

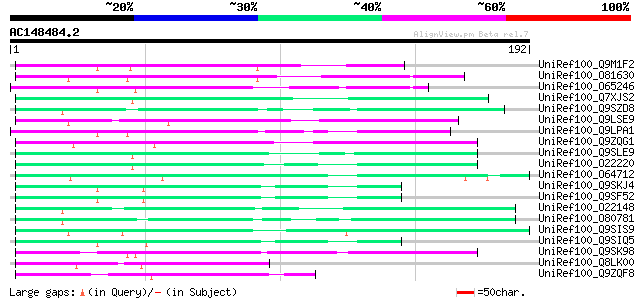
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148484.2 - phase: 0
(192 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9M1F2 Hypothetical protein F9K21.130 [Arabidopsis tha... 63 4e-09
UniRef100_O81630 F8M12.22 protein [Arabidopsis thaliana] 62 6e-09
UniRef100_O65246 F21E10.8 protein [Arabidopsis thaliana] 61 2e-08
UniRef100_Q7XJS2 Putative non-LTR retroelement reverse transcrip... 60 3e-08
UniRef100_Q9SZD8 Hypothetical protein F19B15.120 [Arabidopsis th... 59 5e-08
UniRef100_Q9LSE9 Reverse transcriptase-like protein [Arabidopsis... 58 1e-07
UniRef100_Q9LPA1 T32E20.19 [Arabidopsis thaliana] 57 2e-07
UniRef100_Q9ZQG1 Putative non-LTR retroelement reverse transcrip... 56 6e-07
UniRef100_Q9SLE9 Putative non-LTR retroelement reverse transcrip... 55 1e-06
UniRef100_O22220 Putative non-LTR retroelement reverse transcrip... 54 2e-06
UniRef100_O64712 Putative reverse transcriptase [Arabidopsis tha... 54 2e-06
UniRef100_Q9SKJ4 Putative non-LTR retroelement reverse transcrip... 54 3e-06
UniRef100_Q9SF52 Putative non-LTR reverse transcriptase [Arabido... 54 3e-06
UniRef100_O22148 Putative non-LTR retroelement reverse transcrip... 52 6e-06
UniRef100_O80781 Putative non-LTR retroelement reverse transcrip... 52 8e-06
UniRef100_Q9SIS9 Putative non-LTR retroelement reverse transcrip... 50 2e-05
UniRef100_Q9SIQ5 Putative non-LTR retroelement reverse transcrip... 50 2e-05
UniRef100_Q9SK98 Very similar to retrotransposon reverse transcr... 48 1e-04
UniRef100_Q8LK00 Putative reverse transcriptase [Sorghum bicolor] 48 1e-04
UniRef100_Q9ZQF8 Putative non-LTR retroelement reverse transcrip... 46 5e-04
>UniRef100_Q9M1F2 Hypothetical protein F9K21.130 [Arabidopsis thaliana]
Length = 851
Score = 63.2 bits (152), Expect = 4e-09
Identities = 45/148 (30%), Positives = 63/148 (42%), Gaps = 20/148 (13%)
Query: 3 HLFFACSFAIQVWRLSGLWSDVSAANSND-DSNIGTVFALLD--MLTTKQRVVFAATVWS 59
H F C FA WRLS S+ SN+ + NI + LL +T Q+++ +W
Sbjct: 717 HALFTCPFATMAWRLSDTPLYRSSILSNNIEDNISNILLLLQNTTITDSQKLIPFWLLWR 776
Query: 60 LWKHRNLKVWEDVTEVAAVVVDRARVMITEW-QIANTKTPAESRAATTAPPTQLLSNSAE 118
+WK RN V+ ++ E ++ V RA+ EW T+ P TTA
Sbjct: 777 IWKARNNVVFNNLRESPSITVVRAKAETNEWLNATQTQGPRRLPKRTTAA---------- 826
Query: 119 VNSSDDGLVLWQKPTSGRYKCNVDAVFS 146
G W KP KCN DA F+
Sbjct: 827 ------GNTTWVKPQMPYIKCNFDASFN 848
>UniRef100_O81630 F8M12.22 protein [Arabidopsis thaliana]
Length = 1662
Score = 62.4 bits (150), Expect = 6e-09
Identities = 49/170 (28%), Positives = 70/170 (40%), Gaps = 21/170 (12%)
Query: 3 HLFFACSFAIQVWRLSGL-WSDVSAANSNDDSNIGTVFALL--DMLTTKQRVVFAATVWS 59
H+ F C +A +W LS W + + + NI + + L T+QR+ +W
Sbjct: 1412 HVLFTCPYAASIWGLSNFPWLPGHTFSQDTEENISFLINSFSNNTLNTEQRLAPFWLIWR 1471
Query: 60 LWKHRNLKVWEDVTEVAAVVVDRARVMITEW-QIANTKTPAESRAATTAPPTQLLSNSAE 118
LWK RN V+ +E + VV + + EW Q N + A S
Sbjct: 1472 LWKARNNLVFNKFSESCSRVVTQTEAEVNEWLQSVNRREDA----------------SVL 1515
Query: 119 VNSSDDGLVLWQKPTSGRYKCNVDAVFSSNFNRTGIGICIHDEEGAFVLA 168
SS V W+KP KCN DA F+ N N G I E A ++A
Sbjct: 1516 TRSSHRANVRWKKPVFPLVKCNFDAGFTGN-NTQSTGGWIIPETKALLIA 1564
>UniRef100_O65246 F21E10.8 protein [Arabidopsis thaliana]
Length = 301
Score = 60.8 bits (146), Expect = 2e-08
Identities = 42/157 (26%), Positives = 71/157 (44%), Gaps = 18/157 (11%)
Query: 1 MAHLFFACSFAIQVWRLSGLWSDVSAANSND-DSNIGTVFALLDML-TTKQRVVFAATVW 58
+ H+ F C +A+++WRLS L + ++D + NI + A + TT+Q+V+ +W
Sbjct: 37 ICHVLFTCPYAMRIWRLSNLPQLTGLSITHDVEENISLLDAYYNNNHTTQQKVIHFWLIW 96
Query: 59 SLWKHRNLKVWEDVTEVAAVVVDRARVMITEWQIANTKTPAESRAATTAPPTQLLSNSAE 118
+WK N V + E ++V+ + + +W T Q +S A
Sbjct: 97 KIWKAHNNLVLNNYRECPSMVIQQTVAEVYDW-------------TTIINGMQAMSLPAR 143
Query: 119 VNSSDDGLVLWQKPTSGRYKCNVDAVFSSNFNRTGIG 155
+ + G W K + KCN DA F N N G+G
Sbjct: 144 IQTK--GKTSWMKHAAPFVKCNFDAGFQEN-NEQGMG 177
>UniRef100_Q7XJS2 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1344
Score = 60.1 bits (144), Expect = 3e-08
Identities = 43/177 (24%), Positives = 70/177 (39%), Gaps = 22/177 (12%)
Query: 3 HLFFACSFAIQVWRLSGLWSDVSAANSNDDSNIGTVFALLDM--LTTKQRVVFAATVWSL 60
H+ F C A QVW ++ L S S +++ +N+ + L L R V +W L
Sbjct: 1074 HILFECPLARQVWAITHLSSAGSEFSNSVYTNMSRLIDLTQQNDLPHHLRFVSPWILWFL 1133
Query: 61 WKHRNLKVWEDVTEVAAVVVDRARVMITEWQIANTKTPAESRAATTAPPTQLLSNSAEVN 120
WK+RN ++E + +VD+A EW A T + +
Sbjct: 1134 WKNRNALLFEGKGSITTTLVDKAYEAYHEWFSAQTHMQNDEKHLK--------------- 1178
Query: 121 SSDDGLVLWQKPTSGRYKCNVDAVFSSNFNRTGIGICIHDEEGAFVLAKMTSFPCLH 177
+ W P G KCN+ +S + +G + D +G +L SF +H
Sbjct: 1179 -----ITKWCPPLPGELKCNIGFAWSKQHHFSGASWVVRDSQGKVLLHSRRSFNEVH 1230
>UniRef100_Q9SZD8 Hypothetical protein F19B15.120 [Arabidopsis thaliana]
Length = 575
Score = 59.3 bits (142), Expect = 5e-08
Identities = 54/188 (28%), Positives = 76/188 (39%), Gaps = 27/188 (14%)
Query: 3 HLFFACSFAIQVWRLS-------GLWSDVSAANSNDDSNIGTVFALLDMLTTKQRVVFAA 55
HL F C+FA W +S G W+D N N+G + K +
Sbjct: 307 HLLFKCTFARLTWAISSIPIPLGGEWADSIYVNLYWVFNLGNGNPQWE----KASQLVPW 362
Query: 56 TVWSLWKHRNLKVWEDVTEVAAVVVDRARVMITEWQIANTKTPAESRAATTAPPTQLLSN 115
+W LWK+RN V+ A V+ RA + EW+I +T AES
Sbjct: 363 LLWRLWKNRNELVFRGREFNAQEVLRRAEDDLEEWRI---RTEAES-----------CGT 408
Query: 116 SAEVNSSDDGLVLWQKPTSGRYKCNVDAVFSSNFNRTGIGICIHDEEGAFVLAKMTSFPC 175
+VN S G W+ P KCN DA ++ + R GIG + +E+G + P
Sbjct: 409 KPQVNRSSCGR--WRPPPHQWVKCNTDATWNRDNERCGIGWVLRNEKGEVKWMGARALPK 466
Query: 176 LHQVTVGE 183
L V E
Sbjct: 467 LKSVLEAE 474
>UniRef100_Q9LSE9 Reverse transcriptase-like protein [Arabidopsis thaliana]
Length = 343
Score = 58.2 bits (139), Expect = 1e-07
Identities = 43/168 (25%), Positives = 70/168 (41%), Gaps = 16/168 (9%)
Query: 3 HLFFACSFAIQVWRLSGL-WSDVSAANSNDDSNIGTVFALLDMLTTKQRVVFAATV---W 58
HLFF C +A QVWR SG+ ++ ++ + + L L +Q +F + W
Sbjct: 68 HLFFDCFYAQQVWRASGIPHQELRTTGITMETKMELL--LSSCLANRQPQLFNLAIWILW 125
Query: 59 SLWKHRNLKVWEDVTEVAAVVVDRARVMITEWQIANTKTPAESRAATTAPPTQLLSNSAE 118
LWK RN V++ + + RAR + EW+ NT + ++ +S+
Sbjct: 126 RLWKSRNQLVFQQKSISWQNTLQRARNDVQEWEDTNTYVQSLNQQV----------HSSR 175
Query: 119 VNSSDDGLVLWQKPTSGRYKCNVDAVFSSNFNRTGIGICIHDEEGAFV 166
WQ+P S K N D F+ G + DE G ++
Sbjct: 176 HQQPTMARTKWQRPPSTWIKYNYDGAFNHQTRNAKAGWLMRDENGVYM 223
>UniRef100_Q9LPA1 T32E20.19 [Arabidopsis thaliana]
Length = 957
Score = 57.4 bits (137), Expect = 2e-07
Identities = 47/166 (28%), Positives = 73/166 (43%), Gaps = 18/166 (10%)
Query: 1 MAHLFFACSFAIQVWRLSGLWSDVSAANSND-DSNIGTVFALL--DMLTTKQRVVFAATV 57
+ H+ F +A W+L+ L SND + NI + + + LT +Q+++ +
Sbjct: 791 ICHVLFNYPYATTTWKLAKLSYLSGHTFSNDVEENISVLIDSISNNNLTEQQKLLPFWLI 850
Query: 58 WSLWKHRNLKVWEDVTEVAAVVVDRARVMITEWQIANTKTPAESRAATTAPPTQLLSNSA 117
+ +WK RN V+ + ++ VV ++ V + EW I ATT PP LS A
Sbjct: 851 FRIWKARNNYVFNKFRKCSSKVVSQSEVDVNEWII--NMCERRGNTATTRPP---LSQKA 905
Query: 118 EVNSSDDGLVLWQKPTSGRYKCNVDAVFSSNFNRTGIGICIHDEEG 163
W KPT KCN +A N ++ G I D EG
Sbjct: 906 S----------WTKPTVSFVKCNFNAGLHKNTTQSTGGWIIRDPEG 941
>UniRef100_Q9ZQG1 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1138
Score = 55.8 bits (133), Expect = 6e-07
Identities = 44/174 (25%), Positives = 73/174 (41%), Gaps = 17/174 (9%)
Query: 3 HLFFACSFAIQVWRLSGLWS-DVSAANSNDDSNIGTVFALLDMLTTKQRVV--FAATVWS 59
H+ F CS A QVW LSG+ + + N++ +NI +F L M+ V + +W
Sbjct: 872 HVLFTCSMARQVWALSGVPTPEFGFQNASIFANIQFLFELKKMILVPDLVKRSWPWVLWR 931
Query: 60 LWKHRNLKVWEDVTEVAAVVVDRARVMITEWQIANTKTPAESRAATTAPPTQLLSNSAEV 119
LWK+RN ++ +T + + + EW A ++ +S S E
Sbjct: 932 LWKNRNKLFFDGITFCPLNSIVKIQEDTLEWFQAQSQIR--------------VSESEEG 977
Query: 120 NSSDDGLVLWQKPTSGRYKCNVDAVFSSNFNRTGIGICIHDEEGAFVLAKMTSF 173
W+ P G KCN+ + +S G + DE G+ +L +F
Sbjct: 978 QRVIPFTHSWEPPPEGWVKCNIGSAWSGKKKVCGGAWVLRDEHGSVILHSRRAF 1031
>UniRef100_Q9SLE9 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1319
Score = 54.7 bits (130), Expect = 1e-06
Identities = 42/173 (24%), Positives = 68/173 (39%), Gaps = 23/173 (13%)
Query: 3 HLFFACSFAIQVWRLSGLWSDVSAANSNDDSNIGTVFALLDM--LTTKQRVVFAATVWSL 60
HL F C FA QVW LS + + + ++ SNI V + R V +W +
Sbjct: 1048 HLLFQCPFARQVWALSLIQAPATGFGTSIFSNINHVIQNSQNFGIPRHMRTVSPWLLWEI 1107
Query: 61 WKHRNLKVWEDVTEVAAVVVDRARVMITEWQIANTKTPAESRAATTAPPTQLLSNSAEVN 120
WK+RN +++ ++ +V +A W A K +S V+
Sbjct: 1108 WKNRNKTLFQGTGLTSSEIVAKAYEECNLWINAQEK------------------SSGGVS 1149
Query: 121 SSDDGLVLWQKPTSGRYKCNVDAVFSSNFNRTGIGICIHDEEGAFVLAKMTSF 173
S+ W P +G KCN+ +S G+ + D G +L S+
Sbjct: 1150 PSEH---KWNPPPAGELKCNIGVAWSRQKQLAGVSWVLRDSMGQVLLHSRRSY 1199
>UniRef100_O22220 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1094
Score = 53.9 bits (128), Expect = 2e-06
Identities = 41/173 (23%), Positives = 68/173 (38%), Gaps = 23/173 (13%)
Query: 3 HLFFACSFAIQVWRLSGLWSDVSAANSNDDSNIGTVFALLDM--LTTKQRVVFAATVWSL 60
H+ F C A QVW L+ + S + +N+ V L+ R V +W L
Sbjct: 825 HILFQCPLARQVWALTPMQSPNHGFGDSIFTNVNHVIGNCHNTELSPHLRYVSPWIIWIL 884
Query: 61 WKHRNLKVWEDVTEVAAVVVDRARVMITEWQIANTKTPAESRAATTAPPTQLLSNSAEVN 120
WK+RN +++E + V+ +V +A EW A+ + PT+ L+
Sbjct: 885 WKNRNKRLFEGIGSVSLSIVGKALEDCKEWLKAH-------ELICSKEPTKDLT------ 931
Query: 121 SSDDGLVLWQKPTSGRYKCNVDAVFSSNFNRTGIGICIHDEEGAFVLAKMTSF 173
W P KCN+ +S G+ + + +G +L SF
Sbjct: 932 --------WIPPLMNELKCNIGIAWSKKHQMAGVSWVVRNWKGRVLLHSRCSF 976
>UniRef100_O64712 Putative reverse transcriptase [Arabidopsis thaliana]
Length = 365
Score = 53.9 bits (128), Expect = 2e-06
Identities = 51/197 (25%), Positives = 77/197 (38%), Gaps = 21/197 (10%)
Query: 3 HLFFACSFAIQVWRLSGLW-SDVSAANSNDDSNIGTVFALLDMLTTKQRVVFAA--TVWS 59
H+ F C + VWR + + + S+ + N+ + L TT F +W
Sbjct: 87 HIMFNCPYTQSVWRSANIIIGNQWGPPSSFEDNLNRLIQLSKTQTTNSLDRFLPFWIMWR 146
Query: 60 LWKHRNLKVWEDVTEVAAVVVDRARVMITEWQIANTKTPAESRAATTAPPTQLLSNSAEV 119
LWK RN+ +++ + + TEW AN T + T P +S++
Sbjct: 147 LWKSRNVFLFQQKCQSPDYEARKGIQDATEWLNANETTENTNVHVATNPIQTSRRDSSQ- 205
Query: 120 NSSDDGLVLWQKPTSGRYKCNVDAVFSSNFNRTGIGICIHDEEGAFVL---AKMTSFPC- 175
W P G KCN D+ ++ T G I + G VL AK+ S C
Sbjct: 206 ---------WNPPPEGWVKCNFDSGYTQGSPYTRSGWTIRECNGHIVLCGNAKLQSSTCS 256
Query: 176 LHQVTVGEAMGLFEAFQ 192
LH EA+G A Q
Sbjct: 257 LH----AEALGFLHALQ 269
>UniRef100_Q9SKJ4 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1750
Score = 53.5 bits (127), Expect = 3e-06
Identities = 40/146 (27%), Positives = 58/146 (39%), Gaps = 18/146 (12%)
Query: 3 HLFFACSFAIQVWRLSGLWSDVSAANSND-DSNIGTVFALLDMLTTK--QRVVFAATVWS 59
H F C FA WRLS + SND + NI + + T +++ +W
Sbjct: 1478 HALFTCPFATMAWRLSDSSLIRNQLMSNDFEENISNILNFVQDTTMSDFHKLLPVWLIWR 1537
Query: 60 LWKHRNLKVWEDVTEVAAVVVDRARVMITEWQIANTKTPAESRAATTAPPTQLLSNSAEV 119
+WK RN V+ E + V A+ +W A +S T +P Q+ N E
Sbjct: 1538 IWKARNNVVFNKFRESPSKTVLSAKAETHDWLNA-----TQSHKKTPSPTRQIAENKIE- 1591
Query: 120 NSSDDGLVLWQKPTSGRYKCNVDAVF 145
W+ P + KCN DA F
Sbjct: 1592 ---------WRNPPATYVKCNFDAGF 1608
>UniRef100_Q9SF52 Putative non-LTR reverse transcriptase [Arabidopsis thaliana]
Length = 484
Score = 53.5 bits (127), Expect = 3e-06
Identities = 40/146 (27%), Positives = 58/146 (39%), Gaps = 18/146 (12%)
Query: 3 HLFFACSFAIQVWRLSGLWSDVSAANSND-DSNIGTVFALLDMLTTK--QRVVFAATVWS 59
H F C FA WRLS + SND + NI + + T +++ +W
Sbjct: 212 HALFTCPFATMAWRLSDSSLIRNQLMSNDFEENISNILNFVQDTTMSDFHKLLPVWLIWR 271
Query: 60 LWKHRNLKVWEDVTEVAAVVVDRARVMITEWQIANTKTPAESRAATTAPPTQLLSNSAEV 119
+WK RN V+ E + V A+ +W A +S T +P Q+ N E
Sbjct: 272 IWKARNNVVFNKFRESPSKTVLSAKAETHDWLNA-----TQSHKKTPSPTRQIAENKIE- 325
Query: 120 NSSDDGLVLWQKPTSGRYKCNVDAVF 145
W+ P + KCN DA F
Sbjct: 326 ---------WRNPPATYVKCNFDAGF 342
>UniRef100_O22148 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1374
Score = 52.4 bits (124), Expect = 6e-06
Identities = 44/192 (22%), Positives = 70/192 (35%), Gaps = 29/192 (15%)
Query: 3 HLFFACSFAIQVWRLS-------GLWSDVSAANSNDDSNIGTVFALLDMLTTKQRVVFAA 55
HL F C FA W +S G W++ N + ++ + +
Sbjct: 1104 HLLFKCPFARLTWAISPLPAPPGGEWAESLFRNMHHVLSVHKS----QPEESDHHALIPW 1159
Query: 56 TVWSLWKHRNLKVWEDVTEVAAVVVDRARVMITEWQIANTKTPAESRAATTAPPTQLLSN 115
+W LWK+RN V++ A V+ +A + W N K P ++T
Sbjct: 1160 ILWRLWKNRNDLVFKGREFTAPQVILKATEDMDAWN--NRKEPQPQVTSSTR-------- 1209
Query: 116 SAEVNSSDDGLVLWQKPTSGRYKCNVDAVFSSNFNRTGIGICIHDEEGAFVLAKMTSFPC 175
D V WQ P+ G KCN D +S + G+G + + G + + + P
Sbjct: 1210 --------DRCVKWQPPSHGWVKCNTDGAWSKDLGNCGVGWVLRNHTGRLLWLGLRALPS 1261
Query: 176 LHQVTVGEAMGL 187
V E L
Sbjct: 1262 QQSVLETEVEAL 1273
>UniRef100_O80781 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 292
Score = 52.0 bits (123), Expect = 8e-06
Identities = 52/192 (27%), Positives = 72/192 (37%), Gaps = 27/192 (14%)
Query: 3 HLFFACSFAIQVWRLS-------GLWSDVSAANSNDDSNIGTVFALLDMLTTKQRVVFAA 55
HL F C FA VW +S G W+D AN N+ L + +
Sbjct: 24 HLLFKCCFARLVWAISPIPAYPEGEWTDSLYANLYWVLNLEVEIPKLGKIGN----LVPW 79
Query: 56 TVWSLWKHRNLKVWEDVTEVAAVVVDRARVMITEWQIANTKTPAESRAATTAPPTQLLSN 115
+W LWK RN +++ A V+ RA EW +T+ E +A+
Sbjct: 80 LLWRLWKSRNELMFKGKEYDAPEVLRRAMEDFEEW---STRRELEGKAS---------GP 127
Query: 116 SAEVNSSDDGLVLWQKPTSGRYKCNVDAVFSSNFNRTGIGICIHDEEGAFVLAKMTSFPC 175
E N S V W+ P KCN DA + R GIG + +E G + + P
Sbjct: 128 QVERNLS----VQWKAPPYQWVKCNTDATWQLENPRCGIGWILRNESGGVLWMGARALPR 183
Query: 176 LHQVTVGEAMGL 187
V E L
Sbjct: 184 TKNVLEAELEAL 195
>UniRef100_Q9SIS9 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1715
Score = 50.4 bits (119), Expect = 2e-05
Identities = 45/194 (23%), Positives = 70/194 (35%), Gaps = 16/194 (8%)
Query: 3 HLFFACSFAIQVWRLSGL-WSDVSAANSNDDSNIGTVFA--LLDMLTTKQRVVFAATVWS 59
H+ F CS+A VWR + S+ N + NI + L ++ +W
Sbjct: 1438 HIIFTCSYAQVVWRSANFSGSNRLCFTDNLEENIRLILQGKKNQNLPILNGLMPFWIMWR 1497
Query: 60 LWKHRNLKVWEDVTEVAAVVVDRARVMITEWQIANTKTPAESRAATTAPPTQLLSNSAEV 119
LWK RN +++ + V +A TEW T T + N+A+
Sbjct: 1498 LWKSRNEYLFQQLDRFPWKVAQKAEQEATEW------------VETMVNDTAISHNTAQS 1545
Query: 120 NSSD-DGLVLWQKPTSGRYKCNVDAVFSSNFNRTGIGICIHDEEGAFVLAKMTSFPCLHQ 178
N W P G KCN D+ + + T G + D G + + +
Sbjct: 1546 NDRPLSRSKQWSSPPEGFLKCNFDSGYVQGRDYTSTGWILRDCNGRVLHSGCAKLQQSYS 1605
Query: 179 VTVGEAMGLFEAFQ 192
EA+G A Q
Sbjct: 1606 ALQAEALGFLHALQ 1619
>UniRef100_Q9SIQ5 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1524
Score = 50.4 bits (119), Expect = 2e-05
Identities = 39/146 (26%), Positives = 57/146 (38%), Gaps = 18/146 (12%)
Query: 3 HLFFACSFAIQVWRLSGLWSDVSAANSND-DSNIGTVFALLDMLTTKQ--RVVFAATVWS 59
H F C FA W LS + SND + NI + + T +++ +W
Sbjct: 1252 HALFTCPFATMAWWLSDSSLIRNQLMSNDFEENISNILNFVQDTTMSDFHKLLPVWLIWR 1311
Query: 60 LWKHRNLKVWEDVTEVAAVVVDRARVMITEWQIANTKTPAESRAATTAPPTQLLSNSAEV 119
+WK RN V+ E + V A+ +W A +S T +P Q+ N E
Sbjct: 1312 IWKARNNVVFNKFRESPSKTVLSAKAETHDWLNA-----TQSHKKTPSPTRQIAENKIE- 1365
Query: 120 NSSDDGLVLWQKPTSGRYKCNVDAVF 145
W+ P + KCN DA F
Sbjct: 1366 ---------WRNPPATYVKCNFDAGF 1382
>UniRef100_Q9SK98 Very similar to retrotransposon reverse transcriptase [Arabidopsis
thaliana]
Length = 1231
Score = 48.1 bits (113), Expect = 1e-04
Identities = 42/180 (23%), Positives = 73/180 (40%), Gaps = 21/180 (11%)
Query: 3 HLFFACSFAIQVWRLSGLWSDVSAANSNDDSNIGTVFALL-------DML--TTKQRVVF 53
H+ F+CSF QVW +S + + G+V+A L D L + R F
Sbjct: 944 HVLFSCSFPRQVWAMSNIHVPLLG------FECGSVYANLYHFLINRDNLKWPVELRRSF 997
Query: 54 AATVWSLWKHRNLKVWEDVTEVAAVVVDRARVMITEWQIANTKTPAESRAATTAPPTQLL 113
+W +WK+RNL +E + + R + +W A + + R A Q +
Sbjct: 998 PWIIWRIWKNRNLFFFEGKRFTVLETILKVRKDVEDWFAA--QVVEKERRAEVGQSDQQV 1055
Query: 114 SNSAEVNSSDDGLVLWQKPTSGRYKCNVDAVFSSNFNRTGIGICIHDEEGAFVLAKMTSF 173
+ V+ +V W P + KCNV +S G+ + ++ G ++ +F
Sbjct: 1056 FSPRNVSP----VVRWLPPPTDWVKCNVGLSWSRRNRLAGVAWVLRNDRGNVLMHSRRAF 1111
>UniRef100_Q8LK00 Putative reverse transcriptase [Sorghum bicolor]
Length = 1323
Score = 48.1 bits (113), Expect = 1e-04
Identities = 29/100 (29%), Positives = 46/100 (46%), Gaps = 8/100 (8%)
Query: 3 HLFFACSFAIQVWRLSGLWSD---VSAANSNDDSNIGTVFALLDMLTT---KQRVVFAAT 56
HLFF C ++ VW+ WS + A+ D S + F+ LTT K + T
Sbjct: 1213 HLFFECPYSRLVWQAVSSWSSCPSLQPASWEDRSRLEDCFS--QTLTTGEKKAHTLAILT 1270
Query: 57 VWSLWKHRNLKVWEDVTEVAAVVVDRARVMITEWQIANTK 96
+WS+W RN V+ + + +V + M +W +A K
Sbjct: 1271 LWSIWNRRNAVVFREDRKTTISLVVEIKDMARQWSLAGCK 1310
>UniRef100_Q9ZQF8 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1225
Score = 46.2 bits (108), Expect = 5e-04
Identities = 34/120 (28%), Positives = 54/120 (44%), Gaps = 20/120 (16%)
Query: 3 HLFFACSFAIQVWRLSGLWSDVSAANSNDDSNIGTVFALLDMLTTKQRV---------VF 53
HL F C + Q+W LS + S N ++F D L ++ + +F
Sbjct: 976 HLLFLCPPSRQIWALSPIPSSEYIFPRN------SLFYNFDFLLSRGKEFDIAEDIMEIF 1029
Query: 54 AATVWSLWKHRNLKVWEDVTEVAAVVVDRARVMITEWQIANTKTPAESRAATTAPPTQLL 113
+W +WK RN ++E+V E V++D A W+ AN+K AT PP Q++
Sbjct: 1030 PWILWYIWKSRNRFIFENVIESPQVILDFAIQEANVWKQANSK-----EVATEYPPPQVV 1084
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.321 0.130 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 306,171,602
Number of Sequences: 2790947
Number of extensions: 10975151
Number of successful extensions: 27444
Number of sequences better than 10.0: 90
Number of HSP's better than 10.0 without gapping: 29
Number of HSP's successfully gapped in prelim test: 62
Number of HSP's that attempted gapping in prelim test: 27333
Number of HSP's gapped (non-prelim): 94
length of query: 192
length of database: 848,049,833
effective HSP length: 120
effective length of query: 72
effective length of database: 513,136,193
effective search space: 36945805896
effective search space used: 36945805896
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 71 (32.0 bits)
Medicago: description of AC148484.2


