

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148481.8 + phase: 0 /pseudo
(339 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
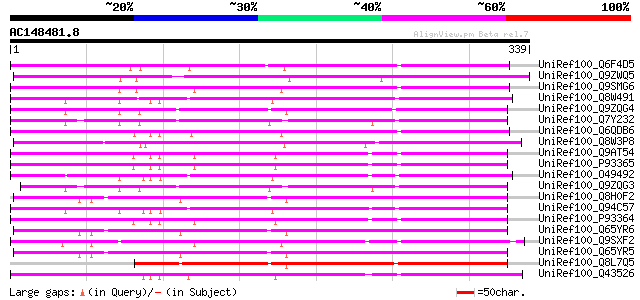
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q6F4D5 UDP-glucose glucosyltransferase [Catharanthus r... 256 8e-67
UniRef100_Q9ZWQ5 UDP-glycose:flavonoid glycosyltransferase [Vign... 254 3e-66
UniRef100_Q9SMG6 Betanidin-5-O-glucosyltransferase [Dorotheanthu... 246 5e-64
UniRef100_Q8W491 Hypothetical protein At4g34120; F28A23.120 [Ara... 244 2e-63
UniRef100_Q9ZQG4 Putative glucosyltransferase [Arabidopsis thali... 240 4e-62
UniRef100_Q7Y232 At2g15490 [Arabidopsis thaliana] 239 1e-61
UniRef100_Q6QDB6 UDP-glucose glucosyltransferase [Rhodiola sacha... 237 4e-61
UniRef100_Q8W3P8 ABA-glucosyltransferase [Phaseolus angularis] 236 9e-61
UniRef100_Q9AT54 Phenylpropanoid:glucosyltransferase 1 [Nicotian... 234 2e-60
UniRef100_P93365 Immediate-early salicylate-induced glucosyltran... 234 3e-60
UniRef100_O49492 Glucosyltransferase-like protein [Arabidopsis t... 234 3e-60
UniRef100_Q9ZQG3 Putative glucosyltransferase [Arabidopsis thali... 232 1e-59
UniRef100_Q8H0F2 Anthocyanin 3'-glucosyltransferase [Gentiana tr... 231 2e-59
UniRef100_Q94C57 Putative glucosyltransferase [Arabidopsis thali... 230 4e-59
UniRef100_P93364 Immediate-early salicylate-induced glucosyltran... 226 9e-58
UniRef100_Q65YR6 UDP-glucose:anthocyanin 3'-O-glucosyltransferas... 223 6e-57
UniRef100_Q9SXF2 UDP-glucose: flavonoid 7-O-glucosyltransferase ... 222 1e-56
UniRef100_Q65YR5 UDP-glucose:anthocyanin 3'-O-glucosyltransferas... 222 1e-56
UniRef100_Q8L7Q5 Putative glucosyltransferase [Arabidopsis thali... 221 2e-56
UniRef100_Q43526 Twi1 protein [Lycopersicon esculentum] 221 2e-56
>UniRef100_Q6F4D5 UDP-glucose glucosyltransferase [Catharanthus roseus]
Length = 487
Score = 256 bits (653), Expect = 8e-67
Identities = 154/397 (38%), Positives = 211/397 (52%), Gaps = 74/397 (18%)
Query: 1 MNPMIDTARLFAKHGVDVTIITTQANALLFKKPIDNDLFSGYSIKACVIQFPAAQVGLPD 60
M P +D A LF GV VT+ITT + +F K I+ SG+ I I+FPA++VGLP+
Sbjct: 18 MLPALDMANLFTSRGVKVTLITTHQHVPMFTKSIERSRNSGFDISIQSIKFPASEVGLPE 77
Query: 61 GVENIKDAT-SREMLAHF--------------IKKQKPH--------------------- 84
G+E++ + EML F +++ +PH
Sbjct: 78 GIESLDQVSGDDEMLPKFMRGVNLLQQPLEQLLQESRPHCLLSDMFFPWTTESAAKFGIP 137
Query: 85 ------------------------ENLVSDSQKFSIPGLPHNIEITSLQLQEYVRE--WS 118
EN+ +D+++F +P LPH I++T Q+ Y RE S
Sbjct: 138 RLLFHGSCSFALSAAESVRRNKPFENVSTDTEEFVVPDLPHQIKLTRTQISTYERENIES 197
Query: 119 EFSDYFDAVYESEGKSYGTLCNSFHELEGDYENLYKSTMGIKAWSVGPVSAWLKKEQNED 178
+F+ V +SE SYG + NSF+ELE DY + Y + +G KAW +GP K Q ED
Sbjct: 198 DFTKMLKKVRDSESTSYGVVVNSFYELEPDYADYYINVLGRKAWHIGPF-LLCNKLQAED 256
Query: 179 --------VIVESELLNWLNSKPNDSVLYVSFGSLTRLSHSQIVEIAHGLENSGHNFIWV 230
I E LNWL+SK +SV+Y+ FGS+ L+ +Q+ EIA LE+SG NFIWV
Sbjct: 257 KAQRGKKSAIDADECLNWLDSKQPNSVIYLCFGSMANLNSAQLHEIATALESSGQNFIWV 316
Query: 231 VRK-KDGEGDEDGFLDDFKQRMKENKKGYIIWNWAPQLLILGHPATAGVVTHCGWNSILE 289
VRK D E F + F++R KE KG II WAPQ LIL H + VTHCGWNS LE
Sbjct: 317 VRKCVDEENSSKWFPEGFEERTKE--KGLIIKGWAPQTLILEHESVGAFVTHCGWNSTLE 374
Query: 290 SLSVSLPIITWPMFAEQFYNEKLLVFVLKIVVSVGSK 326
+ +P++TWP FAEQF+NEKL+ VLK VG++
Sbjct: 375 GICAGVPLVTWPFFAEQFFNEKLITEVLKTGYGVGAR 411
>UniRef100_Q9ZWQ5 UDP-glycose:flavonoid glycosyltransferase [Vigna mungo]
Length = 477
Score = 254 bits (648), Expect = 3e-66
Identities = 147/402 (36%), Positives = 216/402 (53%), Gaps = 72/402 (17%)
Query: 3 PMIDTARLFAKHGVDVTIITTQANALLFKKPIDNDLFSGYSIKACVIQFPAAQVGLPDGV 62
PM+ ARL A G +TI+TT NA LF+K ID+D+ SG+ I+ +++FP Q+GLP+GV
Sbjct: 20 PMVQLARLIASRGQHITILTTSGNAQLFQKTIDDDIASGHHIRLHLLKFPGTQLGLPEGV 79
Query: 63 ENIKDATSR------EMLAHFIKKQ----------------------------------- 81
EN+ AT+ M AHFI+ Q
Sbjct: 80 ENLVSATNNITAGKIHMAAHFIQPQVESVLKESPPDVFIPDIIFTWSKDMSKRLQIPRLV 139
Query: 82 ---------------KPH-ENLVSDSQKFSIPGLPHNIEITSLQLQEYVREWSEFSDYFD 125
K H E +SDS + IPGLPH + + V+ F+ +
Sbjct: 140 FNPISIFDVCMIQAIKAHPEAFLSDSGPYQIPGLPHPLTLP-------VKPSPGFAVLTE 192
Query: 126 AVYESEGKSYGTLCNSFHELEGDYENLYKSTMGIKAWSVGPVSAWLKKEQNEDVIV---E 182
++ E E S+G + NSF EL+ +Y Y+ G K W VGP S +++ + IV
Sbjct: 193 SLLEGEDDSHGVIVNSFAELDAEYTQYYEKLTGRKVWHVGPSSLMVEQIVKKPAIVSEIR 252
Query: 183 SELLNWLNSKPNDSVLYVSFGSLTRLSHSQIVEIAHGLENSGHNFIWVVRKKDGEGDED- 241
+E L WL+SK DSVLY+ FGSL LS Q+ E+A+GL+ SGH+FIWVV +K EG E+
Sbjct: 253 NECLTWLDSKERDSVLYICFGSLVLLSDKQLYELANGLDASGHSFIWVVHRKKKEGQEEE 312
Query: 242 ----GFLDDFKQRMKENKKGYIIWNWAPQLLILGHPATAGVVTHCGWNSILESLSVSLPI 297
+ F+++++ K+G +I WAPQ LIL HPA G +THCGWN+++E++S +P+
Sbjct: 313 EEEKWLPEGFEEKIEREKRGMLIKGWAPQPLILNHPAVGGFLTHCGWNAVVEAISAGVPM 372
Query: 298 ITWPMFAEQFYNEKLLVFVLKIVVSVGSKVNTFWSNEGEVAV 339
+T P F++Q++NEKL+ V V VG+ + EG+ V
Sbjct: 373 VTMPGFSDQYFNEKLITEVHGFGVEVGAAEWSISPYEGKKTV 414
>UniRef100_Q9SMG6 Betanidin-5-O-glucosyltransferase [Dorotheanthus bellidiformis]
Length = 489
Score = 246 bits (629), Expect = 5e-64
Identities = 150/395 (37%), Positives = 215/395 (53%), Gaps = 71/395 (17%)
Query: 1 MNPMIDTARLFAKHGVDVTIITTQANALLFKKPIDNDLFSGYS-IKACVIQFPAAQVGLP 59
M P +D A+LFA GV TIITT NA +F K I+ + + ++ V FP+ + GLP
Sbjct: 23 MIPSLDIAKLFAARGVKTTIITTPLNASMFTKAIEKTRKNTETQMEIEVFSFPSEEAGLP 82
Query: 60 DGVENIKDATS--------------REMLAHFIKKQ------------------------ 81
G EN++ A + +E L +F+ K
Sbjct: 83 LGCENLEQAMAIGANNEFFNAANLLKEQLENFLVKTRPNCLVADMFFTWAADSTAKFNIP 142
Query: 82 ---------------------KPHENLVSDSQKFSIPGLPHNIEITSLQLQEYVREWSE- 119
KP++ + SD++ FS+P LPH +++T LQ+ E +R+ E
Sbjct: 143 TLVFHGFSFFAQCAKEVMWRYKPYKAVSSDTEVFSLPFLPHEVKMTRLQVPESMRKGEET 202
Query: 120 -FSDYFDAVYESEGKSYGTLCNSFHELEGDYENLYKSTMGIKAWSVGPVSAWLKKEQN-- 176
F+ + + E E KSYG + NSF+ELE DY + + +G +AW +GPVS + ++
Sbjct: 203 HFTKRTERIRELERKSYGVIVNSFYELEPDYADFLRKELGRRAWHIGPVSLCNRSIEDKA 262
Query: 177 ----EDVIVESELLNWLNSKPNDSVLYVSFGSLTRLSHSQIVEIAHGLENSGHNFIWVVR 232
+ I E E L WLNSK DSV+Y+ FGS L Q+ EIA LE SG +FIW VR
Sbjct: 263 QRGRQTSIDEDECLKWLNSKKPDSVIYICFGSTGHLIAPQLHEIATALEASGQDFIWAVR 322
Query: 233 KKDGEGDEDGFLDD-FKQRMKENKKGYIIWNWAPQLLILGHPATAGVVTHCGWNSILESL 291
G+G+ + +L ++ R++ KG II WAPQ+LIL H AT G +THCGWNS LE +
Sbjct: 323 GDHGQGNSEEWLPPGYEHRLQG--KGLIIRGWAPQVLILEHEATGGFLTHCGWNSALEGI 380
Query: 292 SVSLPIITWPMFAEQFYNEKLLVFVLKIVVSVGSK 326
S +P++TWP FAEQF+NE+LL +LK+ V+VGSK
Sbjct: 381 SAGVPMVTWPTFAEQFHNEQLLTQILKVGVAVGSK 415
>UniRef100_Q8W491 Hypothetical protein At4g34120; F28A23.120 [Arabidopsis thaliana]
Length = 481
Score = 244 bits (623), Expect = 2e-63
Identities = 154/402 (38%), Positives = 215/402 (53%), Gaps = 78/402 (19%)
Query: 1 MNPMIDTARLFAKHGVDVTIITTQANALLFKKPID--NDLFSGYSIKACVIQFPAAQVGL 58
M P +D A+LF+ G TI+TT N+ +F+KPI+ +L + I + FP +GL
Sbjct: 22 MIPTLDMAKLFSSRGAKSTILTTPLNSKIFQKPIERFKNLNPSFEIDIQIFDFPCVDLGL 81
Query: 59 PDGVENIKDATS--------------------REMLAHFIKKQKPHENLVSD-------- 90
P+G EN+ TS ++ L ++ +P + L++D
Sbjct: 82 PEGCENVDFFTSNNNDDRQYLTLKFFKSTRFFKDQLEKLLETTRP-DCLIADMFFPWATE 140
Query: 91 -SQKFSI-------------------------------------PGLPHNIEITSLQLQE 112
++KF++ P LP NI IT Q+ +
Sbjct: 141 AAEKFNVPRLVFHGTGYFSLCSEYCIRVHNPQNIVASRYEPFVIPDLPGNIVITQEQIAD 200
Query: 113 YVREWSEFSDYFDAVYESEGKSYGTLCNSFHELEGDYENLYKSTMGIKAWSVGPVSAW-- 170
E SE + V ES+ KS G + NSF+ELE DY + YKS + +AW +GP+S +
Sbjct: 201 RDEE-SEMGKFMIEVKESDVKSSGVIVNSFYELEPDYADFYKSVVLKRAWHIGPLSVYNR 259
Query: 171 ---LKKEQNEDV-IVESELLNWLNSKPNDSVLYVSFGSLTRLSHSQIVEIAHGLENSGHN 226
K E+ + I E E L WL+SK DSV+Y+SFGS+ + Q+ EIA GLE SG N
Sbjct: 260 GFEEKAERGKKASINEVECLKWLDSKKPDSVIYISFGSVACFKNEQLFEIAAGLETSGAN 319
Query: 227 FIWVVRKKDGEGDEDGFLDDFKQRMKENKKGYIIWNWAPQLLILGHPATAGVVTHCGWNS 286
FIWVVRK G E+ + F++R+K KG II WAPQ+LIL H AT G VTHCGWNS
Sbjct: 320 FIWVVRKNIGIEKEEWLPEGFEERVK--GKGMIIRGWAPQVLILDHQATCGFVTHCGWNS 377
Query: 287 ILESLSVSLPIITWPMFAEQFYNEKLLVFVLKIVVSVGSKVN 328
+LE ++ LP++TWP+ AEQFYNEKL+ VL+ VSVG+K N
Sbjct: 378 LLEGVAAGLPMVTWPVAAEQFYNEKLVTQVLRTGVSVGAKKN 419
>UniRef100_Q9ZQG4 Putative glucosyltransferase [Arabidopsis thaliana]
Length = 484
Score = 240 bits (613), Expect = 4e-62
Identities = 150/400 (37%), Positives = 207/400 (51%), Gaps = 79/400 (19%)
Query: 1 MNPMIDTARLFAKHGVDVTIITTQANALLFKKPID--NDLFSGYSIKACVIQFPAAQVGL 58
M P++D A+LF++ G T++TT NA +F+KPI+ + I + FP ++GL
Sbjct: 22 MIPILDMAKLFSRRGAKSTLLTTPINAKIFEKPIEAFKNQNPDLEIGIKIFNFPCVELGL 81
Query: 59 PDGVENIKDATS--------------------REMLAHFIKKQKP--------------- 83
P+G EN S ++ L FI+ KP
Sbjct: 82 PEGCENADFINSYQKSDSGDLFLKFLFSTKYMKQQLESFIETTKPSALVADMFFPWATES 141
Query: 84 ------------------------------HENLVSDSQKFSIPGLPHNIEITSLQLQEY 113
H+ + + S F IPGLP +I IT Q
Sbjct: 142 AEKLGVPRLVFHGTSFFSLCCSYNMRIHKPHKKVATSSTPFVIPGLPGDIVITEDQ-ANV 200
Query: 114 VREWSEFSDYFDAVYESEGKSYGTLCNSFHELEGDYENLYKSTMGIKAWSVGPVSAWLKK 173
+E + + V ESE S+G L NSF+ELE Y + Y+S + +AW +GP+S +
Sbjct: 201 AKEETPMGKFMKEVRESETNSFGVLVNSFYELESAYADFYRSFVAKRAWHIGPLSL-SNR 259
Query: 174 EQNEDV-------IVESELLNWLNSKPNDSVLYVSFGSLTRLSHSQIVEIAHGLENSGHN 226
E E I E E L WL+SK SV+Y+SFGS T ++ Q++EIA GLE SG +
Sbjct: 260 ELGEKARRGKKANIDEQECLKWLDSKTPGSVVYLSFGSGTNFTNDQLLEIAFGLEGSGQS 319
Query: 227 FIWVVRKKDGEGDEDGFL-DDFKQRMKENKKGYIIWNWAPQLLILGHPATAGVVTHCGWN 285
FIWVVRK + +GD + +L + FK+R KG II WAPQ+LIL H A G VTHCGWN
Sbjct: 320 FIWVVRKNENQGDNEEWLPEGFKER--TTGKGLIIPGWAPQVLILDHKAIGGFVTHCGWN 377
Query: 286 SILESLSVSLPIITWPMFAEQFYNEKLLVFVLKIVVSVGS 325
S +E ++ LP++TWPM AEQFYNEKLL VL+I V+VG+
Sbjct: 378 SAIEGIAAGLPMVTWPMGAEQFYNEKLLTKVLRIGVNVGA 417
>UniRef100_Q7Y232 At2g15490 [Arabidopsis thaliana]
Length = 484
Score = 239 bits (609), Expect = 1e-61
Identities = 160/409 (39%), Positives = 208/409 (50%), Gaps = 94/409 (22%)
Query: 1 MNPMIDTARLFAKHGVDVTIITTQANALLFKKPID------NDLFSGYSIKACVIQFPAA 54
M P++D A+LFA+ G T++TT NA + +KPI+ DL G I + FP
Sbjct: 19 MIPLLDMAKLFARRGAKSTLLTTPINAKILEKPIEAFKVQNPDLEIGIKI----LNFPCV 74
Query: 55 QVGLPDGVENIKDATS--------------------REMLAHFIKKQKP----------- 83
++GLP+G EN S ++ L FI+ KP
Sbjct: 75 ELGLPEGCENRDFINSYQKSDSFDLFLKFLFSTKYMKQQLESFIETTKPSALVADMFFPW 134
Query: 84 ----------------------------------HENLVSDSQKFSIPGLPHNIEITSLQ 109
H+ + S S F IPGLP +I IT Q
Sbjct: 135 ATESAEKIGVPRLVFHGTSSFALCCSYNMRIHKPHKKVASSSTPFVIPGLPGDIVITEDQ 194
Query: 110 LQEYVREWSEFSDYFDAVYESEGKSYGTLCNSFHELEGDYENLYKSTMGIKAWSVGPVS- 168
E + F ++ V ESE S+G L NSF+ELE Y + Y+S + KAW +GP+S
Sbjct: 195 -ANVTNEETPFGKFWKEVRESETSSFGVLVNSFYELESSYADFYRSFVAKKAWHIGPLSL 253
Query: 169 --------AWLKKEQNEDVIVESELLNWLNSKPNDSVLYVSFGSLTRLSHSQIVEIAHGL 220
A K+ N D E E L WL+SK SV+Y+SFGS T L + Q++EIA GL
Sbjct: 254 SNRGIAEKAGRGKKANID---EQECLKWLDSKTPGSVVYLSFGSGTGLPNEQLLEIAFGL 310
Query: 221 ENSGHNFIWVVRKKD---GEGD-EDGFLDDFKQRMKENKKGYIIWNWAPQLLILGHPATA 276
E SG NFIWVV K + G G+ ED F++R K KG II WAPQ+LIL H A
Sbjct: 311 EGSGQNFIWVVSKNENQVGTGENEDWLPKGFEERNK--GKGLIIRGWAPQVLILDHKAIG 368
Query: 277 GVVTHCGWNSILESLSVSLPIITWPMFAEQFYNEKLLVFVLKIVVSVGS 325
G VTHCGWNS LE ++ LP++TWPM AEQFYNEKLL VL+I V+VG+
Sbjct: 369 GFVTHCGWNSTLEGIAAGLPMVTWPMGAEQFYNEKLLTKVLRIGVNVGA 417
>UniRef100_Q6QDB6 UDP-glucose glucosyltransferase [Rhodiola sachalinensis]
Length = 480
Score = 237 bits (604), Expect = 4e-61
Identities = 143/394 (36%), Positives = 200/394 (50%), Gaps = 70/394 (17%)
Query: 1 MNPMIDTARLFAKHGVDVTIITTQANALLFKKPIDNDLFSGYSIKACVIQFPAAQVGLPD 60
M PM+D ARLFA GV TI+TT N L + I G+ I I F + GLPD
Sbjct: 21 MIPMVDMARLFASQGVRCTIVTTPGNQPLIARSIGKVQLLGFEIGVTTIPFRGTEFGLPD 80
Query: 61 GVENIKDATSREMLAHFIKK----QKPHENLVSD------------------SQKFSI-- 96
G EN+ S + + HF + ++P E L+ + + KF I
Sbjct: 81 GCENLDSVPSPQHVFHFFEAAGSLREPFEQLLEEHKPDCVVGDMFFPWSTDSAAKFGIPR 140
Query: 97 -----------------------------------PGLPHNIEITSLQLQEYVREW---S 118
PGLP I++T QL ++ E S
Sbjct: 141 LVFHGTSYFALCAGEAVRIHKPYLSVSSDDEPFVIPGLPDEIKLTKSQLPMHLLEGKKDS 200
Query: 119 EFSDYFDAVYESEGKSYGTLCNSFHELEGDYENLYKSTMGIKAWSVGPVSAWLKKEQN-- 176
+ D V E+E SYG + NS +ELE Y + +++ + +AW +GP+S + +
Sbjct: 201 VLAQLLDEVKETEVSSYGVIVNSIYELEPAYADYFRNVLKRRAWEIGPLSLCNRDVEEKA 260
Query: 177 ----EDVIVESELLNWLNSKPNDSVLYVSFGSLTRLSHSQIVEIAHGLENSGHNFIWVVR 232
+ I + E L WL+SK DSV+YV FGS + Q+ EIA GLE SG FIWV+R
Sbjct: 261 MRGMQAAIDQHECLKWLDSKEPDSVVYVCFGSTCKFPDDQLAEIASGLEASGQQFIWVIR 320
Query: 233 KKDGEGDEDGFLDDFKQRMKENKKGYIIWNWAPQLLILGHPATAGVVTHCGWNSILESLS 292
+ + ED F++R+K+ + +I WAPQ+LIL H + G V+HCGWNS LE +S
Sbjct: 321 RMSDDSKEDYLPKGFEERVKD--RALLIRGWAPQVLILDHQSVGGFVSHCGWNSTLEGIS 378
Query: 293 VSLPIITWPMFAEQFYNEKLLVFVLKIVVSVGSK 326
LP++TWP+FAEQFYNEKLL VLKI V+VG++
Sbjct: 379 AGLPMVTWPVFAEQFYNEKLLTEVLKIGVAVGAR 412
>UniRef100_Q8W3P8 ABA-glucosyltransferase [Phaseolus angularis]
Length = 478
Score = 236 bits (601), Expect = 9e-61
Identities = 151/391 (38%), Positives = 202/391 (51%), Gaps = 62/391 (15%)
Query: 3 PMIDTARLFAKHGVDVTIITTQANALLFKKPIDNDLFSGYSIKACVIQFPAAQVGLPDGV 62
PMID AR+FA HG TI+ T + LF+K I D G I + Q + G
Sbjct: 23 PMIDAARMFASHGASSTILATPSTTPLFQKCITRDQKFGLPISIHTLSADVPQSDISVG- 81
Query: 63 ENIKDATSREMLAHFIKKQKPH------------------------------------EN 86
+ + E L + +++PH EN
Sbjct: 82 PFLDTSALLEPLRQLLLQRRPHCIVVDMFHRWSGDVVYELGIPRTLFNGIGCFALCVQEN 141
Query: 87 L--------VSDSQKFSIPGLPHNIEITSLQLQEYVREWSEFSDYFDAVYESEGKSYGTL 138
L +DS+ F +P +P IE+T QL ++R S + + + + E KS+GTL
Sbjct: 142 LRHVAFKSVSTDSEPFLVPNIPDRIEMTMSQLPPFLRNPSGIPERWRGMKQLEEKSFGTL 201
Query: 139 CNSFHELEGDYENLYKSTMGIKAWSVGPVSAWLKKEQNED------VIVESELLNWLNSK 192
NSF++LE Y +L KS G KAW VGPVS + ++++ I E LNWLNSK
Sbjct: 202 INSFYDLEPAYADLIKSKWGNKAWIVGPVSFCNRSKEDKTERGKPPTIDEQNCLNWLNSK 261
Query: 193 PNDSVLYVSFGSLTRLSHSQIVEIAHGLENSGHNFIWVV---------RKKDGEGDEDGF 243
SVLY SFGSL RL Q+ EIA+GLE S +FIWVV K++G G+
Sbjct: 262 KPSSVLYASFGSLARLPPEQLKEIAYGLEASEQSFIWVVGNILHNPSENKENGSGN--WL 319
Query: 244 LDDFKQRMKENKKGYIIWNWAPQLLILGHPATAGVVTHCGWNSILESLSVSLPIITWPMF 303
+ F+QRMKE KG ++ WAPQLLIL H A G +THCGWNS LE +S +P+ITWP+
Sbjct: 320 PEGFEQRMKETGKGLVLRGWAPQLLILEHAAIKGFMTHCGWNSTLEGVSAGVPMITWPLT 379
Query: 304 AEQFYNEKLLVFVLKIVVSVGSKVNTFWSNE 334
AEQF NEKL+ VLK V VG++ W+ E
Sbjct: 380 AEQFSNEKLITEVLKTGVQVGNREWWPWNAE 410
>UniRef100_Q9AT54 Phenylpropanoid:glucosyltransferase 1 [Nicotiana tabacum]
Length = 476
Score = 234 bits (598), Expect = 2e-60
Identities = 147/392 (37%), Positives = 201/392 (50%), Gaps = 71/392 (18%)
Query: 1 MNPMIDTARLFAKHGVDVTIITTQANALLFKKPIDNDLFSGYSIKACVIQFPAAQVGLPD 60
M P +D A+LFA GV TIITT N +F K I + G I+ +I+FPA + GLP+
Sbjct: 17 MIPTLDMAKLFASRGVKATIITTPLNEFVFSKAIQRNKHLGIEIEIRLIKFPAVENGLPE 76
Query: 61 GVENIKDATSREMLAHFIK----KQKPHENLVSD------------------SQKFSI-- 96
E + S E L +F K Q+P E L+ + + KF+I
Sbjct: 77 ECERLDQIPSDEKLPNFFKAVAMMQEPLEQLIEECRPDCLISDMFLPWTTDTAAKFNIPR 136
Query: 97 -----------------------------------PGLPHNIEITSLQLQEYVREWSE-- 119
P LPH I++T Q+ + R E
Sbjct: 137 IVFHGTSFFALCVENSVRLNKPFKNVSSDSETFVVPDLPHEIKLTRTQVSPFERSGEETA 196
Query: 120 FSDYFDAVYESEGKSYGTLCNSFHELEGDYENLYKSTMGIKAWSVGPVSAWLK------K 173
+ V ES+ KSYG + NSF+ELE DY Y +G +AW++GP+S + +
Sbjct: 197 MTRMIKTVRESDSKSYGVVFNSFYELETDYVEHYTKVLGRRAWAIGPLSMCNRDIEDKAE 256
Query: 174 EQNEDVIVESELLNWLNSKPNDSVLYVSFGSLTRLSHSQIVEIAHGLENSGHNFIWVVRK 233
+ I + E L WL+SK SV+YV FGS+ + SQ+ E+A G+E SG FIWVVR
Sbjct: 257 RGKKSSIDKHECLKWLDSKKPSSVVYVCFGSVANFTASQLHELAMGIEASGQEFIWVVRT 316
Query: 234 KDGEGDEDGFLDDFKQRMKENKKGYIIWNWAPQLLILGHPATAGVVTHCGWNSILESLSV 293
+ +ED + F++R KE KG II WAPQ+LIL H + VTHCGWNS LE +S
Sbjct: 317 E--LDNEDWLPEGFEERTKE--KGLIIRGWAPQVLILDHESVGAFVTHCGWNSTLEGVSG 372
Query: 294 SLPIITWPMFAEQFYNEKLLVFVLKIVVSVGS 325
+P++TWP+FAEQF+NEKL+ VLK VGS
Sbjct: 373 GVPMVTWPVFAEQFFNEKLVTEVLKTGAGVGS 404
>UniRef100_P93365 Immediate-early salicylate-induced glucosyltransferase [Nicotiana
tabacum]
Length = 476
Score = 234 bits (596), Expect = 3e-60
Identities = 146/392 (37%), Positives = 201/392 (51%), Gaps = 71/392 (18%)
Query: 1 MNPMIDTARLFAKHGVDVTIITTQANALLFKKPIDNDLFSGYSIKACVIQFPAAQVGLPD 60
M P +D A+LFA GV TIITT N +F K I + G I+ +I+FPA + GLP+
Sbjct: 17 MIPTLDMAKLFASRGVKATIITTPLNEFVFSKAIQRNKHLGIEIEIRLIKFPAVENGLPE 76
Query: 61 GVENIKDATSREMLAHFIK----KQKPHENLVSD------------------SQKFSI-- 96
E + S E L +F K Q+P E L+ + + KF+I
Sbjct: 77 ECERLDQIPSDEKLPNFFKAVAMMQEPLEQLIEECRPDCLISDMFLPWTTDTAAKFNIPR 136
Query: 97 -----------------------------------PGLPHNIEITSLQLQEYVREWSE-- 119
P LPH I++T Q+ + R E
Sbjct: 137 IVFHGTSFFALCVENSVRLNKPFKNVSSDSETFVVPDLPHEIKLTRTQVSPFERSGEETA 196
Query: 120 FSDYFDAVYESEGKSYGTLCNSFHELEGDYENLYKSTMGIKAWSVGPVSAWLK------K 173
+ V ES+ KSYG + NSF+ELE DY Y +G +AW++GP+S + +
Sbjct: 197 MTRMIKTVRESDSKSYGVVFNSFYELETDYVEHYTKVLGRRAWAIGPLSMCNRDIEDKAE 256
Query: 174 EQNEDVIVESELLNWLNSKPNDSVLYVSFGSLTRLSHSQIVEIAHGLENSGHNFIWVVRK 233
+ I + E L WL+SK SV+Y+ FGS+ + SQ+ E+A G+E SG FIWVVR
Sbjct: 257 RGKKSSIDKHECLKWLDSKKPSSVVYICFGSVANFTASQLHELAMGVEASGQEFIWVVRT 316
Query: 234 KDGEGDEDGFLDDFKQRMKENKKGYIIWNWAPQLLILGHPATAGVVTHCGWNSILESLSV 293
+ +ED + F++R KE KG II WAPQ+LIL H + VTHCGWNS LE +S
Sbjct: 317 E--LDNEDWLPEGFEERTKE--KGLIIRGWAPQVLILDHESVGAFVTHCGWNSTLEGVSG 372
Query: 294 SLPIITWPMFAEQFYNEKLLVFVLKIVVSVGS 325
+P++TWP+FAEQF+NEKL+ VLK VGS
Sbjct: 373 GVPMVTWPVFAEQFFNEKLVTEVLKTGAGVGS 404
>UniRef100_O49492 Glucosyltransferase-like protein [Arabidopsis thaliana]
Length = 478
Score = 234 bits (596), Expect = 3e-60
Identities = 158/400 (39%), Positives = 214/400 (53%), Gaps = 78/400 (19%)
Query: 1 MNPMIDTARLFAKHGVDVTIITTQANA-LLFKKPIDNDLFSGYSIKACVIQFPAAQVGLP 59
M P +D A+LFA G TI+TT NA L F+KPI N L G I + FP ++GLP
Sbjct: 23 MIPTLDMAKLFATKGAKSTILTTPLNAKLFFEKPIKN-LNPGLEIDIQIFNFPCVELGLP 81
Query: 60 DGVENIKDATS------REMLAHFIKKQKPHEN-------------LVSD---------S 91
+G EN+ TS EM+ F + ++ L++D +
Sbjct: 82 EGCENVDFFTSNNNDDKNEMIVKFFFSTRFFKDQLEKLLGTTRPDCLIADMFFPWATEAA 141
Query: 92 QKFSIP-------------------------------------GLPHNIEITSLQLQEYV 114
KF++P LP NI IT Q+ +
Sbjct: 142 GKFNVPRLVFHGTGYFSLCAGYCIGVHKPQKRVASSSEPFVIPELPGNIVITEEQIIDGD 201
Query: 115 REWSEFSDYFDAVYESEGKSYGTLCNSFHELEGDYENLYKSTMGIKAWSVGPVSAW---- 170
E S+ + V ESE KS G + NSF+ELE DY + YKS + +AW +GP+S +
Sbjct: 202 GE-SDMGKFMTEVRESEVKSSGVVLNSFYELEHDYADFYKSCVQKRAWHIGPLSVYNRGF 260
Query: 171 -LKKEQNEDV-IVESELLNWLNSKPNDSVLYVSFGSLTRLSHSQIVEIAHGLENSGHNFI 228
K E+ + I E+E L WL+SK +SV+YVSFGS+ + Q+ EIA GLE SG +FI
Sbjct: 261 EEKAERGKKANIDEAECLKWLDSKKPNSVIYVSFGSVAFFKNEQLFEIAAGLEASGTSFI 320
Query: 229 WVVRKKDGEGDEDGFLDDFKQRMKENKKGYIIWNWAPQLLILGHPATAGVVTHCGWNSIL 288
WVVRK + E+ + F++R+K KG II WAPQ+LIL H AT G VTHCGWNS+L
Sbjct: 321 WVVRKT--KEKEEWLPEGFEERVK--GKGMIIRGWAPQVLILDHQATCGFVTHCGWNSLL 376
Query: 289 ESLSVSLPIITWPMFAEQFYNEKLLVFVLKIVVSVGSKVN 328
E ++ LP++TWP+ AEQFYNEKL+ VL+ VSVG+K N
Sbjct: 377 EGVAAGLPMVTWPVAAEQFYNEKLVTQVLRTGVSVGAKKN 416
>UniRef100_Q9ZQG3 Putative glucosyltransferase [Arabidopsis thaliana]
Length = 460
Score = 232 bits (591), Expect = 1e-59
Identities = 157/402 (39%), Positives = 203/402 (50%), Gaps = 94/402 (23%)
Query: 8 ARLFAKHGVDVTIITTQANALLFKKPID------NDLFSGYSIKACVIQFPAAQVGLPDG 61
A+LFA+ G T++TT NA + +KPI+ DL G I + FP ++GLP+G
Sbjct: 2 AKLFARRGAKSTLLTTPINAKILEKPIEAFKVQNPDLEIGIKI----LNFPCVELGLPEG 57
Query: 62 VENIKDATS--------------------REMLAHFIKKQKP------------------ 83
EN S ++ L FI+ KP
Sbjct: 58 CENRDFINSYQKSDSFDLFLKFLFSTKYMKQQLESFIETTKPSALVADMFFPWATESAEK 117
Query: 84 ---------------------------HENLVSDSQKFSIPGLPHNIEITSLQLQEYVRE 116
H+ + S S F IPGLP +I IT Q E
Sbjct: 118 IGVPRLVFHGTSSFALCCSYNMRIHKPHKKVASSSTPFVIPGLPGDIVITEDQ-ANVTNE 176
Query: 117 WSEFSDYFDAVYESEGKSYGTLCNSFHELEGDYENLYKSTMGIKAWSVGPVS-------- 168
+ F ++ V ESE S+G L NSF+ELE Y + Y+S + KAW +GP+S
Sbjct: 177 ETPFGKFWKEVRESETSSFGVLVNSFYELESSYADFYRSFVAKKAWHIGPLSLSNRGIAE 236
Query: 169 -AWLKKEQNEDVIVESELLNWLNSKPNDSVLYVSFGSLTRLSHSQIVEIAHGLENSGHNF 227
A K+ N D E E L WL+SK SV+Y+SFGS T L + Q++EIA GLE SG NF
Sbjct: 237 KAGRGKKANID---EQECLKWLDSKTPGSVVYLSFGSGTGLPNEQLLEIAFGLEGSGQNF 293
Query: 228 IWVVRKKD---GEGD-EDGFLDDFKQRMKENKKGYIIWNWAPQLLILGHPATAGVVTHCG 283
IWVV K + G G+ ED F++R K KG II WAPQ+LIL H A G VTHCG
Sbjct: 294 IWVVSKNENQVGTGENEDWLPKGFEERNK--GKGLIIRGWAPQVLILDHKAIGGFVTHCG 351
Query: 284 WNSILESLSVSLPIITWPMFAEQFYNEKLLVFVLKIVVSVGS 325
WNS LE ++ LP++TWPM AEQFYNEKLL VL+I V+VG+
Sbjct: 352 WNSTLEGIAAGLPMVTWPMGAEQFYNEKLLTKVLRIGVNVGA 393
>UniRef100_Q8H0F2 Anthocyanin 3'-glucosyltransferase [Gentiana triflora]
Length = 482
Score = 231 bits (589), Expect = 2e-59
Identities = 147/396 (37%), Positives = 207/396 (52%), Gaps = 77/396 (19%)
Query: 3 PMIDTARLFAKHGVDVTIITTQANALLFKKPIDNDLFSGYSI------------------ 44
P ID A+LF+ GV T+ITT N+ +F K I+ G+ I
Sbjct: 19 PTIDMAKLFSSRGVKATLITTHNNSAIFLKAINRSKILGFDISVLTIKFPSAEFGLPEGY 78
Query: 45 ----------------KACVI-QFP-------------------------AAQVGLPDGV 62
+AC++ Q P AA+ G+P +
Sbjct: 79 ETADQARSIDMMDEFFRACILLQEPLEELLKEHRPQALVADLFFYWANDAAAKFGIPRLL 138
Query: 63 ENIKDATSREMLA-HFIKKQKPHENLVSDSQKFSIPGLPHNIEITSLQL---QEYVREWS 118
++S M+A +++ KP++NL SDS F +P +P I +T Q+ E +
Sbjct: 139 --FHGSSSFAMIAAESVRRNKPYKNLSSDSDPFVVPDIPDKIILTKSQVPTPDETEENNT 196
Query: 119 EFSDYFDAVYESEGKSYGTLCNSFHELEGDYENLYKSTMGIKAWSVGPVSAWLKKEQNED 178
++ + + ESE YG + NSF+ELE DY + K+ +G +AW +GP+S L + ED
Sbjct: 197 HITEMWKNISESENDCYGVIVNSFYELEPDYVDYCKNVLGRRAWHIGPLS--LCNNEGED 254
Query: 179 V--------IVESELLNWLNSKPNDSVLYVSFGSLTRLSHSQIVEIAHGLENSGHNFIWV 230
V I E LNWL+SK DSV+YV FGS+ + +Q+ E+A GLE SG FIWV
Sbjct: 255 VAERGKKSDIDAHECLNWLDSKNPDSVVYVCFGSMANFNAAQLHELAMGLEESGQEFIWV 314
Query: 231 VRKKDGEGDEDG-FLDDFKQRMKENKKGYIIWNWAPQLLILGHPATAGVVTHCGWNSILE 289
VR E DE F D F++R++EN KG II WAPQ+LIL H A V+HCGWNS LE
Sbjct: 315 VRTCVDEEDESKWFPDGFEKRVQENNKGLIIKGWAPQVLILEHEAVGAFVSHCGWNSTLE 374
Query: 290 SLSVSLPIITWPMFAEQFYNEKLLVFVLKIVVSVGS 325
+ + ++TWP+FAEQFYNEKL+ +L+ VSVGS
Sbjct: 375 GICGGVAMVTWPLFAEQFYNEKLMTDILRTGVSVGS 410
>UniRef100_Q94C57 Putative glucosyltransferase [Arabidopsis thaliana]
Length = 483
Score = 230 bits (587), Expect = 4e-59
Identities = 152/398 (38%), Positives = 212/398 (53%), Gaps = 77/398 (19%)
Query: 1 MNPMIDTARLFAKHGVDVTIITTQANALLFKKPID--NDLFSGYSIKACVIQFPAAQVGL 58
M P +D A+LF+ G TI+TT N+ + +KPID +L G I + FP ++GL
Sbjct: 23 MIPTLDMAKLFSSRGAKSTILTTSLNSKILQKPIDTFKNLNPGLEIDIQIFNFPCVELGL 82
Query: 59 PDGVENIKDATS------REMLAHFIKKQKPHEN-------------LVSD--------- 90
P+G EN+ TS EM+ F + ++ L++D
Sbjct: 83 PEGCENVDFFTSNNNDDKNEMIVKFFFSTRFFKDQLEKLLGTTRPDCLIADMFFPWATEA 142
Query: 91 SQKFSIP-------------------------------------GLPHNIEITSLQLQEY 113
+ KF++P LP NI IT Q+ +
Sbjct: 143 AGKFNVPRLVFHGTGYFSLCAGYCIGVHKPQKRVASSSEPFVIPELPGNIVITEEQIIDG 202
Query: 114 VREWSEFSDYFDAVYESEGKSYGTLCNSFHELEGDYENLYKSTMGIKAWSVGPVSAW--- 170
E S+ + V ESE KS G + NSF+ELE DY + YKS + +AW +GP+S +
Sbjct: 203 DGE-SDMGKFMTEVRESEVKSSGVVLNSFYELEHDYADFYKSCVQKRAWHIGPLSVYNRG 261
Query: 171 --LKKEQNEDV-IVESELLNWLNSKPNDSVLYVSFGSLTRLSHSQIVEIAHGLENSGHNF 227
K E+ + I E+E L WL+SK +SV+YVSFGS+ + Q+ EIA GLE SG +F
Sbjct: 262 FEEKAERGKKANIDEAECLKWLDSKKPNSVIYVSFGSVAFFKNEQLFEIAAGLEASGTSF 321
Query: 228 IWVVRKKDGEGDEDGFLDDFKQRMKENKKGYIIWNWAPQLLILGHPATAGVVTHCGWNSI 287
IWVVRK + E+ + F++R+K KG II WAPQ+LIL H AT G VTHCGWNS+
Sbjct: 322 IWVVRKTK-DDREEWLPEGFEERVK--GKGMIIRGWAPQVLILDHQATGGFVTHCGWNSL 378
Query: 288 LESLSVSLPIITWPMFAEQFYNEKLLVFVLKIVVSVGS 325
LE ++ LP++TWP+ AEQFYNEKL+ VL+ VSVG+
Sbjct: 379 LEGVAAGLPMVTWPVGAEQFYNEKLVTQVLRTGVSVGA 416
>UniRef100_P93364 Immediate-early salicylate-induced glucosyltransferase [Nicotiana
tabacum]
Length = 476
Score = 226 bits (575), Expect = 9e-58
Identities = 143/392 (36%), Positives = 200/392 (50%), Gaps = 71/392 (18%)
Query: 1 MNPMIDTARLFAKHGVDVTIITTQANALLFKKPIDNDLFSGYSIKACVIQFPAAQVGLPD 60
M P +D A+L A GV TIITT N +F K I + G I+ +I+FPA + GLP+
Sbjct: 17 MIPTLDMAKLVASRGVKATIITTPLNESVFSKSIQRNKHLGIEIEIRLIKFPAVENGLPE 76
Query: 61 GVENIKDATSREMLAHFIK----KQKPHENLVSD------------------SQKFSI-- 96
E + S + L +F K Q+P E L+ + + KF++
Sbjct: 77 ECERLDLIPSDDKLPNFFKAVAMMQEPLEQLIEECRPNCLVSDMFLPWTTDTAAKFNMPR 136
Query: 97 -----------------------------------PGLPHNIEITSLQLQEYVREWSE-- 119
P LPH I++T QL + + E
Sbjct: 137 IVFHGTSFFALCVENSIRLNKPFKNVSSDSETFVVPNLPHEIKLTRTQLSPFEQSGEETT 196
Query: 120 FSDYFDAVYESEGKSYGTLCNSFHELEGDYENLYKSTMGIKAWSVGPVSAWLK------K 173
+ +V ES+ KSYG + NSF+ELE DY Y +G +AW++GP+S + +
Sbjct: 197 MTRMIKSVRESDSKSYGVIFNSFNELEHDYVEHYTKVLGRRAWAIGPLSMCNRDIEDKAE 256
Query: 174 EQNEDVIVESELLNWLNSKPNDSVLYVSFGSLTRLSHSQIVEIAHGLENSGHNFIWVVRK 233
+ I + E L WL+SK SV+YV FGS+ + SQ+ E+A G+E SG FIWVVR
Sbjct: 257 RGKQSSIDKHECLKWLDSKKPSSVVYVCFGSVANFTASQLHELAMGIEASGQEFIWVVRT 316
Query: 234 KDGEGDEDGFLDDFKQRMKENKKGYIIWNWAPQLLILGHPATAGVVTHCGWNSILESLSV 293
+ +ED + ++R KE KG II WAPQ+LIL H + VTHCGWNS LE +S
Sbjct: 317 E--LDNEDWLPEGLEERTKE--KGLIIRGWAPQVLILDHESVGAFVTHCGWNSTLEGVSG 372
Query: 294 SLPIITWPMFAEQFYNEKLLVFVLKIVVSVGS 325
+P++TWP+FAEQF+NEKL+ VLK VGS
Sbjct: 373 GVPMVTWPVFAEQFFNEKLVTEVLKTGAGVGS 404
>UniRef100_Q65YR6 UDP-glucose:anthocyanin 3'-O-glucosyltransferase [Gentiana scabra
var. buergeri]
Length = 482
Score = 223 bits (568), Expect = 6e-57
Identities = 144/396 (36%), Positives = 203/396 (50%), Gaps = 77/396 (19%)
Query: 3 PMIDTARLFAKHGVDVTIITTQANALLFKKPIDNDLFSGYSI------------------ 44
P ID A+LF+ GV T+ITT N+ +F K I G+ I
Sbjct: 19 PTIDMAKLFSSRGVKATLITTHNNSAIFLKAISRSKILGFDISVLTIKFPSAEFGLPEGY 78
Query: 45 ----------------KACVI-QFP-------------------------AAQVGLPDGV 62
+AC++ Q P AA+ G+P +
Sbjct: 79 ETADQARSIDMMDEFFRACILLQEPLEELLKEHRPQALVADLFFYWANDAAAKFGIPRLL 138
Query: 63 ENIKDATSREML-AHFIKKQKPHENLVSDSQKFSIPGLPHNIEITSLQL---QEYVREWS 118
++S M+ A +++ KP++NL SDS F +P +P I +T Q+ E +
Sbjct: 139 --FHGSSSFAMISAESVRRNKPYKNLSSDSDPFVVPDIPDKIILTKSQVPTPDETEENNT 196
Query: 119 EFSDYFDAVYESEGKSYGTLCNSFHELEGDYENLYKSTMGIKAWSVGPVSAWLKKEQNED 178
++ + + ESE YG + NSF+ELE DY + K+ +G +AW +GP+ L + ED
Sbjct: 197 HITEMWKNISESENDCYGVIVNSFYELEPDYVDYCKNVLGRRAWHIGPLL--LCNNEGED 254
Query: 179 V--------IVESELLNWLNSKPNDSVLYVSFGSLTRLSHSQIVEIAHGLENSGHNFIWV 230
V I E LNWL+SK SV+YV FGS+ + +Q+ E+A GLE SG FIWV
Sbjct: 255 VAQRGKKSDIDAHECLNWLDSKNPYSVVYVCFGSMANFNAAQLHELAMGLEESGQEFIWV 314
Query: 231 VRKKDGEGDEDG-FLDDFKQRMKENKKGYIIWNWAPQLLILGHPATAGVVTHCGWNSILE 289
VR E DE F D F++R++EN KG II WAPQ+LIL H A V+HCGWNS LE
Sbjct: 315 VRTCVDEKDESKWFPDGFEKRVQENNKGLIIKGWAPQVLILEHEAVGAFVSHCGWNSTLE 374
Query: 290 SLSVSLPIITWPMFAEQFYNEKLLVFVLKIVVSVGS 325
+ + ++TWP+FAEQFYNEKL+ +L+ V VGS
Sbjct: 375 GICGGVAMVTWPLFAEQFYNEKLMTDILRTGVPVGS 410
>UniRef100_Q9SXF2 UDP-glucose: flavonoid 7-O-glucosyltransferase [Scutellaria
baicalensis]
Length = 476
Score = 222 bits (566), Expect = 1e-56
Identities = 144/401 (35%), Positives = 208/401 (50%), Gaps = 73/401 (18%)
Query: 1 MNPMIDTARLFAKHGVDVTIITTQANALLFKKP--------------------------- 33
M PM+D A+LF+ GV TII T A A +K
Sbjct: 17 MIPMLDMAKLFSSRGVKTTIIATPAFAEPIRKARESGHDIGLTTTKFPPKGSSLPDNIRS 76
Query: 34 ---IDNDLFSGYSIKACVIQFP-------------------------AAQVGLPDGVENI 65
+ +DL + ++Q P AA+ G+P + +
Sbjct: 77 LDQVTDDLLPHFFRALELLQEPVEEIMEDLKPDCLVSDMFLPWTTDSAAKFGIPRLLFHG 136
Query: 66 KDATSREMLAHFIKKQKPHENLVSDSQKFSIPGLPHNIEITSLQLQEY-VREWSE--FSD 122
+R A + QKP++N+ SDS+ F + GLPH + Q+ +Y ++E + FS
Sbjct: 137 TSLFAR-CFAEQMSIQKPYKNVSSDSEPFVLRGLPHEVSFVRTQIPDYELQEGGDDAFSK 195
Query: 123 YFDAVYESEGKSYGTLCNSFHELEGDYENLYKSTMGIKAWSVGPVSAWLKKEQN------ 176
+ +++ KSYG + NSF ELE +Y + K+ G KAW +GP+ + + +
Sbjct: 196 MAKQMRDADKKSYGDVINSFEELESEYADYNKNVFGKKAWHIGPLKLFNNRAEQKSSQRG 255
Query: 177 -EDVIVESELLNWLNSKPNDSVLYVSFGSLTRLSHSQIVEIAHGLENSGHNFIWVVRKKD 235
E I + E L WLNSK +SV+Y+ FGS+ + +Q+ E A GLE+SG +FIWVVR +
Sbjct: 256 KESAIDDHECLAWLNSKKPNSVVYMCFGSMATFTPAQLHETAVGLESSGQDFIWVVR--N 313
Query: 236 GEGDEDGFLDDFKQRMKENKKGYIIWNWAPQLLILGHPATAGVVTHCGWNSILESLSVSL 295
G +ED F++R+K KG +I WAPQ++IL HP+T VTHCGWNS LE + L
Sbjct: 314 GGENEDWLPQGFEERIKG--KGLMIRGWAPQVMILDHPSTGAFVTHCGWNSTLEGICAGL 371
Query: 296 PIITWPMFAEQFYNEKLLVFVLKIVVSVGSKVNTFWSNEGE 336
P++TWP+FAEQFYNEKL+ VLK VSVG+K W GE
Sbjct: 372 PMVTWPVFAEQFYNEKLVTEVLKTGVSVGNKK---WQRVGE 409
>UniRef100_Q65YR5 UDP-glucose:anthocyanin 3'-O-glucosyltransferase [Gentiana scabra
var. buergeri]
Length = 482
Score = 222 bits (565), Expect = 1e-56
Identities = 143/396 (36%), Positives = 203/396 (51%), Gaps = 77/396 (19%)
Query: 3 PMIDTARLFAKHGVDVTIITTQANALLFKKPIDNDLFSGYSI------------------ 44
P ID A+LF+ GV T+ITT N+ +F K I G+ I
Sbjct: 19 PTIDMAKLFSSRGVKATLITTHNNSAIFLKAISRSKILGFDISVLTIKFPSAEFGLPEGY 78
Query: 45 ----------------KACVI-QFP-------------------------AAQVGLPDGV 62
+AC++ Q P AA+ G+P +
Sbjct: 79 ETADQARSIDLMDEFFRACILLQEPLEELLKEHRPQALVADLFFYWANDAAAKFGIPRLL 138
Query: 63 ENIKDATSREML-AHFIKKQKPHENLVSDSQKFSIPGLPHNIEITSLQL---QEYVREWS 118
++S M+ A +++ KP++NL SDS F +P +P I +T Q+ + +
Sbjct: 139 --FHGSSSFAMISAESVRRNKPYKNLSSDSDPFVVPDIPDKIILTKSQVPTPDDTEENNT 196
Query: 119 EFSDYFDAVYESEGKSYGTLCNSFHELEGDYENLYKSTMGIKAWSVGPVSAWLKKEQNED 178
++ + + ESE YG + NSF+ELE DY + K+ +G +AW +GP+ L + ED
Sbjct: 197 HITEMWKNISESENDCYGVIVNSFYELEPDYVDYCKNVLGRRAWHIGPLL--LCNNEGED 254
Query: 179 V--------IVESELLNWLNSKPNDSVLYVSFGSLTRLSHSQIVEIAHGLENSGHNFIWV 230
V I E LNWL+SK SV+YV FGS+ + +Q+ E+A GLE SG FIWV
Sbjct: 255 VAQRGEKSDIDAHEYLNWLDSKNPYSVVYVCFGSMANFNAAQLHELAMGLEESGQEFIWV 314
Query: 231 VRKKDGEGDEDG-FLDDFKQRMKENKKGYIIWNWAPQLLILGHPATAGVVTHCGWNSILE 289
VR E DE F D F++R++EN KG II WAPQ+LIL H A V+HCGWNS LE
Sbjct: 315 VRTCVDEKDESKWFPDGFEKRVQENNKGLIIKGWAPQVLILEHEAVGAFVSHCGWNSTLE 374
Query: 290 SLSVSLPIITWPMFAEQFYNEKLLVFVLKIVVSVGS 325
+ + ++TWP+FAEQFYNEKL+ +L+ V VGS
Sbjct: 375 GICGGVAMVTWPLFAEQFYNEKLMTDILRTGVPVGS 410
>UniRef100_Q8L7Q5 Putative glucosyltransferase [Arabidopsis thaliana]
Length = 372
Score = 221 bits (564), Expect = 2e-56
Identities = 121/252 (48%), Positives = 160/252 (63%), Gaps = 12/252 (4%)
Query: 82 KPHENLVSDSQKFSIPGLPHNIEITSLQLQEYVREWSEFSDYFDAVYESEGKSYGTLCNS 141
KPH+ + + S F IPGLP +I IT Q +E + + V ESE S+G L NS
Sbjct: 58 KPHKKVATSSTPFVIPGLPGDIVITEDQAN-VAKEETPMGKFMKEVRESETNSFGVLVNS 116
Query: 142 FHELEGDYENLYKSTMGIKAWSVGPVSAWLKKEQNEDV-------IVESELLNWLNSKPN 194
F+ELE Y + Y+S + +AW +GP+S +E E I E E L WL+SK
Sbjct: 117 FYELESAYADFYRSFVAKRAWHIGPLSL-SNRELGEKARRGKKANIDEQECLKWLDSKTP 175
Query: 195 DSVLYVSFGSLTRLSHSQIVEIAHGLENSGHNFIWVVRKKDGEGDEDGFLDD-FKQRMKE 253
SV+Y+SFGS T ++ Q++EIA GLE SG +FIWVVRK + +GD + +L + FK+R
Sbjct: 176 GSVVYLSFGSGTNFTNDQLLEIAFGLEGSGQSFIWVVRKNENQGDNEEWLPEGFKERT-- 233
Query: 254 NKKGYIIWNWAPQLLILGHPATAGVVTHCGWNSILESLSVSLPIITWPMFAEQFYNEKLL 313
KG II WAPQ+LIL H A G VTHCGWNS +E ++ LP++TWPM AEQFYNEKLL
Sbjct: 234 TGKGLIIPGWAPQVLILDHKAIGGFVTHCGWNSAIEGIAAGLPMVTWPMGAEQFYNEKLL 293
Query: 314 VFVLKIVVSVGS 325
VL+I V+VG+
Sbjct: 294 TKVLRIGVNVGA 305
>UniRef100_Q43526 Twi1 protein [Lycopersicon esculentum]
Length = 466
Score = 221 bits (563), Expect = 2e-56
Identities = 144/402 (35%), Positives = 202/402 (49%), Gaps = 73/402 (18%)
Query: 1 MNPMIDTARLFAKHGVDVTIITTQANALLFKKPIDNDLFSGYSIKACVIQFPAAQVGLPD 60
M P +D A + A GV TIITT N +F K I+ + G I +++FPA + LP+
Sbjct: 13 MIPTLDMANVVACRGVKATIITTPLNESVFSKAIERNKHLGIEIDIRLLKFPAKENDLPE 72
Query: 61 GVENIKDATSREMLAHFIKKQKPHEN-------------LVSD---------SQKFSI-- 96
E + S + L +F+K ++ LVSD + KFSI
Sbjct: 73 DCERLDLVPSDDKLPNFLKAAAMMKDEFEELIGECRPDCLVSDMFLPWTTDSAAKFSIPR 132
Query: 97 -----------------------------------PGLPHNIEITSLQLQEYVR--EWSE 119
P LPH I +T QL + + E +
Sbjct: 133 IVFHGTSYFALCVGDTIRRNKPFKNVSSDTETFVVPDLPHEIRLTRTQLSPFEQSDEETG 192
Query: 120 FSDYFDAVYESEGKSYGTLCNSFHELEGDYENLYKSTMGIKAWSVGPVSAWLK------K 173
+ AV ES+ KSYG + NSF+ELE DY Y +G K W++GP+S + +
Sbjct: 193 MAPMIKAVRESDAKSYGVIFNSFYELESDYVEHYTKVVGRKNWAIGPLSLCNRDIEDKAE 252
Query: 174 EQNEDVIVESELLNWLNSKPNDSVLYVSFGSLTRLSHSQIVEIAHGLENSGHNFIWVVRK 233
+ I E L WL+SK + S++YV FGS + +Q+ E+A GLE SG +FIWV+R
Sbjct: 253 RGRKSSIDEHACLKWLDSKKSSSIVYVCFGSTADFTTAQMQELAMGLEASGQDFIWVIR- 311
Query: 234 KDGEGDEDGFLDDFKQRMKENKKGYIIWNWAPQLLILGHPATAGVVTHCGWNSILESLSV 293
G+ED + F++R KE KG II WAPQ +IL H A VTHCGWNS LE +S
Sbjct: 312 ---TGNEDWLPEGFEERTKE--KGLIIRGWAPQSVILDHEAIGAFVTHCGWNSTLEGISA 366
Query: 294 SLPIITWPMFAEQFYNEKLLVFVLKIVVSVGSKVNTFWSNEG 335
+P++TWP+FAEQF+NEKL+ V++ VGSK ++EG
Sbjct: 367 GVPMVTWPVFAEQFFNEKLVTEVMRSGAGVGSKQWKRTASEG 408
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.317 0.135 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 606,805,749
Number of Sequences: 2790947
Number of extensions: 26199447
Number of successful extensions: 70895
Number of sequences better than 10.0: 1068
Number of HSP's better than 10.0 without gapping: 983
Number of HSP's successfully gapped in prelim test: 86
Number of HSP's that attempted gapping in prelim test: 68954
Number of HSP's gapped (non-prelim): 1288
length of query: 339
length of database: 848,049,833
effective HSP length: 128
effective length of query: 211
effective length of database: 490,808,617
effective search space: 103560618187
effective search space used: 103560618187
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 75 (33.5 bits)
Medicago: description of AC148481.8


