

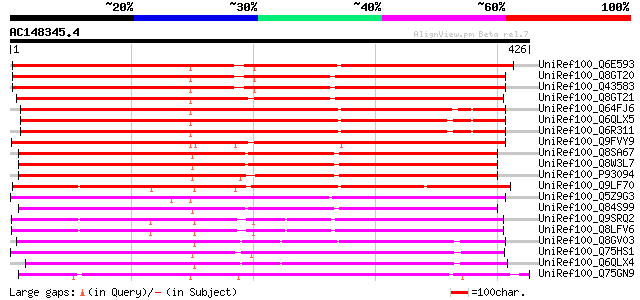
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148345.4 - phase: 0
(426 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q6E593 Benzoyl coenzyme A: benzyl alcohol benzoyl tran... 460 e-128
UniRef100_Q8GT20 Benzoyl coenzyme A: benzyl alcohol benzoyl tran... 459 e-128
UniRef100_Q43583 Hsr201 protein [Nicotiana tabacum] 456 e-127
UniRef100_Q8GT21 Benzoyl coenzyme A: benzyl alcohol benzoyl tran... 434 e-120
UniRef100_Q64FJ6 Alcohol acyl transferase [Malus domestica] 392 e-108
UniRef100_Q6QLX5 Alcohol acyl transferase [Pyrus communis] 387 e-106
UniRef100_Q6R311 Alcohol acyl transferase [Malus domestica] 384 e-105
UniRef100_Q9FVY9 Putative hypersensitivity-related (Hsr)protein ... 379 e-103
UniRef100_Q8SA67 Putative acyltransferase [Cucumis melo] 363 7e-99
UniRef100_Q8W3L7 Alcohol acetyltransferase [Cucumis melo] 362 9e-99
UniRef100_P93094 Hypothetical protein [Cucumis melo] 359 7e-98
UniRef100_Q9LF70 Hypothetical protein K10A8_20 [Arabidopsis thal... 352 9e-96
UniRef100_Q5Z9G3 Putative benzoyl coenzyme A, benzyl alcohol ben... 346 6e-94
UniRef100_Q84S99 Alcohol acyltransferase [Cucumis melo] 322 1e-86
UniRef100_Q9SRQ2 Putative hypersensitivity-related gene [Arabido... 320 4e-86
UniRef100_Q8LFV6 Putative hypersensitivity-related gene [Arabido... 319 8e-86
UniRef100_Q8GV03 Acyltransferase 2 [Capsicum chinense] 307 4e-82
UniRef100_Q75HS1 Hypothetical protein P0628H02.3 [Oryza sativa] 304 3e-81
UniRef100_Q6QLX4 Alcohol acyl transferase [Lycopersicon esculentum] 299 1e-79
UniRef100_Q75GN9 Putative hypersensitivity-related protein [Oryz... 277 4e-73
>UniRef100_Q6E593 Benzoyl coenzyme A: benzyl alcohol benzoyl transferase [Petunia
hybrida]
Length = 460
Score = 460 bits (1183), Expect = e-128
Identities = 238/452 (52%), Positives = 302/452 (66%), Gaps = 51/452 (11%)
Query: 3 SSPLLFTVRRCQPELVPPAAPTPREVKLLSDIDDQECLRFNMPIIFIYRHEPSMIEKDPA 62
SS L+FTVRR +PEL+ PA PTPRE K LSDIDDQE LRF +P+I YR + SM KDP
Sbjct: 6 SSELVFTVRRQEPELIAPAKPTPRETKFLSDIDDQEGLRFQIPVINFYRKDSSMGGKDPV 65
Query: 63 KVLKRALSQTLVYYYPLAGRIREGARDKLMVDCTGEGVMFIEAETDVTLDEFGDALHPPF 122
+V+K+A+++TLV+YYP AGR+REG KLMVDCTGEGVMF+EA DVTL+EFGD L PPF
Sbjct: 66 EVIKKAIAETLVFYYPFAGRLREGNDRKLMVDCTGEGVMFVEANADVTLEEFGDELQPPF 125
Query: 123 PCFQELIYDVPGTNQIIDHPILLIQ----------------------------------M 148
PC +EL+YDVPG+ ++ P+LLIQ M
Sbjct: 126 PCLEELLYDVPGSAGVLHCPLLLIQVTRLRCGGFIFALRLNHTMSDAPGLVQFMTAVGEM 185
Query: 149 ARGAHQPSIQPVWRREILMARDPPRVTCNHHEYEQILSSNINKEEDVAATI------VHH 202
ARGA PS PVW RE+L AR+PP+VTC HHEYE++ D T+ VH
Sbjct: 186 ARGATAPSTLPVWCRELLNARNPPQVTCTHHEYEEV--------PDTKGTLIPLDDMVHR 237
Query: 203 SFFFTLTRIAEIRRLIPFHLRQCSTFDLITACFWCCRTKALQLEPDEEVRMMCIVNARSR 262
SFFF T ++ +RR +P HL CSTF+++TA W CRT +++ +P+EEVR++CIVNARSR
Sbjct: 238 SFFFGPTEVSALRRFVPPHLHNCSTFEVLTAALWRCRTISIKPDPEEEVRVLCIVNARSR 297
Query: 263 FRADHSPLVGYYGNCFAFPAAVTTAGKLCGNPLGYGVELIRKAKAQVTEEYMHSLADLMV 322
F GYYGN FAFP AVTTA KLC NPLGY +EL++K K+ VTEEYM S+ADLMV
Sbjct: 298 FNPQLPS--GYYGNAFAFPVAVTTAEKLCKNPLGYALELVKKTKSDVTEEYMKSVADLMV 355
Query: 323 IKERCLFTTIRSCVVSDLTRAKYAEVNFGWGEAVYGGVVNCVAGPLPR-ATFIIPHKNAE 381
IK R FT +R+ +VSD+TRA + EV+FGWG+AVYGG G +P A+F IP +N +
Sbjct: 356 IKGRPHFTVVRTYLVSDVTRAGFGEVDFGWGKAVYGGPAKGGVGAIPGVASFYIPFRNKK 415
Query: 382 GEEGLILLIFLTSEVMKRFAEELDKMFGNQNQ 413
GE G+++ I L M++F +ELD M Q
Sbjct: 416 GENGIVVPICLPGFAMEKFVKELDSMLKGDAQ 447
>UniRef100_Q8GT20 Benzoyl coenzyme A: benzyl alcohol benzoyl transferase [Nicotiana
tabacum]
Length = 460
Score = 459 bits (1180), Expect = e-128
Identities = 237/447 (53%), Positives = 304/447 (67%), Gaps = 53/447 (11%)
Query: 3 SSPLLFTVRRCQPELVPPAAPTPREVKLLSDIDDQECLRFNMPIIFIYRHEPSMIEKDPA 62
SS L+FTVRR +PEL+ PA PTPRE+K LSDIDDQE LRF +P+I Y + SM KDP
Sbjct: 6 SSELVFTVRRQKPELIAPAKPTPREIKFLSDIDDQEGLRFQIPVIQFYHKDSSMGRKDPV 65
Query: 63 KVLKRALSQTLVYYYPLAGRIREGARDKLMVDCTGEGVMFIEAETDVTLDEFGDALHPPF 122
KV+K+A+++TLV+YYP AGR+REG KLMVDCTGEG+MF+EA+ DVTL++FGD L PPF
Sbjct: 66 KVIKKAIAETLVFYYPFAGRLREGNGRKLMVDCTGEGIMFVEADADVTLEQFGDELQPPF 125
Query: 123 PCFQELIYDVPGTNQIIDHPILLIQ----------------------------------M 148
PC +EL+YDVP + +++ P+LLIQ M
Sbjct: 126 PCLEELLYDVPDSAGVLNCPLLLIQVTRLRCGGFIFALRLNHTMSDAPGLVQFMTAVGEM 185
Query: 149 ARGAHQPSIQPVWRREILMARDPPRVTCNHHEYEQILSSNINKEEDVAATI------VHH 202
ARGA PSI PVW RE+L AR+PP+VTC HHEY+++ D TI VH
Sbjct: 186 ARGASAPSILPVWCRELLNARNPPQVTCTHHEYDEV--------RDTKGTIIPLDDMVHK 237
Query: 203 SFFFTLTRIAEIRRLIPFHLRQCSTFDLITACFWCCRTKALQLEPDEEVRMMCIVNARSR 262
SFFF + ++ +RR +P HLR+CSTF+L+TA W CRT +L+ +P+EEVR +CIVNARSR
Sbjct: 238 SFFFGPSEVSALRRFVPHHLRKCSTFELLTAVLWRCRTMSLKPDPEEEVRALCIVNARSR 297
Query: 263 FRADHSPL-VGYYGNCFAFPAAVTTAGKLCGNPLGYGVELIRKAKAQVTEEYMHSLADLM 321
F + PL GYYGN FAFP AVTTA KL NPLGY +EL++K K+ VTEEYM S+ADLM
Sbjct: 298 F---NPPLPTGYYGNAFAFPVAVTTAAKLSKNPLGYALELVKKTKSDVTEEYMKSVADLM 354
Query: 322 VIKERCLFTTIRSCVVSDLTRAKYAEVNFGWGEAVYGGVVNCVAGPLPR-ATFIIPHKNA 380
V+K R FT +R+ +VSD+TR + EV+FGWG+AVYGG G +P A+F IP KN
Sbjct: 355 VLKGRPHFTVVRTFLVSDVTRGGFGEVDFGWGKAVYGGPAKGGVGAIPGVASFYIPFKNK 414
Query: 381 EGEEGLILLIFLTSEVMKRFAEELDKM 407
+GE G+++ I L M+ F +ELD M
Sbjct: 415 KGENGIVVPICLPGFAMETFVKELDGM 441
>UniRef100_Q43583 Hsr201 protein [Nicotiana tabacum]
Length = 460
Score = 456 bits (1173), Expect = e-127
Identities = 236/447 (52%), Positives = 302/447 (66%), Gaps = 53/447 (11%)
Query: 3 SSPLLFTVRRCQPELVPPAAPTPREVKLLSDIDDQECLRFNMPIIFIYRHEPSMIEKDPA 62
SS L+FTVRR +PEL+ PA PTPRE K LSDIDDQE LRF +P+I Y + SM KDP
Sbjct: 6 SSELVFTVRRQKPELIAPAKPTPRETKFLSDIDDQEGLRFQIPVIQFYHKDSSMGRKDPV 65
Query: 63 KVLKRALSQTLVYYYPLAGRIREGARDKLMVDCTGEGVMFIEAETDVTLDEFGDALHPPF 122
KV+K+A+++TLV+YYP AGR+REG KLMVDCTGEG+MF+EA+ DVTL++FGD L PPF
Sbjct: 66 KVIKKAIAETLVFYYPFAGRLREGNGRKLMVDCTGEGIMFVEADADVTLEQFGDELQPPF 125
Query: 123 PCFQELIYDVPGTNQIIDHPILLIQ----------------------------------M 148
PC +EL+YDVP + +++ P+LLIQ M
Sbjct: 126 PCLEELLYDVPDSAGVLNCPLLLIQVTRLRCGGFIFALRLNHTMSDAPGLVQFMTAVGEM 185
Query: 149 ARGAHQPSIQPVWRREILMARDPPRVTCNHHEYEQILSSNINKEEDVAATI------VHH 202
ARG PSI PVW RE+L AR+PP+VTC HHEY+++ D TI VH
Sbjct: 186 ARGGSAPSILPVWCRELLNARNPPQVTCTHHEYDEV--------RDTKGTIIPLDDMVHK 237
Query: 203 SFFFTLTRIAEIRRLIPFHLRQCSTFDLITACFWCCRTKALQLEPDEEVRMMCIVNARSR 262
SFFF + ++ +RR +P HLR+CSTF+L+TA W CRT +L+ +P+EEVR +CIVNARSR
Sbjct: 238 SFFFGPSEVSALRRFVPHHLRKCSTFELLTAVLWRCRTMSLKPDPEEEVRALCIVNARSR 297
Query: 263 FRADHSPL-VGYYGNCFAFPAAVTTAGKLCGNPLGYGVELIRKAKAQVTEEYMHSLADLM 321
F + PL GYYGN FAFP AVTTA KL NPLGY +EL++K K+ VTEEYM S+ADLM
Sbjct: 298 F---NPPLPTGYYGNAFAFPVAVTTAAKLSKNPLGYALELVKKTKSDVTEEYMKSVADLM 354
Query: 322 VIKERCLFTTIRSCVVSDLTRAKYAEVNFGWGEAVYGGVVNCVAGPLPR-ATFIIPHKNA 380
V+K R FT +R+ +VSD+TR + EV+FGWG+AVYGG G +P A+F IP KN
Sbjct: 355 VLKGRPHFTVVRTFLVSDVTRGGFGEVDFGWGKAVYGGPAKGGVGAIPGVASFYIPFKNK 414
Query: 381 EGEEGLILLIFLTSEVMKRFAEELDKM 407
+GE G+++ I L M+ F +ELD M
Sbjct: 415 KGENGIVVPICLPGFAMETFVKELDGM 441
>UniRef100_Q8GT21 Benzoyl coenzyme A: benzyl alcohol benzoyl transferase [Clarkia
breweri]
Length = 456
Score = 434 bits (1115), Expect = e-120
Identities = 233/438 (53%), Positives = 294/438 (66%), Gaps = 45/438 (10%)
Query: 6 LLFTVRRCQPELVPPAAPTPREVKLLSDIDDQECLRFNMPIIFIYRHE-PSMIEKDPAKV 64
L F V R +PEL+ PA TP E K LSD++DQE LRF +P+I Y+H SM E+DP +V
Sbjct: 7 LSFEVCRRKPELIRPAKQTPHEFKKLSDVEDQEGLRFQIPVIQFYKHNNESMQERDPVQV 66
Query: 65 LKRALSQTLVYYYPLAGRIREGARDKLMVDCTGEGVMFIEAETDVTLDEFGDALHPPFPC 124
++ +++ LVYYYP AGR+RE KL+V+CTGEGVMFIEA+ DVTL++FGDAL PPFPC
Sbjct: 67 IREGIARALVYYYPFAGRLREVDGRKLVVECTGEGVMFIEADADVTLEQFGDALQPPFPC 126
Query: 125 FQELIYDVPGTNQIIDHPILLIQ----------------------------------MAR 150
F +L++DVPG+ I+D P+LLIQ MAR
Sbjct: 127 FDQLLFDVPGSGGILDSPLLLIQVTRLKCGSFIFALRLNHTMADAAGIVLFMKAVGEMAR 186
Query: 151 GAHQPSIQPVWRREILMARDPPRVTCNHHEYEQILSSNINKEEDVAATIVHHSFFFTLTR 210
GA PS PVW R IL AR PP+VT NH EYE++ + +D+A H SFFF T
Sbjct: 187 GAATPSTLPVWDRHILNARVPPQVTFNHREYEEVKGTIFTPFDDLA----HRSFFFGSTE 242
Query: 211 IAEIRRLIPFHLRQCST-FDLITACFWCCRTKALQLEPDEEVRMMCIVNARSRFRADHSP 269
I+ +R+ IP HLR CST +++TAC W CRT A++ PDEEVRM+CIVNARS+F + P
Sbjct: 243 ISAMRKQIPPHLRSCSTTIEVLTACLWRCRTLAIKPNPDEEVRMICIVNARSKF---NPP 299
Query: 270 LV-GYYGNCFAFPAAVTTAGKLCGNPLGYGVELIRKAKAQVTEEYMHSLADLMVIKERCL 328
L GYYGN FA PAAVTTAGKLC NPLG+ +ELIRKAK +VTEEYMHS+ADLMV R
Sbjct: 300 LPDGYYGNAFAIPAAVTTAGKLCNNPLGFALELIRKAKREVTEEYMHSVADLMVATGRPH 359
Query: 329 FTTIRSCVVSDLTRAKYAEVNFGWGEAVYGGVVNCVAGPLPRAT-FIIPHKNAEGEEGLI 387
FT + + +VSD+TRA + EV+FGWGEAVYGG G +P T F IP +N +GE+G++
Sbjct: 360 FTVVNTYLVSDVTRAGFGEVDFGWGEAVYGGPAKGGVGVIPGVTSFYIPLRNRQGEKGIV 419
Query: 388 LLIFLTSEVMKRFAEELD 405
L I L S M+ FAE L+
Sbjct: 420 LPICLPSAAMEIFAEALN 437
>UniRef100_Q64FJ6 Alcohol acyl transferase [Malus domestica]
Length = 455
Score = 392 bits (1008), Expect = e-108
Identities = 207/436 (47%), Positives = 279/436 (63%), Gaps = 44/436 (10%)
Query: 10 VRRCQPELVPPAAPTPREVKLLSDIDDQECLRFNMPIIFIYRHEPSMIE-KDPAKVLKRA 68
V+R QPEL+ PA TP+E K LSDIDDQE LR +PII Y+ PS+ + ++P K ++ A
Sbjct: 9 VKRLQPELITPAKSTPQETKFLSDIDDQESLRVQIPIIMCYKDNPSLNKNRNPVKAIREA 68
Query: 69 LSQTLVYYYPLAGRIREGARDKLMVDCTGEGVMFIEAETDVTLDEFGDALHPPFPCFQEL 128
LS+ LVYYYPLAGR+REG KL+VDC GEG++F+EA DVTL++ GD + PP P +E
Sbjct: 69 LSRALVYYYPLAGRLREGPNRKLVVDCNGEGILFVEASADVTLEQLGDKILPPCPLLEEF 128
Query: 129 IYDVPGTNQIIDHPILLIQ----------------------------------MARGAHQ 154
+Y+ PG++ IID P+LLIQ MARGAH
Sbjct: 129 LYNFPGSDGIIDCPLLLIQVTCLTCGGFILALRLNHTMCDAAGLLLFLTAIAEMARGAHA 188
Query: 155 PSIQPVWRREILMARDPPRVTCNHHEYEQILS-SNINKEEDVAATIVHHSFFFTLTRIAE 213
PSI PVW RE+L ARDPPR+TC HHEYE ++ S+ + + +V SF+F +
Sbjct: 189 PSILPVWERELLFARDPPRITCAHHEYEDVIGHSDGSYASSNQSNMVQRSFYFGAKEMRV 248
Query: 214 IRRLIPFHL-RQCSTFDLITACFWCCRTKALQLEPDEEVRMMCIVNARSRFRADHSPLVG 272
+R+ IP HL CSTFDLITAC W CRT AL + P E VR+ CIVNAR + PL G
Sbjct: 249 LRKQIPPHLISTCSTFDLITACLWKCRTLALNINPKEAVRVSCIVNARGKHNNVRLPL-G 307
Query: 273 YYGNCFAFPAAVTTAGKLCGNPLGYGVELIRKAKAQVTEEYMHSLADLMVIKERCLFTTI 332
YYGN FAFPAA++ A LC NPLGY +EL++KAKA + EEY+ S+ADL+V++ R +++
Sbjct: 308 YYGNAFAFPAAISKAEPLCKNPLGYALELVKKAKATMNEEYLRSVADLLVLRGRPQYSST 367
Query: 333 RS-CVVSDLTRAKYAEVNFGWGEAVYGGVVNCVAGPLPRATFIIPHKNAEGEEGLILLIF 391
S +VSD TR + +VNFGWG+ V+ G V L +F + HKN E+G+++ +
Sbjct: 368 GSYLIVSDNTRVGFGDVNFGWGQPVFAGPVKA----LDLISFYVQHKN-NTEDGILVPMC 422
Query: 392 LTSEVMKRFAEELDKM 407
L S M+RF +EL+++
Sbjct: 423 LPSSAMERFQQELERI 438
>UniRef100_Q6QLX5 Alcohol acyl transferase [Pyrus communis]
Length = 442
Score = 387 bits (994), Expect = e-106
Identities = 205/436 (47%), Positives = 281/436 (64%), Gaps = 44/436 (10%)
Query: 10 VRRCQPELVPPAAPTPREVKLLSDIDDQECLRFNMPIIFIYRHEPSMIE-KDPAKVLKRA 68
V+R QPEL+ PA PTP+E K LSDIDDQE LRF +P+I Y+ PS+ + ++P KV+K A
Sbjct: 9 VKRLQPELITPAKPTPQETKFLSDIDDQEGLRFQLPVIMCYKDNPSLNKNRNPIKVIKEA 68
Query: 69 LSQTLVYYYPLAGRIREGARDKLMVDCTGEGVMFIEAETDVTLDEFGDALHPPFPCFQEL 128
LS+ LVYYYPLAGR+REG KLMV+C GEG++F+EA DVTL++ GD + PP P +E
Sbjct: 69 LSRALVYYYPLAGRLREGPNRKLMVNCNGEGILFVEASADVTLEQLGDKILPPCPLLEEF 128
Query: 129 IYDVPGTNQIIDHPILLIQ----------------------------------MARGAHQ 154
+++ PG++ II P+LL+Q M RGA
Sbjct: 129 LFNFPGSDGIIGCPLLLVQVTCLTCGGFILALRLNHTMCDATGLLMFLTAITEMGRGADA 188
Query: 155 PSIQPVWRREILMARDPPRVTCNHHEYEQILS-SNINKEEDVAATIVHHSFFFTLTRIAE 213
PSI PVW RE+L ARDPPR+TC H+EYE ++ S+ + + +V SF+F +
Sbjct: 189 PSILPVWERELLFARDPPRITCAHYEYEDVIDHSDGSYAFSNQSNMVQRSFYFGAKEMRV 248
Query: 214 IRRLIPFHL-RQCSTFDLITACFWCCRTKALQLEPDEEVRMMCIVNARSRFRADHSPLVG 272
+R+ IP HL CSTFDLITAC W CRT L++ P + VR+ CIVNAR + H PL G
Sbjct: 249 LRKQIPPHLISTCSTFDLITACLWKCRTLVLKINPKQAVRVSCIVNARGKHNNVHIPL-G 307
Query: 273 YYGNCFAFPAAVTTAGKLCGNPLGYGVELIRKAKAQVTEEYMHSLADLMVIKERCLFTTI 332
YYGN FAFPAAV+ A LC NPLGY +EL++KAKA + EEY+ S+ADL+V++ R +++
Sbjct: 308 YYGNAFAFPAAVSKAEPLCKNPLGYALELVKKAKATMNEEYLRSVADLLVLRGRPQYSST 367
Query: 333 RS-CVVSDLTRAKYAEVNFGWGEAVYGGVVNCVAGPLPRATFIIPHKNAEGEEGLILLIF 391
S +VSD TRA + +VNFGWG+ V+ G A L +F + HKN E+G+++ +
Sbjct: 368 GSYLIVSDNTRAGFGDVNFGWGQPVFAG----PAKALDLISFYVQHKN-NIEDGILVPMC 422
Query: 392 LTSEVMKRFAEELDKM 407
L S M+RF +EL+++
Sbjct: 423 LPSSAMERFQQELERI 438
>UniRef100_Q6R311 Alcohol acyl transferase [Malus domestica]
Length = 459
Score = 384 bits (985), Expect = e-105
Identities = 204/436 (46%), Positives = 278/436 (62%), Gaps = 44/436 (10%)
Query: 10 VRRCQPELVPPAAPTPREVKLLSDIDDQECLRFNMPIIFIYRHEPSMIEK-DPAKVLKRA 68
V+R Q EL+ PA PT +E K LSDIDDQE LRF +P+I Y+ PS+ + +P KV++ A
Sbjct: 9 VKRLQLELITPAKPTLQEAKFLSDIDDQEGLRFQVPVIMCYKDNPSLNKNCNPVKVIREA 68
Query: 69 LSQTLVYYYPLAGRIREGARDKLMVDCTGEGVMFIEAETDVTLDEFGDALHPPFPCFQEL 128
LS+ LVYYYPLAGR++EG KLMVDC GEG++F+EA DVTL++ GD + PP P +E
Sbjct: 69 LSRALVYYYPLAGRLKEGPNRKLMVDCNGEGILFVEASADVTLEQLGDKILPPCPLLEEF 128
Query: 129 IYDVPGTNQIIDHPILLIQ----------------------------------MARGAHQ 154
+++ PG++ II P+LL+Q MARGAH
Sbjct: 129 LFNFPGSDGIIGCPLLLVQVTCLTCGGFILALRVNHTMCDAPGLLLFLTAIAEMARGAHA 188
Query: 155 PSIQPVWRREILMARDPPRVTCNHHEYEQILS-SNINKEEDVAATIVHHSFFFTLTRIAE 213
PSI PVW RE+L +RDPPR+TC HHEYE ++ S+ + +V SF+F +
Sbjct: 189 PSILPVWERELLFSRDPPRITCAHHEYEDVIDHSDGLYASSNQSNMVQRSFYFGAKEMRV 248
Query: 214 IRRLIPFHL-RQCSTFDLITACFWCCRTKALQLEPDEEVRMMCIVNARSRFRADHSPLVG 272
+R+ IP HL CSTFDLITAC W CRT AL + P E VR+ CIVNAR + PL G
Sbjct: 249 LRKQIPPHLISTCSTFDLITACLWKCRTLALNINPKEAVRVSCIVNARGKHNNVRLPL-G 307
Query: 273 YYGNCFAFPAAVTTAGKLCGNPLGYGVELIRKAKAQVTEEYMHSLADLMVIKERCLFTTI 332
YYGN FAFPAA++ A LC NPLGY +EL++KAKA + EEY+ S+ADL+V++ R +++
Sbjct: 308 YYGNAFAFPAAISKAEPLCKNPLGYALELVKKAKATMNEEYLRSVADLLVLRGRPQYSST 367
Query: 333 RS-CVVSDLTRAKYAEVNFGWGEAVYGGVVNCVAGPLPRATFIIPHKNAEGEEGLILLIF 391
S +VSD TRA + +VNFGWG+ V+ G A L +F + HKN E+G+++ +
Sbjct: 368 GSYLIVSDNTRAGFGDVNFGWGQPVFAG----PAKALDLISFYVQHKN-NTEDGILVPMC 422
Query: 392 LTSEVMKRFAEELDKM 407
L S M+RF +EL+++
Sbjct: 423 LPSSAMERFQQELERI 438
>UniRef100_Q9FVY9 Putative hypersensitivity-related (Hsr)protein [Oryza sativa]
Length = 553
Score = 379 bits (972), Expect = e-103
Identities = 206/449 (45%), Positives = 281/449 (61%), Gaps = 47/449 (10%)
Query: 2 TSSPLLFTVRRCQPELVPPAAPTPREVKLLSDIDDQECLRFNMPIIFIYRHEPSMIEKDP 61
T++ L FTVRR ELV PA PTPRE+K LSDIDDQ+ LRF++P+I YR +M +DP
Sbjct: 5 TAAALKFTVRRKPAELVAPAGPTPRELKKLSDIDDQDGLRFHIPVIQFYRRSAAMGGRDP 64
Query: 62 AKVLKRALSQTLVYYYPLAGRIREGARDKLMVDCTGEGVMFIEAETDVTLDEFGDALHPP 121
A V++ A+++ LV YYP AGR+RE KL VDCTGEGV+FIEA+ DV L+ FG AL PP
Sbjct: 65 APVIRAAVARALVSYYPFAGRLRELEGRKLAVDCTGEGVLFIEADADVRLEHFGGALQPP 124
Query: 122 FPCFQELIYDVPGTNQIIDHPILLIQ---------------------------------- 147
FPC +EL++DVPG+++++ P+LL Q
Sbjct: 125 FPCLEELVFDVPGSSEVLGSPLLLFQVTRLACGGFILAVRLHHTMADAQGLVQFLGAVAE 184
Query: 148 MARG--AHQPSIQPVWRREILMARDPPRVTCNHHEYEQI--LSSNINKEEDVAATIVHHS 203
MARG A PS+ PVW RE+L AR PPR H EY+++ I +D+A H S
Sbjct: 185 MARGGAAAAPSVAPVWGREMLEARSPPRPAFAHREYDEVPDTKGTIIPLDDMA----HRS 240
Query: 204 FFFTLTRIAEIR-RLIPFHLRQCSTFDLITACFWCCRTKALQLEPDEEVRMMCIVNARSR 262
FFF +A +R L P + +TF+++T C W CRT AL + DE +RM+CIVNAR
Sbjct: 241 FFFGAREVAAVRSHLAPGIRERATTFEVLTGCLWRCRTAALAPDDDEVMRMICIVNARGG 300
Query: 263 FRADHSPLV---GYYGNCFAFPAAVTTAGKLCGNPLGYGVELIRKAKAQVTEEYMHSLAD 319
++ + GYYGN FAFP AV TAG+L PLGY VEL+R AK +V+ EYM S+AD
Sbjct: 301 GKSGGGAGMIPEGYYGNAFAFPVAVATAGELRARPLGYAVELVRAAKGEVSVEYMRSVAD 360
Query: 320 LMVIKERCLFTTIRSCVVSDLTRAKYAEVNFGWGEAVYGGVVNCVAGPLPR-ATFIIPHK 378
LMV + R FT +R+ +VSD+T+A + +++FGWG+ YGG G +P A+F+IP K
Sbjct: 361 LMVQRGRPHFTVVRAYLVSDVTKAGFGDLDFGWGKPAYGGPAKGGVGAIPGVASFLIPFK 420
Query: 379 NAEGEEGLILLIFLTSEVMKRFAEELDKM 407
NA+GE+G+++ + L M +F EE+ K+
Sbjct: 421 NAKGEDGIVVPMCLPGPAMDKFVEEMGKL 449
>UniRef100_Q8SA67 Putative acyltransferase [Cucumis melo]
Length = 461
Score = 363 bits (931), Expect = 7e-99
Identities = 186/429 (43%), Positives = 269/429 (62%), Gaps = 42/429 (9%)
Query: 8 FTVRRCQPELVPPAAPTPREVKLLSDIDDQECLRFNMPIIFIYRHEPSMIEKDPAKVLKR 67
F VR+CQPEL+ PA PTP E K LSD+DDQ+ LRF +P++ IY H PS+ +DP KV+K
Sbjct: 11 FQVRKCQPELIAPANPTPYEFKQLSDVDDQQSLRFQLPLVNIYHHNPSLEGRDPVKVIKE 70
Query: 68 ALSQTLVYYYPLAGRIREGARDKLMVDCTGEGVMFIEAETDVTLDEFGDALHPPFPCFQE 127
A+++ LV+YYPLAGR+REG KL V+CTGEG++FIEA+ DV+L++F D L +
Sbjct: 71 AIAKALVFYYPLAGRLREGPGRKLFVECTGEGILFIEADADVSLEQFRDTLPYSLSSMEN 130
Query: 128 -LIYDVPGTNQIIDHPILLIQM----------------------------------ARGA 152
+I++ ++ +++ P+LLIQ+ ARGA
Sbjct: 131 NIIHNSLNSDGVLNSPLLLIQVTRLKCGGFIFGIHFDHTMADGFGIAQFMKAIAEIARGA 190
Query: 153 HQPSIQPVWRREILMARDPPRVTCNHHEYEQILSSNINKEEDVAATIVHHSFFFTLTRIA 212
PSI PVW+R +L ARDPPR+T H+EY+Q++ + A ++ FFFT +I+
Sbjct: 191 FAPSILPVWQRALLTARDPPRITVRHYEYDQVVDTKSTLIP--ANNMIDRLFFFTQRQIS 248
Query: 213 EIRRLIPFHLRQCSTFDLITACFWCCRTKALQLEPDEEVRMMCIVNARSRFRADHSPLVG 272
+R+ +P HL CS+F+++ A W RT A QL+P+EEVR +C+VN RS+ +G
Sbjct: 249 TLRQTLPAHLHDCSSFEVLAAYVWRLRTIAFQLKPEEEVRFLCVVNLRSKIDIP----LG 304
Query: 273 YYGNCFAFPAAVTTAGKLCGNPLGYGVELIRKAKAQVTEEYMHSLADLMVIKERCLFTTI 332
+YGN FPA +TT KLCGNPLGY V+LIRKAKA+ T+EY+ S+ D MVIK R FT I
Sbjct: 305 FYGNAIVFPAVITTVAKLCGNPLGYAVDLIRKAKAKATKEYIKSMVDFMVIKGRPRFTEI 364
Query: 333 RSCVVSDLTRAKYAEVNFGWGEAVYGGVVNCVAGPL-PRATFIIPHKNAEGEEGLILLIF 391
++SD+TR + V+FGWG+A++GG + G + ++ I N GE+G+++ +
Sbjct: 365 GPFMMSDITRIGFENVDFGWGKAIFGGPIIGGCGIIRGMISYSIAFMNRNGEKGIVVPLC 424
Query: 392 LTSEVMKRF 400
L M+RF
Sbjct: 425 LPPPAMERF 433
>UniRef100_Q8W3L7 Alcohol acetyltransferase [Cucumis melo]
Length = 461
Score = 362 bits (930), Expect = 9e-99
Identities = 187/429 (43%), Positives = 267/429 (61%), Gaps = 42/429 (9%)
Query: 8 FTVRRCQPELVPPAAPTPREVKLLSDIDDQECLRFNMPIIFIYRHEPSMIEKDPAKVLKR 67
F VR+CQPEL+ PA PTP E K LSD+DDQ+ LR +P + IY H PS+ +DP KV+K
Sbjct: 11 FHVRKCQPELIAPANPTPYEFKQLSDVDDQQSLRLQLPFVNIYPHNPSLEGRDPVKVIKE 70
Query: 68 ALSQTLVYYYPLAGRIREGARDKLMVDCTGEGVMFIEAETDVTLDEFGDALHPPFPCFQE 127
A+ + LV+YYPLAGR+REG KL V+CTGEG++FIEA+ DV+L+EF D L Q
Sbjct: 71 AIGKALVFYYPLAGRLREGPGRKLFVECTGEGILFIEADADVSLEEFWDTLPYSLSSMQN 130
Query: 128 -LIYDVPGTNQIIDHPILLIQM----------------------------------ARGA 152
+I++ ++++++ P+LLIQ+ ARGA
Sbjct: 131 NIIHNALNSDEVLNSPLLLIQVTRLKCGGFIFGLCFNHTMADGFGIVQFMKATAEIARGA 190
Query: 153 HQPSIQPVWRREILMARDPPRVTCNHHEYEQILSSNINKEEDVAATIVHHSFFFTLTRIA 212
PSI PVW+R +L ARDPPR+T H+EY+Q++ + A ++ FFFT +I+
Sbjct: 191 FAPSILPVWQRALLTARDPPRITVRHYEYDQVVDTKSTLIP--ANNMIDRLFFFTQRQIS 248
Query: 213 EIRRLIPFHLRQCSTFDLITACFWCCRTKALQLEPDEEVRMMCIVNARSRFRADHSPLVG 272
+R+ +P HL CS+F+++ A W RT A QL+P+EEVR +C+VN RS+ +G
Sbjct: 249 TLRQTLPAHLHDCSSFEVLAAYVWRLRTIAFQLKPEEEVRFLCVVNLRSKIDIP----LG 304
Query: 273 YYGNCFAFPAAVTTAGKLCGNPLGYGVELIRKAKAQVTEEYMHSLADLMVIKERCLFTTI 332
+YGN FPA +TT KLCGNPLGY V+LIRKAKA+ T+EY+ S+ D MVIK R FT I
Sbjct: 305 FYGNAIVFPAVITTIAKLCGNPLGYAVDLIRKAKAKATKEYIKSMVDFMVIKGRPRFTEI 364
Query: 333 RSCVVSDLTRAKYAEVNFGWGEAVYGGVVNCVAGPL-PRATFIIPHKNAEGEEGLILLIF 391
++SD+TR + V+FGWG+A++GG + G + ++ I N GE+G+++ +
Sbjct: 365 GPFMMSDITRIGFENVDFGWGKAIFGGPIIGGCGIIRGMISYSIAFMNRNGEKGIVVPLC 424
Query: 392 LTSEVMKRF 400
L M+RF
Sbjct: 425 LPPPAMERF 433
>UniRef100_P93094 Hypothetical protein [Cucumis melo]
Length = 455
Score = 359 bits (922), Expect = 7e-98
Identities = 191/434 (44%), Positives = 263/434 (60%), Gaps = 52/434 (11%)
Query: 8 FTVRRCQPELVPPAAPTPREVKLLSDIDDQECLRFNMPIIFIYRHEPSMIEKDPAKVLKR 67
F VR+CQPEL+ PA PTP E K LSD+DDQ+ LR +P + IY H PS+ +DP KV+K
Sbjct: 4 FHVRKCQPELIAPANPTPYEFKQLSDVDDQQSLRLQLPFVNIYPHNPSLEGRDPVKVIKE 63
Query: 68 ALSQTLVYYYPLAGRIREGARDKLMVDCTGEGVMFIEAETDVTLDEFGDALHPPFPCFQE 127
A+ + LV+YYPLAGR+REG KL V+CTGEG++FIEA+ DV+L+EF D L Q
Sbjct: 64 AIGKALVFYYPLAGRLREGPGRKLFVECTGEGILFIEADADVSLEEFWDTLPYSLSSMQN 123
Query: 128 -LIYDVPGTNQIIDHPILLIQM----------------------------------ARGA 152
+I++ ++++++ P+LLIQ+ ARGA
Sbjct: 124 NIIHNALNSDEVLNSPLLLIQVTRLKCGGFIFGLCFNHTMADGFGIVQFMKATAEIARGA 183
Query: 153 HQPSIQPVWRREILMARDPPRVTCNHHEYEQILSSN-----INKEEDVAATIVHHSFFFT 207
PSI PVW+R +L ARDPPR+T H+EY+Q++ +N + D FFF+
Sbjct: 184 FAPSILPVWQRALLTARDPPRITFRHYEYDQVVDMKSGLIPVNSKID-------QLFFFS 236
Query: 208 LTRIAEIRRLIPFHLRQCSTFDLITACFWCCRTKALQLEPDEEVRMMCIVNARSRFRADH 267
+I+ +R+ +P HL C +F+++TA W RT ALQ +P+EEVR +C++N RS+
Sbjct: 237 QLQISTLRQTLPAHLHDCPSFEVLTAYVWRLRTIALQFKPEEEVRFLCVMNLRSKIDIP- 295
Query: 268 SPLVGYYGNCFAFPAAVTTAGKLCGNPLGYGVELIRKAKAQVTEEYMHSLADLMVIKERC 327
+GYYGN PA +TTA KLCGNPLGY V+LIRKAKA+ T EY+ S DLMVIK R
Sbjct: 296 ---LGYYGNAVVVPAVITTAAKLCGNPLGYAVDLIRKAKAKATMEYIKSTVDLMVIKGRP 352
Query: 328 LFTTIRSCVVSDLTRAKYAEVNFGWGEAVYGGVVNCVAG-PLPRATFIIPHKNAEGEEGL 386
FT + S ++SDLTR V+FGWG+A++GG A +F +P N GE+G
Sbjct: 353 YFTVVGSFMMSDLTRIGVENVDFGWGKAIFGGPTTTGARITRGLVSFCVPFMNRNGEKGT 412
Query: 387 ILLIFLTSEVMKRF 400
L + L M+RF
Sbjct: 413 ALSLCLPPPAMERF 426
>UniRef100_Q9LF70 Hypothetical protein K10A8_20 [Arabidopsis thaliana]
Length = 461
Score = 352 bits (904), Expect = 9e-96
Identities = 199/449 (44%), Positives = 280/449 (62%), Gaps = 47/449 (10%)
Query: 3 SSPLLFTVRRCQPELVPPAAPTPREVKLLSDIDDQECLRFNMPIIFIYRHEPSMIEKDPA 62
S L F + R +PELV PA PTPRE+K LSDIDDQE LRF++P IF YRH P+ DP
Sbjct: 2 SGSLTFKIYRQKPELVSPAKPTPRELKPLSDIDDQEGLRFHIPTIFFYRHNPTT-NSDPV 60
Query: 63 KVLKRALSQTLVYYYPLAGRIREGARDKLMVDCTGEGVMFIEAETDVTLDEFG--DALHP 120
V++RAL++TLVYYYP AGR+REG KL VDCTGEGV+FIEA+ DVTL EF DAL P
Sbjct: 61 AVIRRALAETLVYYYPFAGRLREGPNRKLAVDCTGEGVLFIEADADVTLVEFEEKDALKP 120
Query: 121 PFPCFQELIYDVPGTNQIIDHPILLIQMAR------------------------------ 150
PFPCF+EL+++V G+ ++++ P++L+Q+ R
Sbjct: 121 PFPCFEELLFNVEGSCEMLNTPLMLMQVTRLKCGGFIFAVRINHAMSDAGGLTLFLKTMC 180
Query: 151 ----GAHQPSIQPVWRREILMARDPPRVTCNHHEYEQI--LSSNINKEEDVAATIVHHSF 204
G H P++ PVW R +L AR RVT H EY+++ + + + D +V S
Sbjct: 181 EFVRGYHAPTVAPVWERHLLSARVLLRVTHAHREYDEMPAIGTELGSRRD---NLVGRSL 237
Query: 205 FFTLTRIAEIRRLIPFHLRQCST-FDLITACFWCCRTKALQLEPDEEVRMMCIVNARSRF 263
FF ++ IRRL+P +L ST +++T+ W RT AL+ + D+E+R++ IVNARS+
Sbjct: 238 FFGPCEMSAIRRLLPPNLVNSSTNMEMLTSFLWRYRTIALRPDQDKEMRLILIVNARSKL 297
Query: 264 RADHSPLVGYYGNCFAFPAAVTTAGKLCGNPLGYGVELIRKAKAQVTEEYMHSLADLMVI 323
+ P GYYGN FAFP A+ TA +L PL + + LI++AK+ VTEEYM SLADLMVI
Sbjct: 298 KNPPLPR-GYYGNAFAFPVAIATANELTKKPLEFALRLIKEAKSSVTEEYMRSLADLMVI 356
Query: 324 KERCLFTTIRSCVVSDLTRAKYAEVNFG-WGEAVYGGVVNCVAGPLPRATFIIPHKNAEG 382
K R F++ + +VSD+ +A+++FG WG+ VYGG+ LP A+F + + G
Sbjct: 357 KGRPSFSSDGAYLVSDV--RIFADIDFGIWGKPVYGGIGTAGVEDLPGASFYVSFEKRNG 414
Query: 383 EEGLILLIFLTSEVMKRFAEELDKMFGNQ 411
E G+++ + L + M+RF EEL+ +F Q
Sbjct: 415 EIGIVVPVCLPEKAMQRFVEELEGVFNGQ 443
>UniRef100_Q5Z9G3 Putative benzoyl coenzyme A, benzyl alcohol benzoyl transferase
[Oryza sativa]
Length = 450
Score = 346 bits (888), Expect = 6e-94
Identities = 200/447 (44%), Positives = 260/447 (57%), Gaps = 42/447 (9%)
Query: 1 MTSSPL-LFTVRRCQPELVPPAAPTPREVKLLSDIDDQECLRFNMPIIFIYRHEPSMIEK 59
M SSPL FTVRR +P LV PAAPTPREVK LSDIDD E +RF I +YR+ P+ +
Sbjct: 1 MASSPLPAFTVRRGEPVLVTPAAPTPREVKALSDIDDGEGMRFYSSGIHLYRNNPAKKGQ 60
Query: 60 DPAKVLKRALSQTLVYYYPLAGRIREGARDKLMVDCTGEGVMFIEAETDVTLDEFGDALH 119
DPA V++ AL++ LV YYPLAGR+RE A KL+V+C G+GVMF EA+ D+T D+FGD
Sbjct: 61 DPAMVIREALARALVPYYPLAGRLREEAGRKLVVECAGQGVMFAEADADLTADDFGDVQS 120
Query: 120 PPFPCFQELIYD---VPGTNQIIDHPILLIQ----------------------------- 147
PPFPCF+ I + V G ++ P+L IQ
Sbjct: 121 PPFPCFERFILESTTVAGVEPVVGRPLLYIQVTRLRCGGFIFGQRFCHCVVDAPGGMQFE 180
Query: 148 -----MARGAHQPSIQPVWRREILMARDPPRVTCNHHEYEQILSSNINKEEDVAATIVHH 202
+ARGA PS+ P W RE+ MARDPPR + H EY + +V
Sbjct: 181 KAVCELARGAAAPSVSPSWGREMFMARDPPRPSYPHLEYREPAGGADRLLATPPEDMVRV 240
Query: 203 SFFFTLTRIAEIRRLIPFHLR-QCSTFDLITACFWCCRTKALQLEPDEEVRMMCIVNARS 261
FFF IA +R+ P +R CS F+L+ AC W RT AL P EEVR+ IVNAR
Sbjct: 241 PFFFGPREIAGLRQHAPASVRGACSRFELVAACIWRSRTAALGYAPGEEVRLSFIVNARG 300
Query: 262 RFRADHSPLVGYYGNCFAFPAAVTTAGKLCGNPLGYGVELIRKAKAQVTEEYMHSLADLM 321
R AD G+YGN FA+ A TTAG+LCG LGY + L++KAK+ VT EY+ S+ADLM
Sbjct: 301 R--ADVPLPEGFYGNAFAYSVAATTAGELCGGDLGYALGLVKKAKSAVTYEYLQSVADLM 358
Query: 322 VIKERCLFTTIRSCVVSDLTRAKYAEVNFGWGEAVYGGVVNCVAGPLPRAT-FIIPHKNA 380
V+ R LF R+ +VSD++ A + V+FGWGEAVYGG GPL T + KN
Sbjct: 359 VVAGRPLFALSRTYIVSDVSHAGFKSVDFGWGEAVYGGPAKGGEGPLLGVTNYFSRSKNG 418
Query: 381 EGEEGLILLIFLTSEVMKRFAEELDKM 407
+GE+ +++ I L + M +F E+ +
Sbjct: 419 KGEQSVVVPICLPKDAMDKFQLEVQAL 445
>UniRef100_Q84S99 Alcohol acyltransferase [Cucumis melo]
Length = 461
Score = 322 bits (825), Expect = 1e-86
Identities = 171/429 (39%), Positives = 250/429 (57%), Gaps = 42/429 (9%)
Query: 8 FTVRRCQPELVPPAAPTPREVKLLSDIDDQECLRFNMPIIFIYRHEPSMIEKDPAKVLKR 67
F VR+CQPEL+ PA PTP E K LSD+DDQ+ LR +P + IY H PS+ +DP KV+K
Sbjct: 11 FHVRKCQPELIAPANPTPYEFKQLSDVDDQQSLRLQLPFVNIYPHNPSLEGRDPVKVIKE 70
Query: 68 ALSQTLVYYYPLAGRIREGARDKLMVDCTGEGVMFIEAETDVTLDEFGDALHPPFPCFQE 127
A+ + LV YYPLAGR+REG KL V+CTGEG++FIEA+ DV+L++F D L +
Sbjct: 71 AIGKALVLYYPLAGRLREGPGRKLFVECTGEGILFIEADADVSLEQFRDTLPYSLSSMEN 130
Query: 128 -LIYDVPGTNQIIDHPILLIQM----------------------------------ARGA 152
+I++ ++ +++ +LLIQ+ ARGA
Sbjct: 131 NIIHNALNSDGVLNSQLLLIQVTRLKCGGFIFGIRFNHTMADGFGIAQFMKAIAEIARGA 190
Query: 153 HQPSIQPVWRREILMARDPPRVTCNHHEYEQILSSNINKEEDVAATIVHHSFFFTLTRIA 212
PSI PVW+R +L AR +T H+EY+Q++ + A ++ FFF+ +I+
Sbjct: 191 FAPSILPVWQRALLTARYLSEITVRHYEYDQVVDTKSTLIP--ANNMIDRLFFFSQLQIS 248
Query: 213 EIRRLIPFHLRQCSTFDLITACFWCCRTKALQLEPDEEVRMMCIVNARSRFRADHSPLVG 272
+R+ +P H CS+F+++ C W RT QL+P+E+VR +C+VN RS+ +G
Sbjct: 249 TLRQTLPAHRHDCSSFEVLADCVWRLRTIGFQLKPEEDVRFLCVVNLRSKIDIP----LG 304
Query: 273 YYGNCFAFPAAVTTAGKLCGNPLGYGVELIRKAKAQVTEEYMHSLADLMVIKERCLFTTI 332
YYGN FPA +TTA KLCGNPLGY V+LIRKAKA+ T + D MVIK R FT +
Sbjct: 305 YYGNAVVFPAVITTAAKLCGNPLGYAVDLIRKAKAKATTHLTSPMVDFMVIKGRPRFTEL 364
Query: 333 RSCVVSDLTRAKYAEVNFGWGEAVYGGVVNCVAGPL-PRATFIIPHKNAEGEEGLILLIF 391
++SD+TR + V+FGWG+A++GG + G + ++ I N + L++ +
Sbjct: 365 MRFMMSDITRIGFENVDFGWGKAIFGGPIIGGCGIIRGMISYSIAFMNRHNKNSLVVPLC 424
Query: 392 LTSEVMKRF 400
L M+ F
Sbjct: 425 LPPPDMESF 433
>UniRef100_Q9SRQ2 Putative hypersensitivity-related gene [Arabidopsis thaliana]
Length = 454
Score = 320 bits (821), Expect = 4e-86
Identities = 186/446 (41%), Positives = 258/446 (57%), Gaps = 52/446 (11%)
Query: 2 TSSPLLFTVRRCQPELVPPAAPTPREVKLLSDIDDQECLRFNMPIIFIYRHEPSMIEKDP 61
T++ L F V R Q ELV PA PTPRE+K LSDIDDQ+ LRF +P+IF YR S + DP
Sbjct: 11 TTTGLSFKVHRQQRELVTPAKPTPRELKPLSDIDDQQGLRFQIPVIFFYRPNLSS-DLDP 69
Query: 62 AKVLKRALSQTLVYYYPLAGRIREGARDKLMVDCTGEGVMFIEAETDVTLDEF--GDALH 119
+V+K+AL+ LVYYYP AGR+RE + KL VDCTGEGV+FIEAE DV L E DAL
Sbjct: 70 VQVIKKALADALVYYYPFAGRLRELSNRKLAVDCTGEGVLFIEAEADVALAELEEADALL 129
Query: 120 PPFPCFQELIYDVPGTNQIIDHPILLIQMAR----------------------------- 150
PPFP +EL++DV G++ +++ P+LL+Q+ R
Sbjct: 130 PPFPFLEELLFDVEGSSDVLNTPLLLVQVTRLKCCGFIFALRFNHTMTDGAGLSLFLKSL 189
Query: 151 -----GAHQPSIQPVWRREIL-MARDPPRVTCNHHEYEQILSSNINKEEDVAAT---IVH 201
G H PS+ PVW R +L ++ RVT H EY+ + DV AT +V
Sbjct: 190 CELACGLHAPSVPPVWNRHLLTVSASEARVTHTHREYDDQVGI------DVVATGHPLVS 243
Query: 202 HSFFFTLTRIAEIRRLIPFHLRQCSTFDLITACFWCCRTKALQLEPDEEVRMMCIVNARS 261
SFFF I+ IR+L+P L S F+ +++ W CRT AL +P+ E+R+ CI+N+RS
Sbjct: 244 RSFFFRAEEISAIRKLLPPDLHNTS-FEALSSFLWRCRTIALNPDPNTEMRLTCIINSRS 302
Query: 262 RFRADHSPLV-GYYGNCFAFPAAVTTAGKLCGNPLGYGVELIRKAKAQVTEEYMHSLADL 320
+ R + PL GYYGN F PAA+ TA L PL + + LI++ K+ VTE+Y+ S+ L
Sbjct: 303 KLR--NPPLEPGYYGNVFVIPAAIATARDLIEKPLEFALRLIQETKSSVTEDYVRSVTAL 360
Query: 321 MVIKERCLFTTIRSCVVSDLTRAKYAEVNFG-WGEAVYGGVVNCVAGPLPRATFIIPHKN 379
M + R +F + ++SDL +++FG WG+ VYGG P +F +P KN
Sbjct: 361 MATRGRPMFVASGNYIISDLRHFDLGKIDFGPWGKPVYGGTAKAGIALFPGVSFYVPFKN 420
Query: 380 AEGEEGLILLIFLTSEVMKRFAEELD 405
+GE G ++ I L M+ F EL+
Sbjct: 421 KKGETGTVVAISLPVRAMETFVAELN 446
>UniRef100_Q8LFV6 Putative hypersensitivity-related gene [Arabidopsis thaliana]
Length = 454
Score = 319 bits (818), Expect = 8e-86
Identities = 186/446 (41%), Positives = 258/446 (57%), Gaps = 52/446 (11%)
Query: 2 TSSPLLFTVRRCQPELVPPAAPTPREVKLLSDIDDQECLRFNMPIIFIYRHEPSMIEKDP 61
T++ L F V R Q ELV PA PTPRE+K LSDIDDQ+ LRF +P+IF YR S + DP
Sbjct: 11 TTTGLSFKVHRQQRELVTPAKPTPRELKPLSDIDDQQGLRFQIPVIFFYRPNLSS-DLDP 69
Query: 62 AKVLKRALSQTLVYYYPLAGRIREGARDKLMVDCTGEGVMFIEAETDVTLDEF--GDALH 119
+V+K+AL+ LVYYYP AGR+RE + KL VDCTGEGV+FIEAE DV L E DAL
Sbjct: 70 VQVIKKALADALVYYYPFAGRLRELSNRKLAVDCTGEGVLFIEAEADVALAELEEADALL 129
Query: 120 PPFPCFQELIYDVPGTNQIIDHPILLIQMAR----------------------------- 150
PPFP +EL++DV G++ +++ P+LL+Q+ R
Sbjct: 130 PPFPFLEELLFDVEGSSDVLNTPLLLVQVTRLKCCGFIFALRFNHTMTDGAGLSLFLKSL 189
Query: 151 -----GAHQPSIQPVWRREIL-MARDPPRVTCNHHEYEQILSSNINKEEDVAAT---IVH 201
G H PS+ PVW R +L ++ RVT H EY+ + DV AT +V
Sbjct: 190 CELACGLHAPSVPPVWNRHLLTVSASEARVTHTHREYDDQVGI------DVVATGHPLVS 243
Query: 202 HSFFFTLTRIAEIRRLIPFHLRQCSTFDLITACFWCCRTKALQLEPDEEVRMMCIVNARS 261
SFFF I+ IR+L+P L S F+ +++ W CRT AL +P+ E+R+ CI+N+RS
Sbjct: 244 RSFFFRAEEISAIRKLLPPDLHNTS-FEALSSFLWRCRTIALNPDPNTEMRLTCIINSRS 302
Query: 262 RFRADHSPLV-GYYGNCFAFPAAVTTAGKLCGNPLGYGVELIRKAKAQVTEEYMHSLADL 320
+ R + PL GYYGN F PAA+ TA L PL + + LI++ K+ VTE+Y+ S+ L
Sbjct: 303 KLR--NPPLEPGYYGNVFVIPAAIATARDLIEKPLEFVLRLIQETKSSVTEDYVRSVTAL 360
Query: 321 MVIKERCLFTTIRSCVVSDLTRAKYAEVNFG-WGEAVYGGVVNCVAGPLPRATFIIPHKN 379
M + R +F + ++SDL +++FG WG+ VYGG P +F +P KN
Sbjct: 361 MATRGRPMFVASGNYIISDLRHFDLGKIDFGPWGKPVYGGTAKAGIALFPGVSFYVPFKN 420
Query: 380 AEGEEGLILLIFLTSEVMKRFAEELD 405
+GE G ++ I L M+ F EL+
Sbjct: 421 KKGETGTVVAISLPVRAMETFVAELN 446
>UniRef100_Q8GV03 Acyltransferase 2 [Capsicum chinense]
Length = 453
Score = 307 bits (786), Expect = 4e-82
Identities = 170/436 (38%), Positives = 248/436 (55%), Gaps = 41/436 (9%)
Query: 6 LLFTVRRCQPELVPPAAPTPREVKLLSDIDDQECLRFNMPIIFIYRHEPSMIEKDPAKVL 65
+ +++ +P+LV P+ TPRE K LSDIDDQ R +PI+ Y++ M KDPAK++
Sbjct: 11 MAISIKHHKPKLVVPSIVTPRETKHLSDIDDQGSARLQIPILMFYKYNSLMEGKDPAKLI 70
Query: 66 KRALSQTLVYYYPLAGRIREGARDKLMVDCTGEGVMFIEAETDVTLDEFGDALHPPFPCF 125
K LS+TL +YYPLAGR+ EG KLMV+C EGV+F+EA+ +V L++ GD++ PP P
Sbjct: 71 KDGLSKTLSFYYPLAGRLIEGPNRKLMVNCNSEGVLFVEADANVELEKLGDSIKPPCPYL 130
Query: 126 QELIYDVPGTNQIIDHPILLIQMAR----------------------------------G 151
L+++VPG++ II P+LL+Q+ R G
Sbjct: 131 DLLLHNVPGSDGIIGCPLLLVQVTRFSCGGFAVGLRLNHTMMDGYGLNMFLNALSELIQG 190
Query: 152 AHQPSIQPVWRREILMARDPPRVTCNHHEYEQILSSNINKEEDVAATIVHHSFFFTLTRI 211
A PSI PVW R +L AR P +TC HHE+++ + S I E + ++ SFFF +
Sbjct: 191 ASAPSILPVWERHLLSARSLPSITCTHHEFDEQIESKI-AWESMEDKLIQKSFFFGKKEM 249
Query: 212 AEIRRLIPFHLRQCSTFDLITACFWCCRTKALQLEPDEEVRMMCIVNARSRFRADHSPLV 271
I+ + + + + F+L+ A W CRT AL L P+E VR+ ++N R + + P
Sbjct: 250 EAIKNQVSPNC-ESTKFELLMAFLWKCRTIALGLHPEEIVRLTYLINIRGKLQKFELP-A 307
Query: 272 GYYGNCFAFPAAVTTAGKLCGNPLGYGVELIRKAKAQVTEEYMHSLADLMVIKERCLFTT 331
GYYGN F P AV+ AG LC N L Y VEL++K K + EEY+ SL DLM IK R T
Sbjct: 308 GYYGNAFVTPTAVSKAGLLCSNSLTYAVELVKKVKDHMNEEYIKSLIDLMAIKGRPELTK 367
Query: 332 IRSCVVSDLTRAKYAEVNFGWGEAVYGGVVNCVAGPLPRATFIIPHKNAEGEEGLILLIF 391
+ +VSD EV FGWG ++GGV ++ +F +P KN +G++G+++ I
Sbjct: 368 SWNFIVSDNRSIGLDEVGFGWGRPIFGGVAKAIS----FISFGVPVKNDKGDKGILIAIS 423
Query: 392 LTSEVMKRFAEELDKM 407
L MK+F E + K+
Sbjct: 424 LPPMAMKKFEEVVYKV 439
>UniRef100_Q75HS1 Hypothetical protein P0628H02.3 [Oryza sativa]
Length = 464
Score = 304 bits (779), Expect = 3e-81
Identities = 184/449 (40%), Positives = 255/449 (55%), Gaps = 52/449 (11%)
Query: 1 MTSSPLL-FTVRRCQPELVPPAAPTPREVKLLSDIDDQECLRFNMPIIFIYRHEPSMIEK 59
M +P L F+VRR + ELV PA PTP E K+LSDIDDQ+ LRFN I YRH PS
Sbjct: 1 MAGAPTLAFSVRRRERELVAPAKPTPYEFKMLSDIDDQDILRFNRSGILFYRHSPSKDGL 60
Query: 60 DPAKVLKRALSQTLVYYYPLAGRIRE-GARDKLMVDCTGEGVMFIEAETDVTLDEFGDAL 118
DP KV+K A+S+TLV++YP+AGR RE KL+V+CTGEGV+F+EA+ + +DE G +L
Sbjct: 61 DPVKVIKAAISETLVHFYPVAGRFRELRPTRKLVVECTGEGVVFVEADANFRMDELGTSL 120
Query: 119 HPPFPCFQELIYDVPG-TNQIIDHPILLIQM----------------------------- 148
PP PC+ L+ + T ++D P+L IQ+
Sbjct: 121 APPVPCYDMLLCEPESPTADVVDRPLLFIQVTRLACGGFVFGMHICHCMADGSGIVQFLT 180
Query: 149 -----ARGAH-QPSIQPVWRREILMARDPPRVTCNHHEYEQILSSNINKEEDVAA---TI 199
ARG H P+++PVW RE+L AR PP VT +H EY + N +DV +
Sbjct: 181 ALTEFARGVHGAPTVRPVWEREVLTARWPPTVTRDHVEYTPLP----NPGKDVLSPTDAY 236
Query: 200 VHHSFFFTLTRIAEIRRLIPFHLRQCST-FDLITACFWCCRTKALQLEPDEEVRMMCIVN 258
HH FFF + IA +R P LR S+ FDL+ A W CRT AL+ +P + VR+ VN
Sbjct: 237 AHHVFFFGASEIAALRSQAPPDLRAVSSRFDLVGAFMWRCRTAALRYDPGDVVRLHMFVN 296
Query: 259 ARSRFRADHSPLVGYYGNCFAFPAAVTTAGKLCGNPLGYGVELIRKAKAQVTEE-YMHSL 317
AR R R+ GYYGN F AA AG+L P GY + L+ +AKA+ +EE Y+ S+
Sbjct: 297 ARVRNRSKRPVPRGYYGNAIVFAAASVPAGELWRRPFGYALRLLMQAKARASEEGYVQSV 356
Query: 318 ADLMVIKERCLFTTIRSCVVSDLTRAKYAEVNFGWGEAVYGGVVNCVAGPLPRATFIIPH 377
A+ R F R+ ++SD+T+A ++FGWG+ VYGG + ATF +
Sbjct: 357 ANFNAAHRRPPFPKARTYLISDMTQAGLMAIDFGWGKPVYGGPATTML-----ATFHLEG 411
Query: 378 KNAEGEEGLILLIFLTSEVMKRFAEELDK 406
+N GE G+I+ I L + V++R +E++K
Sbjct: 412 RNEVGEAGVIVPIRLPNPVIERLIQEVNK 440
>UniRef100_Q6QLX4 Alcohol acyl transferase [Lycopersicon esculentum]
Length = 442
Score = 299 bits (765), Expect = 1e-79
Identities = 164/429 (38%), Positives = 244/429 (56%), Gaps = 40/429 (9%)
Query: 14 QPELVPPAAPTPREVKLLSDIDDQECLRFNMPIIFIYRHEPSMIEKDPAKVLKRALSQTL 73
+P+LV P++ T E K LS+IDDQ +R +PI+ Y++ SM KD AK++K LS+TL
Sbjct: 13 KPKLVVPSSVTSHETKRLSEIDDQGFIRLQIPILMFYKYNSSMKGKDLAKIIKDGLSKTL 72
Query: 74 VYYYPLAGRIREGARDKLMVDCTGEGVMFIEAETDVTLDEFGDALHPPFPCFQELIYDVP 133
V+YYPLAGR+ EG KLMV+C GEGV+FIE + ++ L++ G+++ PP P L+++V
Sbjct: 73 VFYYPLAGRLIEGPNKKLMVNCNGEGVLFIEGDANIELEKLGESIKPPCPYLDLLLHNVH 132
Query: 134 GTNQIIDHPILLIQMAR----------------------------------GAHQPSIQP 159
G++ II P+LLIQ+ R GA PSI P
Sbjct: 133 GSDGIIGSPLLLIQVTRFTCGGFAVGFRFNHTMMDAYGFKMFLNALSELIQGASTPSILP 192
Query: 160 VWRREILMARDPPRVTCNHHEYEQILSSNINKEEDVAATIVHHSFFFTLTRIAEIRRLIP 219
VW R +L AR P +TC HHE+++ + S I E + ++ SFFF + I+ +P
Sbjct: 193 VWERHLLSARSSPSITCIHHEFDEEIESKI-AWESMEDKLIQQSFFFGNEEMEVIKNQVP 251
Query: 220 FHLRQCSTFDLITACFWCCRTKALQLEPDEEVRMMCIVNARSRFRADHSPLVGYYGNCFA 279
+ +C+ F+L+ A W CRT AL L DE VR+ ++N R + + +GYYGN F
Sbjct: 252 PNY-ECTKFELLMAFLWKCRTIALNLHSDEIVRLTYVINIRGKKSLNIELPIGYYGNAFI 310
Query: 280 FPAAVTTAGKLCGNPLGYGVELIRKAKAQVTEEYMHSLADLMVIKERCLFTTIRSCVVSD 339
P V+ AG LC NP+ Y VELI+K K + EEY+ SL DLMV K R T + +VSD
Sbjct: 311 TPVVVSKAGLLCSNPVTYAVELIKKVKDHINEEYIKSLIDLMVTKGRPELTKSWNFLVSD 370
Query: 340 LTRAKYAEVNFGWGEAVYGGVVNCVAGPLPRATFIIPHKNAEGEEGLILLIFLTSEVMKR 399
+ E +FGWG ++GG++ ++ +F + KN +GE+G+++ I L MK+
Sbjct: 371 NRYIGFDEFDFGWGNPIFGGILKAIS----FTSFGVSVKNDKGEKGVLIAISLPPLAMKK 426
Query: 400 FAEELDKMF 408
+ + F
Sbjct: 427 LQDIYNMTF 435
>UniRef100_Q75GN9 Putative hypersensitivity-related protein [Oryza sativa]
Length = 456
Score = 277 bits (709), Expect = 4e-73
Identities = 168/465 (36%), Positives = 248/465 (53%), Gaps = 58/465 (12%)
Query: 8 FTVRRCQPELVPPAAPTPREVKLLSDIDDQECLRFNMPIIFIYR------HEPSMIEKDP 61
FT RR + ELV PA TP E K LSDID+Q LRF + ++ ++P DP
Sbjct: 4 FTARRGKSELVAPARATPNERKYLSDIDNQHSLRFYATAVEFFQLCTFDGYKPH----DP 59
Query: 62 AKVLKRALSQTLVYYYPLAGRIREGARDKLMVDCTGEGVMFIEAETDVTLDEFGDALHPP 121
K ++ AL++ LV+YYP+AGR+RE + KL+VDCT EGV+F+EA DV L+E G L P
Sbjct: 60 VKAIRSALAEALVHYYPIAGRLRELPQGKLVVDCTVEGVVFVEAYADVRLEELGKPLLLP 119
Query: 122 FPCFQELIYDVPGTNQIIDHPILLIQ---------------------------------- 147
+PC +E + D T ++ P+L +Q
Sbjct: 120 YPCVEEFLCDPGDTKVVVGKPLLFLQVTRLKCGGFVIGLHMCHNISDGFGMAHFIKAVGD 179
Query: 148 MARGAHQPSIQPVWRREILMARDPPRVTCNHHEYEQILS---SNINKEEDVAATIVHHSF 204
+ARG +I P+W RE+L PP++T H YE + +N + T+V F
Sbjct: 180 IARGEALLTISPLWNREMLTMCYPPQITHTHLAYEPLRDGDPTNDIMQSTTPDTMVGQYF 239
Query: 205 FFTLTRIAEIRRLIPFHLRQC-STFDLITACFWCCRTKALQLEPDEEVRMMCIVNARSRF 263
F I+ +R +P HLRQ +TF+LI A W CRT AL D+ VR+M +N+R +
Sbjct: 240 LFGPREISAMRNHVPVHLRQSYTTFELIAAAVWKCRTAALGYSLDQHVRLMFTLNSRGNW 299
Query: 264 RADHSPLVGYYGNCFAFPAAVTTAGKLCGNPLGYGVELIRKAKAQVTEEYMHSLADLMVI 323
+ + GYYG C FP A TT LCGNPLGY ++L+RKAK +VT+EY+ S D +
Sbjct: 300 KRNPPIPQGYYGCCLVFPVAETTVADLCGNPLGYALDLVRKAKLEVTDEYVKSTVDFLAS 359
Query: 324 KERCLFTTIRSCVVSDLTRAKYAEVNFGWGEAVYGGVVNCVAGPLPR--ATFIIPHKNAE 381
++ R+ +VSD+T +++FGWG+ + GG+ +AG + ++ KNA+
Sbjct: 360 RKWPSLVVDRTYIVSDITSVGDDKLDFGWGKRMGGGIP--MAGDIMSRLISYFTKCKNAD 417
Query: 382 GEEGLILLIFLTSEVMKRFAEELDKMFGNQNQPTTIGPSFVISTL 426
GE+ +++ ++L S M RFA E+ Q G F++S L
Sbjct: 418 GEDCIVVPMYLPSITMDRFAAEISVWSMKQ------GSKFIVSAL 456
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.325 0.140 0.433
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 709,916,721
Number of Sequences: 2790947
Number of extensions: 28842648
Number of successful extensions: 70179
Number of sequences better than 10.0: 226
Number of HSP's better than 10.0 without gapping: 138
Number of HSP's successfully gapped in prelim test: 88
Number of HSP's that attempted gapping in prelim test: 69415
Number of HSP's gapped (non-prelim): 395
length of query: 426
length of database: 848,049,833
effective HSP length: 130
effective length of query: 296
effective length of database: 485,226,723
effective search space: 143627110008
effective search space used: 143627110008
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 76 (33.9 bits)
Medicago: description of AC148345.4


