

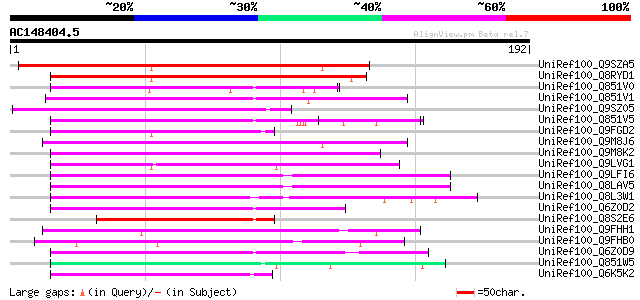
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148404.5 - phase: 0
(192 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9SZA5 Hypothetical protein F17M5.40 [Arabidopsis thal... 119 4e-26
UniRef100_Q8RYD1 Auxin response factor 36 [Arabidopsis thaliana] 113 2e-24
UniRef100_Q851V0 Putative auxin response factor [Oryza sativa] 77 2e-13
UniRef100_Q851V1 Putative auxin response factor [Oryza sativa] 73 5e-12
UniRef100_Q9SZ05 Hypothetical protein AT4g34400 [Arabidopsis tha... 72 6e-12
UniRef100_Q851V5 Putative auxin response factor [Oryza sativa] 67 2e-10
UniRef100_Q9FGD2 Gb|AAF30309.1 [Arabidopsis thaliana] 64 2e-09
UniRef100_Q9M8J6 F28L1.16 protein [Arabidopsis thaliana] 64 2e-09
UniRef100_Q9M8K2 F28L1.10 protein [Arabidopsis thaliana] 63 4e-09
UniRef100_Q9LVG1 Arabidopsis thaliana genomic DNA, chromosome 5,... 62 6e-09
UniRef100_Q9LFI6 Hypothetical protein F4P12_10 [Arabidopsis thal... 60 4e-08
UniRef100_Q8LAV5 Hypothetical protein [Arabidopsis thaliana] 60 4e-08
UniRef100_Q8L3W1 Reduced vernalization response 1 [Arabidopsis t... 59 7e-08
UniRef100_Q6Z0D2 Hypothetical protein OSJNBa0078D03.39 [Oryza sa... 57 2e-07
UniRef100_Q8S2E6 P0022F10.13 protein [Oryza sativa] 57 3e-07
UniRef100_Q9FHH1 Gb|AAF30309.1 [Arabidopsis thaliana] 55 1e-06
UniRef100_Q9FHB0 Arabidopsis thaliana genomic DNA, chromosome 5,... 54 2e-06
UniRef100_Q6Z0D9 Auxin response factor-like [Oryza sativa] 54 3e-06
UniRef100_Q851W5 Putative auxin response factor [Oryza sativa] 54 3e-06
UniRef100_Q6K5K2 Hypothetical protein OJ1212_D02.14 [Oryza sativa] 54 3e-06
>UniRef100_Q9SZA5 Hypothetical protein F17M5.40 [Arabidopsis thaliana]
Length = 461
Score = 119 bits (298), Expect = 4e-26
Identities = 62/138 (44%), Positives = 88/138 (62%), Gaps = 8/138 (5%)
Query: 4 ILIWKFILLLQALPKTFSDNLNKKLPENVTLKGPSGVTWNIRLTTRDG-FVYFVDGWQQF 62
I I F LLQ +P+ FS + +KLP+ VTLK PSGVT+N+ + D + F GW +F
Sbjct: 96 IKITTFCNLLQVIPRKFSTHCKRKLPQIVTLKSPSGVTYNVGVEEDDEKTMAFRFGWDKF 155
Query: 63 MNDHSLKANDFLVCKYNGESHFEVLIFDGESFCEKEASYFVEKCGHAQTAQG-------G 115
+ DHSL+ ND LV K++G S FEVL+FDG++ CEK SYFV KCGHA+ +G
Sbjct: 156 VKDHSLEENDLLVFKFHGVSEFEVLVFDGQTLCEKPTSYFVRKCGHAEKTKGIIDFNATS 215
Query: 116 SNASETNNSIEEVDTDSN 133
S + + + + ++V+T N
Sbjct: 216 SRSPKRHFNPDDVETTPN 233
>UniRef100_Q8RYD1 Auxin response factor 36 [Arabidopsis thaliana]
Length = 337
Score = 113 bits (283), Expect = 2e-24
Identities = 56/119 (47%), Positives = 77/119 (64%), Gaps = 2/119 (1%)
Query: 16 LPKTFSDNLNKKLPENVTLKGPSGVTWNIRLTTRDG-FVYFVDGWQQFMNDHSLKANDFL 74
+P+ FS + +KLP+ VTLK PSGVT+N+ + D + F GW +F+ DHSL+ ND L
Sbjct: 39 IPRKFSTHCKRKLPQIVTLKSPSGVTYNVGVEEDDEKTMAFRFGWDKFVKDHSLEENDLL 98
Query: 75 VCKYNGESHFEVLIFDGESFCEKEASYFVEKCGHAQTAQGGSNASETNNSI-EEVDTDS 132
V K++G S FEVL+FDG++ CEK SYFV KCGHA+ + E I ++DT S
Sbjct: 99 VFKFHGVSEFEVLVFDGQTLCEKPTSYFVRKCGHAEKTKASHTGYEQEEHINSDIDTAS 157
>UniRef100_Q851V0 Putative auxin response factor [Oryza sativa]
Length = 750
Score = 77.0 bits (188), Expect = 2e-13
Identities = 43/113 (38%), Positives = 66/113 (58%), Gaps = 7/113 (6%)
Query: 16 LPKTFSDNLNKKLPENVTLKGPSGVTWNIRLTTRD-GFVYFVDGWQQFMNDHSLKANDFL 74
+P F++N N + E V LK PSG TW+I + D G + GW++F++ + ++ ND L
Sbjct: 46 VPSRFANNFNGHISEVVNLKSPSGKTWSIGVAYSDTGELVLRSGWKEFVDANGVQENDCL 105
Query: 75 VCKYNGESHFEVLIFDGESFCEKEASYFVEKCGHAQT-----AQGGSNASETN 122
+ +Y+G S F+VLIFD S CEK + +FVE G + A+GG + N
Sbjct: 106 LFRYSGVSSFDVLIFD-PSGCEKASPHFVENRGFGREEKSAGAEGGGRDGDKN 157
Score = 70.5 bits (171), Expect = 2e-11
Identities = 42/114 (36%), Positives = 63/114 (54%), Gaps = 9/114 (7%)
Query: 16 LPKTFSDNLNKKLPENVTLKGPSGVTWNIRLTTRD-GFVYFVDGWQQFMNDHSLKANDFL 74
+P F++N N + E V L+ PSG TW+I + D G + GW++F++ + ++ D L
Sbjct: 424 VPARFANNFNGHISEEVNLRSPSGETWSIGVANSDAGELVLQPGWKEFVDGNGIEEGDCL 483
Query: 75 VCKYNG-ESHFEVLIFDGESFCEKEASYFVEKCG------HAQTAQGGSNASET 121
+ +Y+G S F+VLIFD S CEK + +FV G A QGG N T
Sbjct: 484 LFRYSGVSSSFDVLIFD-PSGCEKASPHFVGSHGFGRAENSAGAEQGGRNGRRT 536
>UniRef100_Q851V1 Putative auxin response factor [Oryza sativa]
Length = 306
Score = 72.8 bits (177), Expect = 5e-12
Identities = 41/135 (30%), Positives = 66/135 (48%), Gaps = 2/135 (1%)
Query: 14 QALPKTFSDNLNKKLPENVTLKGPSGVTWNIRLTTRDGFVYFVDGWQQFMNDHSLKANDF 73
+ +P F N K+P+++ L+ SG+T+++++T G V GW +++ H LK DF
Sbjct: 44 KTIPNEFLHNFGGKIPKSIKLETRSGLTFDVQVTKNSGRVVLQSGWASYVSAHDLKIGDF 103
Query: 74 LVCKYNGESHFEVLIFDGESFCEKEASYFVEKCGHA-QTAQGGSNASETNNSIEEVDTDS 132
LV KY+G+S + LIFD S CEK V+ G + A S + ++
Sbjct: 104 LVFKYSGDSQLKTLIFD-SSGCEKVCEKPVDMSGRSYDIAMRNSQDEKKKRKQRDISRQG 162
Query: 133 NGGDSPEQFTDDAVP 147
S E + VP
Sbjct: 163 TVKPSEEGLKAELVP 177
>UniRef100_Q9SZ05 Hypothetical protein AT4g34400 [Arabidopsis thaliana]
Length = 389
Score = 72.4 bits (176), Expect = 6e-12
Identities = 36/103 (34%), Positives = 53/103 (50%), Gaps = 1/103 (0%)
Query: 2 FLILIWKFILLLQALPKTFSDNLNKKLPENVTLKGPSGVTWNIRLTTRDGFVYFVDGWQQ 61
F + + F +P ++ D++ + P+ V L+GP G +W + +D V F GW +
Sbjct: 17 FTVFVSHFSSEFMVIPVSYYDHIPHRFPKTVILRGPGGCSWKVATEIKDDEVLFSQGWPK 76
Query: 62 FMNDHSLKANDFLVCKYNGESHFEVLIFDGESFCEKEASYFVE 104
F+ D++L DFL YNG FEV IF G C KE S E
Sbjct: 77 FVRDNTLNDGDFLTFAYNGAHIFEVSIFRGYDAC-KEISEVTE 118
>UniRef100_Q851V5 Putative auxin response factor [Oryza sativa]
Length = 1029
Score = 67.4 bits (163), Expect = 2e-10
Identities = 37/100 (37%), Positives = 52/100 (52%), Gaps = 2/100 (2%)
Query: 16 LPKTFSDNLNKKLPENVTLKGPSGVTWNIRLTTRDGFVYFVDGWQQFMNDHSLKANDFLV 75
+P F++N ++ + LK +G T ++ + + GW +F N H +K DFLV
Sbjct: 164 IPYKFAENFRDQIQGTIKLKARNGNTCSVLVDKCSNKLVLTKGWAEFANSHDIKMGDFLV 223
Query: 76 CKYNGESHFEVLIFDGESFCEKEASYFVEKCG-HAQTAQG 114
+Y G S FEV IFD S C K AS+ G HAQ QG
Sbjct: 224 FRYTGNSQFEVKIFD-PSGCVKAASHNAVNIGQHAQNMQG 262
Score = 64.3 bits (155), Expect = 2e-09
Identities = 41/154 (26%), Positives = 72/154 (46%), Gaps = 18/154 (11%)
Query: 16 LPKTFSDNLNKKLPENVTLKGPSGVTWNIRLTTRDGFVYFVDGWQQFMNDHSLKANDFLV 75
+P F+ + + + + L+ SG T+++++T + + GW+ F+N H L DFLV
Sbjct: 467 IPDKFARHFKGVISKTIKLEPRSGYTFDVQVTKKLNILVLGSGWESFVNAHDLNMGDFLV 526
Query: 76 CKYNGESHFEVLIFDGESFCEKEASYFVEKC---------GHAQTAQGGSNASETNNSIE 126
KYNG+ +VLIFD S CEK S +E H + + + N
Sbjct: 527 FKYNGDFLLQVLIFD-PSGCEKSTSCSMENAIDHVGQGWKEHNDISTSYHDQPKGNKHWM 585
Query: 127 EVDTDSNG--------GDSPEQFTDDAVPKTTAI 152
+ D+ S G ++P +F+ +P+ T +
Sbjct: 586 QKDSSSKGNKIGNTRSSNTPSKFSGCILPRGTCL 619
Score = 62.4 bits (150), Expect = 6e-09
Identities = 46/145 (31%), Positives = 67/145 (45%), Gaps = 8/145 (5%)
Query: 16 LPKTFSDNLNKKLPENVTLKGPSGVTWNIRLTTRDGFVYFVDGWQQFMNDHSLKANDFLV 75
+P F K+P + L+ G T+++++T G + GW+ F+ H L+ DFLV
Sbjct: 748 IPNEFLQYFRGKIPRTIKLQLRDGCTYDVQVTKNLGKISLQSGWKAFVTAHDLQMGDFLV 807
Query: 76 CKYNGESHFEVLIFDGESFCEKEASYFVEK----CGH--AQTAQGGSNASETN-NSIEEV 128
Y+G S +VLIF G S CEK S K CG + SN+ + S + V
Sbjct: 808 FSYDGISKLKVLIF-GPSGCEKVHSRSTLKNATHCGEKWEEPLHISSNSHDLPVKSPQNV 866
Query: 129 DTDSNGGDSPEQFTDDAVPKTTAIQ 153
DS EQ D A + A+Q
Sbjct: 867 SKSEKQWDSSEQENDTANIEEVALQ 891
>UniRef100_Q9FGD2 Gb|AAF30309.1 [Arabidopsis thaliana]
Length = 334
Score = 64.3 bits (155), Expect = 2e-09
Identities = 35/87 (40%), Positives = 45/87 (51%), Gaps = 5/87 (5%)
Query: 16 LPKTFSDNLNKKLPENVTLKGPSGVTWNIRLTTRDG----FVYFVDGWQQFMNDHSLKAN 71
+P F D L K LP+ L G W + T D V+F GWQ F ND SL+
Sbjct: 25 IPPAFIDMLEKPLPKEAFLVDEIGRLWCVETKTEDTEERFCVFFTKGWQSFANDQSLEFG 84
Query: 72 DFLVCKYNGESHFEVLIFDGESFCEKE 98
DFLV Y+G+S F V IF + C+K+
Sbjct: 85 DFLVFSYDGDSRFSVTIFANDG-CKKD 110
>UniRef100_Q9M8J6 F28L1.16 protein [Arabidopsis thaliana]
Length = 175
Score = 63.9 bits (154), Expect = 2e-09
Identities = 37/138 (26%), Positives = 63/138 (44%), Gaps = 3/138 (2%)
Query: 13 LQALPKTFSDNLNKKLPENVTLKGPSGVTWNIRLTTRDGFVYFVDGWQQFMNDHSLKAND 72
L+ +P+++ ++L ++LP+ L G G W + + ++ VY GW+ F+ D+ LK +
Sbjct: 23 LKLIPRSYYEHLPRRLPKTAILTGTGGRVWKVAMMSKREQVYLARGWENFVADNELKDGE 82
Query: 73 FLVCKYNGESHFEVLIFDGESFCEKEASYFVEKCGHAQTAQG---GSNASETNNSIEEVD 129
FL ++G +EV I+ S E A VE+ + GS T EE
Sbjct: 83 FLTFVFDGYKSYEVSIYGRGSCKETRAVVHVEEISDESESDNDSLGSLVDVTPMPAEENS 142
Query: 130 TDSNGGDSPEQFTDDAVP 147
D+ G + D P
Sbjct: 143 DDTEGDNDSCDSVVDVTP 160
>UniRef100_Q9M8K2 F28L1.10 protein [Arabidopsis thaliana]
Length = 330
Score = 63.2 bits (152), Expect = 4e-09
Identities = 35/122 (28%), Positives = 59/122 (47%)
Query: 16 LPKTFSDNLNKKLPENVTLKGPSGVTWNIRLTTRDGFVYFVDGWQQFMNDHSLKANDFLV 75
+P+++ + L + LP+ L G G W + +T++ VYF GW F+ D+ LK +FL
Sbjct: 40 IPRSYYNLLPRPLPKTAILIGTGGRFWKVAMTSKQEQVYFEQGWGNFVADNQLKEGEFLT 99
Query: 76 CKYNGESHFEVLIFDGESFCEKEASYFVEKCGHAQTAQGGSNASETNNSIEEVDTDSNGG 135
++G +EV I+ E A VE+ S S +N S++ + DS+
Sbjct: 100 FVFDGHKSYEVSIYGRGDCKETRAVIQVEEISDDTEDDNVSLHSPSNVSLDSLSNDSHHS 159
Query: 136 DS 137
S
Sbjct: 160 TS 161
>UniRef100_Q9LVG1 Arabidopsis thaliana genomic DNA, chromosome 5, P1 clone:MGO3
[Arabidopsis thaliana]
Length = 326
Score = 62.4 bits (150), Expect = 6e-09
Identities = 43/137 (31%), Positives = 69/137 (49%), Gaps = 9/137 (6%)
Query: 16 LPKTFSDNLNKKLPENVTLKGPSGVTWNIRLTTRDG------FVYFVDGWQQFMNDHSLK 69
LP +F+ L K LPE VT++ SG W + L G FV V+GW++ + D LK
Sbjct: 29 LPISFTRFLPKSLPETVTVRSISGNIWKLELKKCCGDDDTEKFV-MVNGWKRIVKDEDLK 87
Query: 70 ANDFLVCKYNGESHFEVLIFDGESFCEK--EASYFVEKCGHAQTAQGGSNASETNNSIEE 127
DFL +++G S F I++ + C+K +S E+ + + AS+ S++E
Sbjct: 88 GGDFLEFEFDGSSCFHFCIYEHRTMCKKIRRSSDQSEEIKVESDSDEQNQASDDVLSLDE 147
Query: 128 VDTDSNGGDSPEQFTDD 144
D DS+ + +DD
Sbjct: 148 DDDDSDYNCGEDNDSDD 164
>UniRef100_Q9LFI6 Hypothetical protein F4P12_10 [Arabidopsis thaliana]
Length = 283
Score = 59.7 bits (143), Expect = 4e-08
Identities = 37/148 (25%), Positives = 60/148 (40%), Gaps = 3/148 (2%)
Query: 16 LPKTFSDNLNKKLPENVTLKGPSGVTWNIRLTTRDGFVYFVDGWQQFMNDHSLKANDFLV 75
+P F L LP+ V L+G G W + L G Y GW +F +H LK +F+
Sbjct: 26 IPLPFMAFLADPLPKTVKLQGLGGKLWTVSLKKISGAAYLTRGWPKFAEEHELKNGEFMT 85
Query: 76 CKYNGESHFEVLIFDGESFCEKEASYFVEKCGHAQTAQGGSNASETNNSIEEVDTDSNGG 135
Y+G FEV +FD E A + + S + +S + V+ D +
Sbjct: 86 FVYDGHRTFEVSVFDRWGSKEVRAEI---QAIPLSDSDSDSVVEDEKDSTDVVEDDDDED 142
Query: 136 DSPEQFTDDAVPKTTAIQSPFIPTGKRT 163
+ ++ D + + I P + T
Sbjct: 143 EDEDEDDDGSFDEDEEISQSLYPIDEET 170
>UniRef100_Q8LAV5 Hypothetical protein [Arabidopsis thaliana]
Length = 286
Score = 59.7 bits (143), Expect = 4e-08
Identities = 37/148 (25%), Positives = 60/148 (40%), Gaps = 3/148 (2%)
Query: 16 LPKTFSDNLNKKLPENVTLKGPSGVTWNIRLTTRDGFVYFVDGWQQFMNDHSLKANDFLV 75
+P F L LP+ V L+G G W + L G Y GW +F +H LK +F+
Sbjct: 26 IPLPFMAFLADPLPKTVKLQGLGGKLWTVSLKKISGAAYLTRGWPKFAEEHELKNGEFMT 85
Query: 76 CKYNGESHFEVLIFDGESFCEKEASYFVEKCGHAQTAQGGSNASETNNSIEEVDTDSNGG 135
Y+G FEV +FD E A + + S + +S + V+ D +
Sbjct: 86 FVYDGHRTFEVSVFDRWGSKEVRAEI---QAIPLSDSDSDSVVEDEKDSTDVVEDDDDED 142
Query: 136 DSPEQFTDDAVPKTTAIQSPFIPTGKRT 163
+ ++ D + + I P + T
Sbjct: 143 EDEDEDDDGSFDEDEEISQSLYPIDEET 170
>UniRef100_Q8L3W1 Reduced vernalization response 1 [Arabidopsis thaliana]
Length = 341
Score = 58.9 bits (141), Expect = 7e-08
Identities = 47/174 (27%), Positives = 72/174 (41%), Gaps = 21/174 (12%)
Query: 16 LPKTFSDNLNKKLPENVTLKGPSGVTWNIRLTTRDGFVYFVDGWQQFMNDHSLKANDFLV 75
+P F +L V L P G W + L D ++F DGWQ+F++ +S++ L+
Sbjct: 22 VPDKFVSKFKDELSVAVALTVPDGHVWRVGLRKADNKIWFQDGWQEFVDRYSIRIGYLLI 81
Query: 76 CKYNGESHFEVLIFDGESFCEKEASYFVEKCGHAQTAQGGSNASETNNSIEEVDTDSNGG 135
+Y G S F V IF + E +Y G +A + +E+ D +
Sbjct: 82 FRYEGNSAFSVYIF---NLSHSEINY--HSTGLMDSAHNHFKRARLFEDLEDEDAEVIFP 136
Query: 136 DS--PEQFTDDAVP-----KTTAIQSPF---------IPTGKRTKRRRRSPKAA 173
S P + VP ++AIQ+ F PT K K+R R K A
Sbjct: 137 SSVYPSPLPESTVPANKGYASSAIQTLFTGPVKAEEPTPTPKIPKKRGRKKKNA 190
>UniRef100_Q6Z0D2 Hypothetical protein OSJNBa0078D03.39 [Oryza sativa]
Length = 391
Score = 57.4 bits (137), Expect = 2e-07
Identities = 33/109 (30%), Positives = 55/109 (50%), Gaps = 1/109 (0%)
Query: 16 LPKTFSDNLNKKLPENVTLKGPSGVTWNIRLTTRDGFVYFVDGWQQFMNDHSLKANDFLV 75
+PK F L +PE + L+ + ++ +R+ V F GW QF+ L D ++
Sbjct: 49 IPKEFVQRLKGDIPEEIQLETHNRNSYTVRVDKSQEKVIFAAGWAQFVKTFDLHMGDSMM 108
Query: 76 CKYNGESHFEVLIFDGESFCEKEASYFVEKCGHAQTAQGGSNASETNNS 124
++ G S F+V+IFD + EK S V+ +G ++A+ET NS
Sbjct: 109 FRFKGNSQFDVIIFD-QVGREKVCSVAVDDYLDPNVQEGRTDATETLNS 156
>UniRef100_Q8S2E6 P0022F10.13 protein [Oryza sativa]
Length = 402
Score = 56.6 bits (135), Expect = 3e-07
Identities = 24/66 (36%), Positives = 41/66 (61%), Gaps = 1/66 (1%)
Query: 33 TLKGPSGVTWNIRLTTRDGFVYFVDGWQQFMNDHSLKANDFLVCKYNGESHFEVLIFDGE 92
TL+GPSG TW + + +F GW +F+ D +L+ +F+V +Y+G + F ++FD
Sbjct: 62 TLEGPSGRTWLVVIRRTAEGTFFTSGWPKFVQDQALRELEFVVFRYDGNTRFTAMVFD-R 120
Query: 93 SFCEKE 98
+ CE+E
Sbjct: 121 TACERE 126
>UniRef100_Q9FHH1 Gb|AAF30309.1 [Arabidopsis thaliana]
Length = 300
Score = 55.1 bits (131), Expect = 1e-06
Identities = 41/147 (27%), Positives = 63/147 (41%), Gaps = 10/147 (6%)
Query: 13 LQALPKTFSDNLNKKLPENVTLKGPSGVTWNIRLT-TRDGFVYFVDGWQQFMNDHSLKAN 71
L +P++++ LP+ LK P G WN++ T +++ + +GW +F+ D+ L
Sbjct: 25 LLVIPRSYNRYYPNPLPQTAVLKNPEGRFWNVQWTKSQEVIISLQEGWVKFVKDNGLIDR 84
Query: 72 DFLVCKYNGESHFEVLIFDGESFCEKEASYFVEKCGHAQTAQGGSNASETNNSIEEVDTD 131
DFL+ Y+G F V I E A +++ + G EE DTD
Sbjct: 85 DFLLFTYDGSRSFWVRIHRNGLPLEPTAPIKIQEISDDEDETNGDGDPHME---EEGDTD 141
Query: 132 SNG------GDSPEQFTDDAVPKTTAI 152
N G S E DD TAI
Sbjct: 142 ENMIVSLSLGSSDEVGDDDDDDYDTAI 168
>UniRef100_Q9FHB0 Arabidopsis thaliana genomic DNA, chromosome 5, P1 clone:MGO3
[Arabidopsis thaliana]
Length = 312
Score = 53.9 bits (128), Expect = 2e-06
Identities = 41/150 (27%), Positives = 67/150 (44%), Gaps = 16/150 (10%)
Query: 10 ILLLQALPKTFSDN------LNKKLPENVTLKGPSGVTWNIRLTTRDGFV---YFVDGWQ 60
+LL +L +TF N L K LP+ VT++ SG W + G V V GW+
Sbjct: 2 VLLQSSLNRTFLVNPVMICCLPKSLPKTVTVRSISGNIWKLGFKKCGGEVERFVMVSGWK 61
Query: 61 QFMNDHSLKANDFLVCKYNGESHFEVLIFDGESFCEKEASYFVEKCGHAQTAQGGSNASE 120
+ + D +L D L +++G F IFD E+ C++ + + GS+ E
Sbjct: 62 KIVRDENLNGGDLLSFEFDGSHFFNFSIFDHETTCKRLKRSSEQS---KDIIKVGSDCEE 118
Query: 121 TNNSIEEV----DTDSNGGDSPEQFTDDAV 146
+ + ++V DS+ D+ DD V
Sbjct: 119 ESQASDDVIVLNSDDSDDSDNDYSVEDDNV 148
>UniRef100_Q6Z0D9 Auxin response factor-like [Oryza sativa]
Length = 270
Score = 53.5 bits (127), Expect = 3e-06
Identities = 37/140 (26%), Positives = 65/140 (46%), Gaps = 6/140 (4%)
Query: 16 LPKTFSDNLNKKLPENVTLKGPSGVTWNIRLTTRDGFVYFVDGWQQFMNDHSLKANDFLV 75
+PK F L +P + L+ + + +R+ V F +GW QF+ L+ D ++
Sbjct: 40 IPKEFVQRLKGDIPGEIQLETRNRNSHTVRVDKTQEKVIFTEGWAQFVKTFDLQMGDSMM 99
Query: 76 CKYNGESHFEVLIFDGESFCEKEASYFVEKCGHAQTAQGGSNASETNNSIEEVDTDSNGG 135
++NG S F+V+I D + EK S V+ + + +A+ET NS + ++
Sbjct: 100 FRFNGNSQFDVIIVD-QIGREKACSAVVDDSQNPNVQERRVDATETLNS-----SRAHSQ 153
Query: 136 DSPEQFTDDAVPKTTAIQSP 155
P Q T + V + A P
Sbjct: 154 PMPMQSTTETVNHSHARPCP 173
>UniRef100_Q851W5 Putative auxin response factor [Oryza sativa]
Length = 519
Score = 53.5 bits (127), Expect = 3e-06
Identities = 38/175 (21%), Positives = 71/175 (39%), Gaps = 30/175 (17%)
Query: 16 LPKTFSDNLNKKLPENVTLKGPSGVTWNIRLTTRDGFVYFVDGWQQFMNDHSLKANDFLV 75
+P F D+ L + L P G+ + +++T GW+ F++ H ++ ND L+
Sbjct: 43 IPNKFLDHFGGTLSRTIELVSPKGIVYIVKVTEHMNKTILQCGWEAFVDAHHIEENDSLL 102
Query: 76 CKYNGESHFEVLIFDGESFCEK-----------------EASYFVEKCGHAQTAQGGSN- 117
++ S FEVLI D + CEK A + + H +T Q +
Sbjct: 103 FRHIENSRFEVLILDSDG-CEKVFTCAGIKKTSSVQERNAAPVDISRSTHDETTQSSGSK 161
Query: 118 --------ASETNNSIEEVDTDSNGGDSPEQFTDDAVPKTTA---IQSPFIPTGK 161
+ ++ S+ G+S E+ TD + + + + P +P G+
Sbjct: 162 KFVRCQRASDSQRGKTAKLAETSSSGESGEEGTDSSTSEDESSYELDDPQMPPGR 216
>UniRef100_Q6K5K2 Hypothetical protein OJ1212_D02.14 [Oryza sativa]
Length = 1048
Score = 53.5 bits (127), Expect = 3e-06
Identities = 27/82 (32%), Positives = 45/82 (53%), Gaps = 1/82 (1%)
Query: 16 LPKTFSDNLNKKLPENVTLKGPSGVTWNIRLTTRDGFVYFVDGWQQFMNDHSLKANDFLV 75
+P + +L + +V L+ G IRL+ DG + F +GW F+++H +K +FL+
Sbjct: 392 IPPAVAPSLKDLIDRHVYLEDSEGKCSKIRLSVVDGSLAFYEGWNSFVSEHCIKWGEFLL 451
Query: 76 CKYNGESHFEVLIFDGESFCEK 97
+Y ES F V +F G CE+
Sbjct: 452 FEYTPESTFSVRVF-GIDSCER 472
Score = 32.3 bits (72), Expect = 6.8
Identities = 20/58 (34%), Positives = 28/58 (47%), Gaps = 2/58 (3%)
Query: 32 VTLKGPSGVTWNIRLTTRDGFVYFVDGWQQFMNDHSLKANDFLVCKYNGESHFEVLIF 89
V LK P W + + FV F GW+ F ++L+A D VCK+ E + L F
Sbjct: 986 VILKDPMRRLWPVFYHDKPVFVGFTAGWKPFAAANNLQAGD--VCKFVKEMDEDELAF 1041
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.317 0.134 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 354,341,198
Number of Sequences: 2790947
Number of extensions: 14903946
Number of successful extensions: 42406
Number of sequences better than 10.0: 89
Number of HSP's better than 10.0 without gapping: 53
Number of HSP's successfully gapped in prelim test: 36
Number of HSP's that attempted gapping in prelim test: 42304
Number of HSP's gapped (non-prelim): 123
length of query: 192
length of database: 848,049,833
effective HSP length: 120
effective length of query: 72
effective length of database: 513,136,193
effective search space: 36945805896
effective search space used: 36945805896
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 71 (32.0 bits)
Medicago: description of AC148404.5


