

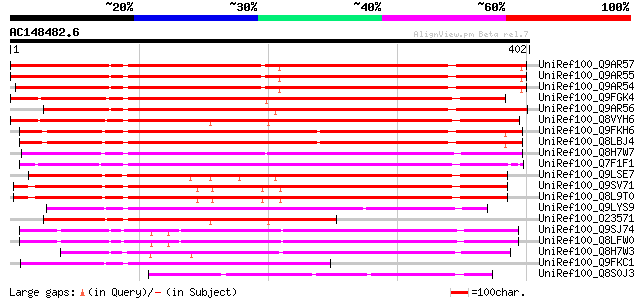
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148482.6 - phase: 0
(402 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9AR57 Putative membrane protein [Solanum tuberosum] 411 e-113
UniRef100_Q9AR55 Putative membrane protein [Solanum tuberosum] 410 e-113
UniRef100_Q9AR54 Putative membrane protein [Solanum tuberosum] 410 e-113
UniRef100_Q9FGK4 Arabidopsis thaliana genomic DNA, chromosome 5,... 405 e-111
UniRef100_Q9AR56 Putative membrane protein [Solanum tuberosum] 400 e-110
UniRef100_Q8VYH6 AT4g17280/dl4675c [Arabidopsis thaliana] 394 e-108
UniRef100_Q9FKH6 Arabidopsis thaliana genomic DNA, chromosome 5,... 364 3e-99
UniRef100_Q8LBJ4 Putative membrane protein [Arabidopsis thaliana] 361 2e-98
UniRef100_Q8H7W7 Putative membrane protein [Oryza sativa] 341 2e-92
UniRef100_Q7F1F1 Putative auxin-induced protein [Oryza sativa] 335 1e-90
UniRef100_Q9LSE7 Emb|CAB45497.1 [Arabidopsis thaliana] 319 8e-86
UniRef100_Q9SV71 Hypothetical protein At4g12980 [Arabidopsis tha... 310 6e-83
UniRef100_Q8L9T0 Hypothetical protein [Arabidopsis thaliana] 308 2e-82
UniRef100_Q9LYS9 Hypothetical protein F17J16_120 [Arabidopsis th... 268 3e-70
UniRef100_O23571 Hypothetical protein dl4675c [Arabidopsis thali... 228 3e-58
UniRef100_Q9SJ74 Expressed protein [Arabidopsis thaliana] 226 9e-58
UniRef100_Q8LFW0 Hypothetical protein [Arabidopsis thaliana] 226 9e-58
UniRef100_Q8H7W3 Hypothetical protein OSJNBa0064E16.14 [Oryza sa... 208 2e-52
UniRef100_Q9FKC1 Emb|CAB86935.1 [Arabidopsis thaliana] 169 2e-40
UniRef100_Q8S0J3 B1078G07.42 protein [Oryza sativa] 138 3e-31
>UniRef100_Q9AR57 Putative membrane protein [Solanum tuberosum]
Length = 400
Score = 411 bits (1057), Expect = e-113
Identities = 210/405 (51%), Positives = 274/405 (66%), Gaps = 16/405 (3%)
Query: 1 MVKNLRFMFLISILMFVTTTLAQTTTQTCKSQNFTNNAIFTSCRDLPQLTSYLHWTYDQT 60
M K L + SIL+ + + Q C + F NN IF +C LP L S LHW+Y
Sbjct: 1 MNKLLTTLLFSSILISTLFSTSYGQNQNCSAFAFRNNQIFATCNALPLLNSVLHWSYHPD 60
Query: 61 TGKLDIAFRHKGITDTNRWVAWAINPNNDLASSMNGAQALVAILQSSGTPKAYTSSIANS 120
+D+A+RH G+ +T+ WVAW +N + + M G+Q LVA SSG AYTS +++
Sbjct: 61 NHTVDLAYRHGGVPNTD-WVAWGLNIDG---TRMVGSQCLVAFRNSSGEIHAYTSPVSSY 116
Query: 121 RTQLAESNISYPHSGLIATHENNEVTIYASITLPVGTPSLVHLWQDGAMSGSTPQMHDMT 180
TQLAE +S+ + A + NNE I+A++ LP G + WQ+GA+SG H +
Sbjct: 117 GTQLAEGALSFNVPRIGAEYSNNEFIIFATLELPAGRTNFNQAWQNGAVSGQALTAHVQS 176
Query: 181 SANTQSKESLDLRSGASEQGSGGGSLS---RRRNTHGVLNAISWGILMPLGAVIARYLKV 237
N +S S+D +G E G GG S++ RRRN HGVLNA+SWG+LMP+GAV ARYLKV
Sbjct: 177 GDNMRSFGSVDFANG--ELGGGGSSVTSRQRRRNVHGVLNAVSWGVLMPMGAVFARYLKV 234
Query: 238 FKSADPAWFYLHVTCQSAAYIVGVAGWGTGLKLGSDSAGVTYSTHRTLGIVIFCLGTLQV 297
FK+A+PAWFY+HV CQ++AYIVGVAGWGTGLKLGSDS G+ ++THR +GI +FCLGTLQV
Sbjct: 235 FKAANPAWFYIHVACQTSAYIVGVAGWGTGLKLGSDSTGIEFTTHRNIGITLFCLGTLQV 294
Query: 298 FALLLRPKKDHKIRFYWNLYHWGVGYATIIISIINIFKGFEALEVSAADRYDNWKHAYTG 357
FALLLRPK DHK R YWN+YH VGYA I ++I N+F+GF+AL + NWK AYTG
Sbjct: 295 FALLLRPKPDHKYRLYWNIYHHAVGYAVISLAIANVFEGFDAL-----NGQKNWKRAYTG 349
Query: 358 IIAALGGVAVLLEAYTWIIVIKRKKSENKLQGMNGTNG--NGYGS 400
+I A+G +AVLLEA+TW IVIKRKK+++ NGTNG N YG+
Sbjct: 350 VIIAIGAIAVLLEAFTWFIVIKRKKTDSNKHTQNGTNGTVNPYGN 394
>UniRef100_Q9AR55 Putative membrane protein [Solanum tuberosum]
Length = 400
Score = 410 bits (1055), Expect = e-113
Identities = 209/405 (51%), Positives = 273/405 (66%), Gaps = 16/405 (3%)
Query: 1 MVKNLRFMFLISILMFVTTTLAQTTTQTCKSQNFTNNAIFTSCRDLPQLTSYLHWTYDQT 60
M K L + SIL+ + + Q C + F NN IF +C LP L S LHW+Y
Sbjct: 1 MNKLLTTLLFSSILISTLFSTSYGQNQNCSAFAFRNNQIFATCNALPLLNSVLHWSYHPD 60
Query: 61 TGKLDIAFRHKGITDTNRWVAWAINPNNDLASSMNGAQALVAILQSSGTPKAYTSSIANS 120
+D+A+RH G+ +T+ WVAW +N + + M G+Q LVA SSG AYTS +++
Sbjct: 61 NHTVDLAYRHGGVPNTD-WVAWGLNIDG---TRMVGSQCLVAFRNSSGEIHAYTSPVSSY 116
Query: 121 RTQLAESNISYPHSGLIATHENNEVTIYASITLPVGTPSLVHLWQDGAMSGSTPQMHDMT 180
TQLA+ +S+ + A + NNE I+A++ LP G S WQ+GA+SG H +
Sbjct: 117 GTQLAKGALSFNVPRIGAEYSNNEFIIFATLELPAGRTSFNQAWQNGAVSGQALTAHVQS 176
Query: 181 SANTQSKESLDLRSGASEQGSGGGSLS---RRRNTHGVLNAISWGILMPLGAVIARYLKV 237
N +S S+D +G E G GG S++ RRRN HGVLNA+SWG+LMP+GAV ARYLKV
Sbjct: 177 GDNMRSFGSIDFANG--ELGGGGSSVTSRQRRRNVHGVLNAVSWGVLMPMGAVFARYLKV 234
Query: 238 FKSADPAWFYLHVTCQSAAYIVGVAGWGTGLKLGSDSAGVTYSTHRTLGIVIFCLGTLQV 297
FK+A+PAWFY+HV CQ++AYIVG+AGWGTGLKLGSDS G+ ++THR +GI +FCLGTLQV
Sbjct: 235 FKAANPAWFYIHVACQTSAYIVGIAGWGTGLKLGSDSTGIEFTTHRNIGITLFCLGTLQV 294
Query: 298 FALLLRPKKDHKIRFYWNLYHWGVGYATIIISIINIFKGFEALEVSAADRYDNWKHAYTG 357
FALLLRPK DHK R YWN+YH VGYA I ++I N+F+GF+AL + NWK AYTG
Sbjct: 295 FALLLRPKPDHKYRLYWNIYHHAVGYAVISLAIANVFQGFDAL-----NGQKNWKRAYTG 349
Query: 358 IIAALGGVAVLLEAYTWIIVIKRKKSENKLQGMNGTNG--NGYGS 400
+I A+G +AVLLEA+TW IVIKRKK++ NGTNG N YG+
Sbjct: 350 VIIAIGAIAVLLEAFTWFIVIKRKKTDTNKHTQNGTNGTVNPYGN 394
>UniRef100_Q9AR54 Putative membrane protein [Solanum tuberosum]
Length = 400
Score = 410 bits (1055), Expect = e-113
Identities = 208/401 (51%), Positives = 272/401 (66%), Gaps = 16/401 (3%)
Query: 5 LRFMFLISILMFVTTTLAQTTTQTCKSQNFTNNAIFTSCRDLPQLTSYLHWTYDQTTGKL 64
L + SIL+ + + Q C + F NN IF +C LP L S LHW+Y +
Sbjct: 5 LTTLLFSSILISTLFSTSYGQNQNCSAFAFRNNQIFATCNALPLLNSVLHWSYHPDNHTV 64
Query: 65 DIAFRHKGITDTNRWVAWAINPNNDLASSMNGAQALVAILQSSGTPKAYTSSIANSRTQL 124
D+A+RH G+ +T+ WVAW +N + + M G+Q LVA SSG AYTS +++ TQL
Sbjct: 65 DLAYRHGGVPNTD-WVAWGLNIDG---TRMVGSQCLVAFRNSSGEIHAYTSPVSSYGTQL 120
Query: 125 AESNISYPHSGLIATHENNEVTIYASITLPVGTPSLVHLWQDGAMSGSTPQMHDMTSANT 184
AE +S+ + A + NNE I+A++ LP G S WQ+GA+SG H + N
Sbjct: 121 AEGALSFNVPRIGAEYSNNEFIIFATLELPAGRTSFNQAWQNGAVSGQALTAHVQSGDNM 180
Query: 185 QSKESLDLRSGASEQGSGGGSLS---RRRNTHGVLNAISWGILMPLGAVIARYLKVFKSA 241
+S S+D +G E G GG S++ RRRN HG+LNA+SWG+LMP+GAV ARYLKVFK+A
Sbjct: 181 RSFGSVDFANG--ELGGGGSSVTSRQRRRNVHGILNAVSWGVLMPMGAVFARYLKVFKAA 238
Query: 242 DPAWFYLHVTCQSAAYIVGVAGWGTGLKLGSDSAGVTYSTHRTLGIVIFCLGTLQVFALL 301
+PAWFY+HV CQ++AYIVGVAGWGTGLKLGSDS G+ ++THR +GI +FCLGTLQVFALL
Sbjct: 239 NPAWFYIHVACQTSAYIVGVAGWGTGLKLGSDSTGIEFTTHRNIGITLFCLGTLQVFALL 298
Query: 302 LRPKKDHKIRFYWNLYHWGVGYATIIISIINIFKGFEALEVSAADRYDNWKHAYTGIIAA 361
LRPK DHK R YWN+YH VGYA I ++I N+F+GF+AL + NWK AYTG+I A
Sbjct: 299 LRPKPDHKYRLYWNIYHHAVGYAVISLAITNVFQGFDAL-----NGQKNWKRAYTGVIIA 353
Query: 362 LGGVAVLLEAYTWIIVIKRKKSENKLQGMNGTNG--NGYGS 400
+G +AVLLEA+TW IVIKRKK+++ NGTNG N YG+
Sbjct: 354 IGAIAVLLEAFTWFIVIKRKKTDSNKHTQNGTNGTVNPYGN 394
>UniRef100_Q9FGK4 Arabidopsis thaliana genomic DNA, chromosome 5, P1 clone:MNJ7
[Arabidopsis thaliana]
Length = 395
Score = 405 bits (1040), Expect = e-111
Identities = 197/388 (50%), Positives = 273/388 (69%), Gaps = 15/388 (3%)
Query: 1 MVKNLRFMFLISILMFVTTTLAQTTTQTCKSQNFTNNAIFTSCRDLPQLTSYLHWTYDQT 60
M + + +S+ +F+ T A Q C + F+ N +F SC DLP L S+LH+TYD +
Sbjct: 1 MAISSNLLLCLSLFIFIITKSA--LAQKCSNYKFSTNRLFESCNDLPVLDSFLHYTYDSS 58
Query: 61 TGKLDIAFRHKGITDTNRWVAWAINPNNDLASSMNGAQALVAILQSSGTPKAYTSSIANS 120
+G L IA+RH +T +WVAWA+NP + + M GAQA+VA QS GT +AYTS I++
Sbjct: 59 SGNLQIAYRHTKLTP-GKWVAWAVNPTS---TGMVGAQAIVAYPQSDGTVRAYTSPISSY 114
Query: 121 RTQLAESNISYPHSGLIATHENNEVTIYASITLPVGTPSLVH-LWQDGAMSGSTPQMHDM 179
+T L E+ +S+ S L AT++NNE+ IYA + LP+ +++ +WQDG++SG+ P H
Sbjct: 115 QTSLLEAELSFNVSQLSATYQNNEMVIYAILNLPLANGGIINTVWQDGSLSGNNPLPHPT 174
Query: 180 TSANTQSKESLDLRSGAS---EQGSGGGSLSRRRNTHGVLNAISWGILMPLGAVIARYLK 236
+ N +S +L+L SGAS G+GG S R+RN HG+LN +SWGI+MP+GA+IARYLK
Sbjct: 175 SGNNVRSVSTLNLVSGASGSTSTGAGGASKLRKRNIHGILNGVSWGIMMPIGAIIARYLK 234
Query: 237 VFKSADPAWFYLHVTCQSAAYIVGVAGWGTGLKLGSDSAGVTYSTHRTLGIVIFCLGTLQ 296
V KSADPAWFYLHV CQS+AYI+GVAGW TGLKLG++SAG+ ++ HR +GI +FCL T+Q
Sbjct: 235 VSKSADPAWFYLHVFCQSSAYIIGVAGWATGLKLGNESAGIQFTFHRAVGIALFCLATIQ 294
Query: 297 VFALLLRPKKDHKIRFYWNLYHWGVGYATIIISIINIFKGFEALEVSAADRYDNWKHAYT 356
VFA+ LRPK +HK R YWN+YH VGY+ II++++N+FKG + L W++AYT
Sbjct: 295 VFAMFLRPKPEHKYRVYWNIYHHTVGYSVIILAVVNVFKGLDILSPE-----KQWRNAYT 349
Query: 357 GIIAALGGVAVLLEAYTWIIVIKRKKSE 384
II LG VAV+LE +TW +VIKR K+E
Sbjct: 350 AIIVVLGIVAVVLEGFTWYVVIKRGKAE 377
>UniRef100_Q9AR56 Putative membrane protein [Solanum tuberosum]
Length = 402
Score = 400 bits (1029), Expect = e-110
Identities = 197/379 (51%), Positives = 256/379 (66%), Gaps = 13/379 (3%)
Query: 27 QTCKSQNFTNNAIFTSCRDLPQLTSYLHWTYDQTTGKLDIAFRHKGITDTNRWVAWAINP 86
Q C + FTNN +F++C LP L S+LHWTY +D+A+RH G+T+++ WVAWA+N
Sbjct: 27 QNCSTHQFTNNNLFSTCNPLPVLNSFLHWTYHPDNHTVDLAYRHGGVTESS-WVAWALNL 85
Query: 87 NNDLASSMNGAQALVAILQSSGTPKAYTSSIANSRTQLAESNISYPHSGLIATHENNEVT 146
+ + M G Q+L+A SSG AYTS IA T L E +S+ + A +E+
Sbjct: 86 DG---TGMAGCQSLIAFRNSSGQIHAYTSPIAGYGTTLTEGALSFGVPRISAEFVRSEMI 142
Query: 147 IYASITLPVGTPSLVHLWQDGAMSGSTPQMHDMTSANTQSKESLDLRSGASEQGSGGG-- 204
I+A++ LP+ S +WQ+G +S ++H + N +S ++D SG + G+GGG
Sbjct: 143 IFATLELPINRTSFTQVWQNGQVSEQALRVHQTSGDNMRSVGTVDFASGQTSAGAGGGIS 202
Query: 205 --SLSRRRNTHGVLNAISWGILMPLGAVIARYLKVFKSADPAWFYLHVTCQSAAYIVGVA 262
+ RRRN HGVLNA+SWG+LMP+GA+ ARYLKVFKSA+PAWFYLH CQ+ AY VGVA
Sbjct: 203 ASARQRRRNIHGVLNAVSWGVLMPMGAIFARYLKVFKSANPAWFYLHAGCQTVAYAVGVA 262
Query: 263 GWGTGLKLGSDSAGVTYSTHRTLGIVIFCLGTLQVFALLLRPKKDHKIRFYWNLYHWGVG 322
GWGTGLKLGSDS G+ + THR +GI +FCLGTLQVFALLLRPK DHK R YWN+YH G
Sbjct: 263 GWGTGLKLGSDSVGIRFDTHRNIGITLFCLGTLQVFALLLRPKPDHKFRLYWNIYHHVTG 322
Query: 323 YATIIISIINIFKGFEALEVSAADRYDNWKHAYTGIIAALGGVAVLLEAYTWIIVIKRKK 382
Y II+SIIN+F+GF+AL + NWK AY G+I LG +AVLLEA TW IVIKRKK
Sbjct: 323 YTVIILSIINVFEGFDAL-----NGQKNWKKAYIGVIIFLGAIAVLLEAITWFIVIKRKK 377
Query: 383 SENKLQGMNGTNGNGYGSR 401
+ + +G NGY SR
Sbjct: 378 TSVSDKYPHGNGTNGYASR 396
>UniRef100_Q8VYH6 AT4g17280/dl4675c [Arabidopsis thaliana]
Length = 402
Score = 394 bits (1013), Expect = e-108
Identities = 198/400 (49%), Positives = 273/400 (67%), Gaps = 15/400 (3%)
Query: 1 MVKNLRFMFLISILMFVTTTLAQTTTQTCKSQNFTNNAIFTSCRDLPQLTSYLHWTYDQT 60
++K L + +S+++ ++ T + QTC F++N +F SC DLP L S+LH+TY+ +
Sbjct: 7 IMKFLNQILCLSLILSISMTTL-SFAQTCSKYKFSSNNVFDSCNDLPFLDSFLHYTYESS 65
Query: 61 TGKLDIAFRHKGITDTNRWVAWAINPNNDLASSMNGAQALVAILQSSGTPKAYTSSIANS 120
TG L IA+RH +T + +WVAWA+NP + + M GAQA+VA QS GT + YTS I +
Sbjct: 66 TGSLHIAYRHTKLT-SGKWVAWAVNPTS---TGMVGAQAIVAYPQSDGTVRVYTSPIRSY 121
Query: 121 RTQLAESNISYPHSGLIATHENNEVTIYASITLP--VGTPSLVH-LWQDGAMSGSTPQMH 177
+T L E ++S+ SGL AT++NNE+ + AS+ L +G ++ +WQDG+MSG++ H
Sbjct: 122 QTSLLEGDLSFNVSGLSATYQNNEIVVLASLKLAQDLGNGGTINTVWQDGSMSGNSLLPH 181
Query: 178 DMTSANTQSKESLDLRSGASEQ--GSGGGSLSRRRNTHGVLNAISWGILMPLGAVIARYL 235
+ N +S +L+L SG S G+GG S R+RN HG+LN +SWGI+MPLGA+IARYL
Sbjct: 182 PTSGNNVRSVSTLNLVSGVSAAAGGAGGSSKLRKRNIHGILNGVSWGIMMPLGAIIARYL 241
Query: 236 KVFKSADPAWFYLHVTCQSAAYIVGVAGWGTGLKLGSDSAGVTYSTHRTLGIVIFCLGTL 295
+V KSADPAWFY+HV CQ++AYI+GVAGW TGLKLG DS G+ YSTHR +GI +F L T+
Sbjct: 242 RVAKSADPAWFYIHVFCQASAYIIGVAGWATGLKLGGDSPGIQYSTHRAIGIALFSLATV 301
Query: 296 QVFALLLRPKKDHKIRFYWNLYHWGVGYATIIISIINIFKGFEALEVSAADRYDNWKHAY 355
QVFA+ LRPK +HK R YWN+YH +GY II+ ++N+FKG L WK+AY
Sbjct: 302 QVFAMFLRPKPEHKHRLYWNIYHHTIGYTIIILGVVNVFKGLGILSPK-----KQWKNAY 356
Query: 356 TGIIAALGGVAVLLEAYTWIIVIKRKKSENKLQGMNGTNG 395
GII L VA LLEA+TW +VIKR+K E K +NG
Sbjct: 357 IGIIVVLAIVATLLEAFTWYVVIKRRKLEAKTAQHGASNG 396
>UniRef100_Q9FKH6 Arabidopsis thaliana genomic DNA, chromosome 5, P1 clone:MXH1
[Arabidopsis thaliana]
Length = 404
Score = 364 bits (934), Expect = 3e-99
Identities = 189/402 (47%), Positives = 252/402 (62%), Gaps = 26/402 (6%)
Query: 8 MFLISILMFVTTTLAQTTTQTCKSQNFTNNAIFTSCRDLPQLTSYLHWTYDQTTGKLDIA 67
+F + + V T Q+ C + FTNN F C DL L S+LHWTY++ G + IA
Sbjct: 11 LFAVLATLLVLTVNGQSL---CNTHRFTNNLAFADCSDLSALGSFLHWTYNEQNGTVSIA 67
Query: 68 FRHKGITDTNRWVAWAINPNNDLASSMNGAQALVAILQSSGTP-KAYTSSIANSRTQLAE 126
+RH G T + WVAW +NP++ + M G QALVA ++ +AYTSS+++ T+L
Sbjct: 68 YRHPG-TSASSWVAWGLNPSS---TQMVGTQALVAFTNTTTNQFQAYTSSVSSYGTRLER 123
Query: 127 SNISYPHSGLIATHENNEVTIYASITLPVGTPSLVHLWQDGAMSGSTPQMHDMTSANTQS 186
S++S+ SGL AT + EVTI+A++ L + LWQ G + P H + N +S
Sbjct: 124 SSLSFGVSGLSATLVSGEVTIFATLELSPNLITANQLWQVGPVVNGVPASHQTSGDNMRS 183
Query: 187 KESLDLRSGASEQGSGG-GSLSRRRNTHGVLNAISWGILMPLGAVIARYLKVFKSADPAW 245
+D R+G + G GG G R+RNTHGVLNA+SWG+LMP+GA++ARY+KVF ADP W
Sbjct: 184 SGRIDFRTGQASAGGGGSGDRLRKRNTHGVLNAVSWGVLMPMGAMMARYMKVF--ADPTW 241
Query: 246 FYLHVTCQSAAYIVGVAGWGTGLKLGSDSAGVTYSTHRTLGIVIFCLGTLQVFALLLRPK 305
FYLH+ Q + Y++GVAGW TG+KLG+DS G +YSTHR LGI +F TLQVFALL+RPK
Sbjct: 242 FYLHIAFQVSGYVIGVAGWATGIKLGNDSPGTSYSTHRNLGIALFTFATLQVFALLVRPK 301
Query: 306 KDHKIRFYWNLYHWGVGYATIIISIINIFKGFEALEVSAADRYDNWKHAYTGIIAALGGV 365
DHK R YWN+YH VGY TII+SI+NIFKGF+ L D D W+ AY GI+ LG
Sbjct: 302 PDHKYRTYWNVYHHTVGYTTIILSIVNIFKGFDIL-----DPEDKWRWAYIGILIFLGAC 356
Query: 366 AVLLEAYTWIIVIKRKK----------SENKLQGMNGTNGNG 397
++LE TW IV++RK S G+NGT G
Sbjct: 357 VLILEPLTWFIVLRRKSRGGNTVAAPTSSKYSNGVNGTTTTG 398
>UniRef100_Q8LBJ4 Putative membrane protein [Arabidopsis thaliana]
Length = 404
Score = 361 bits (926), Expect = 2e-98
Identities = 188/402 (46%), Positives = 251/402 (61%), Gaps = 26/402 (6%)
Query: 8 MFLISILMFVTTTLAQTTTQTCKSQNFTNNAIFTSCRDLPQLTSYLHWTYDQTTGKLDIA 67
+F + + V T Q+ C + FTNN F C DL L S+LHWTY++ G + IA
Sbjct: 11 LFAVLATLLVLTVNGQSL---CNTHRFTNNLAFADCSDLSALGSFLHWTYNEQNGTVSIA 67
Query: 68 FRHKGITDTNRWVAWAINPNNDLASSMNGAQALVAILQSSGTP-KAYTSSIANSRTQLAE 126
+RH G T + WVAW +NP++ + M G QALVA ++ +AYTSS+++ T+L
Sbjct: 68 YRHPG-TSASSWVAWGLNPSS---TQMVGTQALVAFTNTTTNQFQAYTSSVSSYGTRLER 123
Query: 127 SNISYPHSGLIATHENNEVTIYASITLPVGTPSLVHLWQDGAMSGSTPQMHDMTSANTQS 186
S++S+ SGL AT + EVTI+A++ L + LWQ G + P H + N +S
Sbjct: 124 SSLSFGVSGLSATLVSGEVTIFATLELSPNLITANQLWQVGPVVNGVPASHQTSGDNMRS 183
Query: 187 KESLDLRSGASEQGSGG-GSLSRRRNTHGVLNAISWGILMPLGAVIARYLKVFKSADPAW 245
+D R+G + G GG G R+RNTHGVLNA+SWG+LMP+GA++ARY+KVF ADP W
Sbjct: 184 SGRIDFRTGQASAGGGGSGDRLRKRNTHGVLNAVSWGVLMPMGAMMARYMKVF--ADPTW 241
Query: 246 FYLHVTCQSAAYIVGVAGWGTGLKLGSDSAGVTYSTHRTLGIVIFCLGTLQVFALLLRPK 305
FYLH+ Q + Y++GVAGW T +KLG+DS G +YSTHR LGI +F TLQVFALL+RPK
Sbjct: 242 FYLHIAFQVSGYVIGVAGWATRIKLGNDSPGTSYSTHRNLGIALFTFATLQVFALLVRPK 301
Query: 306 KDHKIRFYWNLYHWGVGYATIIISIINIFKGFEALEVSAADRYDNWKHAYTGIIAALGGV 365
DHK R YWN+YH VGY TII+SI+NIFKGF+ L D D W+ AY GI+ LG
Sbjct: 302 PDHKYRTYWNVYHHTVGYTTIILSIVNIFKGFDIL-----DPEDKWRWAYIGILIFLGAC 356
Query: 366 AVLLEAYTWIIVIKRKK----------SENKLQGMNGTNGNG 397
++LE TW IV++RK S G+NGT G
Sbjct: 357 VLILEPLTWFIVLRRKSRGGNTVAAPTSSKYSNGVNGTTTTG 398
>UniRef100_Q8H7W7 Putative membrane protein [Oryza sativa]
Length = 384
Score = 341 bits (874), Expect = 2e-92
Identities = 175/389 (44%), Positives = 237/389 (59%), Gaps = 12/389 (3%)
Query: 10 LISILMFVTTTLAQTTTQTCKSQNFTNNAIFTSCRDLPQLTSYLHWTYDQTTGKLDIAFR 69
L++ ++ + + Q C S F N F C LP L + LHWT+ G D+AFR
Sbjct: 5 LLAGVLLLQLAASAAGQQQCLSATFQNGQTFLKCNPLPVLGASLHWTHHAENGTADVAFR 64
Query: 70 HKGITDTNRWVAWAINPNNDLASSMNGAQALVAILQSSGTPKAYTSSIANSRTQLAESNI 129
++ WVAW IN ++M G+ +A SG+ + + N+ L ++
Sbjct: 65 AP--QQSSGWVAWGINTRG---TTMPGSSVFIASQDGSGSVSVLMTVLENTSPSLTNGSL 119
Query: 130 SYPH-SGLIATHENNEVTIYASITLPVGTPSLVHLWQDGAMSGSTPQMHDMTSANTQSKE 188
S+ S A + N TI+A+I LP + + +WQ G S H + N QS
Sbjct: 120 SFDVLSPPTADYTNGVYTIFATIALPNNSTTQNTVWQAGPGSTGNVGQHATSGPNVQSML 179
Query: 189 SLDLRSGASEQGSGGGSLSRRRNTHGVLNAISWGILMPLGAVIARYLKVFKSADPAWFYL 248
LD SG S G+ S RRN HG+LNA+SWGIL+P+GA+IARYL+VF++ADPAWFYL
Sbjct: 180 RLDFSSGQST-GTASNSRLHRRNIHGILNAVSWGILIPMGAMIARYLRVFEAADPAWFYL 238
Query: 249 HVTCQSAAYIVGVAGWGTGLKLGSDSAGVTYSTHRTLGIVIFCLGTLQVFALLLRPKKDH 308
H+TCQ + YI+GVAGW GLKLGS+S G+TYS HR +GI IFCL TLQVFALLLRP K +
Sbjct: 239 HITCQLSGYILGVAGWALGLKLGSESKGITYSAHRNIGIAIFCLATLQVFALLLRPDKKN 298
Query: 309 KIRFYWNLYHWGVGYATIIISIINIFKGFEALEVSAADRYDNWKHAYTGIIAALGGVAVL 368
K RFYWN+YH VGY+ I+++ +NIFKG + L+ ++ WK +Y I+A L GVA+L
Sbjct: 299 KYRFYWNIYHHSVGYSAIVLAAVNIFKGLDILKPAS-----GWKRSYIAILATLAGVALL 353
Query: 369 LEAYTWIIVIKRKKSENKLQGMNGTNGNG 397
LEA TW+IV++RKKS+ TNGNG
Sbjct: 354 LEAITWVIVLRRKKSDKSSSPYGATNGNG 382
>UniRef100_Q7F1F1 Putative auxin-induced protein [Oryza sativa]
Length = 390
Score = 335 bits (859), Expect = 1e-90
Identities = 176/393 (44%), Positives = 236/393 (59%), Gaps = 15/393 (3%)
Query: 8 MFLISILMFVTTTLAQTTTQTCKSQNFTNNAIFTSCRDLPQLTSYLHWTYDQT-TGKLDI 66
+FL S+L+ A+ C ++ F++N ++ +C DLP L + +HWTYD + L +
Sbjct: 6 VFLASLLLAAVAP-ARAAGGGCAAERFSSNRVYAACSDLPHLGASVHWTYDAAASASLSV 64
Query: 67 AFRHKGITDTNRWVAWAINPNNDLASSMNGAQALVAILQSSGTPKAYTSSIANSRTQLAE 126
AF WVAW +NP M G QALVA+ + G + + A
Sbjct: 65 AFV-AAPPSPGGWVAWGLNPTG---GGMAGTQALVALPKGGGGGYEVQTFDIEGYSLSAP 120
Query: 127 SNISYPHSGLIA-THENNEVTIYASITLPVGTPSLVHLWQDGAMSGSTPQMHDMTSANTQ 185
+ YP + L A + V+++ + L GT + +WQ G +S + H M+S N
Sbjct: 121 GKLKYPATDLAAEVAADGRVSVFGKLALQNGTAEVNQVWQVGPVSSGSMVPHAMSSDNKA 180
Query: 186 SKESLDLRSGASEQGSGGGSLSRRRNTHGVLNAISWGILMPLGAVIARYLKVFKSADPAW 245
+ L+L +GA+ GGGS R++NTHG+LNA+SWG+L+P+GA+ ARYLK FKSADPAW
Sbjct: 181 AMGKLNLLTGAATSSGGGGSNLRKKNTHGILNAVSWGLLLPMGAIFARYLKTFKSADPAW 240
Query: 246 FYLHVTCQSAAYIVGVAGWGTGLKLGSDSAGVTYSTHRTLGIVIFCLGTLQVFALLLRPK 305
FYLHV CQ Y VGV+GW TG+ LG+ S G+TYS HR +GI +F LGTLQ+FAL LRPK
Sbjct: 241 FYLHVACQLIGYGVGVSGWATGIHLGNLSKGITYSLHRNIGITVFALGTLQIFALFLRPK 300
Query: 306 KDHKIRFYWNLYHWGVGYATIIISIINIFKGFEALEVSAADRYDNWKHAYTGIIAALGGV 365
KDHK R YWN YH VGY II+ I+NIFKG L V WK Y I+ LGG+
Sbjct: 301 KDHKYRIYWNAYHHSVGYTIIILGIVNIFKGMSILNVE-----QKWKTGYIITISILGGI 355
Query: 366 AVLLEAYTWIIVIKRKKSENKLQGMNGTNGNGY 398
AV+LEA TW IV+KR+K ENK NG + NG+
Sbjct: 356 AVILEAVTWSIVLKRRKEENK--SYNGAS-NGH 385
>UniRef100_Q9LSE7 Emb|CAB45497.1 [Arabidopsis thaliana]
Length = 393
Score = 319 bits (818), Expect = 8e-86
Identities = 166/383 (43%), Positives = 234/383 (60%), Gaps = 21/383 (5%)
Query: 15 MFVTTTLAQTTTQTCKSQNFTNNAIFTSCRDLPQLTSYLHWTYDQTTGKLDIAFRHKGIT 74
+F+ ++ +QTCKSQ F+ + + C DLPQL ++LH++YD + L + F
Sbjct: 14 IFLALLISPAVSQTCKSQTFSGDKTYPHCLDLPQLKAFLHYSYDASNTTLAVVFSAPP-A 72
Query: 75 DTNRWVAWAINPNNDLASSMNGAQALVAILQSSGTPKAYTSSIANSRTQLAESNISYPHS 134
W+AWAINP A+ M G+Q LVA + +S + L S +++
Sbjct: 73 KPGGWIAWAINPK---ATGMVGSQTLVAYKDPGNGVAVVKTLNISSYSSLIPSKLAFDVW 129
Query: 135 GLIA---THENNEVTIYASITLP---VGTPSLVHLWQDGAMSGSTPQM--HDMTSANTQS 186
+ A + + I+A + +P V + +WQ G G + H SAN S
Sbjct: 130 DMKAEEAARDGGSLRIFARVKVPADLVAKGKVNQVWQVGPELGPGGMIGRHAFDSANLAS 189
Query: 187 KESLDLRSGASEQGSGGG----SLSRRRNTHGVLNAISWGILMPLGAVIARYLKVFKSAD 242
SLDL+ S GG + + RN HG+LNA+SWGIL P+GA+IARY++VF SAD
Sbjct: 190 MSSLDLKGDNSGGTISGGDEVNAKIKNRNIHGILNAVSWGILFPIGAIIARYMRVFDSAD 249
Query: 243 PAWFYLHVTCQSAAYIVGVAGWGTGLKLGSDSAGVTYSTHRTLGIVIFCLGTLQVFALLL 302
PAWFYLHV+CQ +AY++GVAGW TGLKLG++S G+ +S HR +GI +F L T+Q+FA+LL
Sbjct: 250 PAWFYLHVSCQFSAYVIGVAGWATGLKLGNESEGIRFSAHRNIGIALFTLATIQMFAMLL 309
Query: 303 RPKKDHKIRFYWNLYHWGVGYATIIISIINIFKGFEALEVSAADRYDNWKHAYTGIIAAL 362
RPKKDHK RFYWN+YH GVGYA + + IIN+FKG L+ D +K AY +IA L
Sbjct: 310 RPKKDHKYRFYWNIYHHGVGYAILTLGIINVFKGLNILKPQ-----DTYKTAYIAVIAVL 364
Query: 363 GGVAVLLEAYTWIIVIKRKKSEN 385
GG+A+LLEA TW++V+KRK + +
Sbjct: 365 GGIALLLEAITWVVVLKRKSNNS 387
>UniRef100_Q9SV71 Hypothetical protein At4g12980 [Arabidopsis thaliana]
Length = 394
Score = 310 bits (793), Expect = 6e-83
Identities = 167/394 (42%), Positives = 239/394 (60%), Gaps = 26/394 (6%)
Query: 4 NLRFMFLISILMFVTTTLAQTTTQTCKSQNFTNNAIFTSCRDLPQLTSYLHWTYDQTTGK 63
+L F+F +L + A + + +C SQ F+ + C DLP L + LH++YD +
Sbjct: 9 SLSFLFWALLL-----SPAVSQSSSCSSQTFSGVKSYPHCLDLPDLKAILHYSYDASNTT 63
Query: 64 LDIAFRHKGITDTNRWVAWAINPNNDLASSMNGAQALVAILQSSGTPKAYTSSIANSRTQ 123
L + F + W+AWAINP + + M G+QALVA S + T+ S +
Sbjct: 64 LAVVFSAPP-SKPGGWIAWAINPKS---TGMAGSQALVASKDPSTGVASVTTLNIVSYSS 119
Query: 124 LAESNISYPHSGLIATHENNE---VTIYASITLPV---GTPSLVHLWQDG-AMSGSTPQM 176
L S +S+ + A N+ + I+A + +P + + +WQ G +S Q
Sbjct: 120 LVPSKLSFDVWDVKAEEAANDGGALRIFAKVKVPADLAASGKVNQVWQVGPGVSNGRIQA 179
Query: 177 HDMTSANTQSKESLDLRS---GASEQGSGGGSLSR--RRNTHGVLNAISWGILMPLGAVI 231
HD + N S SLDL G G GG SR +RN HG+LNA+SWG+L P+GA+I
Sbjct: 180 HDFSGPNLNSVGSLDLTGTTPGVPVSGGGGAGNSRIHKRNIHGILNAVSWGLLFPIGAMI 239
Query: 232 ARYLKVFKSADPAWFYLHVTCQSAAYIVGVAGWGTGLKLGSDSAGVTYSTHRTLGIVIFC 291
ARY+++F+SADPAWFYLHV+CQ +AY++GVAGW TGLKLGS+S G+ Y+THR +GI +F
Sbjct: 240 ARYMRIFESADPAWFYLHVSCQFSAYVIGVAGWATGLKLGSESKGIQYNTHRNIGICLFS 299
Query: 292 LGTLQVFALLLRPKKDHKIRFYWNLYHWGVGYATIIISIINIFKGFEALEVSAADRYDNW 351
+ TLQ+FA+LLRP+KDHK RF WN+YH GVGY+ +I+ IIN+FKG L +
Sbjct: 300 IATLQMFAMLLRPRKDHKFRFVWNIYHHGVGYSILILGIINVFKGLSILNPK-----HTY 354
Query: 352 KHAYTGIIAALGGVAVLLEAYTWIIVIKRKKSEN 385
K AY +I LGG+ +LLE TW+IV+KRK +++
Sbjct: 355 KTAYIAVIGTLGGITLLLEVVTWVIVLKRKSAKS 388
>UniRef100_Q8L9T0 Hypothetical protein [Arabidopsis thaliana]
Length = 394
Score = 308 bits (788), Expect = 2e-82
Identities = 167/394 (42%), Positives = 238/394 (60%), Gaps = 26/394 (6%)
Query: 4 NLRFMFLISILMFVTTTLAQTTTQTCKSQNFTNNAIFTSCRDLPQLTSYLHWTYDQTTGK 63
+L F+F +L + A + + +C SQ F+ + C DLP L + LH++YD +
Sbjct: 9 SLSFLFWALLL-----SPAVSQSSSCSSQTFSGVKSYPHCLDLPDLKAILHYSYDASNTT 63
Query: 64 LDIAFRHKGITDTNRWVAWAINPNNDLASSMNGAQALVAILQSSGTPKAYTSSIANSRTQ 123
L + F + W+AWAINP + + M G+QALVA S + T+ S +
Sbjct: 64 LAVVFSAPP-SKPGGWIAWAINPKS---TGMAGSQALVASKDPSTGVASVTTLNIVSYSS 119
Query: 124 LAESNISYPHSGLIATHENNE---VTIYASITLPV---GTPSLVHLWQDG-AMSGSTPQM 176
L S +S+ + A N+ + I+A + +P + + +WQ G +S Q
Sbjct: 120 LVPSKLSFDVWDVKAEEAANDGGALRIFAKVKVPADLAASGKVNQVWQVGPGVSNGRIQA 179
Query: 177 HDMTSANTQSKESLDLRS---GASEQGSGGGSLSR--RRNTHGVLNAISWGILMPLGAVI 231
HD + N S SLDL G G GG SR +RN HG+LNA+SWG+L P+GA+I
Sbjct: 180 HDFSGPNLNSVGSLDLTGTTPGVPVSGGGGAGNSRIHKRNIHGILNAVSWGLLFPIGAMI 239
Query: 232 ARYLKVFKSADPAWFYLHVTCQSAAYIVGVAGWGTGLKLGSDSAGVTYSTHRTLGIVIFC 291
ARY+++F+SADPAWFYLHV+CQ +AY +GVAGW TGLKLGS+S G+ Y+THR +GI +F
Sbjct: 240 ARYMRIFESADPAWFYLHVSCQFSAYAIGVAGWATGLKLGSESKGIQYNTHRNIGISLFS 299
Query: 292 LGTLQVFALLLRPKKDHKIRFYWNLYHWGVGYATIIISIINIFKGFEALEVSAADRYDNW 351
+ TLQ+FA+LLRP+KDHK RF WN+YH GVGY+ +I+ IIN+FKG L +
Sbjct: 300 IATLQMFAMLLRPRKDHKFRFVWNIYHHGVGYSILILGIINVFKGLSILNPK-----HTY 354
Query: 352 KHAYTGIIAALGGVAVLLEAYTWIIVIKRKKSEN 385
K AY +I LGG+ +LLE TW+IV+KRK +++
Sbjct: 355 KTAYIAVIGTLGGITLLLEVVTWVIVLKRKSAKS 388
>UniRef100_Q9LYS9 Hypothetical protein F17J16_120 [Arabidopsis thaliana]
Length = 466
Score = 268 bits (684), Expect = 3e-70
Identities = 146/346 (42%), Positives = 207/346 (59%), Gaps = 14/346 (4%)
Query: 29 CKSQNFTNNAIFTSCRDLPQLTSYLHWTYDQTTGKLDIAFRHKGITDTNRWVAWAINPNN 88
C+S +F N F SC DL L SYLH+ Y Q TG L+IA+ H + +++ W++WAINP +
Sbjct: 37 CESYSFNNGKSFRSCTDLLVLNSYLHFNYAQETGVLEIAYHHSNL-ESSSWISWAINPTS 95
Query: 89 DLASSMNGAQALVAILQS-SGTPKAYTSSIANSRTQLAESNISYPHSGLIATHENNEVTI 147
M GAQALVA S SG +AYTSSI + L ES +S + + A + N E+ I
Sbjct: 96 ---KGMVGAQALVAYRNSTSGVMRAYTSSINSYSPMLQESPLSLRVTQVSAEYSNGEMMI 152
Query: 148 YASITLPVGTPSLVHLWQDGAMS-GSTPQMHDMTSANTQSKESLDLRSG-ASEQGSGGGS 205
+A++ LP T + HLWQDG + G MH M+ N +S SLDL SG + S +
Sbjct: 153 FATLVLPPNTTVVNHLWQDGPLKEGDRLGMHAMSGDNLKSMASLDLLSGQVTTTKSVNRN 212
Query: 206 LSRRRNTHGVLNAISWGILMPLGAVIARYLKVFKSADPAWFYLHVTCQSAAYIVG-VAGW 264
+ + H ++NA+SWGILMP+G + ARY+K ++ DP WFY+HV CQ+ Y G + G
Sbjct: 213 MLLVKQIHAIVNALSWGILMPIGVMAARYMKNYEVLDPTWFYIHVVCQTTGYFSGLIGGL 272
Query: 265 GTGLKLGSDSAGVTYSTHRTLGIVIFCLGTLQVFALLLRPKKDHKIRFYWNLYHWGVGYA 324
GT + + + G+ + H +G+++F LG LQ+ +L RP KDHK R YWN YH +GY
Sbjct: 273 GTAIYMARHT-GMRTTLHTVIGLLLFALGFLQILSLKARPNKDHKYRKYWNWYHHTMGYI 331
Query: 325 TIIISIINIFKGFEALEVSAADRYDNWKHAYTGIIAALGGVAVLLE 370
I++SI NI+KG L+ + WK AYT II + AV++E
Sbjct: 332 VIVLSIYNIYKGLSILQPGSI-----WKIAYTTIICCIAAFAVVME 372
>UniRef100_O23571 Hypothetical protein dl4675c [Arabidopsis thaliana]
Length = 273
Score = 228 bits (580), Expect = 3e-58
Identities = 115/232 (49%), Positives = 162/232 (69%), Gaps = 9/232 (3%)
Query: 27 QTCKSQNFTNNAIFTSCRDLPQLTSYLHWTYDQTTGKLDIAFRHKGITDTNRWVAWAINP 86
QTC F++N +F SC DLP L S+LH+TY+ +TG L IA+RH +T + +WVAWA+NP
Sbjct: 8 QTCSKYKFSSNNVFDSCNDLPFLDSFLHYTYESSTGSLHIAYRHTKLT-SGKWVAWAVNP 66
Query: 87 NNDLASSMNGAQALVAILQSSGTPKAYTSSIANSRTQLAESNISYPHSGLIATHENNEVT 146
+ + M GAQA+VA QS GT + YTS I + +T L E ++S+ SGL AT++NNE+
Sbjct: 67 TS---TGMVGAQAIVAYPQSDGTVRVYTSPIRSYQTSLLEGDLSFNVSGLSATYQNNEIV 123
Query: 147 IYASITLP--VGTPSLVH-LWQDGAMSGSTPQMHDMTSANTQSKESLDLRSGASEQ--GS 201
+ AS+ L +G ++ +WQDG+MSG++ H + N +S +L+L SG S G+
Sbjct: 124 VLASLKLAQDLGNGGTINTVWQDGSMSGNSLLPHPTSGNNVRSVSTLNLVSGVSAAAGGA 183
Query: 202 GGGSLSRRRNTHGVLNAISWGILMPLGAVIARYLKVFKSADPAWFYLHVTCQ 253
GG S R+RN HG+LN +SWGI+MPLGA+IARYL+V KSADPAWFY+HV+ +
Sbjct: 184 GGSSKLRKRNIHGILNGVSWGIMMPLGAIIARYLRVAKSADPAWFYIHVSAR 235
>UniRef100_Q9SJ74 Expressed protein [Arabidopsis thaliana]
Length = 404
Score = 226 bits (576), Expect = 9e-58
Identities = 137/405 (33%), Positives = 205/405 (49%), Gaps = 29/405 (7%)
Query: 8 MFLISILMFVTTTLAQTTTQTCKSQNFTNNAIFTSCRDLPQLTSYLHWTYDQTTGKLDIA 67
+ L +L+ + T L ++ C + T + F C LP + + WTY LD+
Sbjct: 4 LILSFLLLLLATKLPESLAGHCTTTTATKS--FEKCISLPTQQASIAWTYHPHNATLDLC 61
Query: 68 FRHKGITDTNRWVAWAINPNNDLASSMNGAQALVAILQSSG-----TPKAYTSSIANSR- 121
F I+ + WV W INP D + M G++ L+A + P SS+ +
Sbjct: 62 FFGTFISPSG-WVGWGINP--DSPAQMTGSRVLIAFPDPNSGQLILLPYVLDSSVKLQKG 118
Query: 122 ---------TQLAESNISYPHSGLIATHENN-EVTIYASITLPVGTPSLVHLWQDGA-MS 170
+L+ S+ S + G +AT N V IYAS+ L + H+W G +
Sbjct: 119 PLLSRPLDLVRLSSSSASL-YGGKMATIRNGASVQIYASVKLSSNNTKIHHVWNRGLYVQ 177
Query: 171 GSTPQMHDMTSANTQSKESLDLRSGASEQGSGGGSLSRRRNTHGVLNAISWGILMPLGAV 230
G +P +H TS + S + D+ SG + GS + + THGV+NAISWG L+P GAV
Sbjct: 178 GYSPTIHPTTSTDLSSFSTFDVTSGFATVNQNSGSRALKV-THGVVNAISWGFLLPAGAV 236
Query: 231 IARYLKVFKSADPAWFYLHVTCQSAAYIVGVAGWGTGLKLGSDSAGVTYSTHRTLGIVIF 290
ARYL+ +S P WFY+H Q +++G G+ G+ LG +S GVTY HR+LGI F
Sbjct: 237 TARYLRQMQSIGPTWFYIHAAIQLTGFLLGTIGFSIGIVLGHNSPGVTYGLHRSLGIATF 296
Query: 291 CLGTLQVFALLLRPKKDHKIRFYWNLYHWGVGYATIIISIINIFKGFEALEVSAADRYDN 350
LQ ALL RPK +K R YW YH VGYA +++ ++N+F+GFE L +
Sbjct: 297 TAAALQTLALLFRPKTTNKFRRYWKSYHHFVGYACVVMGVVNVFQGFEVLREGRSYA--- 353
Query: 351 WKHAYTGIIAALGGVAVLLEAYTWIIVIKRKKSEN-KLQGMNGTN 394
K Y ++ L GV V +E +W++ ++ K E K G+ G +
Sbjct: 354 -KLGYCLCLSTLVGVCVAMEVNSWVVFCRKAKEEKMKRDGLTGVD 397
>UniRef100_Q8LFW0 Hypothetical protein [Arabidopsis thaliana]
Length = 404
Score = 226 bits (576), Expect = 9e-58
Identities = 137/405 (33%), Positives = 205/405 (49%), Gaps = 29/405 (7%)
Query: 8 MFLISILMFVTTTLAQTTTQTCKSQNFTNNAIFTSCRDLPQLTSYLHWTYDQTTGKLDIA 67
+ L +L+ + T L ++ C + T + F C LP + + WTY LD+
Sbjct: 4 LILSFLLLLLATKLPESLAGHCTTTTATKS--FEKCISLPTQQTSIAWTYHPHNATLDLC 61
Query: 68 FRHKGITDTNRWVAWAINPNNDLASSMNGAQALVAILQSSG-----TPKAYTSSIANSR- 121
F I+ + WV W INP D + M G++ L+A + P SS+ +
Sbjct: 62 FFGTFISPSG-WVGWGINP--DSPAQMTGSRVLIAFPDPNSGQLILLPYVLDSSVKLQKG 118
Query: 122 ---------TQLAESNISYPHSGLIATHENN-EVTIYASITLPVGTPSLVHLWQDGA-MS 170
+L+ S+ S + G +AT N V IYAS+ L + H+W G +
Sbjct: 119 PLLSRPLDLVRLSSSSASL-YGGKMATIRNGASVQIYASVKLSSNNTKIHHVWNRGLYVQ 177
Query: 171 GSTPQMHDMTSANTQSKESLDLRSGASEQGSGGGSLSRRRNTHGVLNAISWGILMPLGAV 230
G +P +H TS + S + D+ SG + GS + + THGV+NAISWG L+P GAV
Sbjct: 178 GYSPTIHPTTSTDLSSFSTFDVTSGFATVNQNSGSRALKV-THGVVNAISWGFLLPAGAV 236
Query: 231 IARYLKVFKSADPAWFYLHVTCQSAAYIVGVAGWGTGLKLGSDSAGVTYSTHRTLGIVIF 290
ARYL+ +S P WFY+H Q +++G G+ G+ LG +S GVTY HR+LGI F
Sbjct: 237 TARYLRQMQSIGPTWFYIHAAIQLTGFLLGTIGFSIGIVLGHNSPGVTYGLHRSLGIATF 296
Query: 291 CLGTLQVFALLLRPKKDHKIRFYWNLYHWGVGYATIIISIINIFKGFEALEVSAADRYDN 350
LQ ALL RPK +K R YW YH VGYA +++ ++N+F+GFE L +
Sbjct: 297 TAAALQTLALLFRPKTTNKFRRYWKSYHHFVGYACVVMGVVNVFQGFEVLREGRSYA--- 353
Query: 351 WKHAYTGIIAALGGVAVLLEAYTWIIVIKRKKSEN-KLQGMNGTN 394
K Y ++ L GV V +E +W++ ++ K E K G+ G +
Sbjct: 354 -KLGYCLCLSTLVGVCVAMEVNSWVVFCRKAKEEKMKRDGLTGVD 397
>UniRef100_Q8H7W3 Hypothetical protein OSJNBa0064E16.14 [Oryza sativa]
Length = 417
Score = 208 bits (530), Expect = 2e-52
Identities = 122/363 (33%), Positives = 182/363 (49%), Gaps = 24/363 (6%)
Query: 40 FTSCRDLPQLTSYLHWTYDQTTGKLDIAFRHKGITDTNRWVAWAINPNNDLASSMNGAQA 99
+ C LP + L WTYD LD AF I+ + WVAW +N + A +M GA+
Sbjct: 50 YAKCIALPTQGATLAWTYDARNATLDAAFTGSFISPSG-WVAWGVNKD---APAMTGARV 105
Query: 100 LVAILQSS-----GTPKAYTSSIANSRTQLAESNISYPHSGLIAT--------HENNEVT 146
L A S P + + + L + P A+ + VT
Sbjct: 106 LAAFSDPSTGALLALPFLLSPDVKLQASPLVSRPLDIPLLASSASLVDPARTVRDGATVT 165
Query: 147 IYASITLPVGTPSLVHLWQDGA-MSGSTPQMHDMTSANTQSKESLDLRSGASEQGSGGGS 205
I A+I L L +W G + G +P +H +++ S ++D+ + A+E +
Sbjct: 166 IAATIRLSPNRTKLHFVWNRGLYVQGYSPTIHPTDASDLASHATVDILTTATEASPTASA 225
Query: 206 LSRRRNTHGVLNAISWGILMPLGAVIARYLKVFKSADPAWFYLHVTCQSAAYIVGVAGWG 265
+ THG LNA+SWG L+P+GA +ARYL+ S PAWFY H Q+ Y +G AG+
Sbjct: 226 TLQW--THGSLNALSWGFLLPVGAAVARYLRPCASTGPAWFYAHAAIQATGYALGAAGFA 283
Query: 266 TGLKLGSDSAGVTYSTHRTLGIVIFCLGTLQVFALLLRPKKDHKIRFYWNLYHWGVGYAT 325
GL +GS S GVTY HR LGI G+LQ A+L RPK ++ R YW YH VGY
Sbjct: 284 LGLVMGSASPGVTYKLHRGLGIAAATAGSLQTLAMLFRPKTTNRYRKYWKSYHHLVGYGC 343
Query: 326 IIISIINIFKGFEALEVSAADRYDNWKHAYTGIIAALGGVAVLLEAYTWIIVIKRKKSEN 385
+++ ++N+F+GFE + + A+ WK Y +A L G V LE W++ +R++ E
Sbjct: 344 VVVGVVNVFQGFEVMGLGAS----YWKLGYCMALATLAGGCVALEVNAWVVFCRRQQEEK 399
Query: 386 KLQ 388
++
Sbjct: 400 LMR 402
>UniRef100_Q9FKC1 Emb|CAB86935.1 [Arabidopsis thaliana]
Length = 255
Score = 169 bits (427), Expect = 2e-40
Identities = 93/243 (38%), Positives = 138/243 (56%), Gaps = 7/243 (2%)
Query: 9 FLISILMFVTTTLAQTTTQTCKSQNFTNNAIFTSCRDLPQLTSYLHWTYDQTTGKLDIAF 68
FL + T +C S NF N F SC DLP L S+LH++Y + TG L++A+
Sbjct: 14 FLFVLAPCFTRATTNEVQVSCDSHNFNNGKHFRSCVDLPVLDSFLHYSYVRETGVLEVAY 73
Query: 69 RHKGITDTNRWVAWAINPNNDLASSMNGAQALVAILQS-SGTPKAYTSSIANSRTQLAES 127
RH I +++ W+AW INP + M GAQ L+A S SG +AYTSSI L E
Sbjct: 74 RHTNI-ESSSWIAWGINPTS---KGMIGAQTLLAYRNSTSGFMRAYTSSINGYTPMLQEG 129
Query: 128 NISYPHSGLIATHENNEVTIYASITLPVGTPSLVHLWQDGAMS-GSTPQMHDMTSANTQS 186
+S+ + L A + N E+TI+A++ P T + HLWQDG + G MH M+ + +S
Sbjct: 130 PLSFRVTQLSAEYLNREMTIFATMVWPSNTTVVNHLWQDGPLKEGDRLGMHAMSGNHLKS 189
Query: 187 KESLDLRSG-ASEQGSGGGSLSRRRNTHGVLNAISWGILMPLGAVIARYLKVFKSADPAW 245
+LDL SG + ++ ++ HG++NA+ WGI +P+G + ARY++ +K DP W
Sbjct: 190 MANLDLLSGQVMTTKAANDNMLLVKSIHGLVNAVCWGIFIPIGVMAARYMRTYKGLDPTW 249
Query: 246 FYL 248
FY+
Sbjct: 250 FYI 252
>UniRef100_Q8S0J3 B1078G07.42 protein [Oryza sativa]
Length = 685
Score = 138 bits (347), Expect = 3e-31
Identities = 89/269 (33%), Positives = 126/269 (46%), Gaps = 16/269 (5%)
Query: 108 GTPKAYTSSIANSRT-QLAESNISYPHSGLIATHENNEVTIYASITLPVGTPSLVHLWQD 166
G K Y +SR+ + ++S + +++ + + T P TP L++
Sbjct: 163 GVAKQYKLGGTSSRSCPPDQGSLSLVAKNTLVVAQSSRIYVAFQFTAPQPTPYLIYAV-- 220
Query: 167 GAMSGSTPQMHDMTSANTQSKESLDLRSGASEQGSGGGSLSRRRNTHGVLNAISWGILMP 226
S + P + A Q S + A S GG + + HG + + WG+LMP
Sbjct: 221 -GPSNTNPSGNGDYLAQHQVYTSAAVNYAAGTTSSAGGGAADTKKWHGAMAGLGWGVLMP 279
Query: 227 LGAVIARYLKVFKSADPAWFYLHVTCQSAAYIVGVAGWGTGLKLGSDSAGVTYSTHRTLG 286
+G +ARY FK DP WFY H++ Q +++GVAG G KL D G TH+ +G
Sbjct: 280 VGIALARY---FKKHDPFWFYAHISVQGVGFVLGVAGVVAGFKLNDDVPGG--DTHQAIG 334
Query: 287 IVIFCLGTLQVFALLLRPKKDHKIRFYWNLYHWGVGYATIIISIINIFKGFE-ALEVSAA 345
I + LG LQV A L RP K K+R YWN YH VG A + + NIF G A E +AA
Sbjct: 335 ITVLVLGCLQVLAFLARPDKSSKVRRYWNWYHHNVGRAAVACAAANIFIGLNIAHEGNAA 394
Query: 346 DRYDNWKHAYTGIIAALGGVAVLLEAYTW 374
+ Y + L VAV LE W
Sbjct: 395 ------RAGYGIFLVVLALVAVFLEVKLW 417
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.319 0.133 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 667,650,988
Number of Sequences: 2790947
Number of extensions: 27468402
Number of successful extensions: 88341
Number of sequences better than 10.0: 92
Number of HSP's better than 10.0 without gapping: 52
Number of HSP's successfully gapped in prelim test: 41
Number of HSP's that attempted gapping in prelim test: 88111
Number of HSP's gapped (non-prelim): 129
length of query: 402
length of database: 848,049,833
effective HSP length: 130
effective length of query: 272
effective length of database: 485,226,723
effective search space: 131981668656
effective search space used: 131981668656
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 76 (33.9 bits)
Medicago: description of AC148482.6


