

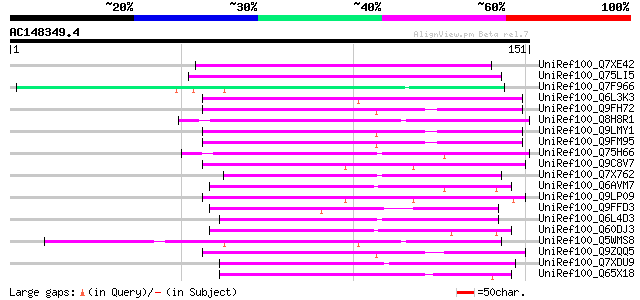
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148349.4 - phase: 0
(151 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q7XE42 Putative mutator-like transposase [Oryza sativa] 75 2e-13
UniRef100_Q75LI5 Putative transposon protein [Oryza sativa] 64 7e-10
UniRef100_Q7F966 OSJNBa0091C07.2 protein [Oryza sativa] 61 4e-09
UniRef100_Q6L3K3 Putative transposon MuDR mudrA-like protein [So... 58 5e-08
UniRef100_Q9FH72 Arabidopsis thaliana genomic DNA, chromosome 5,... 57 1e-07
UniRef100_Q8H8R1 Putative mutator-like transposase [Oryza sativa] 56 2e-07
UniRef100_Q9LMY1 F21F23.10 protein [Arabidopsis thaliana] 55 4e-07
UniRef100_Q9FM95 Similarity to mutator-like transposase [Arabido... 55 4e-07
UniRef100_Q75H66 Hypothetical protein OSJNBb0007E22.22 [Oryza sa... 54 5e-07
UniRef100_Q9C8V7 Mutator-like transposase; 53847-56139 [Arabidop... 53 2e-06
UniRef100_Q7X762 OSJNBb0118P14.3 protein [Oryza sativa] 52 3e-06
UniRef100_Q6AVM7 Putative polyprotein [Oryza sativa] 52 3e-06
UniRef100_Q9LP09 F9C16.9 [Arabidopsis thaliana] 52 3e-06
UniRef100_Q9FFD3 Mutator-like transposase-like protein [Arabidop... 51 6e-06
UniRef100_Q6L4D3 Hypothetical protein OSJNBa0088M05.7 [Oryza sat... 49 2e-05
UniRef100_Q60DJ3 MuDR family transposase protein [Oryza sativa] 49 3e-05
UniRef100_Q5WMS8 Hypothetical protein OJ1333_C12.3 [Oryza sativa] 49 3e-05
UniRef100_Q9ZQQ5 Mutator-like transposase [Arabidopsis thaliana] 47 7e-05
UniRef100_Q7XDU9 Putative mutator protein [Oryza sativa] 47 7e-05
UniRef100_Q65X18 Putative polyprotein [Oryza sativa] 46 2e-04
>UniRef100_Q7XE42 Putative mutator-like transposase [Oryza sativa]
Length = 1005
Score = 75.5 bits (184), Expect = 2e-13
Identities = 34/86 (39%), Positives = 48/86 (55%)
Query: 55 HAWLTLGSYRATYNYFIQPVNSQIYWEPTPYEKSAPPKVRRAAGRPKKNRRKDGNKEPIG 114
H +L Y+ TY + +QPV + W +P K PP+V++ GRPKKNRRKD +K
Sbjct: 769 HECFSLERYKKTYQHVLQPVEHESAWPVSPNPKPLPPRVKKMPGRPKKNRRKDPSKPVKS 828
Query: 115 RSQMKRTYNDTQCGRCGLIGHNLRGC 140
++ + +C RCG GHN R C
Sbjct: 829 GTKSSKVGTKIRCRRCGNYGHNSRTC 854
>UniRef100_Q75LI5 Putative transposon protein [Oryza sativa]
Length = 839
Score = 63.9 bits (154), Expect = 7e-10
Identities = 29/91 (31%), Positives = 46/91 (49%)
Query: 53 HCHAWLTLGSYRATYNYFIQPVNSQIYWEPTPYEKSAPPKVRRAAGRPKKNRRKDGNKEP 112
H + ++ YR TY +QPV + W + + PP+V++ G PK+ RRKD +
Sbjct: 652 HVNMCFSIDQYRNTYQDVLQPVEHESVWPLSTNPRPLPPRVKKMPGSPKRARRKDPTEAA 711
Query: 113 IGRSQMKRTYNDTQCGRCGLIGHNLRGCNKQ 143
++ + +CG C GHN RGC K+
Sbjct: 712 GSSTKSSKRGGSVKCGFCHEKGHNSRGCKKK 742
>UniRef100_Q7F966 OSJNBa0091C07.2 protein [Oryza sativa]
Length = 879
Score = 61.2 bits (147), Expect = 4e-09
Identities = 42/182 (23%), Positives = 72/182 (39%), Gaps = 41/182 (22%)
Query: 3 KMSQNKMKLDGRVGPLCPWQQSRLEKEKNASHNWTPLWSGDTPMQ------RYQIE---- 52
++ QN+ K G +G +CP +L K + S N +W+G + RY ++
Sbjct: 638 RIQQNRSKAKGWLGRICPNILKKLNKYIDLSGNCEAIWNGKDGFEVTDKDKRYTVDLEKR 697
Query: 53 --HCHAWLTLG----------------------------SYRATYNYFIQPVNSQIYWEP 82
C W G Y Y++ + P+ + W
Sbjct: 698 TCSCRYWQLAGIPCAHAITALFVSSKQPEDYIADCYSVEVYNKIYDHCMMPMEGMMQWPI 757
Query: 83 TPYEKSAPPKVRRAAGRPKKNRRKDGNKEPIGRSQMKRTYNDTQCGRCGLIGHNLRGCNK 142
T + K PP + GRP+K RR+D + G+ ++ +T +C +C HN+R C
Sbjct: 758 TGHPKPGPPGYVKMPGRPRKERRRDPLEAKKGK-KLSKTGTKGRCSQCKQTTHNIRTCPM 816
Query: 143 QG 144
+G
Sbjct: 817 KG 818
>UniRef100_Q6L3K3 Putative transposon MuDR mudrA-like protein [Solanum demissum]
Length = 873
Score = 57.8 bits (138), Expect = 5e-08
Identities = 29/95 (30%), Positives = 44/95 (45%), Gaps = 2/95 (2%)
Query: 57 WLTLGSYRATYNYFIQPVNSQIYWEPTPYEKSAPPKVRRAAGRP--KKNRRKDGNKEPIG 114
W + +Y Y + IQPV + +W+ P PP++ + GRP K+ R KD ++ G
Sbjct: 628 WYSREAYMLVYMHKIQPVRGEKFWKVDPSHAMEPPEIHKLVGRPKLKRKREKDEARKREG 687
Query: 115 RSQMKRTYNDTQCGRCGLIGHNLRGCNKQGVARRP 149
R CG C GHN R C +++P
Sbjct: 688 VWSASRKGLKMTCGHCSATGHNQRRCPMLQRSKQP 722
>UniRef100_Q9FH72 Arabidopsis thaliana genomic DNA, chromosome 5, P1 clone:MRD20
[Arabidopsis thaliana]
Length = 733
Score = 56.6 bits (135), Expect = 1e-07
Identities = 33/94 (35%), Positives = 43/94 (45%), Gaps = 4/94 (4%)
Query: 57 WLTLGSYRATYNYFIQPVNSQIYWEPTPYEKSAPPKVRRAAGRPKKNRR-KDGNKEPIGR 115
W ++ ++ YN + PVN WE PP R GRPKKN R KD ++E
Sbjct: 589 WYSVEKWKLCYNSLLFPVNGMELWETHSDVVVMPPPDRIMPGRPKKNDRIKDPSEEASSE 648
Query: 116 SQMKRTYNDTQCGRCGLIGHNLRGCNKQGVARRP 149
+ K C CG IGHN R C + V + P
Sbjct: 649 NSQKAL---VTCSNCGQIGHNKRTCQIELVPKPP 679
>UniRef100_Q8H8R1 Putative mutator-like transposase [Oryza sativa]
Length = 746
Score = 55.8 bits (133), Expect = 2e-07
Identities = 29/102 (28%), Positives = 49/102 (47%), Gaps = 4/102 (3%)
Query: 50 QIEHCHAWLTLGSYRATYNYFIQPVNSQIYWEPTPYEKSAPPKVRRAAGRPKKNRRKDGN 109
++ C+ T+ S++ Y + I PV + +WE PP + GRPK RRK
Sbjct: 608 EVSFCY---TIQSFKQAYMFNIMPVRDKTHWEKMNGVPVNPPVYEKKVGRPKTTRRKQPQ 664
Query: 110 KEPIGRSQMKRTYNDTQCGRCGLIGHNLRGCNKQGVARRPKD 151
+ G +++ + C C +GHN +GC K+ A+ K+
Sbjct: 665 ELDAG-TKISKHGVQIHCSYCKNVGHNKKGCKKRKAAQNGKN 705
>UniRef100_Q9LMY1 F21F23.10 protein [Arabidopsis thaliana]
Length = 753
Score = 54.7 bits (130), Expect = 4e-07
Identities = 32/94 (34%), Positives = 42/94 (44%), Gaps = 4/94 (4%)
Query: 57 WLTLGSYRATYNYFIQPVNSQIYWEPTPYEKSAPPKVRRAAGRPKKNRR-KDGNKEPIGR 115
W ++ ++ YN + PVN WE PP R GRPKKN R KD ++E
Sbjct: 609 WYSVEKWKLCYNSLLFPVNGMELWETHSDVVVMPPPDRIMPGRPKKNDRIKDPSEEASSE 668
Query: 116 SQMKRTYNDTQCGRCGLIGHNLRGCNKQGVARRP 149
+ K C CG GHN R C + V + P
Sbjct: 669 NSQKAL---VTCSNCGQTGHNKRTCQIELVPKPP 699
>UniRef100_Q9FM95 Similarity to mutator-like transposase [Arabidopsis thaliana]
Length = 733
Score = 54.7 bits (130), Expect = 4e-07
Identities = 32/94 (34%), Positives = 42/94 (44%), Gaps = 4/94 (4%)
Query: 57 WLTLGSYRATYNYFIQPVNSQIYWEPTPYEKSAPPKVRRAAGRPKKNRR-KDGNKEPIGR 115
W ++ ++ YN + PVN WE PP R GRPKKN R KD ++E
Sbjct: 589 WYSVEKWKLCYNSLLFPVNGMELWETHSDVVVMPPPDRIMPGRPKKNDRIKDPSEEASSE 648
Query: 116 SQMKRTYNDTQCGRCGLIGHNLRGCNKQGVARRP 149
+ K C CG GHN R C + V + P
Sbjct: 649 NSQKAL---VTCSNCGQTGHNKRTCQIELVPKPP 679
>UniRef100_Q75H66 Hypothetical protein OSJNBb0007E22.22 [Oryza sativa]
Length = 981
Score = 54.3 bits (129), Expect = 5e-07
Identities = 33/103 (32%), Positives = 50/103 (48%), Gaps = 6/103 (5%)
Query: 51 IEHCHAWLTLGSYRATYNYFIQPVNSQIYWEPTPYEKSAPPKVRRAAGRPKKNRRKDGNK 110
+ HC++ + +++A Y I P N + WE + PP + GRPKK+RRK +
Sbjct: 763 LPHCYS---INAFKAVYAENIIPCNDKANWEKMNGPQILPPVYEKKVGRPKKSRRKQ-PQ 818
Query: 111 EPIGRSQMKRTYNDT--QCGRCGLIGHNLRGCNKQGVARRPKD 151
E GR+ K T + C C HN +GC + RPK+
Sbjct: 819 EVQGRNGPKLTKHGVTIHCSYCHEANHNKKGCELRKKGIRPKN 861
>UniRef100_Q9C8V7 Mutator-like transposase; 53847-56139 [Arabidopsis thaliana]
Length = 583
Score = 52.8 bits (125), Expect = 2e-06
Identities = 32/97 (32%), Positives = 40/97 (40%), Gaps = 3/97 (3%)
Query: 57 WLTLGSYRATYNYFIQPVNSQIYWEPTPYEKSAPPKVRRA-AGRPKKNRRKDGNKEPIGR 115
W T ++ TY I PV +I W PP RR GRP R+ G E
Sbjct: 473 WYTTQRWQQTYKDGIAPVQGKILWPRVNRFGVLPPPWRRGNPGRPNNYARRKGRNEAASS 532
Query: 116 S--QMKRTYNDTQCGRCGLIGHNLRGCNKQGVARRPK 150
S + R + C C GHN +GC Q V +PK
Sbjct: 533 SITTLSRLHRVMTCSNCKQEGHNKKGCKNQWVEGQPK 569
>UniRef100_Q7X762 OSJNBb0118P14.3 protein [Oryza sativa]
Length = 939
Score = 52.0 bits (123), Expect = 3e-06
Identities = 27/81 (33%), Positives = 39/81 (47%), Gaps = 1/81 (1%)
Query: 63 YRATYNYFIQPVNSQIYWEPTPYEKSAPPKVRRAAGRPKKNRRKDGNKEPIGRSQMKRTY 122
Y Y+Y I P+ I+WE + PP + GRPKK RRK +E G +++ +
Sbjct: 743 YEQAYSYNIMPLRDSIHWEKMQGIEVKPPVYEKKVGRPKKTRRKQ-PQELEGGTKISKHG 801
Query: 123 NDTQCGRCGLIGHNLRGCNKQ 143
+ C C GHN C K+
Sbjct: 802 VEMHCSYCKNGGHNKTSCKKR 822
>UniRef100_Q6AVM7 Putative polyprotein [Oryza sativa]
Length = 1006
Score = 52.0 bits (123), Expect = 3e-06
Identities = 33/92 (35%), Positives = 45/92 (48%), Gaps = 5/92 (5%)
Query: 59 TLGSYRATYNYFIQPVNSQIYWEPTPYEKSAPPKVRRAAGRPKKNRRKDGNKEPIGRSQM 118
T ++ Y + I P N + WE + PP + AGRPKK+RRK E IG++
Sbjct: 780 TTDAFSKAYGFNIWPCNDKSKWENINGPEIKPPVYEKKAGRPKKSRRK-APYEVIGKNGP 838
Query: 119 KRTYNDT--QCGRCGLIGHNLRGC--NKQGVA 146
K T + C CG HN GC KQG++
Sbjct: 839 KLTKHGVMMHCKYCGEENHNSGGCKLKKQGIS 870
>UniRef100_Q9LP09 F9C16.9 [Arabidopsis thaliana]
Length = 946
Score = 51.6 bits (122), Expect = 3e-06
Identities = 33/103 (32%), Positives = 46/103 (44%), Gaps = 9/103 (8%)
Query: 57 WLTLGSYRATYNYFIQPVNSQIYWEPTPYEKSAPPKVRRA-AGRPKKNRRKDGNKEPIGR 115
W T ++ TYN I PV ++ W PP RR GRPK + R+ G E
Sbjct: 727 WYTTSMWKQTYNDGIGPVQGKLLWPTVNKVGVLPPPWRRGNPGRPKNHARRKGVFESSTA 786
Query: 116 S-----QMKRTYNDTQCGRCGLIGHNLRGCNKQGV---ARRPK 150
S ++ R + C C GHN +GC + V A+RP+
Sbjct: 787 SSSSTTELSRLHRVMTCSNCQGEGHNKQGCKNETVAPPAKRPR 829
>UniRef100_Q9FFD3 Mutator-like transposase-like protein [Arabidopsis thaliana]
Length = 597
Score = 50.8 bits (120), Expect = 6e-06
Identities = 30/92 (32%), Positives = 42/92 (45%), Gaps = 16/92 (17%)
Query: 59 TLGSYRATYNYFIQPVNSQIYWEPTPYEKSA--------PPKVRRAAGRPKKNRRKDGNK 110
T+ SY+ TY+ I + + W+ E PPK RR GRPKK + N
Sbjct: 512 TVSSYQQTYSQMIGEIPDRSLWKDEGEEAGGGVESLRIRPPKTRRPPGRPKKKVVRVEN- 570
Query: 111 EPIGRSQMKRTYNDTQCGRCGLIGHNLRGCNK 142
+KR QCGRC L+GH+ + C +
Sbjct: 571 -------LKRPKRIVQCGRCHLLGHSQKKCTQ 595
>UniRef100_Q6L4D3 Hypothetical protein OSJNBa0088M05.7 [Oryza sativa]
Length = 1092
Score = 48.9 bits (115), Expect = 2e-05
Identities = 25/81 (30%), Positives = 40/81 (48%), Gaps = 1/81 (1%)
Query: 62 SYRATYNYFIQPVNSQIYWEPTPYEKSAPPKVRRAAGRPKKNRRKDGNKEPIGRSQMKRT 121
++R+TY + +QP+ W E P + GRPK RR++ EP ++M +
Sbjct: 871 AFRSTYVHCLQPLEGMSAWPQDDREPLNAPGYIKMPGRPKTERRRE-KHEPPKPTKMPKY 929
Query: 122 YNDTQCGRCGLIGHNLRGCNK 142
+C RC +GHN C+K
Sbjct: 930 GTVIRCTRCKQVGHNKSSCSK 950
>UniRef100_Q60DJ3 MuDR family transposase protein [Oryza sativa]
Length = 1030
Score = 48.5 bits (114), Expect = 3e-05
Identities = 32/92 (34%), Positives = 44/92 (47%), Gaps = 5/92 (5%)
Query: 59 TLGSYRATYNYFIQPVNSQIYWEPTPYEKSAPPKVRRAAGRPKKNRRKDGNKEPIGRSQM 118
T ++ Y + I P N + WE + PP + AGRPKK+RRK E IG++
Sbjct: 804 TTDAFSKAYGFNIWPCNDKSKWENVNGPEIKPPVYEKKAGRPKKSRRK-APYEVIGKNGP 862
Query: 119 KRTYNDTQC--GRCGLIGHNLRGC--NKQGVA 146
K T + CG HN GC KQG++
Sbjct: 863 KLTKHGVMMHYKYCGEENHNSGGCKLKKQGIS 894
>UniRef100_Q5WMS8 Hypothetical protein OJ1333_C12.3 [Oryza sativa]
Length = 892
Score = 48.5 bits (114), Expect = 3e-05
Identities = 35/145 (24%), Positives = 67/145 (46%), Gaps = 16/145 (11%)
Query: 11 LDGRVGPLCPWQQSRLEKEKNASHNWTPLWSGDTPMQRYQIEHCHAWLTLG--------- 61
+D +G +CP +L S N + +G M +++++H + T+
Sbjct: 680 MDKWLGSICPNIHKKLNAYIIDSGNCHAICNG---MDKFEVKHQNHRFTVDLERKTCSCR 736
Query: 62 --SYRATYNYFIQPVNSQIYWEPTPYEKSAPPKVRRAAGRP-KKNRRKDGNKEPIGRSQM 118
S++ TY+ ++PV W + + P R GRP KK R++ +++P +++
Sbjct: 737 IYSFKKTYSCCLEPVEGMESWPTSDRPRPKAPGYVRMPGRPSKKEMRRELHEKPKA-TRV 795
Query: 119 KRTYNDTQCGRCGLIGHNLRGCNKQ 143
R + C C L+GHN C+K+
Sbjct: 796 SRVGSVMTCRLCKLLGHNKASCHKR 820
>UniRef100_Q9ZQQ5 Mutator-like transposase [Arabidopsis thaliana]
Length = 241
Score = 47.4 bits (111), Expect = 7e-05
Identities = 29/101 (28%), Positives = 42/101 (40%), Gaps = 12/101 (11%)
Query: 57 WLTLGSYRATYNYFIQPVNSQIYWEPTPYEKSAPPKVRRAAGRPKKNRR-------KDGN 109
W ++ ++ Y+ PVN W+ PP R GRPK N R +
Sbjct: 117 WYSIEKWKLCYSSLFFPVNGMELWKTHIDVVVMPPPDRIMPGRPKNNDRIRDPTEDRPPQ 176
Query: 110 KEPIGRSQMKRTYNDTQCGRCGLIGHNLRGCNKQGVARRPK 150
K P R +++ T C C IGHN R C ++ V + K
Sbjct: 177 KVPSTREKLQMT-----CSNCQQIGHNKRSCKRKAVPKPSK 212
>UniRef100_Q7XDU9 Putative mutator protein [Oryza sativa]
Length = 929
Score = 47.4 bits (111), Expect = 7e-05
Identities = 23/86 (26%), Positives = 40/86 (45%), Gaps = 1/86 (1%)
Query: 62 SYRATYNYFIQPVNSQIYWEPTPYEKSAPPKVRRAAGRPKKNRRKDGNKEPIGRSQMKRT 121
++++TY + + PV W E P + GRP+ RR++ + EP S+ +
Sbjct: 766 AFKSTYKHCLLPVEGMNAWPEDDREPLTAPGYIKMPGRPRTERRREAH-EPAKPSKASKF 824
Query: 122 YNDTQCGRCGLIGHNLRGCNKQGVAR 147
+C C +GHN C+K A+
Sbjct: 825 GTKVRCRTCKQVGHNKSSCHKHNPAQ 850
>UniRef100_Q65X18 Putative polyprotein [Oryza sativa]
Length = 1040
Score = 45.8 bits (107), Expect = 2e-04
Identities = 28/87 (32%), Positives = 40/87 (45%), Gaps = 4/87 (4%)
Query: 62 SYRATYNYFIQPVNSQIYWEPTPYEKSAPPKVRRAAGRPKKNRRKDGNKEPIGRSQMKRT 121
+Y TY I PV + W T PPK + GRPKK+RR+ ++ P + ++T
Sbjct: 913 AYMRTYTAVIYPVPDEHRWTKTDSPYIDPPKFDKHVGRPKKSRRRGPDEGPRVQGPARKT 972
Query: 122 YNDTQCGRCGLIGHNLRG--CNKQGVA 146
+ C C GH K+GVA
Sbjct: 973 --TSTCSNCRRDGHTCSNIKARKRGVA 997
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.317 0.134 0.442
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 284,896,244
Number of Sequences: 2790947
Number of extensions: 11803969
Number of successful extensions: 34556
Number of sequences better than 10.0: 130
Number of HSP's better than 10.0 without gapping: 39
Number of HSP's successfully gapped in prelim test: 91
Number of HSP's that attempted gapping in prelim test: 34444
Number of HSP's gapped (non-prelim): 144
length of query: 151
length of database: 848,049,833
effective HSP length: 127
effective length of query: 24
effective length of database: 493,599,564
effective search space: 11846389536
effective search space used: 11846389536
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 67 (30.4 bits)
Medicago: description of AC148349.4


