

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148482.4 + phase: 0
(337 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
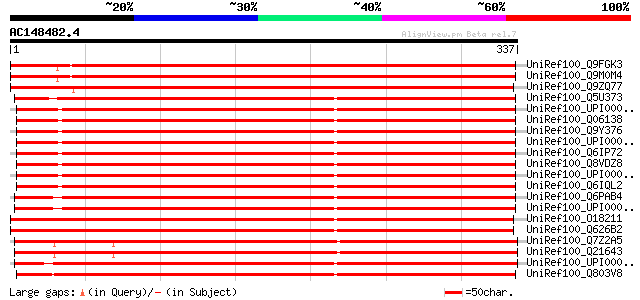
UniRef100_Q9FGK3 Hypothetical MO25-like protein At5g47540 [Arabi... 551 e-156
UniRef100_Q9M0M4 Hypothetical MO25-like protein At4g17270 [Arabi... 516 e-145
UniRef100_Q9ZQ77 Hypothetical MO25-like protein At2g03410 [Arabi... 434 e-120
UniRef100_Q5U373 Zgc:92575 [Brachydanio rerio] 282 1e-74
UniRef100_UPI0000183016 UPI0000183016 UniRef100 entry 280 3e-74
UniRef100_Q06138 Calcium binding protein 39 [Mus musculus] 280 3e-74
UniRef100_Q9Y376 Calcium binding protein 39 [Homo sapiens] 280 3e-74
UniRef100_UPI0000318C3C UPI0000318C3C UniRef100 entry 280 4e-74
UniRef100_Q6IP72 MGC78903 protein [Xenopus laevis] 280 4e-74
UniRef100_Q8VDZ8 Cab39 protein [Mus musculus] 280 4e-74
UniRef100_UPI0000015D96 UPI0000015D96 UniRef100 entry 280 5e-74
UniRef100_Q6IQL2 Zgc:86716 [Brachydanio rerio] 280 5e-74
UniRef100_Q6PAB4 MGC68674 protein [Xenopus laevis] 277 3e-73
UniRef100_UPI0000363038 UPI0000363038 UniRef100 entry 276 4e-73
UniRef100_O18211 Hypothetical MO25-like protein Y53C12A.4 in chr... 276 4e-73
UniRef100_Q626B2 Hypothetical protein CBG01052 [Caenorhabditis b... 275 1e-72
UniRef100_Q7Z2A5 Hypothetical protein R02E12.2 [Caenorhabditis e... 275 1e-72
UniRef100_Q21643 Hypothetical protein R02E12.2 [Caenorhabditis e... 275 1e-72
UniRef100_UPI0000432D61 UPI0000432D61 UniRef100 entry 273 4e-72
UniRef100_Q803V8 Zgc:55451 [Brachydanio rerio] 273 4e-72
>UniRef100_Q9FGK3 Hypothetical MO25-like protein At5g47540 [Arabidopsis thaliana]
Length = 343
Score = 551 bits (1421), Expect = e-156
Identities = 282/339 (83%), Positives = 313/339 (92%), Gaps = 4/339 (1%)
Query: 1 MKGLFKPKPRTPTDIVRQTRDLLLFFDRNT---ESRDSKREEKQMTELCKNIRELKSILY 57
MKGLFK KPRTP D+VRQTRDLLLF DR+T + RDSKR+EK M EL +NIR++KSILY
Sbjct: 1 MKGLFKSKPRTPADLVRQTRDLLLFSDRSTSLPDLRDSKRDEK-MAELSRNIRDMKSILY 59
Query: 58 GNSESEPVSEACAQLTQEFFKENTLRLLIKCIPKLNLEARKDATQVVANLQRQPVQSKLI 117
GNSE+EPV+EACAQLTQEFFKE+TLRLLI C+PKLNLE RKDATQVVANLQRQ V S+LI
Sbjct: 60 GNSEAEPVAEACAQLTQEFFKEDTLRLLITCLPKLNLETRKDATQVVANLQRQQVNSRLI 119
Query: 118 ASDYLENNMDLMDILIVGYENTDMALHYGAMLRECIRHQIVAKYVLNSPHMKKFFDYIQL 177
ASDYLE N+DLMD+LI G+ENTDMALHYGAM RECIRHQIVAKYVL S H+KKFFDYIQL
Sbjct: 120 ASDYLEANIDLMDVLIEGFENTDMALHYGAMFRECIRHQIVAKYVLESDHVKKFFDYIQL 179
Query: 178 PNFDIAADAAATFKELMTRHKSTVAEFLSKNYEWFFADYNSKLLESSNYITRRLAVKLLG 237
PNFDIAADAAATFKEL+TRHKSTVAEFL+KN +WFFADYNSKLLESSNYITRR A+KLLG
Sbjct: 180 PNFDIAADAAATFKELLTRHKSTVAEFLTKNEDWFFADYNSKLLESSNYITRRQAIKLLG 239
Query: 238 DMLLDRSNSAVMTQYVSSRENLRILMNLLRESSKSIQIEAFHVFKLFAANQNKPADIVSI 297
D+LLDRSNSAVMT+YVSSR+NLRILMNLLRESSKSIQIEAFHVFKLFAANQNKPADIV+I
Sbjct: 240 DILLDRSNSAVMTKYVSSRDNLRILMNLLRESSKSIQIEAFHVFKLFAANQNKPADIVNI 299
Query: 298 FVANKSKMLRLLDEFKIDKEDEQFEADKAQVMEEIASLE 336
VAN+SK+LRLL + K DKEDE+FEADK+QV+ EIA+LE
Sbjct: 300 LVANRSKLLRLLADLKPDKEDERFEADKSQVLREIAALE 338
>UniRef100_Q9M0M4 Hypothetical MO25-like protein At4g17270 [Arabidopsis thaliana]
Length = 343
Score = 516 bits (1329), Expect = e-145
Identities = 264/339 (77%), Positives = 303/339 (88%), Gaps = 4/339 (1%)
Query: 1 MKGLFKPKPRTPTDIVRQTRDLLLFFDRNT---ESRDSKREEKQMTELCKNIRELKSILY 57
M+GLFK KPRTP DIVRQTRDLLL+ DR+ + R+SKREEK M EL K+IR+LK ILY
Sbjct: 1 MRGLFKSKPRTPADIVRQTRDLLLYADRSNSFPDLRESKREEK-MVELSKSIRDLKLILY 59
Query: 58 GNSESEPVSEACAQLTQEFFKENTLRLLIKCIPKLNLEARKDATQVVANLQRQPVQSKLI 117
GNSE+EPV+EACAQLTQEFFK +TLR L+ +P LNLEARKDATQVVANLQRQ V S+LI
Sbjct: 60 GNSEAEPVAEACAQLTQEFFKADTLRRLLTSLPNLNLEARKDATQVVANLQRQQVNSRLI 119
Query: 118 ASDYLENNMDLMDILIVGYENTDMALHYGAMLRECIRHQIVAKYVLNSPHMKKFFDYIQL 177
A+DYLE+N+DLMD L+ G+ENTDMALHYG M RECIRHQIVAKYVL+S H+KKFF YIQL
Sbjct: 120 AADYLESNIDLMDFLVDGFENTDMALHYGTMFRECIRHQIVAKYVLDSEHVKKFFYYIQL 179
Query: 178 PNFDIAADAAATFKELMTRHKSTVAEFLSKNYEWFFADYNSKLLESSNYITRRLAVKLLG 237
PNFDIAADAAATFKEL+TRHKSTVAEFL KN +WFFADYNSKLLES+NYITRR A+KLLG
Sbjct: 180 PNFDIAADAAATFKELLTRHKSTVAEFLIKNEDWFFADYNSKLLESTNYITRRQAIKLLG 239
Query: 238 DMLLDRSNSAVMTQYVSSRENLRILMNLLRESSKSIQIEAFHVFKLFAANQNKPADIVSI 297
D+LLDRSNSAVMT+YVSS +NLRILMNLLRESSK+IQIEAFHVFKLF ANQNKP+DI +I
Sbjct: 240 DILLDRSNSAVMTKYVSSMDNLRILMNLLRESSKTIQIEAFHVFKLFVANQNKPSDIANI 299
Query: 298 FVANKSKMLRLLDEFKIDKEDEQFEADKAQVMEEIASLE 336
VAN++K+LRLL + K DKEDE+F+ADKAQV+ EIA+L+
Sbjct: 300 LVANRNKLLRLLADIKPDKEDERFDADKAQVVREIANLK 338
>UniRef100_Q9ZQ77 Hypothetical MO25-like protein At2g03410 [Arabidopsis thaliana]
Length = 348
Score = 434 bits (1116), Expect = e-120
Identities = 213/338 (63%), Positives = 276/338 (81%), Gaps = 3/338 (0%)
Query: 1 MKGLFKPKPRTPTDIVRQTRDLLLFFDRNTESRDSKREEKQ--MTELCKNIRELKSILYG 58
MKGLFK K R P +IVRQTRDL+ + E D++ ++ ELC+NIR+LKSILYG
Sbjct: 1 MKGLFKNKSRLPGEIVRQTRDLIALAESEEEETDARNSKRLGICAELCRNIRDLKSILYG 60
Query: 59 NSESEPVSEACAQLTQEFFKENTLRLLIKCIPKLNLEARKDATQVVANLQRQPVQSKLIA 118
N E+EPV EAC LTQEFF+ +TLR LIK IPKL+LEARKDATQ+VANLQ+Q V+ +L+A
Sbjct: 61 NGEAEPVPEACLLLTQEFFRADTLRPLIKSIPKLDLEARKDATQIVANLQKQQVEFRLVA 120
Query: 119 SDYLENNMDLMDILIVGYENT-DMALHYGAMLRECIRHQIVAKYVLNSPHMKKFFDYIQL 177
S+YLE+N+D++D L+ G ++ ++ALHY ML+EC+RHQ+VAKY+L S +++KFFDY+QL
Sbjct: 121 SEYLESNLDVIDSLVEGIDHDHELALHYTGMLKECVRHQVVAKYILESKNLEKFFDYVQL 180
Query: 178 PNFDIAADAAATFKELMTRHKSTVAEFLSKNYEWFFADYNSKLLESSNYITRRLAVKLLG 237
P FD+A DA+ F+EL+TRHKSTVAE+L+KNYEWFFA+YN+KLLE +Y T+R A KLLG
Sbjct: 181 PYFDVATDASKIFRELLTRHKSTVAEYLAKNYEWFFAEYNTKLLEKGSYFTKRQASKLLG 240
Query: 238 DMLLDRSNSAVMTQYVSSRENLRILMNLLRESSKSIQIEAFHVFKLFAANQNKPADIVSI 297
D+L+DRSNS VM +YVSS +NLRI+MNLLRE +K+IQ+EAFH+FKLF AN+NKP DIV+I
Sbjct: 241 DVLMDRSNSGVMVKYVSSLDNLRIMMNLLREPTKNIQLEAFHIFKLFVANENKPEDIVAI 300
Query: 298 FVANKSKMLRLLDEFKIDKEDEQFEADKAQVMEEIASL 335
VAN++K+LRL + K +KED FE DKA VM EIA+L
Sbjct: 301 LVANRTKILRLFADLKPEKEDVGFETDKALVMNEIATL 338
>UniRef100_Q5U373 Zgc:92575 [Brachydanio rerio]
Length = 334
Score = 282 bits (721), Expect = 1e-74
Identities = 145/334 (43%), Positives = 222/334 (66%), Gaps = 7/334 (2%)
Query: 4 LFKPKPRTPTDIVRQTRDLLLFFDRNTESRDSKREEKQMTELCKNIRELKSILYGNSESE 63
LF ++PT+IV+ +D L + K+ EK E+ K + +K ILYG ++ E
Sbjct: 3 LFGKSHKSPTEIVKTLKDNLSIL-----VKQDKKTEKASEEVSKCLVAMKEILYGTNDKE 57
Query: 64 PVSEACAQLTQEFFKENTLRLLIKCIPKLNLEARKDATQVVANLQRQPVQSKLIASDYLE 123
P +E AQL QE + + L L++ + ++ E +KD Q+ N+ R+ + ++ +Y
Sbjct: 58 PHTETVAQLAQELYNSSLLISLVENLQVIDFEGKKDVCQIFNNILRRQIGTRSPTVEYFC 117
Query: 124 NNMDLMDILIVGYENTDMALHYGAMLRECIRHQIVAKYVLNSPHMKKFFDYIQLPNFDIA 183
++ +++ IL+ GYE +AL+ G MLRECIRH+ +AK VL+S H K FF Y+++ FDIA
Sbjct: 118 SHQEVLFILLKGYETPQVALNCGIMLRECIRHEPLAKIVLHSEHFKDFFSYVEMSTFDIA 177
Query: 184 ADAAATFKELMTRHKSTVAEFLSKNYEWFFADYNSKLLESSNYITRRLAVKLLGDMLLDR 243
+DA ATFK+L+TRHK+ VAEFL +NY+ F +Y KLL S NY+T+R ++KLLG++LLDR
Sbjct: 178 SDAFATFKDLLTRHKALVAEFLEQNYDAVFDNY-EKLLHSENYVTKRQSLKLLGELLLDR 236
Query: 244 SNSAVMTQYVSSRENLRILMNLLRESSKSIQIEAFHVFKLFAANQNKPADIVSIFVANKS 303
N VMT+Y+S ENL+++MNLLR+ S +IQ EAFHVFK+F AN NK IV I + N++
Sbjct: 237 HNFTVMTRYISKPENLKLMMNLLRDKSPNIQFEAFHVFKVFVANPNKTQPIVDILLKNQT 296
Query: 304 KMLRLLDEFKIDK-EDEQFEADKAQVMEEIASLE 336
K++ L F+ D+ +DEQF +K ++++I L+
Sbjct: 297 KLIDFLSNFQKDRTDDEQFNDEKTYLVKQIRDLK 330
>UniRef100_UPI0000183016 UPI0000183016 UniRef100 entry
Length = 341
Score = 280 bits (717), Expect = 3e-74
Identities = 140/333 (42%), Positives = 222/333 (66%), Gaps = 4/333 (1%)
Query: 5 FKPKPRTPTDIVRQTRDLLLFFDRNTESRDSKREEKQMTELCKNIRELKSILYGNSESEP 64
F ++P DIV+ ++ + ++ S K+ EK E+ KN+ +K ILYG +E EP
Sbjct: 5 FGKSHKSPADIVKNLKESMAVLEKQDIS--DKKAEKATEEVSKNLVAMKEILYGTNEKEP 62
Query: 65 VSEACAQLTQEFFKENTLRLLIKCIPKLNLEARKDATQVVANLQRQPVQSKLIASDYLEN 124
+EA AQL QE + L L+ + ++ E +KD Q+ N+ R+ + ++ +Y+
Sbjct: 63 QTEAVAQLAQELYNSGLLSTLVADLQLIDFEGKKDVAQIFNNILRRQIGTRTPTVEYICT 122
Query: 125 NMDLMDILIVGYENTDMALHYGAMLRECIRHQIVAKYVLNSPHMKKFFDYIQLPNFDIAA 184
+++ +L+ GYE+ ++AL+ G MLRECIRH+ +AK +L S FF Y+++ FDIA+
Sbjct: 123 QQNILFMLLKGYESPEIALNCGIMLRECIRHEPLAKIILWSEQFYDFFRYVEMSTFDIAS 182
Query: 185 DAAATFKELMTRHKSTVAEFLSKNYEWFFADYNSKLLESSNYITRRLAVKLLGDMLLDRS 244
DA ATFK+L+TRHK AEFL ++Y+ FF++Y KLL S NY+T+R ++KLLG++LLDR
Sbjct: 183 DAFATFKDLLTRHKLLSAEFLEQHYDRFFSEY-EKLLHSENYVTKRQSLKLLGELLLDRH 241
Query: 245 NSAVMTQYVSSRENLRILMNLLRESSKSIQIEAFHVFKLFAANQNKPADIVSIFVANKSK 304
N +MT+Y+S ENL+++MNLLR+ S++IQ EAFHVFK+F AN NK I+ I + N++K
Sbjct: 242 NFTIMTKYISKPENLKLMMNLLRDKSRNIQFEAFHVFKVFVANPNKTQPILDILLKNQTK 301
Query: 305 MLRLLDEFKIDK-EDEQFEADKAQVMEEIASLE 336
++ L +F+ D+ EDEQF +K ++++I L+
Sbjct: 302 LIEFLSKFQNDRTEDEQFNDEKTYLVKQIRDLK 334
>UniRef100_Q06138 Calcium binding protein 39 [Mus musculus]
Length = 341
Score = 280 bits (717), Expect = 3e-74
Identities = 140/333 (42%), Positives = 223/333 (66%), Gaps = 4/333 (1%)
Query: 5 FKPKPRTPTDIVRQTRDLLLFFDRNTESRDSKREEKQMTELCKNIRELKSILYGNSESEP 64
F ++P DIV+ ++ + ++ S K+ EK E+ KN+ +K ILYG +E EP
Sbjct: 5 FGKSHKSPADIVKNLKESMAVLEKQDIS--DKKAEKATEEVSKNLVAMKEILYGTNEKEP 62
Query: 65 VSEACAQLTQEFFKENTLRLLIKCIPKLNLEARKDATQVVANLQRQPVQSKLIASDYLEN 124
+EA AQL QE + L L+ + ++ E +KD Q+ N+ R+ + ++ +Y+
Sbjct: 63 QTEAVAQLAQELYNSGLLGTLVADLQLIDFEGKKDVAQIFNNILRRQIGTRTPTVEYICT 122
Query: 125 NMDLMDILIVGYENTDMALHYGAMLRECIRHQIVAKYVLNSPHMKKFFDYIQLPNFDIAA 184
+++ +L+ GYE+ ++AL+ G MLRECIRH+ +AK +L S FF Y+++ FDIA+
Sbjct: 123 QQNILFMLLKGYESPEIALNCGIMLRECIRHEPLAKIILWSEQFYDFFRYVEMSTFDIAS 182
Query: 185 DAAATFKELMTRHKSTVAEFLSKNYEWFFADYNSKLLESSNYITRRLAVKLLGDMLLDRS 244
DA ATFK+L+TRHK AEFL ++Y+ FF++Y KLL S NY+T+R ++KLLG++LLDR
Sbjct: 183 DAFATFKDLLTRHKLLSAEFLEQHYDRFFSEY-EKLLHSENYVTKRQSLKLLGELLLDRH 241
Query: 245 NSAVMTQYVSSRENLRILMNLLRESSKSIQIEAFHVFKLFAANQNKPADIVSIFVANKSK 304
N +MT+Y+S ENL+++MNLLR+ S++IQ EAFHVFK+F AN NK I+ I + N++K
Sbjct: 242 NFTIMTKYISKPENLKLMMNLLRDKSRNIQFEAFHVFKVFVANPNKTQPILDILLKNQTK 301
Query: 305 MLRLLDEFKIDK-EDEQFEADKAQVMEEIASLE 336
++ L +F+ D+ EDEQF +K ++++I +L+
Sbjct: 302 LIEFLSKFQNDRTEDEQFNDEKTYLVKQIRNLK 334
>UniRef100_Q9Y376 Calcium binding protein 39 [Homo sapiens]
Length = 341
Score = 280 bits (717), Expect = 3e-74
Identities = 140/333 (42%), Positives = 222/333 (66%), Gaps = 4/333 (1%)
Query: 5 FKPKPRTPTDIVRQTRDLLLFFDRNTESRDSKREEKQMTELCKNIRELKSILYGNSESEP 64
F ++P DIV+ ++ + ++ S K+ EK E+ KN+ +K ILYG +E EP
Sbjct: 5 FGKSHKSPADIVKNLKESMAVLEKQDIS--DKKAEKATEEVSKNLVAMKEILYGTNEKEP 62
Query: 65 VSEACAQLTQEFFKENTLRLLIKCIPKLNLEARKDATQVVANLQRQPVQSKLIASDYLEN 124
+EA AQL QE + L L+ + ++ E +KD Q+ N+ R+ + ++ +Y+
Sbjct: 63 QTEAVAQLAQELYNSGLLSTLVADLQLIDFEGKKDVAQIFNNILRRQIGTRTPTVEYICT 122
Query: 125 NMDLMDILIVGYENTDMALHYGAMLRECIRHQIVAKYVLNSPHMKKFFDYIQLPNFDIAA 184
+++ +L+ GYE+ ++AL+ G MLRECIRH+ +AK +L S FF Y+++ FDIA+
Sbjct: 123 QQNILFMLLKGYESPEIALNCGIMLRECIRHEPLAKIILWSEQFYDFFRYVEMSTFDIAS 182
Query: 185 DAAATFKELMTRHKSTVAEFLSKNYEWFFADYNSKLLESSNYITRRLAVKLLGDMLLDRS 244
DA ATFK+L+TRHK AEFL ++Y+ FF++Y KLL S NY+T+R ++KLLG++LLDR
Sbjct: 183 DAFATFKDLLTRHKLLSAEFLEQHYDRFFSEY-EKLLHSENYVTKRQSLKLLGELLLDRH 241
Query: 245 NSAVMTQYVSSRENLRILMNLLRESSKSIQIEAFHVFKLFAANQNKPADIVSIFVANKSK 304
N +MT+Y+S ENL+++MNLLR+ S++IQ EAFHVFK+F AN NK I+ I + N++K
Sbjct: 242 NFTIMTKYISKPENLKLMMNLLRDKSRNIQFEAFHVFKVFVANPNKTQPILDILLKNQAK 301
Query: 305 MLRLLDEFKIDK-EDEQFEADKAQVMEEIASLE 336
++ L +F+ D+ EDEQF +K ++++I L+
Sbjct: 302 LIEFLSKFQNDRTEDEQFNDEKTYLVKQIRDLK 334
>UniRef100_UPI0000318C3C UPI0000318C3C UniRef100 entry
Length = 341
Score = 280 bits (716), Expect = 4e-74
Identities = 142/333 (42%), Positives = 221/333 (65%), Gaps = 4/333 (1%)
Query: 5 FKPKPRTPTDIVRQTRDLLLFFDRNTESRDSKREEKQMTELCKNIRELKSILYGNSESEP 64
F ++P DIV+ +D + +++ S K+ EK E+ K++ +K ILYG +E EP
Sbjct: 5 FGKSHKSPADIVKNLKDSMTVLEKHDIS--DKKAEKATEEVSKSLVAMKEILYGTNEKEP 62
Query: 65 VSEACAQLTQEFFKENTLRLLIKCIPKLNLEARKDATQVVANLQRQPVQSKLIASDYLEN 124
+EA AQL QE + L LI + ++ E +KD Q+ N+ R+ + ++ +YL
Sbjct: 63 QTEAVAQLAQELYNSGLLSTLIADLQLIDFEGKKDVAQIFNNILRRQIGTRTPTVEYLCT 122
Query: 125 NMDLMDILIVGYENTDMALHYGAMLRECIRHQIVAKYVLNSPHMKKFFDYIQLPNFDIAA 184
+++ +L+ GYE+ ++AL+ G MLRECIRH+ +AK L S FF Y+++ FDIA+
Sbjct: 123 QQNILFMLLKGYESPEIALNCGIMLRECIRHEPLAKITLYSEQFYDFFRYVEMSTFDIAS 182
Query: 185 DAAATFKELMTRHKSTVAEFLSKNYEWFFADYNSKLLESSNYITRRLAVKLLGDMLLDRS 244
DA ATFK+L+TRHK AEFL + Y+ FF++Y KLL S NY+T+R ++KLLG++LLDR
Sbjct: 183 DAFATFKDLLTRHKLLSAEFLEQYYDKFFSEY-EKLLHSENYVTKRQSLKLLGELLLDRH 241
Query: 245 NSAVMTQYVSSRENLRILMNLLRESSKSIQIEAFHVFKLFAANQNKPADIVSIFVANKSK 304
N +MT+Y+S ENL+++MNLLR+ S++IQ EAFHVFK+F AN NK I+ I + N++K
Sbjct: 242 NFTIMTKYISKPENLKLMMNLLRDKSRNIQFEAFHVFKVFVANPNKTQPILDILLKNQTK 301
Query: 305 MLRLLDEFKIDK-EDEQFEADKAQVMEEIASLE 336
++ L +F+ D+ EDEQF +K ++++I L+
Sbjct: 302 LIEFLSKFQNDRTEDEQFNDEKTYLVKQIRDLK 334
>UniRef100_Q6IP72 MGC78903 protein [Xenopus laevis]
Length = 341
Score = 280 bits (716), Expect = 4e-74
Identities = 140/333 (42%), Positives = 222/333 (66%), Gaps = 4/333 (1%)
Query: 5 FKPKPRTPTDIVRQTRDLLLFFDRNTESRDSKREEKQMTELCKNIRELKSILYGNSESEP 64
F ++P DIV+ ++ + ++ S K+ EK E+ KN+ +K ILYG +E EP
Sbjct: 5 FGKSHKSPADIVKNLKESIAVLEKQDIS--DKKAEKATEEVSKNLVAMKEILYGTNEKEP 62
Query: 65 VSEACAQLTQEFFKENTLRLLIKCIPKLNLEARKDATQVVANLQRQPVQSKLIASDYLEN 124
+EA AQL QE + L L+ + ++ E +KD Q+ N+ R+ + ++ +Y+
Sbjct: 63 QTEAVAQLAQELYNSGLLGTLVADLQLIDFEGKKDVAQIFNNILRRQIGTRTPTVEYICT 122
Query: 125 NMDLMDILIVGYENTDMALHYGAMLRECIRHQIVAKYVLNSPHMKKFFDYIQLPNFDIAA 184
+++ +L+ GYE+ ++AL+ G MLRECIRH+ +AK +L S FF Y+++ FDIA+
Sbjct: 123 QQNILFMLLKGYESPEIALNCGIMLRECIRHEPLAKIILWSEPFYDFFRYVEMSTFDIAS 182
Query: 185 DAAATFKELMTRHKSTVAEFLSKNYEWFFADYNSKLLESSNYITRRLAVKLLGDMLLDRS 244
DA ATFK+L+TRHK AEFL ++Y+ FF++Y KLL S NY+T+R ++KLLG++LLDR
Sbjct: 183 DAFATFKDLLTRHKLLSAEFLEQHYDRFFSEY-EKLLHSENYVTKRQSLKLLGELLLDRH 241
Query: 245 NSAVMTQYVSSRENLRILMNLLRESSKSIQIEAFHVFKLFAANQNKPADIVSIFVANKSK 304
N +MT+Y+S ENL+++MNLLR+ S++IQ EAFHVFK+F AN NK ++ I + N+SK
Sbjct: 242 NFTIMTKYISKPENLKLMMNLLRDKSRNIQFEAFHVFKVFVANPNKTQPVLDILLKNQSK 301
Query: 305 MLRLLDEFKIDK-EDEQFEADKAQVMEEIASLE 336
++ L +F+ D+ EDEQF +K ++++I L+
Sbjct: 302 LIEFLSKFQNDRTEDEQFNDEKTYLVKQIRDLK 334
>UniRef100_Q8VDZ8 Cab39 protein [Mus musculus]
Length = 341
Score = 280 bits (716), Expect = 4e-74
Identities = 140/333 (42%), Positives = 222/333 (66%), Gaps = 4/333 (1%)
Query: 5 FKPKPRTPTDIVRQTRDLLLFFDRNTESRDSKREEKQMTELCKNIRELKSILYGNSESEP 64
F ++P DIV+ ++ + ++ S K+ EK E+ KN+ +K ILYG +E EP
Sbjct: 5 FGKSHKSPADIVKNLKESMAVLEKQDIS--DKKAEKATEEVSKNLVAMKEILYGTNEKEP 62
Query: 65 VSEACAQLTQEFFKENTLRLLIKCIPKLNLEARKDATQVVANLQRQPVQSKLIASDYLEN 124
+EA AQL QE + L L+ + ++ E +KD Q+ N+ R+ + ++ +Y+
Sbjct: 63 QTEAVAQLAQELYNSGLLGTLVADLQLIDFEGKKDVAQIFNNILRRQIGTRTPTVEYICT 122
Query: 125 NMDLMDILIVGYENTDMALHYGAMLRECIRHQIVAKYVLNSPHMKKFFDYIQLPNFDIAA 184
+++ +L+ GYE+ ++AL+ G MLRECIRH+ +AK +L S FF Y+++ FDIA+
Sbjct: 123 QQNILFMLLKGYESPEIALNCGIMLRECIRHEPLAKIILWSEQFYDFFRYVEMSTFDIAS 182
Query: 185 DAAATFKELMTRHKSTVAEFLSKNYEWFFADYNSKLLESSNYITRRLAVKLLGDMLLDRS 244
DA ATFK+L+TRHK AEFL ++Y+ FF++Y KLL S NY+T+R ++KLLG++LLDR
Sbjct: 183 DAFATFKDLLTRHKLLSAEFLEQHYDRFFSEY-EKLLHSENYVTKRQSLKLLGELLLDRH 241
Query: 245 NSAVMTQYVSSRENLRILMNLLRESSKSIQIEAFHVFKLFAANQNKPADIVSIFVANKSK 304
N +MT+Y+S ENL+++MNLLR+ S++IQ EAFHVFK+F AN NK I+ I + N++K
Sbjct: 242 NFTIMTKYISKPENLKLMMNLLRDKSRNIQFEAFHVFKVFVANPNKTQPILDILLKNQTK 301
Query: 305 MLRLLDEFKIDK-EDEQFEADKAQVMEEIASLE 336
++ L +F+ D+ EDEQF +K ++++I L+
Sbjct: 302 LIEFLSKFQNDRTEDEQFNDEKTYLVKQIRDLK 334
>UniRef100_UPI0000015D96 UPI0000015D96 UniRef100 entry
Length = 341
Score = 280 bits (715), Expect = 5e-74
Identities = 141/333 (42%), Positives = 221/333 (66%), Gaps = 4/333 (1%)
Query: 5 FKPKPRTPTDIVRQTRDLLLFFDRNTESRDSKREEKQMTELCKNIRELKSILYGNSESEP 64
F ++P DIV+ +D + +++ S K+ EK E+ K++ +K ILYG +E EP
Sbjct: 5 FGKSHKSPADIVKNLKDSMTVLEKHDIS--DKKAEKATEEVSKSLVAMKEILYGTNEKEP 62
Query: 65 VSEACAQLTQEFFKENTLRLLIKCIPKLNLEARKDATQVVANLQRQPVQSKLIASDYLEN 124
+EA AQL QE + L L+ + ++ E +KD Q+ N+ R+ + ++ +YL
Sbjct: 63 QTEAVAQLAQELYNSGLLSTLVADLQLIDFEGKKDVAQIFNNILRRQIGTRTPTVEYLCT 122
Query: 125 NMDLMDILIVGYENTDMALHYGAMLRECIRHQIVAKYVLNSPHMKKFFDYIQLPNFDIAA 184
+++ +L+ GYE+ ++AL+ G MLRECIRH+ +AK L S FF Y+++ FDIA+
Sbjct: 123 QQNILFMLLKGYESAEIALNCGIMLRECIRHEPLAKITLYSEQFYDFFRYVEMSTFDIAS 182
Query: 185 DAAATFKELMTRHKSTVAEFLSKNYEWFFADYNSKLLESSNYITRRLAVKLLGDMLLDRS 244
DA ATFK+L+TRHK AEFL + Y+ FF++Y KLL S NY+T+R ++KLLG++LLDR
Sbjct: 183 DAFATFKDLLTRHKLLSAEFLEQFYDKFFSEY-EKLLHSENYVTKRQSLKLLGELLLDRH 241
Query: 245 NSAVMTQYVSSRENLRILMNLLRESSKSIQIEAFHVFKLFAANQNKPADIVSIFVANKSK 304
N +MT+Y+S ENL+++MNLLR+ S++IQ EAFHVFK+F AN NK I+ I + N++K
Sbjct: 242 NFTIMTKYISKPENLKLMMNLLRDKSRNIQFEAFHVFKVFVANPNKTQPILDILLKNQAK 301
Query: 305 MLRLLDEFKIDK-EDEQFEADKAQVMEEIASLE 336
++ L +F+ D+ EDEQF +K ++++I L+
Sbjct: 302 LIEFLSKFQNDRTEDEQFNDEKTYLVKQIRDLK 334
>UniRef100_Q6IQL2 Zgc:86716 [Brachydanio rerio]
Length = 341
Score = 280 bits (715), Expect = 5e-74
Identities = 141/333 (42%), Positives = 220/333 (65%), Gaps = 4/333 (1%)
Query: 5 FKPKPRTPTDIVRQTRDLLLFFDRNTESRDSKREEKQMTELCKNIRELKSILYGNSESEP 64
F + P DIV+ +D + ++ S K+ EK E+ K++ +K ILYG +E EP
Sbjct: 5 FVKSHKCPADIVKNLKDNMTILEKQDIS--DKKAEKASEEVSKSLLSMKEILYGTNEKEP 62
Query: 65 VSEACAQLTQEFFKENTLRLLIKCIPKLNLEARKDATQVVANLQRQPVQSKLIASDYLEN 124
+EA AQL QE + L L+ + ++ E +KD Q+ ++ R+ + ++ +YL
Sbjct: 63 QTEAVAQLAQELYNSGLLSTLVADLQLIDFEGKKDVAQIFNDILRRQIGTRTPTVEYLCT 122
Query: 125 NMDLMDILIVGYENTDMALHYGAMLRECIRHQIVAKYVLNSPHMKKFFDYIQLPNFDIAA 184
+++ +L+ GYE+ D+AL+ G MLRECIRH+ +AK L S FF Y+++ FDIA+
Sbjct: 123 QQNILFMLLKGYESPDIALNCGIMLRECIRHEPLAKITLCSEQFYDFFRYVEMSTFDIAS 182
Query: 185 DAAATFKELMTRHKSTVAEFLSKNYEWFFADYNSKLLESSNYITRRLAVKLLGDMLLDRS 244
DA ATFK+L+TRHK AEFL ++Y+ FF++Y KLL S NY+T+R ++KLLG++LLDR
Sbjct: 183 DAFATFKDLLTRHKVLSAEFLEQHYDRFFSEY-EKLLHSENYVTKRQSLKLLGELLLDRH 241
Query: 245 NSAVMTQYVSSRENLRILMNLLRESSKSIQIEAFHVFKLFAANQNKPADIVSIFVANKSK 304
N +MT+Y+S ENL+++MNLLR+ S++IQ EAFHVFK+F AN NK I+ I + N++K
Sbjct: 242 NFTIMTKYISKPENLKLMMNLLRDKSRNIQFEAFHVFKVFVANPNKTPPILDILLKNQTK 301
Query: 305 MLRLLDEFKIDK-EDEQFEADKAQVMEEIASLE 336
++ L +F+ D+ EDEQF +K ++++I L+
Sbjct: 302 LIEFLSKFQNDRAEDEQFSDEKTYLIKQIRDLK 334
>UniRef100_Q6PAB4 MGC68674 protein [Xenopus laevis]
Length = 337
Score = 277 bits (708), Expect = 3e-73
Identities = 140/334 (41%), Positives = 217/334 (64%), Gaps = 7/334 (2%)
Query: 4 LFKPKPRTPTDIVRQTRDLLLFFDRNTESRDSKREEKQMTELCKNIRELKSILYGNSESE 63
LF + P +IV+ +D + +R K+ EK E+ K+++ K IL G + E
Sbjct: 6 LFSKSHKNPAEIVKTLKDNMALLERQ-----DKKTEKASEEVSKSLQATKEILCGTGDKE 60
Query: 64 PVSEACAQLTQEFFKENTLRLLIKCIPKLNLEARKDATQVVANLQRQPVQSKLIASDYLE 123
P +E AQL QE + L LI + ++ E +KD +Q+ N+ R+ + ++ +Y+
Sbjct: 61 PQTETVAQLAQELYNSGLLVTLIANLHLIDFEGKKDVSQIFNNILRRQIGTRSPTVEYIS 120
Query: 124 NNMDLMDILIVGYENTDMALHYGAMLRECIRHQIVAKYVLNSPHMKKFFDYIQLPNFDIA 183
++ ++ IL+ GYE+ +ALH G MLREC+RH+ +AK +L S FF Y+++ FDIA
Sbjct: 121 SHQHILFILLKGYESPQVALHCGIMLRECVRHEPLAKVILYSEQFGDFFKYVEMSTFDIA 180
Query: 184 ADAAATFKELMTRHKSTVAEFLSKNYEWFFADYNSKLLESSNYITRRLAVKLLGDMLLDR 243
+DA ATFK+L+TRHK VAEFL +NY+ F DY KLL S NY+T+R ++KLLG+++LDR
Sbjct: 181 SDAFATFKDLLTRHKLMVAEFLEQNYDRIFNDY-EKLLHSENYVTKRQSLKLLGELILDR 239
Query: 244 SNSAVMTQYVSSRENLRILMNLLRESSKSIQIEAFHVFKLFAANQNKPADIVSIFVANKS 303
N ++MT+Y+S ENL+++MNLLR+ S +IQ EAFHVFK+F AN NK IV I + N++
Sbjct: 240 HNFSIMTKYISKPENLKLMMNLLRDKSPNIQFEAFHVFKVFVANPNKTQPIVDILLKNQT 299
Query: 304 KMLRLLDEFKIDK-EDEQFEADKAQVMEEIASLE 336
K++ L F+ D+ +DEQF +K ++++I L+
Sbjct: 300 KLIDFLSSFQKDRTDDEQFTDEKNYLIKQIRDLK 333
>UniRef100_UPI0000363038 UPI0000363038 UniRef100 entry
Length = 333
Score = 276 bits (707), Expect = 4e-73
Identities = 143/334 (42%), Positives = 220/334 (65%), Gaps = 7/334 (2%)
Query: 4 LFKPKPRTPTDIVRQTRDLLLFFDRNTESRDSKREEKQMTELCKNIRELKSILYGNSESE 63
LF ++P DIVR ++ L ++ K+ +K E+ K + +K ILYG+++ E
Sbjct: 3 LFGKSHKSPADIVRTLKENLAILVKH-----DKKTDKASEEVSKCLVSMKEILYGSNDKE 57
Query: 64 PVSEACAQLTQEFFKENTLRLLIKCIPKLNLEARKDATQVVANLQRQPVQSKLIASDYLE 123
P +E AQL QE + L L++ + ++ E +KD Q+ N+ R+ + ++ +Y
Sbjct: 58 PHTETVAQLAQELYNSGLLITLVENLQLIDFEGKKDVCQIFNNILRRQIGTRSPTVEYFC 117
Query: 124 NNMDLMDILIVGYENTDMALHYGAMLRECIRHQIVAKYVLNSPHMKKFFDYIQLPNFDIA 183
++ +++ IL+ GYE +AL+ G MLRECIRH+ +AK VL S FF+Y+++ FDIA
Sbjct: 118 SHQEVLFILLKGYETPQVALNCGIMLRECIRHEPLAKIVLQSDDFPNFFNYVEMSTFDIA 177
Query: 184 ADAAATFKELMTRHKSTVAEFLSKNYEWFFADYNSKLLESSNYITRRLAVKLLGDMLLDR 243
+DA ATFK+L+TRHK VAE L +NY+ FADY KLL S NY+T+R ++KLLG++LLDR
Sbjct: 178 SDAFATFKDLLTRHKVLVAEHLEQNYDAVFADY-EKLLNSENYVTKRQSLKLLGELLLDR 236
Query: 244 SNSAVMTQYVSSRENLRILMNLLRESSKSIQIEAFHVFKLFAANQNKPADIVSIFVANKS 303
N VMT+Y+S ENL+++MNLLR+ S +IQ EAFHVFK+F AN NK IV I + N++
Sbjct: 237 HNFTVMTRYISKPENLKLMMNLLRDKSPNIQFEAFHVFKVFVANPNKTQPIVDILLKNQT 296
Query: 304 KMLRLLDEFKIDK-EDEQFEADKAQVMEEIASLE 336
K++ L+ F+ D+ +DEQF +K ++++I L+
Sbjct: 297 KLIDFLNNFQKDRTDDEQFNDEKTYLIKQIRDLK 330
>UniRef100_O18211 Hypothetical MO25-like protein Y53C12A.4 in chromosome II
[Caenorhabditis elegans]
Length = 338
Score = 276 bits (707), Expect = 4e-73
Identities = 142/336 (42%), Positives = 212/336 (62%), Gaps = 2/336 (0%)
Query: 1 MKGLFKPKPRTPTDIVRQTRDLLLFFDRNTESRDSKREEKQMTELCKNIRELKSILYGNS 60
+K LF +TP D+V+ RD LL DR+ + ++ EK + E K + K+ +YG+
Sbjct: 2 LKPLFGKADKTPADVVKNLRDALLVIDRHGTNTSERKVEKAIEETAKMLALAKTFIYGSD 61
Query: 61 ESEPVSEACAQLTQEFFKENTLRLLIKCIPKLNLEARKDATQVVANLQRQPVQSKLIASD 120
+EP +E QL QE + N L +LIK + K E +KD V NL R+ + ++ +
Sbjct: 62 ANEPNNEQVTQLAQEVYNANVLPMLIKHLHKFEFECKKDVASVFNNLLRRQIGTRSPTVE 121
Query: 121 YLENNMDLMDILIVGYENTDMALHYGAMLRECIRHQIVAKYVLNSPHMKKFFDYIQLPNF 180
YL +++ L++GYE D+AL G+MLRE +RH+ +A+ VL S + ++FF ++Q F
Sbjct: 122 YLAARPEILITLLLGYEQPDIALTCGSMLREAVRHEHLARIVLYSEYFQRFFVFVQSDVF 181
Query: 181 DIAADAAATFKELMTRHKSTVAEFLSKNYEWFFADYNSKLLESSNYITRRLAVKLLGDML 240
DIA DA +TFK+LMT+HK+ AE+L NY+ FF Y S L S NY+TRR ++KLLG++L
Sbjct: 182 DIATDAFSTFKDLMTKHKNMCAEYLDNNYDRFFGQY-SALTNSENYVTRRQSLKLLGELL 240
Query: 241 LDRSNSAVMTQYVSSRENLRILMNLLRESSKSIQIEAFHVFKLFAANQNKPADIVSIFVA 300
LDR N + M +Y++S ENL+ +M LLR+ ++IQ EAFHVFK+F AN NKP I I
Sbjct: 241 LDRHNFSTMNKYITSPENLKTVMELLRDKRRNIQYEAFHVFKIFVANPNKPRPITDILTR 300
Query: 301 NKSKMLRLLDEFKIDK-EDEQFEADKAQVMEEIASL 335
N+ K++ L F D+ DEQF +KA ++++I L
Sbjct: 301 NRDKLVEFLTAFHNDRTNDEQFNDEKAYLIKQIQEL 336
>UniRef100_Q626B2 Hypothetical protein CBG01052 [Caenorhabditis briggsae]
Length = 338
Score = 275 bits (703), Expect = 1e-72
Identities = 141/336 (41%), Positives = 214/336 (62%), Gaps = 2/336 (0%)
Query: 1 MKGLFKPKPRTPTDIVRQTRDLLLFFDRNTESRDSKREEKQMTELCKNIRELKSILYGNS 60
+K LF +TP D+V+ RD LL DR + +++ ++ EK + E K + K+ +YG+
Sbjct: 2 LKPLFGKADKTPADVVKNLRDALLVIDRQSATQNERKVEKAIEETAKMLALAKTFIYGSD 61
Query: 61 ESEPVSEACAQLTQEFFKENTLRLLIKCIPKLNLEARKDATQVVANLQRQPVQSKLIASD 120
+EP +E QL QE + N L +LIK + K E +KD V NL R+ + ++ +
Sbjct: 62 ANEPNNEQVTQLAQEVYNANILPMLIKHLHKFEFECKKDVASVFNNLLRRQIGTRSPTVE 121
Query: 121 YLENNMDLMDILIVGYENTDMALHYGAMLRECIRHQIVAKYVLNSPHMKKFFDYIQLPNF 180
YL +++ L++GYE D+AL G+MLRE +RH+ +A+ VL S + ++FF ++Q F
Sbjct: 122 YLAARPEILITLLLGYEQPDIALTCGSMLREAVRHEHLARIVLYSEYFQRFFVFVQSDVF 181
Query: 181 DIAADAAATFKELMTRHKSTVAEFLSKNYEWFFADYNSKLLESSNYITRRLAVKLLGDML 240
DIA DA +TFK+LMT+HK+ AE+L NY+ FF Y + L S NY+TRR ++KLLG++L
Sbjct: 182 DIATDAFSTFKDLMTKHKNMCAEYLDNNYDRFFTAY-AALTNSENYVTRRQSLKLLGELL 240
Query: 241 LDRSNSAVMTQYVSSRENLRILMNLLRESSKSIQIEAFHVFKLFAANQNKPADIVSIFVA 300
LDR N + M +Y++S ENL+ +M LLR+ ++IQ EAFHVFK+F AN NKP I I
Sbjct: 241 LDRHNFSTMNKYITSPENLKTVMELLRDKRRNIQYEAFHVFKIFVANPNKPRPITDILNR 300
Query: 301 NKSKMLRLLDEFKIDK-EDEQFEADKAQVMEEIASL 335
N+ K++ L F D+ DEQF +KA ++++I L
Sbjct: 301 NRDKLVEFLTAFHNDRTNDEQFNDEKAYLIKQIQEL 336
>UniRef100_Q7Z2A5 Hypothetical protein R02E12.2 [Caenorhabditis elegans]
Length = 377
Score = 275 bits (703), Expect = 1e-72
Identities = 148/346 (42%), Positives = 218/346 (62%), Gaps = 13/346 (3%)
Query: 4 LFKPKPRTPTDIVRQTRDLLLFFDR--------NTESRDSKREEKQMTELCKNIRELKSI 55
LF ++P D+V+ R++L D+ + + K+ +K + E+ KN+ +KS
Sbjct: 4 LFGKSHKSPADVVKTLREVLTILDKLPPPKLDKDGNIQSDKKYDKALDEVSKNVAMIKSF 63
Query: 56 LYGNSESEPVSE---ACAQLTQEFFKENTLRLLIKCIPKLNLEARKDATQVVANLQRQPV 112
+YGN +EP SE AQL QE + N L +LIK +PK E +KD Q+ NL R+ +
Sbjct: 64 IYGNDSAEPSSEHVVQVAQLAQEVYNANILPMLIKMLPKFEFECKKDVGQIFNNLLRRQI 123
Query: 113 QSKLIASDYLENNMDLMDILIVGYENTDMALHYGAMLRECIRHQIVAKYVLNSPHMKKFF 172
++ +YL +++ L+ GY D+AL G MLRE IRH +AK +L S FF
Sbjct: 124 GTRSPTVEYLGARPEILIQLVQGYSVPDIALTCGLMLRESIRHDHLAKIILYSDVFYTFF 183
Query: 173 DYIQLPNFDIAADAAATFKELMTRHKSTVAEFLSKNYEWFFADYNSKLLESSNYITRRLA 232
Y+Q FDI++DA +TFKEL TRHK+ +AEFL NY+ FFA Y + LL S NY+TRR +
Sbjct: 184 LYVQSEVFDISSDAFSTFKELTTRHKAIIAEFLDSNYDTFFAQYQN-LLNSKNYVTRRQS 242
Query: 233 VKLLGDMLLDRSNSAVMTQYVSSRENLRILMNLLRESSKSIQIEAFHVFKLFAANQNKPA 292
+KLLG++LLDR N MT+Y+S+ +NLR++M LLR+ S++IQ EAFHVFK+F AN NKP
Sbjct: 243 LKLLGELLLDRHNFNTMTKYISNPDNLRLMMELLRDKSRNIQYEAFHVFKVFVANPNKPK 302
Query: 293 DIVSIFVANKSKMLRLLDEFKIDK-EDEQFEADKAQVMEEIASLEA 337
I I N+ K++ L EF D+ +DEQF +KA ++++I +++
Sbjct: 303 PISDILNRNREKLVEFLSEFHNDRTDDEQFNDEKAYLIKQIQEMKS 348
>UniRef100_Q21643 Hypothetical protein R02E12.2 [Caenorhabditis elegans]
Length = 636
Score = 275 bits (703), Expect = 1e-72
Identities = 148/346 (42%), Positives = 218/346 (62%), Gaps = 13/346 (3%)
Query: 4 LFKPKPRTPTDIVRQTRDLLLFFDR--------NTESRDSKREEKQMTELCKNIRELKSI 55
LF ++P D+V+ R++L D+ + + K+ +K + E+ KN+ +KS
Sbjct: 263 LFGKSHKSPADVVKTLREVLTILDKLPPPKLDKDGNIQSDKKYDKALDEVSKNVAMIKSF 322
Query: 56 LYGNSESEPVSE---ACAQLTQEFFKENTLRLLIKCIPKLNLEARKDATQVVANLQRQPV 112
+YGN +EP SE AQL QE + N L +LIK +PK E +KD Q+ NL R+ +
Sbjct: 323 IYGNDSAEPSSEHVVQVAQLAQEVYNANILPMLIKMLPKFEFECKKDVGQIFNNLLRRQI 382
Query: 113 QSKLIASDYLENNMDLMDILIVGYENTDMALHYGAMLRECIRHQIVAKYVLNSPHMKKFF 172
++ +YL +++ L+ GY D+AL G MLRE IRH +AK +L S FF
Sbjct: 383 GTRSPTVEYLGARPEILIQLVQGYSVPDIALTCGLMLRESIRHDHLAKIILYSDVFYTFF 442
Query: 173 DYIQLPNFDIAADAAATFKELMTRHKSTVAEFLSKNYEWFFADYNSKLLESSNYITRRLA 232
Y+Q FDI++DA +TFKEL TRHK+ +AEFL NY+ FFA Y + LL S NY+TRR +
Sbjct: 443 LYVQSEVFDISSDAFSTFKELTTRHKAIIAEFLDSNYDTFFAQYQN-LLNSKNYVTRRQS 501
Query: 233 VKLLGDMLLDRSNSAVMTQYVSSRENLRILMNLLRESSKSIQIEAFHVFKLFAANQNKPA 292
+KLLG++LLDR N MT+Y+S+ +NLR++M LLR+ S++IQ EAFHVFK+F AN NKP
Sbjct: 502 LKLLGELLLDRHNFNTMTKYISNPDNLRLMMELLRDKSRNIQYEAFHVFKVFVANPNKPK 561
Query: 293 DIVSIFVANKSKMLRLLDEFKIDK-EDEQFEADKAQVMEEIASLEA 337
I I N+ K++ L EF D+ +DEQF +KA ++++I +++
Sbjct: 562 PISDILNRNREKLVEFLSEFHNDRTDDEQFNDEKAYLIKQIQEMKS 607
>UniRef100_UPI0000432D61 UPI0000432D61 UniRef100 entry
Length = 335
Score = 273 bits (699), Expect = 4e-72
Identities = 138/336 (41%), Positives = 223/336 (66%), Gaps = 8/336 (2%)
Query: 4 LFKPKPRTPTDIVRQTRDLLLFFDRNTESRDSKREEKQMTELCKNIRELKSILYGNSESE 63
LF ++P ++V+ ++ + N R K+ EK ++ KN+ +K++LYG +E+E
Sbjct: 3 LFGKSQKSPAEVVKALKEAV-----NALERGDKKVEKAQEDVSKNLVHIKNMLYGTAETE 57
Query: 64 PVSE-ACAQLTQEFFKENTLRLLIKCIPKLNLEARKDATQVVANLQRQPVQSKLIASDYL 122
P ++ AQL QE + N L LL++ + +++ E +KD QV N+ R+ + ++ +Y+
Sbjct: 58 PQADIVVAQLAQELYNSNLLLLLVQNLSRIDFEGKKDVAQVFNNILRRQIGTRSPTVEYI 117
Query: 123 ENNMDLMDILIVGYENTDMALHYGAMLRECIRHQIVAKYVLNSPHMKKFFDYIQLPNFDI 182
+++ L+ GYE+ D+AL+ G MLREC R++ +AK ++ S FF Y+++ FDI
Sbjct: 118 CTKPEILFTLMSGYEHQDIALNCGTMLRECARYEALAKIMIYSDDFYNFFRYVEVSTFDI 177
Query: 183 AADAAATFKELMTRHKSTVAEFLSKNYEWFFADYNSKLLESSNYITRRLAVKLLGDMLLD 242
A+DA +TFKEL+TRHK AEFL NY+ F+ Y +LL S NY+TRR ++KLLG++LLD
Sbjct: 178 ASDAFSTFKELLTRHKILSAEFLEINYDKVFSHY-QRLLNSENYVTRRQSLKLLGELLLD 236
Query: 243 RSNSAVMTQYVSSRENLRILMNLLRESSKSIQIEAFHVFKLFAANQNKPADIVSIFVANK 302
R N VMT+Y+S+ +NL+++MN+L+E S++IQ EAFHVFK+F AN NKP I+ I + N+
Sbjct: 237 RHNFTVMTRYISNPDNLKLMMNMLKEKSRNIQFEAFHVFKVFVANPNKPKPILDILLRNQ 296
Query: 303 SKMLRLLDEFKIDK-EDEQFEADKAQVMEEIASLEA 337
K++ L F D+ EDEQF +KA ++++I L++
Sbjct: 297 EKLIEFLTRFHTDRSEDEQFNDEKAYLIKQIKELKS 332
>UniRef100_Q803V8 Zgc:55451 [Brachydanio rerio]
Length = 343
Score = 273 bits (699), Expect = 4e-72
Identities = 136/333 (40%), Positives = 223/333 (66%), Gaps = 3/333 (0%)
Query: 5 FKPKPRTPTDIVRQTRDLLLFFDRNTESRDSKREEKQMTELCKNIRELKSILYGNSESEP 64
F+ ++P +IV+ ++ + + ++ ES +SK+ EK E+ KN+ LK +L G + EP
Sbjct: 5 FEKSQKSPAEIVKSLKENVAYLEK-LESSESKKCEKVAEEVSKNLSSLKEVLCGTGDKEP 63
Query: 65 VSEACAQLTQEFFKENTLRLLIKCIPKLNLEARKDATQVVANLQRQPVQSKLIASDYLEN 124
+EA AQL QE + N L LI + +++ E +KD + +N+ R+ + ++ +Y+ +
Sbjct: 64 QTEAVAQLAQELYNTNLLISLIANLQRIDFEGKKDVVHLFSNIVRRQIGARTPTVEYISS 123
Query: 125 NMDLMDILIVGYENTDMALHYGAMLRECIRHQIVAKYVLNSPHMKKFFDYIQLPNFDIAA 184
+ ++ +L+ GYE +++AL+ G MLREC+RH +A+ VL S FF Y+++ FDIA+
Sbjct: 124 HSQILFMLLKGYETSEVALNCGMMLRECLRHDPLARIVLFSEDFYCFFRYVEMSTFDIAS 183
Query: 185 DAAATFKELMTRHKSTVAEFLSKNYEWFFADYNSKLLESSNYITRRLAVKLLGDMLLDRS 244
DA A+FK+L+TRHK A+FL NY+ F +Y KLL S NY+T+R ++KLLG++LLDR
Sbjct: 184 DAFASFKDLLTRHKIMCADFLETNYDRVFTEY-EKLLHSENYVTKRQSLKLLGELLLDRH 242
Query: 245 NSAVMTQYVSSRENLRILMNLLRESSKSIQIEAFHVFKLFAANQNKPADIVSIFVANKSK 304
N V T+Y+S ENL+++MN+LR++S++IQ EAFHVFK+F AN NK ++ I + N+SK
Sbjct: 243 NFTVATKYISRAENLKLMMNMLRDNSRNIQFEAFHVFKVFVANPNKTQPVLDILLKNQSK 302
Query: 305 MLRLLDEFKIDK-EDEQFEADKAQVMEEIASLE 336
++ L F+ D+ EDEQF +K ++++I L+
Sbjct: 303 LVEFLSHFQTDRSEDEQFCDEKNYLIKQIRDLK 335
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.320 0.133 0.364
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 487,538,097
Number of Sequences: 2790947
Number of extensions: 18466813
Number of successful extensions: 64270
Number of sequences better than 10.0: 101
Number of HSP's better than 10.0 without gapping: 68
Number of HSP's successfully gapped in prelim test: 33
Number of HSP's that attempted gapping in prelim test: 64049
Number of HSP's gapped (non-prelim): 113
length of query: 337
length of database: 848,049,833
effective HSP length: 128
effective length of query: 209
effective length of database: 490,808,617
effective search space: 102579000953
effective search space used: 102579000953
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 75 (33.5 bits)
Medicago: description of AC148482.4


