVCE+C
Sbjct: 47 AAAAASAAGSAAGQAAVAAP---PAKKKRTLP---DPDAEVIALSPKTLLATNRFVCEVC 100
Query: 68 NKGFQRDQNLQLHRRGHNLPWKLRQRSSKEI-RKRVYVCPEPTCVHHDPSRALGDLTGIK 126
NKGFQR+QNLQLHRRGHNLPWKL+Q++ + R+RVY+CPEPTCVHHDPSRALGDLTGIK
Sbjct: 101 NKGFQREQNLQLHRRGHNLPWKLKQKNPLQAQRRRVYLCPEPTCVHHDPSRALGDLTGIK 160
Query: 127 KHFCRKHGEKKWKCEKCSKKYAVQSDWKAHSKVCGSREYKCDCGTVFSRRDSFITHRAFC 186
KHFCRKHGEKKWKC+KCSK+YAVQSDWKAHSK+CG+REY+CDCGT+FSRRDSFITHRAFC
Sbjct: 161 KHFCRKHGEKKWKCDKCSKRYAVQSDWKAHSKICGTREYRCDCGTLFSRRDSFITHRAFC 220
Query: 187 DALAEENAKSQNQAVG 202
DALA+E+A+ A G
Sbjct: 221 DALAQESARLPPAAAG 236
Score = 43.5 bits (101), Expect = 0.015
Identities = 45/148 (30%), Positives = 65/148 (43%), Gaps = 19/148 (12%)
Query: 373 PAMSATALLQKAAQMGAAATNASLLRGLGIVSSSASTSSGQQDSLHWGLGPMEPEG---- 428
P MSATALLQKAAQMG++ ++A+ G + + SS H M +G
Sbjct: 452 PQMSATALLQKAAQMGSSTSSAN-GAGASVFGGGFAGSSAPSSIPHGRGTTMVDQGQMHL 510
Query: 429 SSLVSAGLGLGLPCDADSGLKELMLGTPSMFGPKQTTLD----FLGLGMAAGGSAGGGLS 484
SL+++ G G +AD + M G+ SM P+ +D L GG G ++
Sbjct: 511 QSLMNSLAGGG---NAD---HQGMFGSGSMIDPRLYDMDQHEVKFSLQRGGGGGGDGDVT 564
Query: 485 ALITSIGGGSGLDVTAAAASFGNGEFSG 512
+GGG + S GE G
Sbjct: 565 RDFLGVGGGGFM----RGMSMARGEHHG 588
>UniRef100_Q8GYC1 Putative C2H2-type zinc finger protein [Arabidopsis thaliana]
Length = 516
Score = 313 bits (801), Expect = 1e-83
Identities = 203/495 (41%), Positives = 264/495 (53%), Gaps = 84/495 (16%)
Query: 31 PTKKKRNLPGMPDPEAEVIALSPTTLLATNRFVCEICNKGFQRDQNLQLHRRGHNLPWKL 90
P KK+RN PG P+P+AEV+ALSP TL+ATNRF+C++CNKGFQR+QNLQLHRRGHNLPWKL
Sbjct: 52 PPKKRRNQPGNPNPDAEVVALSPKTLMATNRFICDVCNKGFQREQNLQLHRRGHNLPWKL 111
Query: 91 RQRSSKEIRKRVYVCPEPTCVHHDPSRALGDLTGIKKHFCRKHGEKKWKCEKCSKKYAVQ 150
+Q+S+KE++++VY+CPEPTCVHHDPSRALGDLTGIKKH+ RKHGEKKWKCEKCSK+YAVQ
Sbjct: 112 KQKSTKEVKRKVYLCPEPTCVHHDPSRALGDLTGIKKHYYRKHGEKKWKCEKCSKRYAVQ 171
Query: 151 SDWKAHSKVCGSREYKCDCGTVFSRRDSFITHRAFCDALAEENAKSQNQAVGKANSESDS 210
SDWKAHSK CG++EY+CDCGT+FSRRDS+ITHRAFCDAL +E A++
Sbjct: 172 SDWKAHSKTCGTKEYRCDCGTIFSRRDSYITHRAFCDALIQETARN-------------- 217
Query: 211 KVLTGDSLPAVITTTAAAAATTPQSNS-----GVSSALETQKLDLPENPPQIIEEPQVVV 265
P V T+ AA++ S G SAL L + P P V
Sbjct: 218 --------PTVSFTSMTAASSGVGSGGIYGRLGGGSALSHHHL---SDHPNFGFNPLV-- 264
Query: 266 TTTASALNASCSSSTSSKSNGCAATSTGVFASLFASSTASASASLQPQAPAFSDLIRSMG 325
N + +SS ++ + +S F ASS + + +F + G
Sbjct: 265 -----GYNLNIASS-DNRRDFIPQSSNPNFLIQSASSQGMLNTTPNNNNQSF---MNQHG 315
Query: 326 CTDPRPTDFSAPPSSEAISLCLSTSHGSSIFGTG-GQECRQYVPTHQPPAMSATALLQKA 384
P D I+L S+ +S F G QE + T P S L+
Sbjct: 316 LIQFDPVD--------NINL-KSSGTNNSFFNLGFFQENTKNSETSLPSLYSTDVLVHHR 366
Query: 385 AQMGAAATNASLLRGLGIVSSSASTSSGQQDSLHWGLGPMEPEGSSLVSAGLGLG--LPC 442
+ A +N S L + S +S +L GL SS+++ G G +
Sbjct: 367 EENLNAGSNVSATALLQKATQMGSVTSNDPSALFRGLA-SSSNSSSVIANHFGGGRIMEN 425
Query: 443 DADSGLKELMLG---------------------TPSMFGPKQTTLDFLGL-GMAAGGSAG 480
D + L+ LM +M G + TLDFLG+ GM + G
Sbjct: 426 DNNGNLQGLMNSLAAVNGGGGSGGSIFDVQFGDNGNMSGSDKLTLDFLGVGGMVRNVNRG 485
Query: 481 GGLSALITSIGGGSG 495
GG GGG G
Sbjct: 486 GG--------GGGRG 492
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.312 0.127 0.368
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 857,094,283
Number of Sequences: 2790947
Number of extensions: 37185070
Number of successful extensions: 269251
Number of sequences better than 10.0: 8279
Number of HSP's better than 10.0 without gapping: 780
Number of HSP's successfully gapped in prelim test: 7678
Number of HSP's that attempted gapping in prelim test: 161763
Number of HSP's gapped (non-prelim): 49028
length of query: 519
length of database: 848,049,833
effective HSP length: 132
effective length of query: 387
effective length of database: 479,644,829
effective search space: 185622548823
effective search space used: 185622548823
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 77 (34.3 bits)
Medicago: description of AC148484.5


