

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148404.8 - phase: 0
(416 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
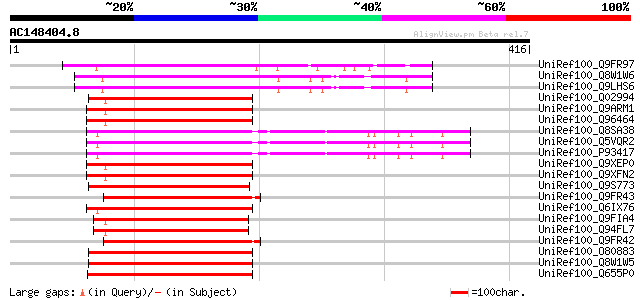
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9FR97 MYB65 [Arabidopsis thaliana] 204 4e-51
UniRef100_Q8W1W6 Putative transcription factor MYB33 [Arabidopsi... 203 8e-51
UniRef100_Q9LHS6 MYB family transcription factor-like [Arabidops... 203 8e-51
UniRef100_Q02994 Protein 3 [Petunia hybrida] 201 4e-50
UniRef100_Q9ARM1 Transcription factor GAMyb [Hordeum vulgare] 199 9e-50
UniRef100_Q96464 GAMyb protein [Hordeum vulgare] 199 9e-50
UniRef100_Q8SA38 Transcription factor GAMyb [Oryza sativa] 199 9e-50
UniRef100_Q5VQR2 Putative transcription factor GAMyb [Oryza sativa] 199 9e-50
UniRef100_P93417 Transcription factor GAMyb [Oryza sativa] 199 9e-50
UniRef100_Q9XEP0 Gibberellin MYB transcription factor [Lolium te... 198 3e-49
UniRef100_Q9XFN2 Myb protein [Avena sativa] 197 5e-49
UniRef100_Q9S773 Putative myb-related protein [Arabidopsis thali... 196 8e-49
UniRef100_Q9FR43 Anther-specific myb-related protein 2 [Nicotian... 196 1e-48
UniRef100_Q6IX76 Transcription factor Myb3 [Triticum aestivum] 195 2e-48
UniRef100_Q9FIA4 Similarity to MYB-related transcription factor ... 192 1e-47
UniRef100_Q94FL7 Putative transcription factor MYB120 [Arabidops... 192 1e-47
UniRef100_Q9FR42 Anther-specific myb-related protein 1 [Nicotian... 192 1e-47
UniRef100_O80883 Putative MYB family transcription factor [Arabi... 191 3e-47
UniRef100_Q8W1W5 Putative transcription factor MYB101 [Arabidops... 191 3e-47
UniRef100_Q655P0 Putative transcription factor GAMyb [Oryza sativa] 189 2e-46
>UniRef100_Q9FR97 MYB65 [Arabidopsis thaliana]
Length = 553
Score = 204 bits (519), Expect = 4e-51
Identities = 130/339 (38%), Positives = 178/339 (52%), Gaps = 50/339 (14%)
Query: 43 SSGDDDRKRQSILSADQETDEGDTTG--GEGGEGGLKKGPWTAAEDEILVAHVQKYGEGN 100
++ D D S + + + + G G LKKGPWT+ ED IL+ +V+K+GEGN
Sbjct: 6 ATADSDDGMHSSIHNESPAPDSISNGCRSRGKRSVLKKGPWTSTEDGILIDYVKKHGEGN 65
Query: 101 WNSVRKCTGLARCGKSCRLRWANHLRPDLRKGAITAEEERRIIELHHKMGNKWAQMAALL 160
WN+V+K T LARCGKSCRLRWANHLRP+L+KGA + EEE+ I+E+H KMGNKWAQMA L
Sbjct: 66 WNAVQKHTSLARCGKSCRLRWANHLRPNLKKGAFSQEEEQLIVEMHAKMGNKWAQMAEHL 125
Query: 161 PGRTDNEIKNFWNTRCKKRGRANLPIYPEDLSSECL-----LNQENADMLTNEASQHD-- 213
PGRTDNEIKN+WNTR K+R RA LP+YP ++ + L + N + D
Sbjct: 126 PGRTDNEIKNYWNTRIKRRQRAGLPLYPPEIYVDDLHWSEEYTKSNIIRVDRRRRHQDFL 185
Query: 214 EAENKVPEVIFKDYKLRPDILPPCFDKLLAGL---------------------LLRPSKR 252
+ N V+F D +LP D L+ L +L K+
Sbjct: 186 QLGNSKDNVLFDDLNFAASLLPAASD--LSDLVACNMLGTGASSSRYESYMPPILPSPKQ 243
Query: 253 PWKSDMNLYSYSNNA----AAPAAFDQ--LRKYPMPSPSSP------PWEDINEPHSYNQ 300
W+S S+N +P F ++K P SP P+E+ HS +
Sbjct: 244 IWESGSRFPMCSSNIKHEFQSPEHFQNTAVQKNPRSCSISPCDVDHHPYEN---QHSSHM 300
Query: 301 FNEYDNPMVGIHMAPKTSSFEPIYGFMKTEPPSLQYSQT 339
D+ V M P + +P++G +K E PS QYS+T
Sbjct: 301 MMVPDSHTVTYGMHPTS---KPLFGAVKLELPSFQYSET 336
>UniRef100_Q8W1W6 Putative transcription factor MYB33 [Arabidopsis thaliana]
Length = 520
Score = 203 bits (516), Expect = 8e-51
Identities = 128/329 (38%), Positives = 180/329 (53%), Gaps = 49/329 (14%)
Query: 53 SILSADQETDEGDTTGGEGGE-------GGLKKGPWTAAEDEILVAHVQKYGEGNWNSVR 105
S S D + +E G + LKKGPW++AED+IL+ +V K+GEGNWN+V+
Sbjct: 2 SYTSTDSDHNESPAADDNGSDCRSRWDGHALKKGPWSSAEDDILIDYVNKHGEGNWNAVQ 61
Query: 106 KCTGLARCGKSCRLRWANHLRPDLRKGAITAEEERRIIELHHKMGNKWAQMAALLPGRTD 165
K T L RCGKSCRLRWANHLRP+L+KGA + EEE+ I+ELH KMGN+WA+MAA LPGRTD
Sbjct: 62 KHTSLFRCGKSCRLRWANHLRPNLKKGAFSQEEEQLIVELHAKMGNRWARMAAHLPGRTD 121
Query: 166 NEIKNFWNTRCKKRGRANLPIYPEDLSSECL-LNQENA-DMLTNEASQHDE--AENKVPE 221
NEIKN+WNTR K+R RA LP+YP ++ E L +QE A + E +H +
Sbjct: 122 NEIKNYWNTRIKRRQRAGLPLYPPEMHVEALEWSQEYAKSRVMGEDRRHQDFLQLGSCES 181
Query: 222 VIFKDYKLRPDILPPCFD-------KLLAGLLLRP------------SKRPWKSDMNLYS 262
+F D D++P FD K + P SKR W+S++ LY
Sbjct: 182 NVFFDTLNFTDMVPGTFDLADMTAYKNMGNCASSPRYENFMTPTIPSSKRLWESEL-LYP 240
Query: 263 YSNNAAAPAAFDQLRKYPMPSPSSPPWEDINEPHSYNQFNEYDNPMVGIHMAPK------ 316
++ F ++ SP + I++ S++ + ++P+ G +P
Sbjct: 241 -GCSSTIKQEFSSPEQFRNTSP-----QTISKTCSFSVPCDVEHPLYGNRHSPVMIPDSH 294
Query: 317 ------TSSFEPIYGFMKTEPPSLQYSQT 339
+P+YG +K E PS QYS+T
Sbjct: 295 TPTDGIVPYSKPLYGAVKLELPSFQYSET 323
>UniRef100_Q9LHS6 MYB family transcription factor-like [Arabidopsis thaliana]
Length = 451
Score = 203 bits (516), Expect = 8e-51
Identities = 128/329 (38%), Positives = 180/329 (53%), Gaps = 49/329 (14%)
Query: 53 SILSADQETDEGDTTGGEGGE-------GGLKKGPWTAAEDEILVAHVQKYGEGNWNSVR 105
S S D + +E G + LKKGPW++AED+IL+ +V K+GEGNWN+V+
Sbjct: 2 SYTSTDSDHNESPAADDNGSDCRSRWDGHALKKGPWSSAEDDILIDYVNKHGEGNWNAVQ 61
Query: 106 KCTGLARCGKSCRLRWANHLRPDLRKGAITAEEERRIIELHHKMGNKWAQMAALLPGRTD 165
K T L RCGKSCRLRWANHLRP+L+KGA + EEE+ I+ELH KMGN+WA+MAA LPGRTD
Sbjct: 62 KHTSLFRCGKSCRLRWANHLRPNLKKGAFSQEEEQLIVELHAKMGNRWARMAAHLPGRTD 121
Query: 166 NEIKNFWNTRCKKRGRANLPIYPEDLSSECL-LNQENA-DMLTNEASQHDE--AENKVPE 221
NEIKN+WNTR K+R RA LP+YP ++ E L +QE A + E +H +
Sbjct: 122 NEIKNYWNTRIKRRQRAGLPLYPPEMHVEALEWSQEYAKSRVMGEDRRHQDFLQLGSCES 181
Query: 222 VIFKDYKLRPDILPPCFD-------KLLAGLLLRP------------SKRPWKSDMNLYS 262
+F D D++P FD K + P SKR W+S++ LY
Sbjct: 182 NVFFDTLNFTDMVPGTFDLADMTAYKNMGNCASSPRYENFMTPTIPSSKRLWESEL-LYP 240
Query: 263 YSNNAAAPAAFDQLRKYPMPSPSSPPWEDINEPHSYNQFNEYDNPMVGIHMAPK------ 316
++ F ++ SP + I++ S++ + ++P+ G +P
Sbjct: 241 -GCSSTIKQEFSSPEQFRNTSP-----QTISKTCSFSVPCDVEHPLYGNRHSPVMIPDSH 294
Query: 317 ------TSSFEPIYGFMKTEPPSLQYSQT 339
+P+YG +K E PS QYS+T
Sbjct: 295 TPTDGIVPYSKPLYGAVKLELPSFQYSET 323
>UniRef100_Q02994 Protein 3 [Petunia hybrida]
Length = 517
Score = 201 bits (510), Expect = 4e-50
Identities = 93/135 (68%), Positives = 111/135 (81%), Gaps = 4/135 (2%)
Query: 64 GDTTGGEGGEGG----LKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRL 119
G +TG G G LKKGPWTAAED IL+ +V+K+GEGNWN+V++ +GL RCGKSCRL
Sbjct: 12 GASTGRSNGAGSSRQVLKKGPWTAAEDSILMEYVKKHGEGNWNAVQRNSGLMRCGKSCRL 71
Query: 120 RWANHLRPDLRKGAITAEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKR 179
RWANHLRP+L+KGA T EEER IIELH K+GNKWA+MAA LPGRTDNEIKN+WNTR K+R
Sbjct: 72 RWANHLRPNLKKGAFTVEEERIIIELHAKLGNKWARMAAQLPGRTDNEIKNYWNTRLKRR 131
Query: 180 GRANLPIYPEDLSSE 194
RA LPIYP++L +
Sbjct: 132 QRAGLPIYPQELQQQ 146
>UniRef100_Q9ARM1 Transcription factor GAMyb [Hordeum vulgare]
Length = 553
Score = 199 bits (507), Expect = 9e-50
Identities = 94/136 (69%), Positives = 111/136 (81%), Gaps = 3/136 (2%)
Query: 62 DEGDTTGGEGGEGG---LKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCR 118
D+G + GG GG LKKGPWT+AED ILV +V+K+GEGNWN+V+K TGL RCGKSCR
Sbjct: 25 DDGSSGGGSPHRGGGPPLKKGPWTSAEDAILVDYVKKHGEGNWNAVQKNTGLFRCGKSCR 84
Query: 119 LRWANHLRPDLRKGAITAEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKK 178
LRWANHLRP+L+KGA T EEER II+LH KMGNKWA+MAA LPGRTDNEIKN+WNTR K+
Sbjct: 85 LRWANHLRPNLKKGAFTPEEERLIIQLHSKMGNKWARMAAHLPGRTDNEIKNYWNTRIKR 144
Query: 179 RGRANLPIYPEDLSSE 194
RA LPIYP + ++
Sbjct: 145 CQRAGLPIYPASVCNQ 160
>UniRef100_Q96464 GAMyb protein [Hordeum vulgare]
Length = 553
Score = 199 bits (507), Expect = 9e-50
Identities = 94/136 (69%), Positives = 111/136 (81%), Gaps = 3/136 (2%)
Query: 62 DEGDTTGGEGGEGG---LKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCR 118
D+G + GG GG LKKGPWT+AED ILV +V+K+GEGNWN+V+K TGL RCGKSCR
Sbjct: 25 DDGSSGGGSPHRGGGPPLKKGPWTSAEDAILVDYVKKHGEGNWNAVQKNTGLFRCGKSCR 84
Query: 119 LRWANHLRPDLRKGAITAEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKK 178
LRWANHLRP+L+KGA T EEER II+LH KMGNKWA+MAA LPGRTDNEIKN+WNTR K+
Sbjct: 85 LRWANHLRPNLKKGAFTPEEERLIIQLHSKMGNKWARMAAHLPGRTDNEIKNYWNTRIKR 144
Query: 179 RGRANLPIYPEDLSSE 194
RA LPIYP + ++
Sbjct: 145 CQRAGLPIYPASVCNQ 160
>UniRef100_Q8SA38 Transcription factor GAMyb [Oryza sativa]
Length = 553
Score = 199 bits (507), Expect = 9e-50
Identities = 138/345 (40%), Positives = 180/345 (52%), Gaps = 44/345 (12%)
Query: 62 DEGDTTGG--EGGEGGLKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRL 119
D+G + G GG LKKGPWT+AED ILV +V+K+GEGNWN+V+K TGL RCGKSCRL
Sbjct: 24 DDGSSGGSPHRGGGPPLKKGPWTSAEDAILVDYVKKHGEGNWNAVQKNTGLFRCGKSCRL 83
Query: 120 RWANHLRPDLRKGAITAEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKR 179
RWANHLRP+L+KGA TAEEER II+LH KMGNKWA+MAA LPGRTDNEIKN+WNTR K+
Sbjct: 84 RWANHLRPNLKKGAFTAEEERLIIQLHSKMGNKWARMAAHLPGRTDNEIKNYWNTRIKRC 143
Query: 180 GRANLPIYPEDLSSECLLNQENADMLTNEASQHDEAENKVPEVIFKDYKLRPDILPPCFD 239
RA LPIYP + ++ N D +S D EN +++ + PD F
Sbjct: 144 QRAGLPIYPTSVCNQ----SSNEDQQC--SSDFDCGENLSNDLLNANGLYLPDFTCDNFI 197
Query: 240 KLLAGLLLRPSKRPWKSDMNLYSYSNNAAAPAAFDQLRKYPMPSPSS---PPWED----- 291
L P S NL S + + + DQ+ + M S P D
Sbjct: 198 ANSEALPYAPHLSA-VSISNLLGQSFASKSCSFMDQVNQTGMLKQSDGVLPGLSDTINGV 256
Query: 292 INEPHSYNQFNEYDNPMVG---IHMAPKTSSF----------------------EPIYGF 326
I+ ++ +E VG +H A TS P G
Sbjct: 257 ISSVDQFSNDSEKLKQAVGFDYLHEANSTSKIIAPFGGALNGSHAFLNGNFSASRPTSGP 316
Query: 327 MKTEPPSLQYSQTQHRSYM--IPYPQVEPVQSVPTSLERSHFLPS 369
+K E PSLQ +++ S++ P ++P + V L+ PS
Sbjct: 317 LKMELPSLQDTESDPNSWLKYTVAPALQPTELVDPYLQSPAATPS 361
>UniRef100_Q5VQR2 Putative transcription factor GAMyb [Oryza sativa]
Length = 474
Score = 199 bits (507), Expect = 9e-50
Identities = 138/345 (40%), Positives = 180/345 (52%), Gaps = 44/345 (12%)
Query: 62 DEGDTTGG--EGGEGGLKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRL 119
D+G + G GG LKKGPWT+AED ILV +V+K+GEGNWN+V+K TGL RCGKSCRL
Sbjct: 24 DDGSSGGSPHRGGGPPLKKGPWTSAEDAILVDYVKKHGEGNWNAVQKNTGLFRCGKSCRL 83
Query: 120 RWANHLRPDLRKGAITAEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKR 179
RWANHLRP+L+KGA TAEEER II+LH KMGNKWA+MAA LPGRTDNEIKN+WNTR K+
Sbjct: 84 RWANHLRPNLKKGAFTAEEERLIIQLHSKMGNKWARMAAHLPGRTDNEIKNYWNTRIKRC 143
Query: 180 GRANLPIYPEDLSSECLLNQENADMLTNEASQHDEAENKVPEVIFKDYKLRPDILPPCFD 239
RA LPIYP + ++ N D +S D EN +++ + PD F
Sbjct: 144 QRAGLPIYPTSVCNQ----SSNEDQQC--SSDFDCGENLSNDLLNANGLYLPDFTCDNFI 197
Query: 240 KLLAGLLLRPSKRPWKSDMNLYSYSNNAAAPAAFDQLRKYPMPSPSS---PPWED----- 291
L P S NL S + + + DQ+ + M S P D
Sbjct: 198 ANSEALPYAPHLSA-VSISNLLGQSFASKSCSFMDQVNQTGMLKQSDGVLPGLSDTINGV 256
Query: 292 INEPHSYNQFNEYDNPMVG---IHMAPKTSSF----------------------EPIYGF 326
I+ ++ +E VG +H A TS P G
Sbjct: 257 ISSVDQFSNDSEKLKQAVGFDYLHEANSTSKIIAPFGGALNGSHAFLNGNFSASRPTSGP 316
Query: 327 MKTEPPSLQYSQTQHRSYM--IPYPQVEPVQSVPTSLERSHFLPS 369
+K E PSLQ +++ S++ P ++P + V L+ PS
Sbjct: 317 LKMELPSLQDTESDPNSWLKYTVAPALQPTELVDPYLQSPAATPS 361
>UniRef100_P93417 Transcription factor GAMyb [Oryza sativa]
Length = 553
Score = 199 bits (507), Expect = 9e-50
Identities = 138/345 (40%), Positives = 180/345 (52%), Gaps = 44/345 (12%)
Query: 62 DEGDTTGG--EGGEGGLKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRL 119
D+G + G GG LKKGPWT+AED ILV +V+K+GEGNWN+V+K TGL RCGKSCRL
Sbjct: 24 DDGSSGGSPHRGGGPPLKKGPWTSAEDAILVDYVKKHGEGNWNAVQKNTGLFRCGKSCRL 83
Query: 120 RWANHLRPDLRKGAITAEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKR 179
RWANHLRP+L+KGA TAEEER II+LH KMGNKWA+MAA LPGRTDNEIKN+WNTR K+
Sbjct: 84 RWANHLRPNLKKGAFTAEEERLIIQLHSKMGNKWARMAAHLPGRTDNEIKNYWNTRIKRC 143
Query: 180 GRANLPIYPEDLSSECLLNQENADMLTNEASQHDEAENKVPEVIFKDYKLRPDILPPCFD 239
RA LPIYP + ++ N D +S D EN +++ + PD F
Sbjct: 144 QRAGLPIYPTSVCNQ----SSNEDQQC--SSDFDCGENLSNDLLNANGLYLPDFTCDNFI 197
Query: 240 KLLAGLLLRPSKRPWKSDMNLYSYSNNAAAPAAFDQLRKYPMPSPSS---PPWED----- 291
L P S NL S + + + DQ+ + M S P D
Sbjct: 198 ANSEALPYAPHLSA-VSISNLLGQSFASKSCSFMDQVNQTGMLKQSDGVLPGLSDTINGV 256
Query: 292 INEPHSYNQFNEYDNPMVG---IHMAPKTSSF----------------------EPIYGF 326
I+ ++ +E VG +H A TS P G
Sbjct: 257 ISSVDQFSNDSEKLKQAVGFDYLHEANSTSKIIAPFGGALNGSHAFLNGNFSASRPTSGP 316
Query: 327 MKTEPPSLQYSQTQHRSYM--IPYPQVEPVQSVPTSLERSHFLPS 369
+K E PSLQ +++ S++ P ++P + V L+ PS
Sbjct: 317 LKMELPSLQDTESDPNSWLKYTVAPALQPTELVDPYLQSPAATPS 361
>UniRef100_Q9XEP0 Gibberellin MYB transcription factor [Lolium temulentum]
Length = 548
Score = 198 bits (503), Expect = 3e-49
Identities = 93/136 (68%), Positives = 111/136 (81%), Gaps = 3/136 (2%)
Query: 62 DEGDTTGGEGGEGG---LKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCR 118
D G ++GG GG LKKGPWT+AED ILV +V+K+GEGNWN+V+K TGL RCGKSCR
Sbjct: 25 DGGGSSGGSPHRGGGPPLKKGPWTSAEDAILVDYVKKHGEGNWNAVQKNTGLFRCGKSCR 84
Query: 119 LRWANHLRPDLRKGAITAEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKK 178
LRWANHLRP+L+KGA T EEER II+LH KMGNKWA+MAA LPGRTDNEIKN+WNTR K+
Sbjct: 85 LRWANHLRPNLKKGAFTPEEERLIIQLHSKMGNKWARMAAHLPGRTDNEIKNYWNTRIKR 144
Query: 179 RGRANLPIYPEDLSSE 194
RA LP+YP + ++
Sbjct: 145 CQRAGLPVYPASVINQ 160
>UniRef100_Q9XFN2 Myb protein [Avena sativa]
Length = 546
Score = 197 bits (501), Expect = 5e-49
Identities = 93/136 (68%), Positives = 111/136 (81%), Gaps = 3/136 (2%)
Query: 62 DEGDTTGGEGGEGG---LKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCR 118
D+G ++GG GG LKKGPWT+AED ILV +V+K+GEGNWN+V+K TGL RCGKSCR
Sbjct: 24 DDGGSSGGSPHRGGGPPLKKGPWTSAEDAILVDYVKKHGEGNWNAVQKNTGLFRCGKSCR 83
Query: 119 LRWANHLRPDLRKGAITAEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKK 178
LRWANHLRP+L+KGA T EEER II+LH KMGNKWA+MAA LPGRTDNEIKN+WNTR K+
Sbjct: 84 LRWANHLRPNLKKGAFTPEEERLIIQLHSKMGNKWARMAANLPGRTDNEIKNYWNTRIKR 143
Query: 179 RGRANLPIYPEDLSSE 194
RA L IYP + ++
Sbjct: 144 CQRAGLTIYPASIINQ 159
>UniRef100_Q9S773 Putative myb-related protein [Arabidopsis thaliana]
Length = 389
Score = 196 bits (499), Expect = 8e-49
Identities = 91/129 (70%), Positives = 106/129 (81%)
Query: 64 GDTTGGEGGEGGLKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWAN 123
G + GEGG LKKGPWT AEDE L A+V++YGEGNWNSV+K T LARCGKSCRLRWAN
Sbjct: 7 GASEDGEGGGVVLKKGPWTVAEDETLAAYVREYGEGNWNSVQKKTWLARCGKSCRLRWAN 66
Query: 124 HLRPDLRKGAITAEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKRGRAN 183
HLRP+LRKG+ T EEER II+LH ++GNKWA+MAA LPGRTDNEIKN+WNTR K+ R
Sbjct: 67 HLRPNLRKGSFTPEEERLIIQLHSQLGNKWARMAAQLPGRTDNEIKNYWNTRLKRFQRQG 126
Query: 184 LPIYPEDLS 192
LP+YP + S
Sbjct: 127 LPLYPPEYS 135
>UniRef100_Q9FR43 Anther-specific myb-related protein 2 [Nicotiana tabacum]
Length = 474
Score = 196 bits (498), Expect = 1e-48
Identities = 89/126 (70%), Positives = 109/126 (85%), Gaps = 2/126 (1%)
Query: 76 LKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKGAIT 135
LKKGPWTAAED IL+ +V+K GEGNWN+V++ +GL RCGKSCRLRWANHLRP+L+KGA +
Sbjct: 20 LKKGPWTAAEDAILMEYVKKNGEGNWNAVQRNSGLMRCGKSCRLRWANHLRPNLKKGAFS 79
Query: 136 AEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKRGRANLPIYPEDLSSEC 195
EEER I+ELH K+GNKWA+MAA +PGRTDNEIKN+WNTR K+R RA LPIYP+D+ +
Sbjct: 80 LEEERFIVELHAKLGNKWARMAAQMPGRTDNEIKNYWNTRLKRRQRAGLPIYPQDIQPQ- 138
Query: 196 LLNQEN 201
LNQ+N
Sbjct: 139 -LNQQN 143
>UniRef100_Q6IX76 Transcription factor Myb3 [Triticum aestivum]
Length = 503
Score = 195 bits (495), Expect = 2e-48
Identities = 92/135 (68%), Positives = 110/135 (81%), Gaps = 2/135 (1%)
Query: 62 DEGDTTGG--EGGEGGLKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRL 119
D+G + G GG LKKGPWT+AED ILV +V+K+GEGNWN+V+K TGL RCGKSCRL
Sbjct: 25 DDGSSGGSPHRGGGPPLKKGPWTSAEDAILVDYVKKHGEGNWNAVQKNTGLFRCGKSCRL 84
Query: 120 RWANHLRPDLRKGAITAEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKR 179
RWANHLRP+L+KGA T EEER II+LH KMGNKWA+MAA LPGRTD+EIKN+WNTR K+
Sbjct: 85 RWANHLRPNLKKGAFTPEEERLIIQLHSKMGNKWARMAAHLPGRTDDEIKNYWNTRIKRC 144
Query: 180 GRANLPIYPEDLSSE 194
RA LPIYP + ++
Sbjct: 145 QRAGLPIYPASVCNQ 159
>UniRef100_Q9FIA4 Similarity to MYB-related transcription factor [Arabidopsis
thaliana]
Length = 581
Score = 192 bits (489), Expect = 1e-47
Identities = 90/135 (66%), Positives = 108/135 (79%), Gaps = 11/135 (8%)
Query: 68 GGEGGEGG-----------LKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKS 116
GG G +GG LKKGPWTAAEDEIL A+V++ GEGNWN+V+K TGLARCGKS
Sbjct: 7 GGAGKDGGSTNHLSDGGVILKKGPWTAAEDEILAAYVRENGEGNWNAVQKNTGLARCGKS 66
Query: 117 CRLRWANHLRPDLRKGAITAEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRC 176
CRLRWANHLRP+L+KG+ T +EER II+LH ++GNKWA+MAA LPGRTDNEIKN+WNTR
Sbjct: 67 CRLRWANHLRPNLKKGSFTGDEERLIIQLHAQLGNKWARMAAQLPGRTDNEIKNYWNTRL 126
Query: 177 KKRGRANLPIYPEDL 191
K+ R LP+YP D+
Sbjct: 127 KRLLRQGLPLYPPDI 141
>UniRef100_Q94FL7 Putative transcription factor MYB120 [Arabidopsis thaliana]
Length = 523
Score = 192 bits (489), Expect = 1e-47
Identities = 90/135 (66%), Positives = 108/135 (79%), Gaps = 11/135 (8%)
Query: 68 GGEGGEGG-----------LKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKS 116
GG G +GG LKKGPWTAAEDEIL A+V++ GEGNWN+V+K TGLARCGKS
Sbjct: 7 GGAGKDGGSTNHLSDGGVILKKGPWTAAEDEILAAYVRENGEGNWNAVQKNTGLARCGKS 66
Query: 117 CRLRWANHLRPDLRKGAITAEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRC 176
CRLRWANHLRP+L+KG+ T +EER II+LH ++GNKWA+MAA LPGRTDNEIKN+WNTR
Sbjct: 67 CRLRWANHLRPNLKKGSFTGDEERLIIQLHAQLGNKWARMAAQLPGRTDNEIKNYWNTRL 126
Query: 177 KKRGRANLPIYPEDL 191
K+ R LP+YP D+
Sbjct: 127 KRLLRQGLPLYPPDI 141
>UniRef100_Q9FR42 Anther-specific myb-related protein 1 [Nicotiana tabacum]
Length = 470
Score = 192 bits (488), Expect = 1e-47
Identities = 88/126 (69%), Positives = 107/126 (84%), Gaps = 1/126 (0%)
Query: 76 LKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKGAIT 135
LKKGPWTA ED IL+ +V+K GEGNWN+V++ +GL RCGKSCRLRWANHLRP+L+KGA +
Sbjct: 20 LKKGPWTATEDAILMEYVKKNGEGNWNAVQRNSGLMRCGKSCRLRWANHLRPNLKKGAFS 79
Query: 136 AEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKRGRANLPIYPEDLSSEC 195
EEER I+ELH K+GNKWA+MAA +PGRTDNEIKN+WNTR K+R RA LPIYP+D+ +
Sbjct: 80 LEEERIIVELHAKLGNKWARMAAQMPGRTDNEIKNYWNTRLKRRQRAGLPIYPQDIQLQ- 138
Query: 196 LLNQEN 201
L QEN
Sbjct: 139 LNQQEN 144
>UniRef100_O80883 Putative MYB family transcription factor [Arabidopsis thaliana]
Length = 490
Score = 191 bits (485), Expect = 3e-47
Identities = 86/132 (65%), Positives = 108/132 (81%), Gaps = 1/132 (0%)
Query: 64 GDTTGGEGGEG-GLKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWA 122
G+TT EG GLKKGPWT ED IL +V+K+GEGNWN+V+K +GL RCGKSCRLRWA
Sbjct: 5 GETTATATMEGRGLKKGPWTTTEDAILTEYVRKHGEGNWNAVQKNSGLLRCGKSCRLRWA 64
Query: 123 NHLRPDLRKGAITAEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKRGRA 182
NHLRP+L+KG+ T +EE+ II+LH K+GNKWA+MA+ LPGRTDNEIKN+WNTR K+R RA
Sbjct: 65 NHLRPNLKKGSFTPDEEKIIIDLHAKLGNKWARMASQLPGRTDNEIKNYWNTRMKRRQRA 124
Query: 183 NLPIYPEDLSSE 194
LP+YP ++ +
Sbjct: 125 GLPLYPHEIQHQ 136
>UniRef100_Q8W1W5 Putative transcription factor MYB101 [Arabidopsis thaliana]
Length = 490
Score = 191 bits (485), Expect = 3e-47
Identities = 86/132 (65%), Positives = 108/132 (81%), Gaps = 1/132 (0%)
Query: 64 GDTTGGEGGEG-GLKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWA 122
G+TT EG GLKKGPWT ED IL +V+K+GEGNWN+V+K +GL RCGKSCRLRWA
Sbjct: 5 GETTATATMEGRGLKKGPWTTTEDAILTEYVRKHGEGNWNAVQKNSGLLRCGKSCRLRWA 64
Query: 123 NHLRPDLRKGAITAEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKRGRA 182
NHLRP+L+KG+ T +EE+ II+LH K+GNKWA+MA+ LPGRTDNEIKN+WNTR K+R RA
Sbjct: 65 NHLRPNLKKGSFTPDEEKIIIDLHAKLGNKWARMASQLPGRTDNEIKNYWNTRMKRRQRA 124
Query: 183 NLPIYPEDLSSE 194
LP+YP ++ +
Sbjct: 125 GLPLYPHEIQHQ 136
>UniRef100_Q655P0 Putative transcription factor GAMyb [Oryza sativa]
Length = 447
Score = 189 bits (479), Expect = 2e-46
Identities = 87/133 (65%), Positives = 106/133 (79%), Gaps = 1/133 (0%)
Query: 63 EGDTTGGEGGEGG-LKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRW 121
EG GG GG GG LKKGPWT AED++LV HV+++GEGNWN+VR+ TGL RCGKSCRLRW
Sbjct: 12 EGGGGGGNGGGGGGLKKGPWTQAEDKLLVDHVRRHGEGNWNAVRRETGLQRCGKSCRLRW 71
Query: 122 ANHLRPDLRKGAITAEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKRGR 181
ANHLRPDLRKG + +EER I+ LH +GNKWA++++ L GRTDNEIKN+WNTR K+R R
Sbjct: 72 ANHLRPDLRKGPFSPDEERLILRLHGLLGNKWARISSYLHGRTDNEIKNYWNTRLKRRVR 131
Query: 182 ANLPIYPEDLSSE 194
A L YP ++ E
Sbjct: 132 AGLTPYPPEIERE 144
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.312 0.131 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 792,987,280
Number of Sequences: 2790947
Number of extensions: 36947126
Number of successful extensions: 94024
Number of sequences better than 10.0: 1332
Number of HSP's better than 10.0 without gapping: 1218
Number of HSP's successfully gapped in prelim test: 114
Number of HSP's that attempted gapping in prelim test: 91666
Number of HSP's gapped (non-prelim): 1703
length of query: 416
length of database: 848,049,833
effective HSP length: 130
effective length of query: 286
effective length of database: 485,226,723
effective search space: 138774842778
effective search space used: 138774842778
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 76 (33.9 bits)
Medicago: description of AC148404.8


