

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148343.5 - phase: 0
(443 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
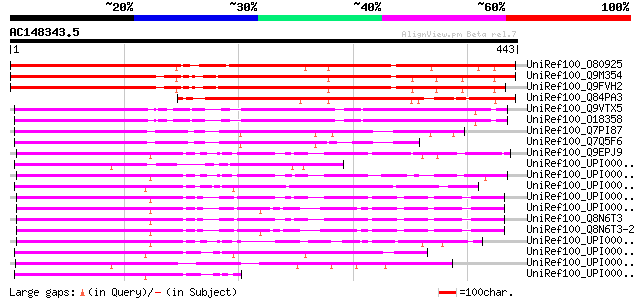
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_O80925 Putative ADP ribosylation factor 1 GTPase activ... 543 e-153
UniRef100_Q9M354 Hypothetical protein F5K20_10 [Arabidopsis thal... 526 e-148
UniRef100_Q9FVH2 ARF GAP-like zinc finger-containing protein ZIG... 509 e-143
UniRef100_Q84PA3 ADP ribosylation GTPase-like protein [Oryza sat... 292 1e-77
UniRef100_Q9VTX5 CG4237-PA [Drosophila melanogaster] 212 2e-53
UniRef100_O18358 Putative ARF1 GTPase activating protein [Drosop... 212 2e-53
UniRef100_Q7PI87 ENSANGP00000024029 [Anopheles gambiae str. PEST] 209 1e-52
UniRef100_Q7Q5F6 ENSANGP00000004294 [Anopheles gambiae str. PEST] 194 4e-48
UniRef100_Q9EPJ9 ADP-ribosylation factor GTPase activating prote... 171 3e-41
UniRef100_UPI000042D5F0 UPI000042D5F0 UniRef100 entry 171 4e-41
UniRef100_UPI00003AC259 UPI00003AC259 UniRef100 entry 167 6e-40
UniRef100_UPI00002BB484 UPI00002BB484 UniRef100 entry 166 1e-39
UniRef100_UPI000036C64D UPI000036C64D UniRef100 entry 165 2e-39
UniRef100_UPI000036C64C UPI000036C64C UniRef100 entry 165 3e-39
UniRef100_Q8N6T3 ADP-ribosylation factor GTPase activating prote... 165 3e-39
UniRef100_Q8N6T3-2 Splice isoform 2 of Q8N6T3 [Homo sapiens] 164 4e-39
UniRef100_UPI00003AC258 UPI00003AC258 UniRef100 entry 161 4e-38
UniRef100_UPI0000364090 UPI0000364090 UniRef100 entry 157 6e-37
UniRef100_UPI00002194E2 UPI00002194E2 UniRef100 entry 157 6e-37
UniRef100_UPI0000364091 UPI0000364091 UniRef100 entry 153 1e-35
>UniRef100_O80925 Putative ADP ribosylation factor 1 GTPase activating protein
[Arabidopsis thaliana]
Length = 456
Score = 543 bits (1400), Expect = e-153
Identities = 287/469 (61%), Positives = 340/469 (72%), Gaps = 41/469 (8%)
Query: 1 MAASRRLRELQSEPSNKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVT 60
MAA+RRLR LQS+P NK+CVDCSQKNPQWAS+SYG+FMCLECSGKHRGLGVHISFVRSVT
Sbjct: 1 MAAARRLRTLQSQPENKVCVDCSQKNPQWASISYGIFMCLECSGKHRGLGVHISFVRSVT 60
Query: 61 MDSWSDLQIKKMEAGGNRNLNTFLSQYGISKETDIITKYNSNAASIYRDRIQAIAEGRSW 120
MDSWS++QIKKM+AGGN LN FL+QYGISKETDII+KYNSNAAS+YRDRIQA+AEGR W
Sbjct: 61 MDSWSEIQIKKMDAGGNERLNNFLAQYGISKETDIISKYNSNAASVYRDRIQALAEGRQW 120
Query: 121 RDPPVVKENASTRAGKGKPPLAAA----SNGGGWDDNWDNDDGDSYGYGSRGGGDIRRNQ 176
RDPP+VKE+ KPPL+ S GGW DNWDNDD S D+RRNQ
Sbjct: 121 RDPPIVKESVGGGLMNKKPPLSQGGGRDSGNGGW-DNWDNDD-------SFRSTDMRRNQ 172
Query: 177 STGDVIGFVGNGVTPSSRSKSTEDIYTRTQLEASAANKEGFFAKKMAENESRPEGLPPSQ 236
S GD G G ++SKS+EDIY+R+QLEASAANKE FFAK+MAENES+PEGLPPSQ
Sbjct: 173 SAGDFRSSGGRGA--PAKSKSSEDIYSRSQLEASAANKESFFAKRMAENESKPEGLPPSQ 230
Query: 237 GGKYVGFGSSPGPAQRISPQN---DYLSVVSEGIGKLSMVAQSA-------TKEITAKVK 286
GGKYVGFGSSPGPA R + Q+ D SV+SEG G+LS+VA SA T E T+KVK
Sbjct: 231 GGKYVGFGSSPGPAPRSNQQSGGGDVFSVMSEGFGRLSLVAASAANVVQTGTMEFTSKVK 290
Query: 287 DGGYDHKVNETVNIVTQKTSEIGQRTWGIMKGVMALASQKVEEFTKDYPDGNSDNWPRNE 346
+GG D V+ETVN+V KT+EIGQRTWGIMKGVMA+ASQKVEEFTK+ + W +
Sbjct: 291 EGGLDQTVSETVNVVASKTTEIGQRTWGIMKGVMAIASQKVEEFTKE----EASTWNQQN 346
Query: 347 NDRHDFNQENKGWNSSTSTRE---GQPSSGGQTNTY-HSNSWDDW-DNQDTRKEVPAKGS 401
+ +N G + T+ Q SS G N+Y +SNSWDDW + +++KE K S
Sbjct: 347 KTEGNGYYQNSGIGNKTANSSFGGSQSSSSGHNNSYRNSNSWDDWGEENNSKKEAAPKVS 406
Query: 402 APHNNDD--WAGWDDAKDDDDEF------DDKSFGNNGTSGSGWTGGGF 442
+++DD WAGWDD DD+F D KS G+NG S + WTGGGF
Sbjct: 407 TSNDDDDGGWAGWDDNDAKDDDFYYQPASDKKSVGHNGKSDTAWTGGGF 455
>UniRef100_Q9M354 Hypothetical protein F5K20_10 [Arabidopsis thaliana]
Length = 459
Score = 526 bits (1356), Expect = e-148
Identities = 288/478 (60%), Positives = 336/478 (70%), Gaps = 56/478 (11%)
Query: 1 MAASRRLRELQSEPSNKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVT 60
MAA+R+LR LQS+P NK+CVDC+QKNPQWASVSYG+FMCLECSGKHRGLGVHISFVRSVT
Sbjct: 1 MAATRQLRTLQSQPENKVCVDCAQKNPQWASVSYGIFMCLECSGKHRGLGVHISFVRSVT 60
Query: 61 MDSWSDLQIKKMEAGGNRNLNTFLSQYGISKETDIITKYNSNAASIYRDRIQAIAEGRSW 120
MDSWS +QIKKMEAGGN LN F +QYGI+KETDII+KYNSNAAS+YRDRIQA+AEGR W
Sbjct: 61 MDSWSAIQIKKMEAGGNERLNKFFAQYGIAKETDIISKYNSNAASVYRDRIQALAEGRPW 120
Query: 121 RDPPVVKENASTRAGKGKPPLAAA-------SNGGGWDDNWDNDDGDSYGYGSRGGGDIR 173
DPPVVKE KPPLA +N GGW D+WDND DSY D+R
Sbjct: 121 NDPPVVKE------ANKKPPLAQGGYGNNNNNNNGGW-DSWDND--DSY------KSDMR 165
Query: 174 RNQSTGDVIGFVGNGVTPSSRSKSTEDIYTRTQLEASAANKEGFFAKKMAENESRPEGLP 233
RNQS D GN +SKS+EDIYTR+QLEASAA KE FFA++MAENES+PEGLP
Sbjct: 166 RNQSAND-FRASGNREGAHVKSKSSEDIYTRSQLEASAAGKESFFARRMAENESKPEGLP 224
Query: 234 PSQGGKYVGFGSSPGPAQRISPQNDYLSVVSEGIGKLSMVAQSA-----------TKEIT 282
PSQGGKYVGFGSS P R + Q+D SVVS+G G+LS+VA SA TKE T
Sbjct: 225 PSQGGKYVGFGSSSAPPPRNNQQDDVFSVVSQGFGRLSLVAASAAQSAASVVQTGTKEFT 284
Query: 283 AKVKDGGYDHKVNETVNIVTQKTSEIGQRTWGIMKGVMALASQKVEEFTKDYPDGNSDNW 342
+KVK+GGYDHKV+ETVN+V KT+EIG RTWGIMKGVMA+A+QKVEEFTK+ S +W
Sbjct: 285 SKVKEGGYDHKVSETVNVVANKTTEIGHRTWGIMKGVMAMATQKVEEFTKE----GSTSW 340
Query: 343 -PRNENDRH----DFNQENKGWNSSTSTREGQPS--SGGQTNTYHSNSWDDWDNQDTRK- 394
++EN+ + +F NK NSS Q S SG N+ +SNSWD W + +K
Sbjct: 341 NQQSENEGNGYYQNFGNGNKAANSSVGGGRPQSSSTSGHYNNSQNSNSWDSWGENENKKT 400
Query: 395 -EVPAKGSAPHNNDD-WAGWDDAKDDDDEF--------DDKSFGNNGTSGSGWTGGGF 442
V KGS+ N+DD W GWDD DD F D KS G+NG S + WTGGGF
Sbjct: 401 EAVAPKGSSASNDDDGWTGWDDHDAKDDGFDGHYQSAGDKKSAGHNGKSDTAWTGGGF 458
>UniRef100_Q9FVH2 ARF GAP-like zinc finger-containing protein ZIGA2 [Arabidopsis
thaliana]
Length = 458
Score = 509 bits (1310), Expect = e-143
Identities = 281/469 (59%), Positives = 328/469 (69%), Gaps = 56/469 (11%)
Query: 1 MAASRRLRELQSEPSNKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVT 60
MAA+R+LR LQS+P NK+CVDC+QKNPQWASVSYG+FMCLECSGKHRGLGVHISFVRSVT
Sbjct: 1 MAATRQLRTLQSQPENKVCVDCAQKNPQWASVSYGIFMCLECSGKHRGLGVHISFVRSVT 60
Query: 61 MDSWSDLQIKKMEAGGNRNLNTFLSQYGISKETDIITKYNSNAASIYRDRIQAIAEGRSW 120
MDSWS +QIKKMEAGGN LN F +QYGI+KETDII+KYNSNAAS+YRDRIQA+AEGR W
Sbjct: 61 MDSWSAIQIKKMEAGGNERLNKFFAQYGIAKETDIISKYNSNAASVYRDRIQALAEGRPW 120
Query: 121 RDPPVVKENASTRAGKGKPPLAAA-------SNGGGWDDNWDNDDGDSYGYGSRGGGDIR 173
DPPVVKE KPPLA +N GGW D+WDND DSY D+R
Sbjct: 121 NDPPVVKE------ANKKPPLAQGGYGNNNNNNNGGW-DSWDND--DSY------KSDMR 165
Query: 174 RNQSTGDVIGFVGNGVTPSSRSKSTEDIYTRTQLEASAANKEGFFAKKMAENESRPEGLP 233
RNQS D GN +SKS+EDIYTR+QLEASAA KE FFA++MAENES+PEGLP
Sbjct: 166 RNQSAND-FRASGNREGAHVKSKSSEDIYTRSQLEASAAGKESFFARRMAENESKPEGLP 224
Query: 234 PSQGGKYVGFGSSPGPAQRISPQNDYLSVVSEGIGKLSMVAQSA-----------TKEIT 282
PSQGGKYVGFGSS P R + Q+D SVVS+G G+LS+VA SA TKE T
Sbjct: 225 PSQGGKYVGFGSSSAPPPRNNQQDDVFSVVSQGFGRLSLVAASAAQSAASVVQTGTKEFT 284
Query: 283 AKVKDGGYDHKVNETVNIVTQKTSEIGQRTWGIMKGVMALASQKVEEFTKDYPDGNSDNW 342
+KVK+GGYDHKV+ETVN+V K +EIG RTWGIMKGVMA+A+QKVEEFTK+ S +W
Sbjct: 285 SKVKEGGYDHKVSETVNVVANKITEIGHRTWGIMKGVMAMATQKVEEFTKE----GSTSW 340
Query: 343 -PRNENDRH----DFNQENKGWNSSTSTREGQPS--SGGQTNTYHSNSWDDWDNQDTRK- 394
++EN+ + +F NK NSS Q S SG N+ +SNSWD W + +K
Sbjct: 341 NQQSENEGNGYYQNFGNGNKAANSSVGGGRPQSSSTSGHYNNSQNSNSWDSWGENENKKT 400
Query: 395 -EVPAKGSAPHNNDD-WAGWDDAKDDDDEF--------DDKSFGNNGTS 433
V KGS+ N+DD W GWDD DD F D KS G+NG S
Sbjct: 401 EAVAPKGSSASNDDDGWTGWDDHDAKDDGFDGHYQSAGDKKSAGHNGKS 449
>UniRef100_Q84PA3 ADP ribosylation GTPase-like protein [Oryza sativa]
Length = 308
Score = 292 bits (748), Expect = 1e-77
Identities = 167/321 (52%), Positives = 204/321 (63%), Gaps = 47/321 (14%)
Query: 147 GGGWDDNWDNDDGDSYGYGSRGGGDIRRNQSTGDVIGFVGNGVTPSSRSKSTEDIYTRTQ 206
GGGWDD WD+D D+RRNQS G RSKST+D+YTR Q
Sbjct: 9 GGGWDD-WDDDFRP----------DMRRNQSVGSFGESGAESGRQPPRSKSTQDMYTRQQ 57
Query: 207 LEASAANKEGFFAKKMAENESRPEGLPPSQGGKYVGFGSSPGPAQR---ISPQNDYLSVV 263
LEASAANK+ FFA++MAENES+PEG+PPSQGGKYVGFGSSP P+ + Q D + VV
Sbjct: 58 LEASAANKDSFFARRMAENESKPEGIPPSQGGKYVGFGSSPAPSANRNGAAAQGDVMQVV 117
Query: 264 SEGIGKLSMVAQSA-----------TKEITAKVKDGGYDHKVNETVNIVTQKTSEIGQRT 312
S+GIG+LS+VA SA TKE +K+++GGYD KVNETVN+V KT+EIG RT
Sbjct: 118 SQGIGRLSLVAASAAQSAASVVQVGTKEFQSKMREGGYDQKVNETVNVVANKTAEIGSRT 177
Query: 313 WGIMKGVMALASQKVEEFTKDYPDGNSDNWPRNENDR---HDFNQE--NKGWNSSTSTRE 367
WGIMKGVMALASQKVEE+ K+ +G D+W R E H F +E GWNSS
Sbjct: 178 WGIMKGVMALASQKVEEYAKEGGNGWGDDWQRREQGSEPYHRFERETNGNGWNSS----- 232
Query: 368 GQPSSGGQTNTYHSNSWDDWDNQDTRKEVPAKGSAPHNNDDWAGWDDAKDDD-DEFD--- 423
S G + Y+SNSWDDWD + +K+ PAK ++D WAGWDD KDD+ D ++
Sbjct: 233 ---SHDGSSKNYNSNSWDDWD-EPVKKDEPAK--ERQSSDSWAGWDDGKDDNFDSYNHST 286
Query: 424 -DKSFGNNGTS-GSGWTGGGF 442
K NGT+ GS WT GGF
Sbjct: 287 PSKGSNQNGTTGGSYWTEGGF 307
>UniRef100_Q9VTX5 CG4237-PA [Drosophila melanogaster]
Length = 468
Score = 212 bits (539), Expect = 2e-53
Identities = 147/451 (32%), Positives = 217/451 (47%), Gaps = 62/451 (13%)
Query: 5 RRLRELQSEPSNKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVTMDSW 64
R L+EL+ + N C +C NPQW SV+YG+++CLECSGKHR LGVH+SFVRSVTMD W
Sbjct: 8 RVLQELKPQDENSKCFECGTHNPQWVSVTYGIWICLECSGKHRSLGVHLSFVRSVTMDKW 67
Query: 65 SDLQIKKMEAGGNRNLNTFL-SQYGISKETDIITKYNSNAASIYRDRIQAIAEGRSWRDP 123
D++++KM+AGGNRN FL Q ++ I +YNS AA++YRD+I +A+G+SW
Sbjct: 68 KDIELEKMKAGGNRNAREFLEDQEDWNERAPITQRYNSKAAALYRDKIATLAQGKSWD-- 125
Query: 124 PVVKENASTRAGKGKPPLAAASNGGGWDDNWDNDDGDSYGYGSRGGGDIRRNQSTGDVIG 183
+KE A R G + S+GG + ++ + + GYG GG
Sbjct: 126 --LKE-AQGRVGSNN----SFSSGGSSNSSYQSRP-SATGYGGNGGYQ------------ 165
Query: 184 FVGNGVTPSSRSKSTEDIYTRTQLEASAANKEGFFAKKMAENESRPEGLPPSQGGKYVGF 243
G G P Y + Q + KE FF+++ EN SRPE LPPSQGGKY GF
Sbjct: 166 -NGGGAEPGG--------YQQYQTQEFKDQKEEFFSRRQVENASRPENLPPSQGGKYAGF 216
Query: 244 GSSPGPAQRISPQNDYLSVVSEGIGKLSMVAQSATK-EITAKVKDGGYDHKVNETVNIVT 302
G + P + Q + S +S S+ + +A+K TAK K TVN+ +
Sbjct: 217 GFTREPPPKTQSQELFDSTLSTLASGWSLFSTNASKLASTAK-------EKAVTTVNLAS 269
Query: 303 QKTSEIGQRTWGIMKGVMALASQKVEEFTKDYPDGNSDNWPRNENDRHDFNQENKGWNSS 362
K E G + GV +AS+ + + + + N + +D N E+
Sbjct: 270 TKIKE-GTLLDSVQCGVTDVASKVTDMGKRGWNNLAGSNISSPQGGYNDPNFEDSSAYQR 328
Query: 363 TSTREGQPSSG-GQTNTYHSNSWDDWDNQDTRKEVPAKGSAPHN---------------- 405
+++ G + G GQ + + W W + K S+ HN
Sbjct: 329 SNSVGGNLAGGLGQQSGVSDSDWGGWQDNGNSKSHMTSSSSYHNQLSSSSGGGTASAGLT 388
Query: 406 -NDDWAGWDDAKDDDDEFDDKSFGNNGTSGS 435
+ DW+G++ E S+ N + GS
Sbjct: 389 RDADWSGFEATNYQSSE---TSYQNASSGGS 416
>UniRef100_O18358 Putative ARF1 GTPase activating protein [Drosophila melanogaster]
Length = 468
Score = 212 bits (539), Expect = 2e-53
Identities = 147/451 (32%), Positives = 217/451 (47%), Gaps = 62/451 (13%)
Query: 5 RRLRELQSEPSNKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVTMDSW 64
R L+EL+ + N C +C NPQW SV+YG+++CLECSGKHR LGVH+SFVRSVTMD W
Sbjct: 8 RVLQELKPQDENSKCFECGTHNPQWVSVTYGIWICLECSGKHRSLGVHLSFVRSVTMDKW 67
Query: 65 SDLQIKKMEAGGNRNLNTFL-SQYGISKETDIITKYNSNAASIYRDRIQAIAEGRSWRDP 123
D++++KM+AGGNRN FL Q ++ I +YNS AA++YRD+I +A+G+SW
Sbjct: 68 KDIELEKMKAGGNRNAREFLEDQEDWNERAPITQRYNSKAAALYRDKIATLAQGKSWD-- 125
Query: 124 PVVKENASTRAGKGKPPLAAASNGGGWDDNWDNDDGDSYGYGSRGGGDIRRNQSTGDVIG 183
+KE A R G + S+GG + ++ + + GYG GG
Sbjct: 126 --LKE-AQGRVGSNN----SFSSGGSSNSSYQSRP-SATGYGGNGGYQ------------ 165
Query: 184 FVGNGVTPSSRSKSTEDIYTRTQLEASAANKEGFFAKKMAENESRPEGLPPSQGGKYVGF 243
G G P Y + Q + KE FF+++ EN SRPE LPPSQGGKY GF
Sbjct: 166 -NGGGAEPGG--------YQQYQTQEFKDQKEEFFSRRQVENASRPENLPPSQGGKYAGF 216
Query: 244 GSSPGPAQRISPQNDYLSVVSEGIGKLSMVAQSATK-EITAKVKDGGYDHKVNETVNIVT 302
G + P + Q + S +S S+ + +A+K TAK K TVN+ +
Sbjct: 217 GFTREPPPKTQSQELFDSTLSTLASGWSLFSTNASKLASTAK-------EKAVTTVNLAS 269
Query: 303 QKTSEIGQRTWGIMKGVMALASQKVEEFTKDYPDGNSDNWPRNENDRHDFNQENKGWNSS 362
K E G + GV +AS+ + + + + N + +D N E+
Sbjct: 270 TKIKE-GTLLDSVQCGVTDVASKVTDMGKRGWNNLAGSNISSRQGGYNDPNFEDSSAYQR 328
Query: 363 TSTREGQPSSG-GQTNTYHSNSWDDWDNQDTRKEVPAKGSAPHN---------------- 405
+++ G + G GQ + + W W + K S+ HN
Sbjct: 329 SNSVGGNLAGGLGQQSGVSDSDWGGWQDNGNSKSHMTSSSSYHNQLSSSSGGGTASAGLT 388
Query: 406 -NDDWAGWDDAKDDDDEFDDKSFGNNGTSGS 435
+ DW+G++ E S+ N + GS
Sbjct: 389 RDADWSGFEATNYQSSE---TSYQNAPSGGS 416
>UniRef100_Q7PI87 ENSANGP00000024029 [Anopheles gambiae str. PEST]
Length = 484
Score = 209 bits (532), Expect = 1e-52
Identities = 140/413 (33%), Positives = 196/413 (46%), Gaps = 57/413 (13%)
Query: 5 RRLRELQSEPSNKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVTMDSW 64
R L EL+ N C +C NPQW SV+YG+++CLECSGKHRGLGVH+SFVRSV+MD W
Sbjct: 8 RVLSELKPSDGNNKCFECGTHNPQWVSVTYGIWICLECSGKHRGLGVHLSFVRSVSMDKW 67
Query: 65 SDLQIKKMEAGGNRNLNTFLSQYGISKET-DIITKYNSNAASIYRDRIQAIAEGRSWRDP 123
D++++KM+ GGNR FL ET I KY++ AA++YRDRI +A+G+ W
Sbjct: 68 KDIELEKMKVGGNRKAREFLDAQDDWDETMPIQRKYSTRAAALYRDRISTLAQGQPW--- 124
Query: 124 PVVKENASTRAGKGKPPLAAASNGGGWDDNWDNDDGDSYGYGSRGGGDIRRNQSTGDVIG 183
E+ + G AA+ + GG + + S GS GGG G
Sbjct: 125 ---DESGALARVNGSGGYAASGSSGGSTGSMTH----SRSSGSMGGGS-----------G 166
Query: 184 FVGNGVTPSSRSKSTED--IYTRTQLEASAANKEGFFAKKMAENESRPEGLPPSQGGKYV 241
GNG + +D Y + Q A KE FF++K EN +RPE LPP+QGGKY
Sbjct: 167 AGGNGGYQNGGDNYYQDGGSYQQYQTPEFKAQKEDFFSRKQEENAARPENLPPNQGGKYA 226
Query: 242 GFGSSPGPAQRISPQNDYLSVVSE---GIGKLSMVAQSATKE-------ITAKVKDGGYD 291
GFG + P R + +V S G S VA A + + KVK+G
Sbjct: 227 GFGYTMDPPPRSQSHELFDTVQSSLATGWNVFSKVANVAKENALKYGSIASQKVKEGSLL 286
Query: 292 HKVNETVNIVTQKTSEIGQRTWGIMKGVMALASQKVEEFTKDYPDGNSDNWPRNENDRHD 351
V V+ + K +E+G++ WG G P S + P ND++
Sbjct: 287 EGVGSQVSNLATKVTEVGRKGWGGFGG----------------PGSGSYSTPSGSNDQYS 330
Query: 352 FNQENKGWNSSTSTR-----EGQPSSGGQTNTYHSNSWDD--WDNQDTRKEVP 397
+ G +S + R + Q S G + +SW + W N +P
Sbjct: 331 NISSDSGVSSPINGRQPGGYQQQRGSTGGSGRRDESSWSNNGWSNDSNSNSLP 383
>UniRef100_Q7Q5F6 ENSANGP00000004294 [Anopheles gambiae str. PEST]
Length = 319
Score = 194 bits (493), Expect = 4e-48
Identities = 129/360 (35%), Positives = 175/360 (47%), Gaps = 56/360 (15%)
Query: 5 RRLRELQSEPSNKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVTMDSW 64
R L EL+ N C +C NPQW SV+YG+++CLECSGKHRGLGVH+SFVRSV+MD W
Sbjct: 8 RVLSELKPSDGNNKCFECGTHNPQWVSVTYGIWICLECSGKHRGLGVHLSFVRSVSMDKW 67
Query: 65 SDLQIKKMEAGGNRNLNTFLSQYGISKET-DIITKYNSNAASIYRDRIQAIAEGRSWRDP 123
D++++KM+ GGNR FL ET I KY++ AA++YRDRI +A+G+ W
Sbjct: 68 KDIELEKMKVGGNRKAREFLDAQDDWDETMPIQRKYSTRAAALYRDRISTLAQGQPW--- 124
Query: 124 PVVKENASTRAGKGKPPLAAASNGGGWDDNWDNDDGDSYGYGSRGGGDIRRNQSTGDVIG 183
E+ + G AA+ + GG + + S GS GGG G
Sbjct: 125 ---DESGALARVNGSGGYAASGSSGGSTGSMTH----SRSSGSMGGGS-----------G 166
Query: 184 FVGNGVTPSSRSKSTED--IYTRTQLEASAANKEGFFAKKMAENESRPEGLPPSQGGKYV 241
GNG + +D Y + Q A KE FF++K EN +RPE LPP+QGGKY
Sbjct: 167 AGGNGGYQNGGDNYYQDGGSYQQYQTPEFKAQKEDFFSRKQEENAARPENLPPNQGGKYA 226
Query: 242 GFGSSPGPAQRISPQNDYLSVVSE---GIGKLSMVAQSATKEITAKVKDGGYDHKVNETV 298
GFG + P R + +V S G S VA A KE K
Sbjct: 227 GFGYTMDPPPRSQSHELFDTVQSSLATGWNVFSKVANVA-KENALKYG------------ 273
Query: 299 NIVTQKTSEIGQRTWGIMKGVMALASQKVEEFTKDYPDGNSDNWPRNENDRHDFNQENKG 358
+I +QK +E+G++ WG G P S + P ND++ + G
Sbjct: 274 SIASQKVTEVGRKGWGGFGG----------------PGSGSYSTPSGSNDQYSNISSDSG 317
>UniRef100_Q9EPJ9 ADP-ribosylation factor GTPase activating protein 1 [Mus musculus]
Length = 414
Score = 171 bits (434), Expect = 3e-41
Identities = 132/451 (29%), Positives = 202/451 (44%), Gaps = 71/451 (15%)
Query: 7 LRELQSEPSNKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVTMDSWSD 66
L+E++++ N +C +C NPQW SV+YG+++CLECSG+HRGLGVH+SFVRSVTMD W D
Sbjct: 10 LKEVRAQDENNVCFECGAFNPQWVSVTYGIWICLECSGRHRGLGVHLSFVRSVTMDKWKD 69
Query: 67 LQIKKMEAGGNRNLNTFL-SQYGISKETDIITKYNSNAASIYRDRIQAIAEGRSWR---- 121
++++KM+AGGN FL +Q + KY+S AA+++RD++ +AEG+ W
Sbjct: 70 IELEKMKAGGNAKFREFLETQDDYEPSWSLQDKYSSRAAALFRDKVATLAEGKEWSLESS 129
Query: 122 -----DPPVVKE-NASTRAGKGKPPLAAASNGGGWDDNWDNDDGDSYGYGSRGGGDIRRN 175
PP K + G+P AAAS ++D W NDD SY +G + R
Sbjct: 130 PAQNWTPPQPKTLQFTAHRASGQPQSAAASGDKAFED-WLNDDLGSY----QGAQENR-- 182
Query: 176 QSTGDVIGFVGNGVTPSSRSKSTEDIYTRTQLEASAANKEGFFAKKMAENESRPEGLPPS 235
+GF GN V P R ED + + + + F + EG
Sbjct: 183 -----YVGF-GNTVPPQKR----EDDFLNNAMSSLYSGWSSFTTGASKFASAAKEGA--- 229
Query: 236 QGGKYVGFGSSPGPAQRISPQNDYLSVVSEGIGKLSMVAQSATKEITAKVKDGGYDHKVN 295
FGS +Q+ S L+ ++ K KVK+G V+
Sbjct: 230 -----TKFGSQ--ASQKASELGHSLN-------------ENVLKPAQEKVKEGRIFDDVS 269
Query: 296 ETVNIVTQKTSEIGQRTWGIMKGVMALASQKVEEFTKDYPDGNSDNWPRNENDRHDFNQE 355
V+ + K +G + W + V S K E+ + +G+S +N ++ N +
Sbjct: 270 SGVSQLASKVQGVGSKGW---RDVTTFFSGKAEDSSDRPLEGHSYQNSSGDNSQNS-NID 325
Query: 356 NKGW---NSSTSTREGQPSS------GGQTNTYHSNSWDDWDNQDTRKEVPAKGSAPHNN 406
W S+ + PSS T S+SWD W + A + N+
Sbjct: 326 QSFWETFGSAEPPKAKSPSSDSWTCADASTGRRSSDSWDVWGSGS------ASNNKNSNS 379
Query: 407 DDWAGWDDAKDDDDEFDDKSFGNNGTSGSGW 437
D W W+ A + K + T+ GW
Sbjct: 380 DGWESWEGASGEGRAKATKKAAPS-TADEGW 409
>UniRef100_UPI000042D5F0 UPI000042D5F0 UniRef100 entry
Length = 416
Score = 171 bits (433), Expect = 4e-41
Identities = 112/307 (36%), Positives = 150/307 (48%), Gaps = 55/307 (17%)
Query: 5 RRLRELQSEPSNKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVTMDSW 64
+ L L + NK+CVDC+ +PQWASVSYG+F+CLECSG HRG GVHISFVRS+TMD W
Sbjct: 8 KELLALMNTGGNKVCVDCNAPSPQWASVSYGIFICLECSGVHRGFGVHISFVRSITMDKW 67
Query: 65 SDLQIKKMEAGGNRNLNTFLSQY----GISKETDIITKYNSNAASIYRDRIQAIAEGRSW 120
SD Q+ KM+ GGN F+ Y G +K + KYNS AA+ YR+++ A G+ W
Sbjct: 68 SDEQLNKMKTGGNEKFKDFMENYGPEGGYTKGMGMQEKYNSWAAAQYREKLTAECAGQPW 127
Query: 121 RDPPVVKENASTRAGKGKPPLAAASNGGGWDDNWDNDDGDSYGYGSRGGGDIRRNQSTGD 180
+S A G P A+S R++++ G
Sbjct: 128 -------SASSPPANFGLPSRPASSQ------------------------TTRKSRAAG- 155
Query: 181 VIGFVGNGVTPSSRSKSTEDIYTRTQLEASAANKEGFFAKKMAENESRPEGLPPSQGGKY 240
G G+ + P SR+ S L + E FF + N +RP+ LPPSQGGKY
Sbjct: 156 --GITGSSLNP-SRTNSPSIPQGSDDLYGQKSANEAFFERMGNANATRPDHLPPSQGGKY 212
Query: 241 VGFGS-------SPGPAQRISP---------QNDYLSVVSEGIGKLSMVAQSATKEITAK 284
GFGS S P+ +S Q + L +++G G S SA +EI
Sbjct: 213 SGFGSTLEPDVASSHPSYSLSSHAAPTLDEFQRNPLGALTKGWGLFSSAIVSAGREINNA 272
Query: 285 VKDGGYD 291
V G D
Sbjct: 273 VVQPGLD 279
>UniRef100_UPI00003AC259 UPI00003AC259 UniRef100 entry
Length = 418
Score = 167 bits (423), Expect = 6e-40
Identities = 130/445 (29%), Positives = 197/445 (44%), Gaps = 90/445 (20%)
Query: 7 LRELQSEPSNKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVTMDSWSD 66
L+E++++ N +C +C NPQW SV+YG+++CLECSGKHRGLGVH+SFVRSVTMD W D
Sbjct: 10 LKEVRAQDENNVCFECGAFNPQWVSVTYGIWICLECSGKHRGLGVHLSFVRSVTMDKWKD 69
Query: 67 LQIKKMEAGGNRNLNTFL-SQYGISKETDIITKYNSNAASIYRDRIQAIAEGRSWR---- 121
++++KM+AGGN FL SQ + KYNS AA+++RD++ +AEG+ W
Sbjct: 70 IELEKMKAGGNSKFREFLESQDDYDPCWSMQEKYNSKAAALFRDQVATVAEGKEWSLETS 129
Query: 122 -----DPPVVKEN-ASTRAGKGKPPLAAASNGGGWDDNWDNDDGDSYGYGSRGGGDIRRN 175
PP K + +ST G P AS+ ++D W NDD +SY +GG + R
Sbjct: 130 PARNWTPPQPKTSLSSTHRSAGPPQNTTASSDKAFED-WLNDDVNSY----QGGQENR-- 182
Query: 176 QSTGDVIGFVGNGVTPSSRSKSTEDIYTRTQLEASAANKEGFFAKKMAENESRPEGLPPS 235
+GF GN VTP + ED + + + + F + EG
Sbjct: 183 -----YVGF-GNTVTPPKK----EDDFLNNAMSSLYSGWSSFTTGASKIASAAKEGA--- 229
Query: 236 QGGKYVGFGSSPGPAQRISPQNDYLSVVSEGIGKLSMVAQSATKEITAKVKDGGYDHKVN 295
FGS +Q+ S L+ ++ K KVK+G +V
Sbjct: 230 -----TRFGSQA--SQKASELGQTLN-------------ENVLKPAQEKVKEGKIFEEVT 269
Query: 296 ETVNIVTQKTSEIGQRTWGIMKGVMALASQKVEEFTKDYPDGNSDNWPRNENDRHDFNQE 355
V+ + K +G + W + V F PD +S+ ++ +
Sbjct: 270 VGVSQLASKVQGVGSKGW-----------RDVTTFFSGKPDDHSERPAEGDS------YQ 312
Query: 356 NKGWNSSTSTREGQPSSGGQTNTYHSNSWDDWDNQDTRKEVPAKGSAPHNNDDWAGWDD- 414
N G G Q N + W+ + N D P K +++ W D+
Sbjct: 313 NSG------------GEGYQNNAVDQSFWETFGNSD-----PPKAHKSPSSESWTYVDNS 355
Query: 415 ----AKDDDDEFDDKSFGNNGTSGS 435
+ D D + + NN S S
Sbjct: 356 TEKKSSDSWDVWGSGTVSNNKNSNS 380
>UniRef100_UPI00002BB484 UPI00002BB484 UniRef100 entry
Length = 414
Score = 166 bits (420), Expect = 1e-39
Identities = 121/424 (28%), Positives = 193/424 (44%), Gaps = 36/424 (8%)
Query: 5 RRLRELQSEPSNKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVTMDSW 64
R L+E++++ N +C +C NPQW SV+YG+++CLECSGKHRGLGVH+SFVRSVTMD W
Sbjct: 8 RVLKEVRTQDENNLCFECGAFNPQWVSVTYGIWICLECSGKHRGLGVHLSFVRSVTMDKW 67
Query: 65 SDLQIKKMEAGGNRNLNTFLS-QYGISKETDIITKYNSNAASIYRDRIQAIAEG------ 117
D++++KM+AGGN FL Q + KYNS AA+++RD++ +AEG
Sbjct: 68 KDIELEKMKAGGNGKFRLFLELQDDYDPNWTLQEKYNSKAAALFRDKVATLAEGKDWSIE 127
Query: 118 ----RSWRDPPVVKENASTRAGKGKPPLAAASNGGGWDDNWDNDDGDSYGYGSRGGGDIR 173
R+W P +S+ G AA+S+ ++D W +DD +SY + GG
Sbjct: 128 TSSARNWTSPQPKTGLSSSHRSGGSGQNAASSSDKAFED-WLSDDVNSY----QSGGGYS 182
Query: 174 RNQSTGDVIGFVGNGVTPSSR-----SKSTEDIYTRTQLEASAANKEGFFAKKMAENESR 228
NQ +GF GN VTP + + + IY+ A+K AK +
Sbjct: 183 GNQE-NRYVGF-GNTVTPEKKEDDFINNAMSSIYSGWSSFTVGASKFASIAKDNTTKLAN 240
Query: 229 PEGLPPSQGGKYVGFGSSPGPAQRISPQNDYLSVVSEGIGKLSMVAQSATKEITAKVKDG 288
L ++ G+ + + P Q L ++ + L+ Q A ++T
Sbjct: 241 QATLKAAELGQTLN-ENVIKPTQEKVKDGKLLDEMAVSVTGLANKVQGAGAKLTNLFMGK 299
Query: 289 GYDHKVNETVNIVTQKTSEIGQRTWGIMKGVMALASQKVEEFTKDYPDGNSDNWPRNEND 348
D E+ + G + + + K P +SD+W +N
Sbjct: 300 AEDGSAGESYQNMDPNQGYQGSSSQSVSRDFWETFGSSSSLKAKKSP--SSDSWTVADNS 357
Query: 349 RHDFNQENKGWNSSTSTREGQPSSGGQTNTYHSNSWD-DWDNQDTR-KEVPAKGSAPH-N 405
+ W S ++ + + +WD +W+N D + K+ P K + N
Sbjct: 358 AKKSSDSWDNWGSEAASNKHSTEESWE-------AWDNNWENADGKSKKSPTKAAEESWN 410
Query: 406 NDDW 409
N +W
Sbjct: 411 NAEW 414
>UniRef100_UPI000036C64D UPI000036C64D UniRef100 entry
Length = 406
Score = 165 bits (418), Expect = 2e-39
Identities = 132/437 (30%), Positives = 191/437 (43%), Gaps = 70/437 (16%)
Query: 7 LRELQSEPSNKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVTMDSWSD 66
L+E++ + N +C +C NPQW SV+YG+++CLECSG+HRGLGVH+SFVRSVTMD W D
Sbjct: 10 LKEVRVQDENNVCFECGAFNPQWVSVTYGIWICLECSGRHRGLGVHLSFVRSVTMDKWKD 69
Query: 67 LQIKKMEAGGNRNLNTFL-SQYGISKETDIITKYNSNAASIYRDRIQAIAEGRSWR---- 121
++++KM+AGGN FL SQ + KYNS AA+++RD++ +A+GR W
Sbjct: 70 IELEKMKAGGNAKFREFLESQEDYDPCWSLQEKYNSRAAALFRDKVATLAKGREWSLESS 129
Query: 122 -----DPPVVKENAST-RAGKGKPPLAAASNGGGWDDNWDNDDGDSYGYGSRGGGDIRRN 175
PP + ST G+P AS+ ++D W NDD SY G++G
Sbjct: 130 PAQNWTPPQPRTLPSTVHRASGQPQSVTASSDKAFED-WLNDDLGSY-QGAQGN------ 181
Query: 176 QSTGDVIGFVGNGVTPSSRSKSTEDIYTRTQLEASAANKEGFFAKKMAENESRPEGLPPS 235
+VG G TP + K ED + + + + F + EG
Sbjct: 182 -------RYVGFGNTPPPQKK--EDDFLNNAMSSLYSGWSSFTTGASRFASAAKEGA--- 229
Query: 236 QGGKYVGFGSSPGPAQRISPQNDYLSVVSEGIGKLSMVAQSATKEITAKVKDGGYDHKVN 295
FGS +Q+ S L+ ++ K KVK+G V+
Sbjct: 230 -----TKFGSQ--ASQKASELGHSLN-------------ENVLKPAQEKVKEGKIFDDVS 269
Query: 296 ETVNIVTQKTSEIGQRTWGIMKGVMALASQKVEEFTKDYPDGNSDNWPRNENDRHDFNQE 355
V+ + K +G + W + V S K E +G+S +N D Q
Sbjct: 270 SGVSQLASKVQGVGSKGW---RDVTTFFSGKAEGPLDSPSEGHS-----YQNSCPDHFQN 321
Query: 356 NKGWNSSTSTREGQPSSGGQTNTYHSNSWDDWDNQDTRKEVPAKGSAPHNNDDWAGWDDA 415
+ S T S T T S S D W DT E ++D W W A
Sbjct: 322 SNIDQSFWETF----GSAEPTKTRKSPSSDSWTCADTSTE-------RRSSDSWEVWGSA 370
Query: 416 KDDDDEFDDKSFGNNGT 432
+ + D G GT
Sbjct: 371 STNRNSNSDGGEGGEGT 387
>UniRef100_UPI000036C64C UPI000036C64C UniRef100 entry
Length = 414
Score = 165 bits (417), Expect = 3e-39
Identities = 135/443 (30%), Positives = 195/443 (43%), Gaps = 74/443 (16%)
Query: 7 LRELQSEPSNKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVTMDSWSD 66
L+E++ + N +C +C NPQW SV+YG+++CLECSG+HRGLGVH+SFVRSVTMD W D
Sbjct: 10 LKEVRVQDENNVCFECGAFNPQWVSVTYGIWICLECSGRHRGLGVHLSFVRSVTMDKWKD 69
Query: 67 LQIKKMEAGGNRNLNTFL-SQYGISKETDIITKYNSNAASIYRDRIQAIAEGRSWR---- 121
++++KM+AGGN FL SQ + KYNS AA+++RD++ +A+GR W
Sbjct: 70 IELEKMKAGGNAKFREFLESQEDYDPCWSLQEKYNSRAAALFRDKVATLAKGREWSLESS 129
Query: 122 -----DPPVVKENAST-RAGKGKPPLAAASNGGGWDDNWDNDDGDSYGYGSRGGGDIRRN 175
PP + ST G+P AS+ ++D W NDD SY G++G
Sbjct: 130 PAQNWTPPQPRTLPSTVHRASGQPQSVTASSDKAFED-WLNDDLGSY-QGAQGN------ 181
Query: 176 QSTGDVIGFVGNGVTPSSRSKSTEDIYTRTQLEASAANKEGF------FAKKMAENESRP 229
+VG G TP + K ED + + + + F FA E ++
Sbjct: 182 -------RYVGFGNTPPPQKK--EDDFLNNAMSSLYSGWSSFTTGASRFASAAKEGATK- 231
Query: 230 EGLPPSQGGKYVGFGSSPGPAQRISPQNDYLSVVSEGIGKLSMVAQSATKEITAKVKDGG 289
G SQ K+ G P PA + + ++ K KVK+G
Sbjct: 232 FGSQASQ--KFWGHKQQPEPASELG----------------HSLNENVLKPAQEKVKEGK 273
Query: 290 YDHKVNETVNIVTQKTSEIGQRTWGIMKGVMALASQKVEEFTKDYPDGNSDNWPRNENDR 349
V+ V+ + K +G + W + V S K E +G+S +N
Sbjct: 274 IFDDVSSGVSQLASK--GVGSKGW---RDVTTFFSGKAEGPLDSPSEGHS-----YQNSC 323
Query: 350 HDFNQENKGWNSSTSTREGQPSSGGQTNTYHSNSWDDWDNQDTRKEVPAKGSAPHNNDDW 409
D Q + S T S T T S S D W DT E ++D W
Sbjct: 324 PDHFQNSNIDQSFWETF----GSAEPTKTRKSPSSDSWTCADTSTE-------RRSSDSW 372
Query: 410 AGWDDAKDDDDEFDDKSFGNNGT 432
W A + + D G GT
Sbjct: 373 EVWGSASTNRNSNSDGGEGGEGT 395
>UniRef100_Q8N6T3 ADP-ribosylation factor GTPase activating protein 1 [Homo sapiens]
Length = 406
Score = 165 bits (417), Expect = 3e-39
Identities = 133/437 (30%), Positives = 191/437 (43%), Gaps = 70/437 (16%)
Query: 7 LRELQSEPSNKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVTMDSWSD 66
L+E++ + N +C +C NPQW SV+YG+++CLECSG+HRGLGVH+SFVRSVTMD W D
Sbjct: 10 LKEVRVQDENNVCFECGAFNPQWVSVTYGIWICLECSGRHRGLGVHLSFVRSVTMDKWKD 69
Query: 67 LQIKKMEAGGNRNLNTFL-SQYGISKETDIITKYNSNAASIYRDRIQAIAEGRSWR---- 121
++++KM+AGGN FL SQ + KYNS AA+++RD++ A+AEGR W
Sbjct: 70 IELEKMKAGGNAKFREFLESQEDYDPCWSLQEKYNSRAAALFRDKVVALAEGREWSLESS 129
Query: 122 -----DPPVVKENAS-TRAGKGKPPLAAASNGGGWDDNWDNDDGDSYGYGSRGGGDIRRN 175
PP + S G+P AS+ ++D W NDD SY G++G
Sbjct: 130 PAQNWTPPQPRTLPSMVHRVSGQPQSVTASSDKAFED-WLNDDLGSY-QGAQGN------ 181
Query: 176 QSTGDVIGFVGNGVTPSSRSKSTEDIYTRTQLEASAANKEGFFAKKMAENESRPEGLPPS 235
+VG G TP + K ED + + + + F + EG
Sbjct: 182 -------RYVGFGNTPPPQKK--EDDFLNNAMSSLYSGWSSFTTGASRFASAAKEGA--- 229
Query: 236 QGGKYVGFGSSPGPAQRISPQNDYLSVVSEGIGKLSMVAQSATKEITAKVKDGGYDHKVN 295
FGS +Q+ S L+ ++ K KVK+G V+
Sbjct: 230 -----TKFGSQ--ASQKASELGHSLN-------------ENVLKPAQEKVKEGKIFDDVS 269
Query: 296 ETVNIVTQKTSEIGQRTWGIMKGVMALASQKVEEFTKDYPDGNSDNWPRNENDRHDFNQE 355
V+ + K +G + W + V S K E +G+S +N D Q
Sbjct: 270 SGVSQLASKVQGVGSKGW---RDVTTFFSGKAEGPLDSPSEGHS-----YQNSGLDHFQN 321
Query: 356 NKGWNSSTSTREGQPSSGGQTNTYHSNSWDDWDNQDTRKEVPAKGSAPHNNDDWAGWDDA 415
+ S T S T T S S D W DT E ++D W W A
Sbjct: 322 SNIDQSFWETF----GSAEPTKTRKSPSSDSWTCADTSTE-------RRSSDSWEVWGSA 370
Query: 416 KDDDDEFDDKSFGNNGT 432
+ + D G GT
Sbjct: 371 STNRNSNSDGGEGGEGT 387
>UniRef100_Q8N6T3-2 Splice isoform 2 of Q8N6T3 [Homo sapiens]
Length = 414
Score = 164 bits (416), Expect = 4e-39
Identities = 136/443 (30%), Positives = 195/443 (43%), Gaps = 74/443 (16%)
Query: 7 LRELQSEPSNKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVTMDSWSD 66
L+E++ + N +C +C NPQW SV+YG+++CLECSG+HRGLGVH+SFVRSVTMD W D
Sbjct: 10 LKEVRVQDENNVCFECGAFNPQWVSVTYGIWICLECSGRHRGLGVHLSFVRSVTMDKWKD 69
Query: 67 LQIKKMEAGGNRNLNTFL-SQYGISKETDIITKYNSNAASIYRDRIQAIAEGRSWR---- 121
++++KM+AGGN FL SQ + KYNS AA+++RD++ A+AEGR W
Sbjct: 70 IELEKMKAGGNAKFREFLESQEDYDPCWSLQEKYNSRAAALFRDKVVALAEGREWSLESS 129
Query: 122 -----DPPVVKENAS-TRAGKGKPPLAAASNGGGWDDNWDNDDGDSYGYGSRGGGDIRRN 175
PP + S G+P AS+ ++D W NDD SY G++G
Sbjct: 130 PAQNWTPPQPRTLPSMVHRVSGQPQSVTASSDKAFED-WLNDDLGSY-QGAQGN------ 181
Query: 176 QSTGDVIGFVGNGVTPSSRSKSTEDIYTRTQLEASAANKEGF------FAKKMAENESRP 229
+VG G TP + K ED + + + + F FA E ++
Sbjct: 182 -------RYVGFGNTPPPQKK--EDDFLNNAMSSLYSGWSSFTTGASRFASAAKEGATK- 231
Query: 230 EGLPPSQGGKYVGFGSSPGPAQRISPQNDYLSVVSEGIGKLSMVAQSATKEITAKVKDGG 289
G SQ K+ G P PA + + ++ K KVK+G
Sbjct: 232 FGSQASQ--KFWGHKQQPEPASELG----------------HSLNENVLKPAQEKVKEGK 273
Query: 290 YDHKVNETVNIVTQKTSEIGQRTWGIMKGVMALASQKVEEFTKDYPDGNSDNWPRNENDR 349
V+ V+ + K +G + W + V S K E +G+S +N
Sbjct: 274 IFDDVSSGVSQLASK--GVGSKGW---RDVTTFFSGKAEGPLDSPSEGHS-----YQNSG 323
Query: 350 HDFNQENKGWNSSTSTREGQPSSGGQTNTYHSNSWDDWDNQDTRKEVPAKGSAPHNNDDW 409
D Q + S T S T T S S D W DT E ++D W
Sbjct: 324 LDHFQNSNIDQSFWETF----GSAEPTKTRKSPSSDSWTCADTSTE-------RRSSDSW 372
Query: 410 AGWDDAKDDDDEFDDKSFGNNGT 432
W A + + D G GT
Sbjct: 373 EVWGSASTNRNSNSDGGEGGEGT 395
>UniRef100_UPI00003AC258 UPI00003AC258 UniRef100 entry
Length = 396
Score = 161 bits (407), Expect = 4e-38
Identities = 128/426 (30%), Positives = 192/426 (45%), Gaps = 89/426 (20%)
Query: 7 LRELQSEPSNKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVTMDSWSD 66
L+E++++ N +C +C NPQW SV+YG+++CLECSGKHRGLGVH+SFVRSVTMD W D
Sbjct: 10 LKEVRAQDENNVCFECGAFNPQWVSVTYGIWICLECSGKHRGLGVHLSFVRSVTMDKWKD 69
Query: 67 LQIKKMEAGGNRNLNTFL-SQYGISKETDIITKYNSNAASIYRDRIQAIAEGRSWR---- 121
++++KM+AGGN FL SQ + KYNS AA+++RD++ +AEG+ W
Sbjct: 70 IELEKMKAGGNSKFREFLESQDDYDPCWSMQEKYNSKAAALFRDQVATVAEGKEWSLETS 129
Query: 122 -----DPPVVKEN-ASTRAGKGKPPLAAASNGGGWDDNWDNDDGDSYGYGSRGGGDIRRN 175
PP K + +ST G P AS+ ++D W NDD +SY +GG + R
Sbjct: 130 PARNWTPPQPKTSLSSTHRSAGPPQNTTASSDKAFED-WLNDDVNSY----QGGQENR-- 182
Query: 176 QSTGDVIGFVGNGVTPSSRSKSTEDIYTRTQLEASAANKEGFFAKKMAENESRPEGLPPS 235
+GF GN VTP + ED + L +
Sbjct: 183 -----YVGF-GNTVTPPKK----EDDF-----------------------------LNNA 203
Query: 236 QGGKYVGFGSSPGPAQRISPQNDYLSVVSEGIGKLSMVAQSATKEITAKVKDGGYDHKVN 295
Y G+ S A +I+ S EG + A E+ +N
Sbjct: 204 MSSLYSGWSSFTTGASKIA------SAAKEGATRFGSQASQKASEL---------GQTLN 248
Query: 296 ETVNIVTQKTSEIGQRTWGIMKGVMALASQKVEEFTKDYPDGNSDNWPRNENDRHDFNQE 355
E V Q+ +G + W + V S K ++ ++ +G D++ + + + N
Sbjct: 249 ENVLKPAQEKG-VGSKGW---RDVTTFFSGKPDDHSERPAEG--DSYQNSGGEGYQNNAV 302
Query: 356 NKGW-----NSSTSTREGQPSSGGQT---NTYHSNSWDDWDNQDTRKEVPAKGSAPHNND 407
++ + NS PSS T N+ S D WD + K S N+D
Sbjct: 303 DQSFWETFGNSDPPKAHKSPSSESWTYVDNSTEKKSSDSWDVWGSGTVSNNKNS---NSD 359
Query: 408 DWAGWD 413
W W+
Sbjct: 360 SWENWE 365
>UniRef100_UPI0000364090 UPI0000364090 UniRef100 entry
Length = 372
Score = 157 bits (397), Expect = 6e-37
Identities = 110/381 (28%), Positives = 176/381 (45%), Gaps = 41/381 (10%)
Query: 5 RRLRELQSEPSNKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVTMDSW 64
R L+E++++ N +C +C NPQW SV+YG+++CLECSGKHRGLGVH+SFVRSVTMD W
Sbjct: 8 RVLKEVRTQDENNLCFECGAFNPQWVSVTYGIWICLECSGKHRGLGVHLSFVRSVTMDKW 67
Query: 65 SDLQIKKMEAGGNRNLNTFLS-QYGISKETDIITKYNSNAASIYRDRIQAIAEG------ 117
D++++KM+AGGN FL Q + + KYNS AA+++RD++ +AEG
Sbjct: 68 KDIELEKMKAGGNGKFRLFLELQDDFNPNWTLQEKYNSKAAALFRDKVATLAEGKEWSME 127
Query: 118 ----RSWRDPPVVKENASTRAGKGKPPLAAASNGGGWDDNWDNDDGDSYGYGSRGGGDIR 173
R+W P +S+ G AASN ++D W +DD +SY G++
Sbjct: 128 TSPARNWTSPQPKTGLSSSHRSGGSGQNTAASNDKAFED-WLSDDVNSYQSGNQ------ 180
Query: 174 RNQSTGDVIGFVGNGVTPSSR-----SKSTEDIYTRTQLEASAANKEGFFAKKMAENESR 228
+GF GN VTP + + + IY+ A+K AK +
Sbjct: 181 ----ENRYVGF-GNTVTPEKKEDDLFNNAMSSIYSGWSSFTVGASKIASIAKDNTAKLAN 235
Query: 229 PEGLPPSQGGKYVGFGSSPGPAQRISPQN--DYLSVVSEGI-GKLSMVAQSATKEITAKV 285
L ++ G+ + +++ D ++V G+ K+ T K
Sbjct: 236 QATLKATELGQTLNENVIKPTQEKVKDGKLLDEMAVSVTGLANKVQGPGTKLTNLFMGKA 295
Query: 286 KDGGYDHKV-NETVNIVTQKTSEIGQRTWGIMKGVMALASQKVEEFTKDYPDGNSDNWPR 344
+DG + N N Q + + + W +L ++K +SD+W
Sbjct: 296 EDGSTEESYQNMDPNQGYQGSQSVSRDFWETFGSNTSLKAKK---------SPSSDSWTM 346
Query: 345 NENDRHDFNQENKGWNSSTST 365
+N + W S ++
Sbjct: 347 ADNSAKKSSDSWDNWGSEAAS 367
>UniRef100_UPI00002194E2 UPI00002194E2 UniRef100 entry
Length = 462
Score = 157 bits (397), Expect = 6e-37
Identities = 120/418 (28%), Positives = 185/418 (43%), Gaps = 66/418 (15%)
Query: 6 RLRELQSEPSNKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVTMDSWS 65
+L +Q E N +C DC +PQWAS +G+F+CL C+G HRGLGVHISFVRS++MD++
Sbjct: 75 KLLAIQKESKNSLCCDCGAPSPQWASPKFGIFICLSCAGVHRGLGVHISFVRSISMDAFK 134
Query: 66 DLQIKKMEAGGNRNLNTFLSQY------GIS-KETDIITKYNSNAASIYRDRIQAIAEGR 118
+I++M GGN F ++ GIS + I +Y A Y++R+ A+ +GR
Sbjct: 135 AAEIERMRLGGNERWREFFEKHADTELRGISWDDATIAERYGGEAGDEYKERLSALVDGR 194
Query: 119 SWRDPPVVKENASTRAGKGKPPLAAASNGGGWDDNWDNDDGDSYGYGSRGGGDIRRNQST 178
+ K + + PL +++G GGG R + S
Sbjct: 195 EYVPGEKKKAAPAENTNRSTAPLGGSADG---------------SKNGGGGGRFRDDPSL 239
Query: 179 GDVIGFVGNGVTPSSRSKSTEDIYTRTQLEASAANKEGFFAKKMAENESRPEGLPPSQGG 238
G +G S +D Y FA+ A+N SR E LPPSQGG
Sbjct: 240 GGSRSESPSGGATSGGKTRVDDKY---------------FARLGADNASRSEDLPPSQGG 284
Query: 239 KYVGFGSSPGPA-----------QRISPQNDYLSVVSEGIGKL-SMVAQSATK------E 280
KY GFG++P PA QN+ ++ +S+G G S V ++AT +
Sbjct: 285 KYAGFGNTPAPAPGGAGGNTALPSMDELQNNPVAALSKGFGWFASTVTKTATTVNNNYIQ 344
Query: 281 ITAK-VKDGGYDHKVNETVNIV---TQKTSEIGQRTWG-IMKGVMALASQKV-----EEF 330
TAK + D + + T V Q+ ++ Q ++ ++G + Q+V +E
Sbjct: 345 PTAKQIADSDFAAQARNTAGTVGRAAQQGAKSAQTSFNRFVEGPDSRQYQQVRQAPIDET 404
Query: 331 TKDYPDGNSDNWPRNENDR-HDFNQENKGWNSSTSTREGQPSSGGQTNTYHSNSWDDW 387
K + D SD + + + + S G P S + T + WDDW
Sbjct: 405 RKGFWDDFSDIPEQQQKQQPKPSSIGTAAMGKGGSGGAGGPGSASASQTAKKDEWDDW 462
>UniRef100_UPI0000364091 UPI0000364091 UniRef100 entry
Length = 350
Score = 153 bits (386), Expect = 1e-35
Identities = 84/209 (40%), Positives = 122/209 (58%), Gaps = 26/209 (12%)
Query: 5 RRLRELQSEPSNKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVTMDSW 64
R L+E++++ N +C +C NPQW SV+YG+++CLECSGKHRGLGVH+SFVRSVTMD W
Sbjct: 8 RVLKEVRTQDENNLCFECGAFNPQWVSVTYGIWICLECSGKHRGLGVHLSFVRSVTMDKW 67
Query: 65 SDLQIKKMEAGGNRNLNTFLS-QYGISKETDIITKYNSNAASIYRDRIQAIAEG------ 117
D++++KM+AGGN FL Q + + KYNS AA+++RD++ +AEG
Sbjct: 68 KDIELEKMKAGGNGKFRLFLELQDDFNPNWTLQEKYNSKAAALFRDKVATLAEGKEWSME 127
Query: 118 ----RSWRDPPVVKENASTRAGKGKPPLAAASNGGGWDDNWDNDDGDSYGYGSRGGGDIR 173
R+W P +S+ G AASN ++D W +DD +SY G++
Sbjct: 128 TSPARNWTSPQPKTGLSSSHRSGGSGQNTAASNDKAFED-WLSDDVNSYQSGNQ------ 180
Query: 174 RNQSTGDVIGFVGNGVTPSSRSKSTEDIY 202
+GF GN VTP K +D++
Sbjct: 181 ----ENRYVGF-GNTVTP---EKKEDDLF 201
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.309 0.129 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 849,052,208
Number of Sequences: 2790947
Number of extensions: 40637985
Number of successful extensions: 133983
Number of sequences better than 10.0: 1100
Number of HSP's better than 10.0 without gapping: 687
Number of HSP's successfully gapped in prelim test: 440
Number of HSP's that attempted gapping in prelim test: 126892
Number of HSP's gapped (non-prelim): 4205
length of query: 443
length of database: 848,049,833
effective HSP length: 130
effective length of query: 313
effective length of database: 485,226,723
effective search space: 151875964299
effective search space used: 151875964299
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 76 (33.9 bits)
Medicago: description of AC148343.5


