

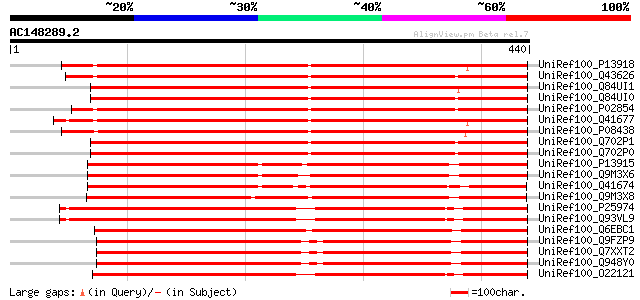
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148289.2 + phase: 0 /pseudo
(440 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_P13918 Vicilin precursor [Pisum sativum] 602 e-171
UniRef100_Q43626 Vicilin 47kD protein [Pisum sativum] 598 e-169
UniRef100_Q84UI1 Allergen Len c 1.0101 [Lens culinaris] 587 e-166
UniRef100_Q84UI0 Allergen Len c 1.0102 [Lens culinaris] 583 e-165
UniRef100_P02854 Provicilin precursor [Pisum sativum] 582 e-165
UniRef100_Q41677 Vicilin precursor [Vicia narbonensis] 577 e-163
UniRef100_P08438 Vicilin precursor [Vicia faba] 575 e-163
UniRef100_Q702P1 Vicilin [Pisum sativum] 575 e-162
UniRef100_Q702P0 Vicilin [Pisum sativum] 570 e-161
UniRef100_P13915 Convicilin precursor [Pisum sativum] 490 e-137
UniRef100_Q9M3X6 Convicilin precursor [Pisum sativum] 487 e-136
UniRef100_Q41674 Convicilin precursor [Vicia narbonensis] 483 e-135
UniRef100_Q9M3X8 Convicilin [Lens culinaris] 471 e-131
UniRef100_P25974 Beta-conglycinin, beta chain precursor [Glycine... 424 e-117
UniRef100_Q93VL9 Beta-conglycinin beta-subunit [Glycine max] 423 e-117
UniRef100_Q6EBC1 Beta-conglutin [Lupinus albus] 417 e-115
UniRef100_Q9FZP9 Alpha' subunit of beta-conglycinin [Glycine max] 407 e-112
UniRef100_Q7XXT2 Prepro beta-conglycinin alpha prime subunit [Gl... 407 e-112
UniRef100_Q948Y0 Beta-conglycinin alpha prime subunit [Glycine max] 407 e-112
UniRef100_O22121 Beta subunit of beta conglycinin [Glycine max] 404 e-111
>UniRef100_P13918 Vicilin precursor [Pisum sativum]
Length = 459
Score = 602 bits (1551), Expect = e-171
Identities = 303/398 (76%), Positives = 356/398 (89%), Gaps = 7/398 (1%)
Query: 45 IKAPCSLLMLLGIVFLASICVSSRSDQDQENPFIFNSNRFQTLFENENGHIRLLQRFDKR 104
+KA LLML+GI FLAS+CVSSRSD +NPFIF SN+FQTLFENENGHIRLLQ+FD+R
Sbjct: 6 MKASFPLLMLMGISFLASVCVSSRSDP--QNPFIFKSNKFQTLFENENGHIRLLQKFDQR 63
Query: 105 SKIFENLQNYRLLEYHSKPHTLFLPQHNDADFILVVVSGKAILTVLNPNNRNSFNLERGD 164
SKIFENLQNYRLLEY SKPHT+FLPQH DAD+ILVV+SGKAILTVL P++RNSFNLERGD
Sbjct: 64 SKIFENLQNYRLLEYKSKPHTIFLPQHTDADYILVVLSGKAILTVLKPDDRNSFNLERGD 123
Query: 165 TIKLPAGTLGYLANRDDNKDLRVLDLAIPVNRPGQFQSFSLSESENQQSFLSGFSKNILE 224
TIKLPAGT+ YL NRDDN++LRVLDLAIPVNRPGQ QSF LS ++NQQ++LSGFSKNILE
Sbjct: 124 TIKLPAGTIAYLVNRDDNEELRVLDLAIPVNRPGQLQSFLLSGNQNQQNYLSGFSKNILE 183
Query: 225 AAFNSNYEEIERVLIEENEQEPQHRRGLRKDERRQQSQEANVIVKVSREQIEELSKNAKS 284
A+FN++YEEIE+VL+EE+E+E QHRR L+ ++RQQSQE NVIVK+SR QIEELSKNAKS
Sbjct: 184 ASFNTDYEEIEKVLLEEHEKETQHRRSLK--DKRQQSQEENVIVKLSRGQIEELSKNAKS 241
Query: 285 SSRRSESSESEPINLRNQKPIYSNKFGNFFEITPEKNPQLKDLDILVNYAEIREGSLLLP 344
+S++S SSESEP NLR++ PIYSN+FG FFEITPEKNPQL+DLDI VN EI+EGSLLLP
Sbjct: 242 TSKKSVSSESEPFNLRSRGPIYSNEFGKFFEITPEKNPQLQDLDIFVNSVEIKEGSLLLP 301
Query: 345 HFNSRATVIVAVEEGKGEFELVGQRNENQQEQREYEEDEQQQ---ERSQQVQRYRARLSP 401
H+NSRA VIV V EGKG+FELVGQRNENQQEQR+ +++E++Q E ++QVQ Y+A+LS
Sbjct: 302 HYNSRAIVIVTVNEGKGDFELVGQRNENQQEQRKEDDEEEEQGEEEINKQVQNYKAKLSS 361
Query: 402 GDVYVIPAGHPIVVTASSDLSLLGFGINAENNQRNFLA 439
GDV+VIPAGHP+ V ASS+L LLGFGINAENNQRNFLA
Sbjct: 362 GDVFVIPAGHPVAVKASSNLDLLGFGINAENNQRNFLA 399
Score = 37.4 bits (85), Expect = 0.86
Identities = 48/222 (21%), Positives = 87/222 (38%), Gaps = 32/222 (14%)
Query: 66 SSRSDQDQENPFIFNSNRFQTLFENENGHIRLLQRFDKRSKIFENLQNYRLLEYHSKPHT 125
S +S + PF S ++ NE G + + +++ ++L + + K +
Sbjct: 243 SKKSVSSESEPFNLRSRG--PIYSNEFG--KFFEITPEKNPQLQDLDIF-VNSVEIKEGS 297
Query: 126 LFLPQHNDADFILVVVS-GKAILTVLNPNNRNSFNLERGDTIKLPAG-------TLGYLA 177
L LP +N ++V V+ GK ++ N N + D + G Y A
Sbjct: 298 LLLPHYNSRAIVIVTVNEGKGDFELVGQRNENQQEQRKEDDEEEEQGEEEINKQVQNYKA 357
Query: 178 NRDDNKDLRVLDLAIPVNRPGQFQ----SFSLSESENQQSFLSGFSKNIL--------EA 225
D+ V+ PV F ++ NQ++FL+G N++ E
Sbjct: 358 KLSSG-DVFVIPAGHPVAVKASSNLDLLGFGINAENNQRNFLAGDEDNVISQIQRPVKEL 416
Query: 226 AFNSNYEEIERVLIEENE------QEPQHRRGLRKDERRQQS 261
AF + +E++R+L + + Q Q RG R+ R S
Sbjct: 417 AFPGSAQEVDRILENQKQSHFADAQPQQRERGSRETRDRLSS 458
>UniRef100_Q43626 Vicilin 47kD protein [Pisum sativum]
Length = 438
Score = 598 bits (1541), Expect = e-169
Identities = 303/392 (77%), Positives = 346/392 (87%), Gaps = 5/392 (1%)
Query: 48 PCSLLMLLGIVFLASICVSSRSDQDQENPFIFNSNRFQTLFENENGHIRLLQRFDKRSKI 107
P LMLL I FLAS+CVSSRSDQ ENPFIF SNRFQTL+ENENGHIRLLQ+FDKRSKI
Sbjct: 5 PIKPLMLLAIAFLASVCVSSRSDQ--ENPFIFKSNRFQTLYENENGHIRLLQKFDKRSKI 62
Query: 108 FENLQNYRLLEYHSKPHTLFLPQHNDADFILVVVSGKAILTVLNPNNRNSFNLERGDTIK 167
FENLQNYRLLEY SKPHTLFLPQ+ DADFILVV+SGKA LTVL N+RNSFNLERGD IK
Sbjct: 63 FENLQNYRLLEYKSKPHTLFLPQYTDADFILVVLSGKATLTVLKSNDRNSFNLERGDAIK 122
Query: 168 LPAGTLGYLANRDDNKDLRVLDLAIPVNRPGQFQSFSLSESENQQSFLSGFSKNILEAAF 227
LPAGT+ YLANRDDN+DLRVLDLAIPVN+PGQ QSF LS ++NQ S LSGFSKNILEAAF
Sbjct: 123 LPAGTIAYLANRDDNEDLRVLDLAIPVNKPGQLQSFLLSGTQNQPSLLSGFSKNILEAAF 182
Query: 228 NSNYEEIERVLIEENEQEPQHRRGLRKDERRQQSQEANVIVKVSREQIEELSKNAKSSSR 287
N+NYEEIE+VL+E+ EQEPQHRR L+ +RRQ+ E NVIVKVSREQIEELSKNAKSSS+
Sbjct: 183 NTNYEEIEKVLLEQQEQEPQHRRSLK--DRRQEINEENVIVKVSREQIEELSKNAKSSSK 240
Query: 288 RSESSESEPINLRNQKPIYSNKFGNFFEITPEKNPQLKDLDILVNYAEIREGSLLLPHFN 347
+S SSES P NLR++ PIYSNKFG FFEITPEKN QL+DLDI VN +I+EGSLLLP++N
Sbjct: 241 KSVSSESGPFNLRSRNPIYSNKFGKFFEITPEKNQQLQDLDIFVNSVDIKEGSLLLPNYN 300
Query: 348 SRATVIVAVEEGKGEFELVGQRNENQQEQREYEEDEQQQERSQQVQRYRARLSPGDVYVI 407
SRA VIV V EGKG+FELVGQRNENQ ++ + +E+EQ++E S+QVQ YRA+LSPGDV+VI
Sbjct: 301 SRAIVIVTVTEGKGDFELVGQRNENQGKEND-KEEEQEEETSKQVQLYRAKLSPGDVFVI 359
Query: 408 PAGHPIVVTASSDLSLLGFGINAENNQRNFLA 439
PAGHP+ + ASSDL+L+GFGINAENN+RNFLA
Sbjct: 360 PAGHPVAINASSDLNLIGFGINAENNERNFLA 391
>UniRef100_Q84UI1 Allergen Len c 1.0101 [Lens culinaris]
Length = 418
Score = 587 bits (1513), Expect = e-166
Identities = 297/373 (79%), Positives = 333/373 (88%), Gaps = 4/373 (1%)
Query: 69 SDQDQENPFIFNSNRFQTLFENENGHIRLLQRFDKRSKIFENLQNYRLLEYHSKPHTLFL 128
S DQENPFIF SNRFQT++ENENGHIRLLQRFDKRSKIFENLQNYRLLEY SKPHT+FL
Sbjct: 1 SRSDQENPFIFKSNRFQTIYENENGHIRLLQRFDKRSKIFENLQNYRLLEYKSKPHTIFL 60
Query: 129 PQHNDADFILVVVSGKAILTVLNPNNRNSFNLERGDTIKLPAGTLGYLANRDDNKDLRVL 188
PQ DADFILVV+SGKAILTVLN N+RNSFNLERGDTIKLPAGT+ YLANRDDN+DLRVL
Sbjct: 61 PQFTDADFILVVLSGKAILTVLNSNDRNSFNLERGDTIKLPAGTIAYLANRDDNEDLRVL 120
Query: 189 DLAIPVNRPGQFQSFSLSESENQQSFLSGFSKNILEAAFNSNYEEIERVLIEENEQEPQH 248
DLAIPVNRPGQ QSF LS ++NQ SFLSGFSKNILEAAFN+ YEEIE+VL+EE EQ+ QH
Sbjct: 121 DLAIPVNRPGQLQSFLLSGTQNQPSFLSGFSKNILEAAFNTEYEEIEKVLLEEQEQKSQH 180
Query: 249 RRGLRKDERRQQSQEANVIVKVSREQIEELSKNAKSSSRRSESSESEPINLRNQKPIYSN 308
RR LR ++RQ+ +VIVKVSREQIEELSKNAKSSS++S SSESEP NLR++ PIYSN
Sbjct: 181 RRSLR--DKRQEITNEDVIVKVSREQIEELSKNAKSSSKKSVSSESEPFNLRSRNPIYSN 238
Query: 309 KFGNFFEITPEKNPQLKDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEGKGEFELVGQ 368
KFG FFEITPEKNPQL+DLDI VN EI+EGSLLLP++NSRA VIV V EGKG+FELVGQ
Sbjct: 239 KFGKFFEITPEKNPQLQDLDIFVNSVEIKEGSLLLPNYNSRAIVIVTVNEGKGDFELVGQ 298
Query: 369 RNENQQEQREY--EEDEQQQERSQQVQRYRARLSPGDVYVIPAGHPIVVTASSDLSLLGF 426
RNENQQEQRE EE+ Q++E ++QVQRYRARLSPGDV VIPAGHP+ + ASSDL+L+GF
Sbjct: 299 RNENQQEQREENDEEEGQEEETTKQVQRYRARLSPGDVLVIPAGHPVAINASSDLNLIGF 358
Query: 427 GINAENNQRNFLA 439
GINA+NNQRNFLA
Sbjct: 359 GINAKNNQRNFLA 371
Score = 35.4 bits (80), Expect = 3.3
Identities = 58/245 (23%), Positives = 92/245 (36%), Gaps = 41/245 (16%)
Query: 24 SQHSSSIASKLSEIISKINMAIKAPCSLLMLLGIVFLASICVSSRSDQDQENPFIFNS-- 81
SQH S+ K EI ++ ++ +K + L +S S +S + PF S
Sbjct: 178 SQHRRSLRDKRQEITNE-DVIVKVSREQIEELSKNAKSS---SKKSVSSESEPFNLRSRN 233
Query: 82 ----NRFQTLFENENGHIRLLQRFDKRSKIFENLQNYRLLEYHSKPHTLFLPQHNDADFI 137
N+F FE LQ D IF N K +L LP +N +
Sbjct: 234 PIYSNKFGKFFEITPEKNPQLQDLD----IFVN-------SVEIKEGSLLLPNYNSRAIV 282
Query: 138 LVVVS-GKAILTVLNPNNRNSFNL------ERGDTIKLPAGTLGYLANRDDNKDLRVLDL 190
+V V+ GK ++ N N E G + Y A D+ V+
Sbjct: 283 IVTVNEGKGDFELVGQRNENQQEQREENDEEEGQEEETTKQVQRYRARLSPG-DVLVIPA 341
Query: 191 AIPV--NRPGQFQ--SFSLSESENQQSFLSGFSKNIL--------EAAFNSNYEEIERVL 238
PV N F ++ NQ++FL+G N++ E AF + E++R+L
Sbjct: 342 GHPVAINASSDLNLIGFGINAKNNQRNFLAGEEDNVISQIQRPVKELAFPGSSREVDRLL 401
Query: 239 IEENE 243
+ +
Sbjct: 402 TNQKQ 406
>UniRef100_Q84UI0 Allergen Len c 1.0102 [Lens culinaris]
Length = 415
Score = 583 bits (1503), Expect = e-165
Identities = 289/371 (77%), Positives = 335/371 (89%), Gaps = 3/371 (0%)
Query: 69 SDQDQENPFIFNSNRFQTLFENENGHIRLLQRFDKRSKIFENLQNYRLLEYHSKPHTLFL 128
S DQENPFIF SNRFQT++ENENGHIRLLQ+FDKRSKIFENLQNYRLLEY SKPHTLFL
Sbjct: 1 SRSDQENPFIFKSNRFQTIYENENGHIRLLQKFDKRSKIFENLQNYRLLEYKSKPHTLFL 60
Query: 129 PQHNDADFILVVVSGKAILTVLNPNNRNSFNLERGDTIKLPAGTLGYLANRDDNKDLRVL 188
PQ+ DADFILVV+SGKA+LTVLN N+RNSFNLERGDTIKLPAGT+ YLANRDDN+DLRVL
Sbjct: 61 PQYTDADFILVVLSGKAVLTVLNSNDRNSFNLERGDTIKLPAGTIAYLANRDDNEDLRVL 120
Query: 189 DLAIPVNRPGQFQSFSLSESENQQSFLSGFSKNILEAAFNSNYEEIERVLIEENEQEPQH 248
DLAIPVN PGQ +SF LS ++NQ SFLSGF+K+ILEAAFN++YEEIE+VL+E+ EQEPQH
Sbjct: 121 DLAIPVNNPGQLESFLLSGTQNQPSFLSGFNKSILEAAFNTDYEEIEKVLLEDQEQEPQH 180
Query: 249 RRGLRKDERRQQSQEANVIVKVSREQIEELSKNAKSSSRRSESSESEPINLRNQKPIYSN 308
RR LR +RRQ+ + NVIVKVSREQI+ELSKNAKSSS++S SSESEP NLR++ PIYSN
Sbjct: 181 RRSLR--DRRQEINKENVIVKVSREQIKELSKNAKSSSKKSVSSESEPFNLRSRNPIYSN 238
Query: 309 KFGNFFEITPEKNPQLKDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEGKGEFELVGQ 368
KFG FFEITPEKNPQL+DLDI VN EI+EGSLLLP++NSRA VIV V EGKG FELVGQ
Sbjct: 239 KFGKFFEITPEKNPQLQDLDIFVNSVEIKEGSLLLPNYNSRAIVIVTVNEGKGYFELVGQ 298
Query: 369 RNENQQEQREYEEDEQQQERSQQVQRYRARLSPGDVYVIPAGHPIVVTASSDLSLLGFGI 428
RNENQ+E+ + +E+EQ++E S QVQRYRA+LSPGDV+V+PAGHP+ + ASSDL+L+GFGI
Sbjct: 299 RNENQREEND-DEEEQEEETSTQVQRYRAKLSPGDVFVVPAGHPVAINASSDLNLIGFGI 357
Query: 429 NAENNQRNFLA 439
NA+NNQRNFLA
Sbjct: 358 NAKNNQRNFLA 368
Score = 35.8 bits (81), Expect = 2.5
Identities = 57/256 (22%), Positives = 90/256 (34%), Gaps = 68/256 (26%)
Query: 25 QHSSSIASKLSEIISKINMAIKAPCSLLMLLGIVFLASICVSSRSDQDQENPFIFNS--- 81
QH S+ + EI +K N+ +K + L +S S +S + PF S
Sbjct: 179 QHRRSLRDRRQEI-NKENVIVKVSREQIKELSKNAKSS---SKKSVSSESEPFNLRSRNP 234
Query: 82 ---NRFQTLFENENGHIRLLQRFDKRSKIFENLQNYRLLEYHSKPHTLFLPQHNDADFIL 138
N+F FE LQ D IF N K +L LP +N ++
Sbjct: 235 IYSNKFGKFFEITPEKNPQLQDLD----IFVN-------SVEIKEGSLLLPNYNSRAIVI 283
Query: 139 VVVS-GKAILTVLNPNNRNS----------------------FNLERGDTIKLPAGTLGY 175
V V+ GK ++ N N L GD +PAG
Sbjct: 284 VTVNEGKGYFELVGQRNENQREENDDEEEQEEETSTQVQRYRAKLSPGDVFVVPAGH--- 340
Query: 176 LANRDDNKDLRVLDLAIPVNRPGQFQSFSLSESENQQSFLSGFSKNIL--------EAAF 227
+AI + F ++ NQ++FL+G N++ E AF
Sbjct: 341 -------------PVAINASSDLNLIGFGINAKNNQRNFLAGEEDNVISQIQRPVKELAF 387
Query: 228 NSNYEEIERVLIEENE 243
+ E++R+L + +
Sbjct: 388 PGSSREVDRLLTNQKQ 403
>UniRef100_P02854 Provicilin precursor [Pisum sativum]
Length = 410
Score = 582 bits (1499), Expect = e-165
Identities = 294/387 (75%), Positives = 341/387 (87%), Gaps = 5/387 (1%)
Query: 53 MLLGIVFLASICVSSRSDQDQENPFIFNSNRFQTLFENENGHIRLLQRFDKRSKIFENLQ 112
MLL I FLAS+CVSSRSDQ ENPFIF SNRFQTL+ENENGHIRLLQ+FDKRSKIFENLQ
Sbjct: 1 MLLAIAFLASVCVSSRSDQ--ENPFIFKSNRFQTLYENENGHIRLLQKFDKRSKIFENLQ 58
Query: 113 NYRLLEYHSKPHTLFLPQHNDADFILVVVSGKAILTVLNPNNRNSFNLERGDTIKLPAGT 172
NYRLLEY SKPHTLFLPQ+ DADFILVV+SGKA LTVL N+RNSFNLERGD IKLPAG+
Sbjct: 59 NYRLLEYKSKPHTLFLPQYTDADFILVVLSGKATLTVLKSNDRNSFNLERGDAIKLPAGS 118
Query: 173 LGYLANRDDNKDLRVLDLAIPVNRPGQFQSFSLSESENQQSFLSGFSKNILEAAFNSNYE 232
+ Y ANRDDN++ RVLDLAIPVN+PGQ QSF LS ++NQ+S LSGFSKNILEAAFN+NYE
Sbjct: 119 IAYFANRDDNEEPRVLDLAIPVNKPGQLQSFLLSGTQNQKSSLSGFSKNILEAAFNTNYE 178
Query: 233 EIERVLIEENEQEPQHRRGLRKDERRQQSQEANVIVKVSREQIEELSKNAKSSSRRSESS 292
EIE+VL+E+ EQEPQHRR L+ +RRQ+ E NVIVKVSR+QIEELSKNAKSSS++S SS
Sbjct: 179 EIEKVLLEQQEQEPQHRRSLK--DRRQEINEENVIVKVSRDQIEELSKNAKSSSKKSVSS 236
Query: 293 ESEPINLRNQKPIYSNKFGNFFEITPEKNPQLKDLDILVNYAEIREGSLLLPHFNSRATV 352
ES P NLR++ PIYSNKFG FFEITPEKN QL+DLDI VN +I+ GSLLLP++NSRA V
Sbjct: 237 ESGPFNLRSRNPIYSNKFGKFFEITPEKNQQLQDLDIFVNSVDIKVGSLLLPNYNSRAIV 296
Query: 353 IVAVEEGKGEFELVGQRNENQQEQREYEEDEQQQERSQQVQRYRARLSPGDVYVIPAGHP 412
IV V EGKG+FELVGQRNENQ ++ + +E+EQ++E S+QVQ YRA+LSPGDV+VIPAGHP
Sbjct: 297 IVTVTEGKGDFELVGQRNENQGKEND-KEEEQEEETSKQVQLYRAKLSPGDVFVIPAGHP 355
Query: 413 IVVTASSDLSLLGFGINAENNQRNFLA 439
+ + ASSDL+L+G GINAENN+RNFLA
Sbjct: 356 VAINASSDLNLIGLGINAENNERNFLA 382
>UniRef100_Q41677 Vicilin precursor [Vicia narbonensis]
Length = 463
Score = 577 bits (1488), Expect = e-163
Identities = 294/405 (72%), Positives = 349/405 (85%), Gaps = 9/405 (2%)
Query: 38 ISKINMAIKAPCSLLMLLGIVFLASICVSSRSDQDQENPFIFNSNRFQTLFENENGHIRL 97
++ I M + P LLMLLGI FLAS+CVSSRSDQ ENPFIF SN+FQTLFEN+NGHIRL
Sbjct: 1 MAAITMKVSFP--LLMLLGISFLASVCVSSRSDQ--ENPFIFKSNKFQTLFENDNGHIRL 56
Query: 98 LQRFDKRSKIFENLQNYRLLEYHSKPHTLFLPQHNDADFILVVVSGKAILTVLNPNNRNS 157
LQ+FD+RSKI ENLQNYRLLEY SKP T+FLPQ +ADFILVV+SGKAILTVL P++RNS
Sbjct: 57 LQKFDERSKILENLQNYRLLEYKSKPRTIFLPQQTNADFILVVLSGKAILTVLKPDDRNS 116
Query: 158 FNLERGDTIKLPAGTLGYLANRDDNKDLRVLDLAIPVNRPGQFQSFSLSESENQQSFLSG 217
FNLERGDTIKLPAGT+ YL N+DDN+DLRVLDLAIPVN P Q QSF LS SENQQS LSG
Sbjct: 117 FNLERGDTIKLPAGTIAYLVNKDDNEDLRVLDLAIPVNGPDQLQSFLLSGSENQQSILSG 176
Query: 218 FSKNILEAAFNSNYEEIERVLIEENEQEPQHRRGLRKDERRQQSQEANVIVKVSREQIEE 277
FSK++LEA+FN+ YEEIE+VL+EE E+E QHRR LR ++RQ SQ+ +VIVK+SR QIEE
Sbjct: 177 FSKSVLEASFNTGYEEIEKVLLEEREKETQHRRSLR--DKRQHSQDEDVIVKLSRGQIEE 234
Query: 278 LSKNAKSSSRRSESSESEPINLRNQKPIYSNKFGNFFEITPEKNPQLKDLDILVNYAEIR 337
LS+NAKSSS++S SSESEP NLR++ PIYSNKFG FFEITPEKNPQL+DLD+LVN EI+
Sbjct: 235 LSRNAKSSSKKSVSSESEPFNLRSRNPIYSNKFGKFFEITPEKNPQLQDLDVLVNSVEIK 294
Query: 338 EGSLLLPHFNSRATVIVAVEEGKGEFELVGQRNENQQEQREYEEDEQQQ---ERSQQVQR 394
EGSLLLPH+NSRA VIV V +GKG+FE+VGQRNEN+Q QR+ +++E++Q + QVQ
Sbjct: 295 EGSLLLPHYNSRAIVIVTVNDGKGDFEIVGQRNENRQGQRKEDDEEEEQGDENTNTQVQN 354
Query: 395 YRARLSPGDVYVIPAGHPIVVTASSDLSLLGFGINAENNQRNFLA 439
Y+A+LS GDV+VIPAGHP+ + ASS+L LLGFGINA+NNQRNFLA
Sbjct: 355 YKAKLSRGDVFVIPAGHPVSIKASSNLDLLGFGINAKNNQRNFLA 399
Score = 38.5 bits (88), Expect = 0.39
Identities = 37/169 (21%), Positives = 68/169 (39%), Gaps = 38/169 (22%)
Query: 122 KPHTLFLPQHNDADFILVVVS-GKAILTVLNPNNRNSFNLERGDTIKLPAGTLGYLANRD 180
K +L LP +N ++V V+ GK ++ N N + D + G D
Sbjct: 294 KEGSLLLPHYNSRAIVIVTVNDGKGDFEIVGQRNENRQGQRKEDDEEEEQG--------D 345
Query: 181 DNKDLRVLD----------LAIPVNRPGQFQS--------FSLSESENQQSFLSGFSKNI 222
+N + +V + IP P ++ F ++ NQ++FL+G N+
Sbjct: 346 ENTNTQVQNYKAKLSRGDVFVIPAGHPVSIKASSNLDLLGFGINAKNNQRNFLAGEEDNV 405
Query: 223 L--------EAAFNSNYEEIERVLIEENEQEPQHRRGLRKDERRQQSQE 263
+ E AF + +E++R+L Q+ H + +R + S E
Sbjct: 406 ISQIDRPVKELAFPGSAQEVDRLL---ENQKQSHFANAQPQQRERGSHE 451
>UniRef100_P08438 Vicilin precursor [Vicia faba]
Length = 463
Score = 575 bits (1483), Expect = e-163
Identities = 292/398 (73%), Positives = 347/398 (86%), Gaps = 7/398 (1%)
Query: 45 IKAPCSLLMLLGIVFLASICVSSRSDQDQENPFIFNSNRFQTLFENENGHIRLLQRFDKR 104
+K LL LLGI FLAS+C+SSRSDQD NPF+F SNRFQTLFENENGHIRLLQ+FD+
Sbjct: 6 LKDSFPLLTLLGIAFLASVCLSSRSDQD--NPFVFESNRFQTLFENENGHIRLLQKFDQH 63
Query: 105 SKIFENLQNYRLLEYHSKPHTLFLPQHNDADFILVVVSGKAILTVLNPNNRNSFNLERGD 164
SK+ ENLQNYRLLEY SKPHT+FLPQ DADFILVV+SGKAILTVL PN+RNSF+LERGD
Sbjct: 64 SKLLENLQNYRLLEYKSKPHTIFLPQQTDADFILVVLSGKAILTVLLPNDRNSFSLERGD 123
Query: 165 TIKLPAGTLGYLANRDDNKDLRVLDLAIPVNRPGQFQSFSLSESENQQSFLSGFSKNILE 224
TIKLPAGT+GYL NRDD +DLRVLDL IPVNRPG+ QSF LS ++NQ S LSGFSKNILE
Sbjct: 124 TIKLPAGTIGYLVNRDDEEDLRVLDLVIPVNRPGEPQSFLLSGNQNQPSILSGFSKNILE 183
Query: 225 AAFNSNYEEIERVLIEENEQEPQHRRGLRKDERRQQSQEANVIVKVSREQIEELSKNAKS 284
A+FN++Y+EIE+VL+EE+ +E HRRGL+ +RRQ+ QE NVIVK+SR+QIEEL+KNAKS
Sbjct: 184 ASFNTDYKEIEKVLLEEHGKEKYHRRGLK--DRRQRGQEENVIVKISRKQIEELNKNAKS 241
Query: 285 SSRRSESSESEPINLRNQKPIYSNKFGNFFEITPEKNPQLKDLDILVNYAEIREGSLLLP 344
SS++S SSESEP NLR+++PIYSNKFG FFEITP++NPQL+DL+I VNY EI EGSLLLP
Sbjct: 242 SSKKSTSSESEPFNLRSREPIYSNKFGKFFEITPKRNPQLQDLNIFVNYVEINEGSLLLP 301
Query: 345 HFNSRATVIVAVEEGKGEFELVGQRNENQQEQR-EYEEDEQQ--QERSQQVQRYRARLSP 401
H+NSRA VIV V EGKG+FELVGQRNENQQ R EY+E+++Q +E +QVQ Y+A+LSP
Sbjct: 302 HYNSRAIVIVTVNEGKGDFELVGQRNENQQGLREEYDEEKEQGEEEIRKQVQNYKAKLSP 361
Query: 402 GDVYVIPAGHPIVVTASSDLSLLGFGINAENNQRNFLA 439
GDV VIPAG+P+ + ASS+L+L+GFGINAENNQR FLA
Sbjct: 362 GDVLVIPAGYPVAIKASSNLNLVGFGINAENNQRYFLA 399
Score = 37.0 bits (84), Expect = 1.1
Identities = 48/218 (22%), Positives = 88/218 (40%), Gaps = 29/218 (13%)
Query: 66 SSRSDQDQENPFIFNSNRFQTLFENENGHIRLLQRFDKRSKIFENLQNYRLLEYHSKPHT 125
S +S + PF S + ++ N+ G + + KR+ ++L N + +
Sbjct: 243 SKKSTSSESEPFNLRSR--EPIYSNKFG--KFFEITPKRNPQLQDL-NIFVNYVEINEGS 297
Query: 126 LFLPQHNDADFILVVVS-GKAILTVLNPNNRNSFNL--------ERGDTIKLPAGTLGYL 176
L LP +N ++V V+ GK ++ N N L E+G+ ++ Y
Sbjct: 298 LLLPHYNSRAIVIVTVNEGKGDFELVGQRNENQQGLREEYDEEKEQGEE-EIRKQVQNYK 356
Query: 177 ANRDDNKDLRV---LDLAIPVNRPGQFQSFSLSESENQQSFLSGFSKNIL--------EA 225
A L + +AI + F ++ NQ+ FL+G N++ E
Sbjct: 357 AKLSPGDVLVIPAGYPVAIKASSNLNLVGFGINAENNQRYFLAGEEDNVISQIHKPVKEL 416
Query: 226 AFNSNYEEIERVLIEENEQEPQHRRGLRKDERRQQSQE 263
AF + +E++ +L Q+ H + ER + SQE
Sbjct: 417 AFPGSAQEVDTLL---ENQKQSHFANAQPRERERGSQE 451
>UniRef100_Q702P1 Vicilin [Pisum sativum]
Length = 415
Score = 575 bits (1481), Expect = e-162
Identities = 288/371 (77%), Positives = 330/371 (88%), Gaps = 3/371 (0%)
Query: 69 SDQDQENPFIFNSNRFQTLFENENGHIRLLQRFDKRSKIFENLQNYRLLEYHSKPHTLFL 128
S DQENPFIF SNRFQTL+ENENGHIRLLQ+FDKRSKIFENLQNYRLLEY SKPHTLFL
Sbjct: 1 SRSDQENPFIFKSNRFQTLYENENGHIRLLQKFDKRSKIFENLQNYRLLEYKSKPHTLFL 60
Query: 129 PQHNDADFILVVVSGKAILTVLNPNNRNSFNLERGDTIKLPAGTLGYLANRDDNKDLRVL 188
PQ+ DADFILVV+SGKA LTVL N+RNSFNLERGD IKLPAGT+ YLANRDDN+DLRVL
Sbjct: 61 PQYTDADFILVVLSGKATLTVLKSNDRNSFNLERGDAIKLPAGTIAYLANRDDNEDLRVL 120
Query: 189 DLAIPVNRPGQFQSFSLSESENQQSFLSGFSKNILEAAFNSNYEEIERVLIEENEQEPQH 248
DLAIPVN+PGQ QSF LS ++NQ S LSGFSKNILEAAFN+NYEEIE+VL+E+ EQEPQH
Sbjct: 121 DLAIPVNKPGQLQSFLLSGTQNQPSLLSGFSKNILEAAFNTNYEEIEKVLLEQQEQEPQH 180
Query: 249 RRGLRKDERRQQSQEANVIVKVSREQIEELSKNAKSSSRRSESSESEPINLRNQKPIYSN 308
RR L+ +RRQ+ E NVIVKVSREQIEELSKNAKSSS++S SSES P NLR++ PIYSN
Sbjct: 181 RRSLK--DRRQEINEENVIVKVSREQIEELSKNAKSSSKKSVSSESGPFNLRSRNPIYSN 238
Query: 309 KFGNFFEITPEKNPQLKDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEGKGEFELVGQ 368
KFG FFEITPEKN QL+DLDI VN +I+EGSLLLP++NSRA VIV V EGKG+FELVGQ
Sbjct: 239 KFGKFFEITPEKNQQLQDLDIFVNSVDIKEGSLLLPNYNSRAIVIVTVTEGKGDFELVGQ 298
Query: 369 RNENQQEQREYEEDEQQQERSQQVQRYRARLSPGDVYVIPAGHPIVVTASSDLSLLGFGI 428
RNENQ ++ + +E+EQ++E S+QVQ YRA+LSPGDV+VIPAGHP+ + ASSDL+L+GFGI
Sbjct: 299 RNENQGKEND-KEEEQEEETSKQVQLYRAKLSPGDVFVIPAGHPVAINASSDLNLIGFGI 357
Query: 429 NAENNQRNFLA 439
NAENN+RNFLA
Sbjct: 358 NAENNERNFLA 368
>UniRef100_Q702P0 Vicilin [Pisum sativum]
Length = 415
Score = 570 bits (1469), Expect = e-161
Identities = 287/371 (77%), Positives = 328/371 (88%), Gaps = 3/371 (0%)
Query: 69 SDQDQENPFIFNSNRFQTLFENENGHIRLLQRFDKRSKIFENLQNYRLLEYHSKPHTLFL 128
S DQENPFIF SNRFQTL+ENENGHIRLLQ+FDKRSKIFENLQNYRLLEY SKP TLFL
Sbjct: 1 SRSDQENPFIFKSNRFQTLYENENGHIRLLQKFDKRSKIFENLQNYRLLEYKSKPRTLFL 60
Query: 129 PQHNDADFILVVVSGKAILTVLNPNNRNSFNLERGDTIKLPAGTLGYLANRDDNKDLRVL 188
PQ DADFILVV+SGKA LTVL N+RNSFNLERGDTIKLPAGT+ YLANRDDN+DLRVL
Sbjct: 61 PQCTDADFILVVLSGKATLTVLKSNDRNSFNLERGDTIKLPAGTIAYLANRDDNEDLRVL 120
Query: 189 DLAIPVNRPGQFQSFSLSESENQQSFLSGFSKNILEAAFNSNYEEIERVLIEENEQEPQH 248
DL IPVN+PGQ QSF LS ++NQ S LSGFSKNILEAAFN+NYEEIE+VL+E+ EQEPQH
Sbjct: 121 DLTIPVNKPGQLQSFLLSGTQNQPSLLSGFSKNILEAAFNTNYEEIEKVLLEQQEQEPQH 180
Query: 249 RRGLRKDERRQQSQEANVIVKVSREQIEELSKNAKSSSRRSESSESEPINLRNQKPIYSN 308
RR L+ +RRQ+ E NVIVKVSREQIEELSKNAKSSS++S SSES P NLR++ PIYSN
Sbjct: 181 RRSLK--DRRQEINEENVIVKVSREQIEELSKNAKSSSKKSVSSESGPFNLRSRNPIYSN 238
Query: 309 KFGNFFEITPEKNPQLKDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEGKGEFELVGQ 368
KFG FFEITPEKN QL+DLDI VN +I+EGSLLLP++NSRA VIV V EGKG+FELVGQ
Sbjct: 239 KFGKFFEITPEKNQQLQDLDIFVNSVDIKEGSLLLPNYNSRAIVIVTVTEGKGDFELVGQ 298
Query: 369 RNENQQEQREYEEDEQQQERSQQVQRYRARLSPGDVYVIPAGHPIVVTASSDLSLLGFGI 428
RNENQ ++ + +E+EQ++E S+QVQ YRA+LSPGDV+VIPAGHP+ + ASSDL+L+GFGI
Sbjct: 299 RNENQGKEND-KEEEQEEETSKQVQLYRAKLSPGDVFVIPAGHPVAINASSDLNLIGFGI 357
Query: 429 NAENNQRNFLA 439
NAENN+RNFLA
Sbjct: 358 NAENNERNFLA 368
>UniRef100_P13915 Convicilin precursor [Pisum sativum]
Length = 571
Score = 490 bits (1261), Expect = e-137
Identities = 246/374 (65%), Positives = 303/374 (80%), Gaps = 13/374 (3%)
Query: 67 SRSDQDQENPFIFNSNRFQTLFENENGHIRLLQRFDKRSKIFENLQNYRLLEYHSKPHTL 126
S Q+ NPF+F SN+F TLFENENGHIR LQRFDKRS +FENLQNYRL+EY +KPHT+
Sbjct: 145 SSESQEHRNPFLFKSNKFLTLFENENGHIRRLQRFDKRSDLFENLQNYRLVEYRAKPHTI 204
Query: 127 FLPQHNDADFILVVVSGKAILTVLNPNNRNSFNLERGDTIKLPAGTLGYLANRDDNKDLR 186
FLPQH DAD ILVV++GKAILTVL+PN+RNS+NLERGDTIK+PAGT YL N+DD +DLR
Sbjct: 205 FLPQHIDADLILVVLNGKAILTVLSPNDRNSYNLERGDTIKIPAGTTSYLVNQDDEEDLR 264
Query: 187 VLDLAIPVNRPGQFQSFSLSESENQQSFLSGFSKNILEAAFNSNYEEIERVLIEENEQEP 246
V+D IPVNRPG+F++F LSE++NQ +L GFSKNILEA+ N+ YE IE+VL+EE E++P
Sbjct: 265 VVDFVIPVNRPGKFEAFGLSENKNQ--YLRGFSKNILEASLNTKYETIEKVLLEEQEKKP 322
Query: 247 QHRRGLRKDERRQQSQEANVIVKVSREQIEELSKNAKSSSRRSESSESEPINLRNQKPIY 306
Q LR +R QQ +E + I+KVSREQIEEL K AKSSS++S SE EP NLR+ KP Y
Sbjct: 323 QQ---LRDRKRTQQGEERDAIIKVSREQIEELRKLAKSSSKKSLPSEFEPFNLRSHKPEY 379
Query: 307 SNKFGNFFEITPEKN-PQLKDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEGKGEFEL 365
SNKFG FEITPEK PQL+DLDILV+ EI +G+L+LPH+NSRA V++ V EGKG EL
Sbjct: 380 SNKFGKLFEITPEKKYPQLQDLDILVSCVEINKGALMLPHYNSRAIVVLLVNEGKGNLEL 439
Query: 366 VGQRNENQQEQREYEEDEQQQERSQQVQRYRARLSPGDVYVIPAGHPIVVTASSDLSLLG 425
+G +NE Q E E ++ER+ +VQRY ARLSPGDV +IPAGHP+ ++ASS+L+LLG
Sbjct: 440 LGLKNEQQ-------EREDRKERNNEVQRYEARLSPGDVVIIPAGHPVAISASSNLNLLG 492
Query: 426 FGINAENNQRNFLA 439
FGINA+NNQRNFL+
Sbjct: 493 FGINAKNNQRNFLS 506
Score = 36.6 bits (83), Expect = 1.5
Identities = 37/162 (22%), Positives = 62/162 (37%), Gaps = 44/162 (27%)
Query: 126 LFLPQHND-ADFILVVVSGKAILTVLNPNN----------------RNSFNLERGDTIKL 168
L LP +N A +L+V GK L +L N R L GD + +
Sbjct: 415 LMLPHYNSRAIVVLLVNEGKGNLELLGLKNEQQEREDRKERNNEVQRYEARLSPGDVVII 474
Query: 169 PAGTLGYLANRDDNKDLRVLDLAIPVNRPGQFQSFSLSESENQQSFLSGFSKNIL----- 223
PAG +AI + F ++ NQ++FLSG N++
Sbjct: 475 PAGH----------------PVAISASSNLNLLGFGINAKNNQRNFLSGSDDNVISQIEN 518
Query: 224 ---EAAFNSNYEEIERVLIEENEQEPQHRRGLRKDERRQQSQ 262
E F + +E+ R++ Q+ H +++ ++SQ
Sbjct: 519 PVKELTFPGSSQEVNRLI---KNQKQSHFASAEPEQKEEESQ 557
>UniRef100_Q9M3X6 Convicilin precursor [Pisum sativum]
Length = 613
Score = 487 bits (1254), Expect = e-136
Identities = 245/374 (65%), Positives = 303/374 (80%), Gaps = 19/374 (5%)
Query: 67 SRSDQDQENPFIFNSNRFQTLFENENGHIRLLQRFDKRSKIFENLQNYRLLEYHSKPHTL 126
S Q++ NPF+F SN+F TLFENENGHIRLLQRFDKRS +FENLQNYRL+EY +KPHT+
Sbjct: 193 SSESQERRNPFLFKSNKFLTLFENENGHIRLLQRFDKRSDLFENLQNYRLVEYRAKPHTI 252
Query: 127 FLPQHNDADFILVVVSGKAILTVLNPNNRNSFNLERGDTIKLPAGTLGYLANRDDNKDLR 186
FLPQH DAD ILVV+SGKAILTVL+PN+RNS+NLERGDTIKLPAGT YL N+DD +DLR
Sbjct: 253 FLPQHIDADLILVVLSGKAILTVLSPNDRNSYNLERGDTIKLPAGTTSYLVNQDDEEDLR 312
Query: 187 VLDLAIPVNRPGQFQSFSLSESENQQSFLSGFSKNILEAAFNSNYEEIERVLIEENEQEP 246
++DL IPVN PG+F++F L++++NQ +L GFSKNILEA++N+ YE IE+VL+EE E++
Sbjct: 313 LVDLVIPVNGPGKFEAFDLAKNKNQ--YLRGFSKNILEASYNTRYETIEKVLLEEQEKD- 369
Query: 247 QHRRGLRKDERRQQSQEANVIVKVSREQIEELSKNAKSSSRRSESSESEPINLRNQKPIY 306
+RRQQ +E + IVKVSREQIEEL K AKSSS++S SE EPINLR+ KP Y
Sbjct: 370 --------RKRRQQGEETDAIVKVSREQIEELKKLAKSSSKKSLPSEFEPINLRSHKPEY 421
Query: 307 SNKFGNFFEITPEKN-PQLKDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEGKGEFEL 365
SNKFG FEITPEK PQL+DLD+ V+ EI EG+L+LPH+NSRA V++ V EGKG EL
Sbjct: 422 SNKFGKLFEITPEKKYPQLQDLDLFVSCVEINEGALMLPHYNSRAIVVLLVNEGKGNLEL 481
Query: 366 VGQRNENQQEQREYEEDEQQQERSQQVQRYRARLSPGDVYVIPAGHPIVVTASSDLSLLG 425
+G +NE Q E E ++ER+ +VQRY ARLSPGDV +IPAGHP+ +TASS+L+LLG
Sbjct: 482 LGLKNEQQ-------EREDRKERNNEVQRYEARLSPGDVVIIPAGHPVAITASSNLNLLG 534
Query: 426 FGINAENNQRNFLA 439
FGINAENN+RNFL+
Sbjct: 535 FGINAENNERNFLS 548
Score = 35.8 bits (81), Expect = 2.5
Identities = 38/162 (23%), Positives = 61/162 (37%), Gaps = 44/162 (27%)
Query: 126 LFLPQHND-ADFILVVVSGKAILTVLNPNN----------------RNSFNLERGDTIKL 168
L LP +N A +L+V GK L +L N R L GD + +
Sbjct: 457 LMLPHYNSRAIVVLLVNEGKGNLELLGLKNEQQEREDRKERNNEVQRYEARLSPGDVVII 516
Query: 169 PAGTLGYLANRDDNKDLRVLDLAIPVNRPGQFQSFSLSESENQQSFLSGFSKNIL----- 223
PAG +AI + F ++ N+++FLSG N++
Sbjct: 517 PAGH----------------PVAITASSNLNLLGFGINAENNERNFLSGSDDNVISQIEN 560
Query: 224 ---EAAFNSNYEEIERVLIEENEQEPQHRRGLRKDERRQQSQ 262
E F + +EI R++ Q+ H +++ Q SQ
Sbjct: 561 PVKELTFPGSVQEINRLI---KNQKQSHFANAEPEQKEQGSQ 599
>UniRef100_Q41674 Convicilin precursor [Vicia narbonensis]
Length = 545
Score = 483 bits (1243), Expect = e-135
Identities = 252/373 (67%), Positives = 297/373 (79%), Gaps = 18/373 (4%)
Query: 67 SRSDQDQENPFIFNSNRFQTLFENENGHIRLLQRFDKRSKIFENLQNYRLLEYHSKPHTL 126
S Q++ NPF+F SN+F TLFENENGHIR LQRFDKRS +FENLQNYRL+EY +KPHT+
Sbjct: 124 SSKSQERRNPFLFKSNKFLTLFENENGHIRRLQRFDKRSDLFENLQNYRLVEYRAKPHTI 183
Query: 127 FLPQHNDADFILVVVSGKAILTVLNPNNRNSFNLERGDTIKLPAGTLGYLANRDDNKDLR 186
FLPQH DAD IL V+SG+AILTVL+PN+RNS+NLERGDTIKLPAGT YL N+DD +DLR
Sbjct: 184 FLPQHIDADLILTVLSGRAILTVLSPNDRNSYNLERGDTIKLPAGTTSYLLNQDDEEDLR 243
Query: 187 VLDLAIPVNRPGQFQSFSLSESENQQSFLSGFSKNILEAAFNSNYEEIERVLIEENEQEP 246
V+DL+I VNRPG+ +SF LS S+NQ +L GFSKNILEA+ N+ YE IE+VL+E EP
Sbjct: 244 VVDLSISVNRPGKVESFGLSGSKNQ--YLRGFSKNILEASLNTKYETIEKVLLE----EP 297
Query: 247 QHRRGLRKDERRQQSQEANVIVKVSREQIEELSKNAKSSSRRSESSESEPINLRNQKPIY 306
Q G +RR Q QE N +VKVSREQ+EEL + AKSSS++ SSE EP NLR+Q P Y
Sbjct: 298 QQSIG---QKRRSQRQETNALVKVSREQVEELKRLAKSSSKKGVSSEFEPFNLRSQNPKY 354
Query: 307 SNKFGNFFEITPEKN-PQLKDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEGKGEFEL 365
SNKFG FEITPEK PQL+DLDI V+ EI EG L+LPH+NSRA VI+ V EGKG EL
Sbjct: 355 SNKFGKLFEITPEKKYPQLQDLDIFVSSVEINEGGLMLPHYNSRAIVILLVNEGKGNLEL 414
Query: 366 VGQRNENQQEQREYEEDEQQQERSQQVQRYRARLSPGDVYVIPAGHPIVVTASSDLSLLG 425
VG +NE QQEQRE E++ QQVQRY ARLSPGDV +IPAGHP+ V+ASS+L+LLG
Sbjct: 415 VGLKNE-QQEQREREDE-------QQVQRYEARLSPGDVVIIPAGHPVAVSASSNLNLLG 466
Query: 426 FGINAENNQRNFL 438
FGINAENNQRNFL
Sbjct: 467 FGINAENNQRNFL 479
Score = 36.6 bits (83), Expect = 1.5
Identities = 37/161 (22%), Positives = 62/161 (37%), Gaps = 43/161 (26%)
Query: 126 LFLPQHND-ADFILVVVSGKAILTVLNPNNRNSFNLER---------------GDTIKLP 169
L LP +N A IL+V GK L ++ N ER GD + +P
Sbjct: 390 LMLPHYNSRAIVILLVNEGKGNLELVGLKNEQQEQREREDEQQVQRYEARLSPGDVVIIP 449
Query: 170 AGTLGYLANRDDNKDLRVLDLAIPVNRPGQFQSFSLSESENQQSFLSGFSKNIL------ 223
AG +A+ + F ++ NQ++FL+G N++
Sbjct: 450 AGH----------------PVAVSASSNLNLLGFGINAENNQRNFLTGSDDNVISQIENP 493
Query: 224 --EAAFNSNYEEIERVLIEENEQEPQHRRGLRKDERRQQSQ 262
E F + +E+ R+L QE H +++ ++SQ
Sbjct: 494 VKELTFPGSAQEVNRLL---KNQEHSHFANAEPEQKGEESQ 531
>UniRef100_Q9M3X8 Convicilin [Lens culinaris]
Length = 518
Score = 471 bits (1211), Expect = e-131
Identities = 238/374 (63%), Positives = 294/374 (77%), Gaps = 13/374 (3%)
Query: 66 SSRSDQDQENPFIFNSNRFQTLFENENGHIRLLQRFDKRSKIFENLQNYRLLEYHSKPHT 125
+S Q++ NPF+F SN+F TLFENENGHIR LQRFDKRS +FENLQNYRL+EY +KPH+
Sbjct: 151 TSSESQERRNPFLFKSNKFLTLFENENGHIRRLQRFDKRSDLFENLQNYRLVEYRAKPHS 210
Query: 126 LFLPQHNDADFILVVVSGKAILTVLNPNNRNSFNLERGDTIKLPAGTLGYLANRDDNKDL 185
+FLPQH DA+FI+VV+SGKAILTVL+PN+RNS+NLERGD IK PAG YL N DD +DL
Sbjct: 211 IFLPQHIDAEFIVVVLSGKAILTVLSPNDRNSYNLERGDAIKSPAGATYYLVNPDDEEDL 270
Query: 186 RVLDLAIPVNRPGQFQSFSLSESENQQSFLSGFSKNILEAAFNSNYEEIERVLIEENEQE 245
RV+D I +NRPG+F++F L S N++ +L GFSK++LEA+ N+ Y+ IE+VL+EE E E
Sbjct: 271 RVVDFVISLNRPGKFEAFDL--SANRRQYLRGFSKSVLEASLNTKYDTIEKVLLEEQENE 328
Query: 246 PQHRRGLRKDERRQQSQEANVIVKVSREQIEELSKNAKSSSRRSESSESEPINLRNQKPI 305
P RR + R Q QE + IVKVSREQIEEL + AKSSS++S SE EP NLR+Q P
Sbjct: 329 PHQRRDRK---GRPQGQEKHAIVKVSREQIEELRRLAKSSSKKSLPSEFEPFNLRSQNPK 385
Query: 306 YSNKFGNFFEITPEKN-PQLKDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEGKGEFE 364
YSNKFG FFE+TPEK PQL+DLD+LV+ EI EG LLLPH+NSRA V++ V EGKG E
Sbjct: 386 YSNKFGKFFEVTPEKKYPQLQDLDLLVSSVEINEGGLLLPHYNSRAIVVLLVNEGKGNLE 445
Query: 365 LVGQRNENQQEQREYEEDEQQQERSQQVQRYRARLSPGDVYVIPAGHPIVVTASSDLSLL 424
LVG +NE Q E E +ER+ +VQRY ARLSPGDV +IPAGHP+ ++ASS+L+LL
Sbjct: 446 LVGFKNEQQ-------EREDNKERNNEVQRYEARLSPGDVVIIPAGHPVSISASSNLNLL 498
Query: 425 GFGINAENNQRNFL 438
GFGINAENN+RNFL
Sbjct: 499 GFGINAENNERNFL 512
Score = 33.9 bits (76), Expect = 9.5
Identities = 15/32 (46%), Positives = 22/32 (67%)
Query: 45 IKAPCSLLMLLGIVFLASICVSSRSDQDQENP 76
IK+ LL+LLGI+FLA +CV+ +D + P
Sbjct: 5 IKSRFPLLLLLGIIFLAFVCVAYANDDEGSEP 36
>UniRef100_P25974 Beta-conglycinin, beta chain precursor [Glycine max]
Length = 439
Score = 424 bits (1091), Expect = e-117
Identities = 221/398 (55%), Positives = 294/398 (73%), Gaps = 26/398 (6%)
Query: 43 MAIKAPCSLLMLLGIVFLASICVSSRSDQDQENPFIF-NSNRFQTLFENENGHIRLLQRF 101
M ++ P LL+LLG VFLAS+CVS + +D+ NPF F +SN FQTLFEN+N IRLLQRF
Sbjct: 2 MRVRFP--LLVLLGTVFLASVCVSLKVREDENNPFYFRSSNSFQTLFENQNVRIRLLQRF 59
Query: 102 DKRSKIFENLQNYRLLEYHSKPHTLFLPQHNDADFILVVVSGKAILTVLNPNNRNSFNLE 161
+KRS ENL++YR++++ SKP+T+ LP H DADF+L V+SG+AILT++N ++R+S+NL
Sbjct: 60 NKRSPQLENLRDYRIVQFQSKPNTILLPHHADADFLLFVLSGRAILTLVNNDDRDSYNLH 119
Query: 162 RGDTIKLPAGTLGYLANRDDNKDLRVLDLAIPVNRPGQFQSFSLSESENQQSFLSGFSKN 221
GD ++PAGT YL N D+++L+++ LAIPVN+PG++ F LS ++ QQS+L GFS N
Sbjct: 120 PGDAQRIPAGTTYYLVNPHDHQNLKIIKLAIPVNKPGRYDDFFLSSTQAQQSYLQGFSHN 179
Query: 222 ILEAAFNSNYEEIERVLIEENEQEPQHRRGLRKDERRQQSQEANVIVKVSREQIEELSKN 281
ILE +F+S +EEI RVL E E +Q Q+ VIV++S+EQI +LS+
Sbjct: 180 ILETSFHSEFEEINRVLFGEEE---------------EQRQQEGVIVELSKEQIRQLSRR 224
Query: 282 AKSSSRRSESSESEPINLRNQKPIYSNKFGNFFEITPEKNPQLKDLDILVNYAEIREGSL 341
AKSSSR++ SSE EP NLR++ PIYSN FG FFEITPEKNPQL+DLDI ++ +I EG+L
Sbjct: 225 AKSSSRKTISSEDEPFNLRSRNPIYSNNFGKFFEITPEKNPQLRDLDIFLSSVDINEGAL 284
Query: 342 LLPHFNSRATVIVAVEEGKGEFELVGQRNENQQEQREYEEDEQQQERSQQVQRYRARLSP 401
LLPHFNS+A VI+ + EG ELVG + E QQ+Q+ Q+E +VQRYRA LS
Sbjct: 285 LLPHFNSKAIVILVINEGDANIELVGIK-EQQQKQK-------QEEEPLEVQRYRAELSE 336
Query: 402 GDVYVIPAGHPIVVTASSDLSLLGFGINAENNQRNFLA 439
DV+VIPA +P VV A+S+L+ L FGINAENNQRNFLA
Sbjct: 337 DDVFVIPAAYPFVVNATSNLNFLAFGINAENNQRNFLA 374
Score = 33.9 bits (76), Expect = 9.5
Identities = 33/131 (25%), Positives = 62/131 (47%), Gaps = 18/131 (13%)
Query: 126 LFLPQHND-ADFILVVVSGKAILTVLNPNNRNSFNLERGDTIKLPAGTLGYLANRDDNKD 184
L LP N A ILV+ G A + ++ + + + +++ Y A ++ D
Sbjct: 284 LLLPHFNSKAIVILVINEGDANIELVGIKEQQQKQKQEEEPLEVQR----YRAELSED-D 338
Query: 185 LRVLDLAIP--VNRPGQ--FQSFSLSESENQQSFLSGFSKNIL--------EAAFNSNYE 232
+ V+ A P VN F +F ++ NQ++FL+G N++ E AF + +
Sbjct: 339 VFVIPAAYPFVVNATSNLNFLAFGINAENNQRNFLAGEKDNVVRQIERQVQELAFPGSAQ 398
Query: 233 EIERVLIEENE 243
++ER+L ++ E
Sbjct: 399 DVERLLKKQRE 409
>UniRef100_Q93VL9 Beta-conglycinin beta-subunit [Glycine max]
Length = 439
Score = 423 bits (1087), Expect = e-117
Identities = 220/398 (55%), Positives = 293/398 (73%), Gaps = 26/398 (6%)
Query: 43 MAIKAPCSLLMLLGIVFLASICVSSRSDQDQENPFIF-NSNRFQTLFENENGHIRLLQRF 101
M ++ P LL+LLG VFLAS+CVS + +D+ NPF F +SN FQTLFEN+NG IRLLQRF
Sbjct: 2 MRVRFP--LLVLLGTVFLASVCVSLKVREDENNPFYFRSSNSFQTLFENQNGRIRLLQRF 59
Query: 102 DKRSKIFENLQNYRLLEYHSKPHTLFLPQHNDADFILVVVSGKAILTVLNPNNRNSFNLE 161
+KRS ENL++YR++++ SKP+T+ LP H DADF+L V+SG+AILT++N ++R+S+NL
Sbjct: 60 NKRSPQLENLRDYRIVQFQSKPNTILLPHHADADFLLFVLSGRAILTLVNNDDRDSYNLH 119
Query: 162 RGDTIKLPAGTLGYLANRDDNKDLRVLDLAIPVNRPGQFQSFSLSESENQQSFLSGFSKN 221
GD ++PAGT YL N D+++L+++ LAIPVN+P ++ F LS ++ QQS+L GFS N
Sbjct: 120 PGDAQRIPAGTTYYLVNPHDHQNLKIIKLAIPVNKPSRYDDFFLSSTQAQQSYLQGFSHN 179
Query: 222 ILEAAFNSNYEEIERVLIEENEQEPQHRRGLRKDERRQQSQEANVIVKVSREQIEELSKN 281
ILE +F+S +EEI RVL E E +Q Q+ VIV++S+EQI +LS+
Sbjct: 180 ILETSFHSEFEEINRVLFGEEE---------------EQRQQEGVIVELSKEQIRQLSRR 224
Query: 282 AKSSSRRSESSESEPINLRNQKPIYSNKFGNFFEITPEKNPQLKDLDILVNYAEIREGSL 341
AKSSSR++ SSE EP NLR++ PIYSN FG FFEITPEKNPQ +DLDI ++ +I EG+L
Sbjct: 225 AKSSSRKTISSEDEPFNLRSRNPIYSNNFGKFFEITPEKNPQPRDLDIFLSSVDINEGAL 284
Query: 342 LLPHFNSRATVIVAVEEGKGEFELVGQRNENQQEQREYEEDEQQQERSQQVQRYRARLSP 401
LLPHFNS+A VI+ + EG ELVG + E QQ+Q+ Q+E +VQRYRA LS
Sbjct: 285 LLPHFNSKAIVILVINEGDANIELVGIK-EQQQKQK-------QEEEPLEVQRYRAELSE 336
Query: 402 GDVYVIPAGHPIVVTASSDLSLLGFGINAENNQRNFLA 439
DV+VIPA +P VV A+S+L+ L FGINAENNQRNFLA
Sbjct: 337 DDVFVIPAAYPFVVNATSNLNFLAFGINAENNQRNFLA 374
Score = 33.9 bits (76), Expect = 9.5
Identities = 33/131 (25%), Positives = 62/131 (47%), Gaps = 18/131 (13%)
Query: 126 LFLPQHND-ADFILVVVSGKAILTVLNPNNRNSFNLERGDTIKLPAGTLGYLANRDDNKD 184
L LP N A ILV+ G A + ++ + + + +++ Y A ++ D
Sbjct: 284 LLLPHFNSKAIVILVINEGDANIELVGIKEQQQKQKQEEEPLEVQR----YRAELSED-D 338
Query: 185 LRVLDLAIP--VNRPGQ--FQSFSLSESENQQSFLSGFSKNIL--------EAAFNSNYE 232
+ V+ A P VN F +F ++ NQ++FL+G N++ E AF + +
Sbjct: 339 VFVIPAAYPFVVNATSNLNFLAFGINAENNQRNFLAGEKDNVVRQIERQVQELAFPGSAQ 398
Query: 233 EIERVLIEENE 243
++ER+L ++ E
Sbjct: 399 DVERLLKKQRE 409
>UniRef100_Q6EBC1 Beta-conglutin [Lupinus albus]
Length = 533
Score = 417 bits (1072), Expect = e-115
Identities = 199/369 (53%), Positives = 290/369 (77%), Gaps = 14/369 (3%)
Query: 73 QENPFIFNSNRFQTLFENENGHIRLLQRFDKRSKIFENLQNYRLLEYHSKPHTLFLPQHN 132
Q NP+ F+S RFQTL++N NG IR+L+RFD+R+ ENLQNYR++E+ SKP+TL LP+H+
Sbjct: 111 QRNPYHFSSQRFQTLYKNRNGKIRVLERFDQRTNRLENLQNYRIVEFQSKPNTLILPKHS 170
Query: 133 DADFILVVVSGKAILTVLNPNNRNSFNLERGDTIKLPAGTLGYLANRDDNKDLRVLDLAI 192
DAD++LVV++G+A +T++NP+ R ++NLE GD +++PAG+ Y+ N DDN+ LRV+ LAI
Sbjct: 171 DADYVLVVLNGRATITIVNPDRRQAYNLEYGDALRIPAGSTSYILNPDDNQKLRVVKLAI 230
Query: 193 PVNRPGQFQSFSLSESENQQSFLSGFSKNILEAAFNSNYEEIERVLI-EENEQE-PQHRR 250
P+N PG F F S +++QQS+ SGFS+N LEA FN+ YEEI+R+++ E+EQE + RR
Sbjct: 231 PINNPGYFYDFYPSSTKDQQSYFSGFSRNTLEATFNTRYEEIQRIILGNEDEQEYEEQRR 290
Query: 251 GLRKDERRQQSQEANVIVKVSREQIEELSKNAKSSSRRSESSESEPINLRNQKPIYSNKF 310
G + Q Q+ VIV VS++QI++L+K+A+SSS + + S+S P NLR+ +PIYSNK+
Sbjct: 291 G-----QEQSDQDEGVIVIVSKKQIQKLTKHAQSSSGKDKPSDSGPFNLRSNEPIYSNKY 345
Query: 311 GNFFEITPEKNPQLKDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEGKGEFELVGQRN 370
GNF+EITP++NPQ++DL+I + Y +I EG+LLLPH+NS+A +V V+EG+G +ELVG R+
Sbjct: 346 GNFYEITPDRNPQVQDLNISLTYIKINEGALLLPHYNSKAIYVVVVDEGEGNYELVGIRD 405
Query: 371 ENQQEQREYEEDEQQQERSQQVQRYRARLSPGDVYVIPAGHPIVVTASSDLSLLGFGINA 430
+ +Q+ ++Q+E+ ++V RY ARLS GD++VIPAG+PI + ASS+L LLGFGINA
Sbjct: 406 QQRQQ-------DEQEEKEEEVIRYSARLSEGDIFVIPAGYPISINASSNLRLLGFGINA 458
Query: 431 ENNQRNFLA 439
+ NQRNFLA
Sbjct: 459 DENQRNFLA 467
Score = 44.7 bits (104), Expect = 0.005
Identities = 50/223 (22%), Positives = 95/223 (42%), Gaps = 48/223 (21%)
Query: 66 SSRSDQDQENPFIFNSNRFQTLFENENGHIRLLQRFDKRSKIFENLQNYRLLEYHSKPHT 125
S + PF SN + ++ N+ G+ + D+ ++ + N L
Sbjct: 321 SGKDKPSDSGPFNLRSN--EPIYSNKYGNFYEITP-DRNPQVQD--LNISLTYIKINEGA 375
Query: 126 LFLPQHND-ADFILVVVSGKAILTVLNPNN----------------RNSFNLERGDTIKL 168
L LP +N A +++VV G+ ++ + R S L GD +
Sbjct: 376 LLLPHYNSKAIYVVVVDEGEGNYELVGIRDQQRQQDEQEEKEEEVIRYSARLSEGDIFVI 435
Query: 169 PAGTLGYLANRDDNKDLRVLDLAIPVNRPGQFQSFSLSESENQQSFLSGFSKNIL----- 223
PAG Y + + + +LR+L F ++ ENQ++FL+G N++
Sbjct: 436 PAG---YPISINASSNLRLL-------------GFGINADENQRNFLAGSKDNVIRQLDR 479
Query: 224 ---EAAFNSNYEEIERVLIEENEQEPQHRRGLRKDERRQQSQE 263
E F + E+IER++ +N+Q+ G + +++QQS++
Sbjct: 480 AVNELTFPGSAEDIERLI--KNQQQSYFANGQPQQQQQQQSEK 520
>UniRef100_Q9FZP9 Alpha' subunit of beta-conglycinin [Glycine max]
Length = 559
Score = 407 bits (1047), Expect = e-112
Identities = 197/366 (53%), Positives = 286/366 (77%), Gaps = 18/366 (4%)
Query: 74 ENPFIFNSNRFQTLFENENGHIRLLQRFDKRSKIFENLQNYRLLEYHSKPHTLFLPQHND 133
+NPF FNS RFQTLF+N+ GH+R+LQRF+KRS+ +NL++YR+LE++SKP+TL LP H D
Sbjct: 147 KNPFHFNSKRFQTLFKNQYGHVRVLQRFNKRSQQLQNLRDYRILEFNSKPNTLLLPHHAD 206
Query: 134 ADFILVVVSGKAILTVLNPNNRNSFNLERGDTIKLPAGTLGYLANRDDNKDLRVLDLAIP 193
AD+++V+++G AILT++N ++R+S+NL+ GD +++PAGT Y+ N D++++LR++ LAIP
Sbjct: 207 ADYLIVILNGTAILTLVNNDDRDSYNLQSGDALRVPAGTTYYVVNPDNDENLRMITLAIP 266
Query: 194 VNRPGQFQSFSLSESENQQSFLSGFSKNILEAAFNSNYEEIERVLIEENEQEPQHRRGLR 253
VN+PG+F+SF LS ++ QQS+L GFSKNILEA++++ +EEI +VL E + Q
Sbjct: 267 VNKPGRFESFFLSSTQAQQSYLQGFSKNILEASYDTKFEEINKVLFGREEGQQQ------ 320
Query: 254 KDERRQQSQEANVIVKVSREQIEELSKNAKSSSRRSESSESEPINLRNQKPIYSNKFGNF 313
+ER Q+S VIV++S++QI ELSK+AKSSSR++ SSE +P NLR++ PIYSNK G
Sbjct: 321 GEERLQES----VIVEISKKQIRELSKHAKSSSRKTISSEDKPFNLRSRDPIYSNKLGKL 376
Query: 314 FEITPEKNPQLKDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEGKGEFELVGQRNENQ 373
FEITPEKNPQL+DLD+ ++ ++ EG+L LPHFNS+A V++ + EG+ ELVG + + Q
Sbjct: 377 FEITPEKNPQLRDLDVFLSVVDMNEGALFLPHFNSKAIVVLVINEGEANIELVGIKEQQQ 436
Query: 374 QEQREYEEDEQQQERSQQVQRYRARLSPGDVYVIPAGHPIVVTASSDLSLLGFGINAENN 433
+ +QQ+E+ +V++YRA LS D++VIPAG+P+VV A+SDL+ FGINAENN
Sbjct: 437 R--------QQQEEQPLEVRKYRAELSEQDIFVIPAGYPVVVNATSDLNFFAFGINAENN 488
Query: 434 QRNFLA 439
QRNFLA
Sbjct: 489 QRNFLA 494
Score = 35.4 bits (80), Expect = 3.3
Identities = 37/153 (24%), Positives = 67/153 (43%), Gaps = 25/153 (16%)
Query: 126 LFLPQHND-ADFILVVVSGKAILTVLNPNNRNSFNLERGDTIKLPAGTLGYLANRDDNKD 184
LFLP N A +LV+ G+A + ++ + +R + P Y A + +D
Sbjct: 404 LFLPHFNSKAIVVLVINEGEANIELVGIKEQQ----QRQQQEEQPLEVRKYRAELSE-QD 458
Query: 185 LRVLDLAIPV----NRPGQFQSFSLSESENQQSFLSGFSKNIL--------EAAFNSNYE 232
+ V+ PV F +F ++ NQ++FL+G N++ E AF + +
Sbjct: 459 IFVIPAGYPVVVNATSDLNFFAFGINAENNQRNFLAGSKDNVISQIPSQVQELAFPGSAK 518
Query: 233 EIERVLIEENEQEPQHRRGLRKDERRQQSQEAN 265
+IE ++ ++E D + QQ +E N
Sbjct: 519 DIENLIKSQSE-------SYFVDAQPQQKEEGN 544
>UniRef100_Q7XXT2 Prepro beta-conglycinin alpha prime subunit [Glycine max]
Length = 621
Score = 407 bits (1047), Expect = e-112
Identities = 197/366 (53%), Positives = 286/366 (77%), Gaps = 18/366 (4%)
Query: 74 ENPFIFNSNRFQTLFENENGHIRLLQRFDKRSKIFENLQNYRLLEYHSKPHTLFLPQHND 133
+NPF FNS RFQTLF+N+ GH+R+LQRF+KRS+ +NL++YR+LE++SKP+TL LP H D
Sbjct: 209 KNPFHFNSKRFQTLFKNQYGHVRVLQRFNKRSQQLQNLRDYRILEFNSKPNTLLLPHHAD 268
Query: 134 ADFILVVVSGKAILTVLNPNNRNSFNLERGDTIKLPAGTLGYLANRDDNKDLRVLDLAIP 193
AD+++V+++G AILT++N ++R+S+NL+ GD +++PAGT Y+ N D++++LR++ LAIP
Sbjct: 269 ADYLIVILNGTAILTLVNNDDRDSYNLQSGDALRVPAGTTYYVVNPDNDENLRMITLAIP 328
Query: 194 VNRPGQFQSFSLSESENQQSFLSGFSKNILEAAFNSNYEEIERVLIEENEQEPQHRRGLR 253
VN+PG+F+SF LS ++ QQS+L GFSKNILEA++++ +EEI +VL E + Q
Sbjct: 329 VNKPGRFESFFLSSTQAQQSYLQGFSKNILEASYDTKFEEINKVLFGREEGQQQ------ 382
Query: 254 KDERRQQSQEANVIVKVSREQIEELSKNAKSSSRRSESSESEPINLRNQKPIYSNKFGNF 313
+ER Q+S VIV++S++QI ELSK+AKSSSR++ SSE +P NLR++ PIYSNK G
Sbjct: 383 GEERLQES----VIVEISKKQIRELSKHAKSSSRKTISSEDKPFNLRSRDPIYSNKLGKL 438
Query: 314 FEITPEKNPQLKDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEGKGEFELVGQRNENQ 373
FEITPEKNPQL+DLD+ ++ ++ EG+L LPHFNS+A V++ + EG+ ELVG + + Q
Sbjct: 439 FEITPEKNPQLRDLDVFLSVVDMNEGALFLPHFNSKAIVVLVINEGEANIELVGIKEQQQ 498
Query: 374 QEQREYEEDEQQQERSQQVQRYRARLSPGDVYVIPAGHPIVVTASSDLSLLGFGINAENN 433
+ +QQ+E+ +V++YRA LS D++VIPAG+P+VV A+SDL+ FGINAENN
Sbjct: 499 R--------QQQEEQPLEVRKYRAELSEQDIFVIPAGYPVVVNATSDLNFFAFGINAENN 550
Query: 434 QRNFLA 439
QRNFLA
Sbjct: 551 QRNFLA 556
Score = 35.4 bits (80), Expect = 3.3
Identities = 37/153 (24%), Positives = 67/153 (43%), Gaps = 25/153 (16%)
Query: 126 LFLPQHND-ADFILVVVSGKAILTVLNPNNRNSFNLERGDTIKLPAGTLGYLANRDDNKD 184
LFLP N A +LV+ G+A + ++ + +R + P Y A + +D
Sbjct: 466 LFLPHFNSKAIVVLVINEGEANIELVGIKEQQ----QRQQQEEQPLEVRKYRAELSE-QD 520
Query: 185 LRVLDLAIPV----NRPGQFQSFSLSESENQQSFLSGFSKNIL--------EAAFNSNYE 232
+ V+ PV F +F ++ NQ++FL+G N++ E AF + +
Sbjct: 521 IFVIPAGYPVVVNATSDLNFFAFGINAENNQRNFLAGSKDNVISQIPSQVQELAFPGSAK 580
Query: 233 EIERVLIEENEQEPQHRRGLRKDERRQQSQEAN 265
+IE ++ ++E D + QQ +E N
Sbjct: 581 DIENLIKSQSE-------SYFVDAQPQQKEEGN 606
>UniRef100_Q948Y0 Beta-conglycinin alpha prime subunit [Glycine max]
Length = 621
Score = 407 bits (1046), Expect = e-112
Identities = 197/366 (53%), Positives = 285/366 (77%), Gaps = 18/366 (4%)
Query: 74 ENPFIFNSNRFQTLFENENGHIRLLQRFDKRSKIFENLQNYRLLEYHSKPHTLFLPQHND 133
+NPF FNS RFQTLF+N+ GH+R+LQRF+KRS+ +NL++YR+LE++SKP+TL LP H D
Sbjct: 209 KNPFHFNSKRFQTLFKNQYGHVRVLQRFNKRSQQLQNLRDYRILEFNSKPNTLLLPHHAD 268
Query: 134 ADFILVVVSGKAILTVLNPNNRNSFNLERGDTIKLPAGTLGYLANRDDNKDLRVLDLAIP 193
AD+++V+++G AILT++N ++R+S+NL+ GD +++PAGT Y+ N D++++LR++ LAIP
Sbjct: 269 ADYLIVILNGTAILTLVNNDDRDSYNLQSGDALRVPAGTTYYVVNPDNDENLRMITLAIP 328
Query: 194 VNRPGQFQSFSLSESENQQSFLSGFSKNILEAAFNSNYEEIERVLIEENEQEPQHRRGLR 253
VN+PG+F+SF LS ++ QQS+L GFSKNILEA++++ +EEI +VL E + Q
Sbjct: 329 VNKPGRFESFFLSSTQAQQSYLQGFSKNILEASYDTKFEEINKVLFGREEGQQQ------ 382
Query: 254 KDERRQQSQEANVIVKVSREQIEELSKNAKSSSRRSESSESEPINLRNQKPIYSNKFGNF 313
+ER Q+S VIV++S++QI ELSK AKSSSR++ SSE +P NLR++ PIYSNK G
Sbjct: 383 GEERLQES----VIVEISKKQIRELSKRAKSSSRKTISSEDKPFNLRSRDPIYSNKLGKL 438
Query: 314 FEITPEKNPQLKDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEGKGEFELVGQRNENQ 373
FEITPEKNPQL+DLD+ ++ ++ EG+L LPHFNS+A V++ + EG+ ELVG + + Q
Sbjct: 439 FEITPEKNPQLRDLDVFLSVVDMNEGALFLPHFNSKAIVVLVINEGEANIELVGIKEQQQ 498
Query: 374 QEQREYEEDEQQQERSQQVQRYRARLSPGDVYVIPAGHPIVVTASSDLSLLGFGINAENN 433
+ +QQ+E+ +V++YRA LS D++VIPAG+P+VV A+SDL+ FGINAENN
Sbjct: 499 R--------QQQEEQPLEVRKYRAELSEQDIFVIPAGYPVVVNATSDLNFFAFGINAENN 550
Query: 434 QRNFLA 439
QRNFLA
Sbjct: 551 QRNFLA 556
Score = 35.0 bits (79), Expect = 4.3
Identities = 37/153 (24%), Positives = 67/153 (43%), Gaps = 25/153 (16%)
Query: 126 LFLPQHND-ADFILVVVSGKAILTVLNPNNRNSFNLERGDTIKLPAGTLGYLANRDDNKD 184
LFLP N A +LV+ G+A + ++ + +R + P Y A + +D
Sbjct: 466 LFLPHFNSKAIVVLVINEGEANIELVGIKEQQ----QRQQQEEQPLEVRKYRAELSE-QD 520
Query: 185 LRVLDLAIPV----NRPGQFQSFSLSESENQQSFLSGFSKNIL--------EAAFNSNYE 232
+ V+ PV F +F ++ NQ++FL+G N++ E AF + +
Sbjct: 521 IFVIPAGYPVVVNATSDLNFFAFGINAENNQRNFLAGSKDNVISQIPSQVQELAFLGSAK 580
Query: 233 EIERVLIEENEQEPQHRRGLRKDERRQQSQEAN 265
+IE ++ ++E D + QQ +E N
Sbjct: 581 DIENLIKSQSE-------SYFVDAQPQQKEEGN 606
>UniRef100_O22121 Beta subunit of beta conglycinin [Glycine max]
Length = 416
Score = 404 bits (1039), Expect = e-111
Identities = 207/370 (55%), Positives = 275/370 (73%), Gaps = 24/370 (6%)
Query: 71 QDQENPFIF-NSNRFQTLFENENGHIRLLQRFDKRSKIFENLQNYRLLEYHSKPHTLFLP 129
+D+ NPF F +SN FQTLFEN+NG IRLLQRF+KRS ENL++YR++++ SKP+T+ LP
Sbjct: 5 EDENNPFYFRSSNSFQTLFENQNGRIRLLQRFNKRSPQLENLRDYRIVQFQSKPNTILLP 64
Query: 130 QHNDADFILVVVSGKAILTVLNPNNRNSFNLERGDTIKLPAGTLGYLANRDDNKDLRVLD 189
H DADF+L V+SG+AILT++N ++R+S+NL GD ++PAGT YL N D+++L+++
Sbjct: 65 HHADADFLLFVLSGRAILTLVNNDDRDSYNLHPGDAQRIPAGTTYYLVNPHDHQNLKIIK 124
Query: 190 LAIPVNRPGQFQSFSLSESENQQSFLSGFSKNILEAAFNSNYEEIERVLIEENEQEPQHR 249
LAIPVN+PG++ F LS ++ QQS+L GFS NILE +F+S +EEI RVL E E
Sbjct: 125 LAIPVNKPGRYDDFFLSSTQAQQSYLQGFSHNILETSFHSEFEEINRVLFGEEE------ 178
Query: 250 RGLRKDERRQQSQEANVIVKVSREQIEELSKNAKSSSRRSESSESEPINLRNQKPIYSNK 309
+Q Q+ VIV++S+EQI +LS+ AKSSSR++ SSE EP NLR++ PIYSN
Sbjct: 179 ---------EQRQQEGVIVELSKEQIRQLSRRAKSSSRKTISSEDEPFNLRSRNPIYSNN 229
Query: 310 FGNFFEITPEKNPQLKDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEGKGEFELVGQR 369
FG FFEITPEKNPQL+DLDI ++ +I EG+LLLPHFNS+A VI+ + EG ELVG +
Sbjct: 230 FGKFFEITPEKNPQLRDLDIFLSSVDINEGALLLPHFNSKAIVILVINEGDANIELVGIK 289
Query: 370 NENQQEQREYEEDEQQQERSQQVQRYRARLSPGDVYVIPAGHPIVVTASSDLSLLGFGIN 429
E QQ+Q+ Q+E +VQRYRA LS DV+VIPA +P VV A+S+L+ L FGIN
Sbjct: 290 -EQQQKQK-------QEEEPLEVQRYRAELSEDDVFVIPAAYPFVVNATSNLNFLAFGIN 341
Query: 430 AENNQRNFLA 439
AENNQRNFLA
Sbjct: 342 AENNQRNFLA 351
Score = 33.9 bits (76), Expect = 9.5
Identities = 33/131 (25%), Positives = 62/131 (47%), Gaps = 18/131 (13%)
Query: 126 LFLPQHND-ADFILVVVSGKAILTVLNPNNRNSFNLERGDTIKLPAGTLGYLANRDDNKD 184
L LP N A ILV+ G A + ++ + + + +++ Y A ++ D
Sbjct: 261 LLLPHFNSKAIVILVINEGDANIELVGIKEQQQKQKQEEEPLEVQR----YRAELSED-D 315
Query: 185 LRVLDLAIP--VNRPGQ--FQSFSLSESENQQSFLSGFSKNIL--------EAAFNSNYE 232
+ V+ A P VN F +F ++ NQ++FL+G N++ E AF + +
Sbjct: 316 VFVIPAAYPFVVNATSNLNFLAFGINAENNQRNFLAGEKDNVVRQIERQVQELAFPGSAQ 375
Query: 233 EIERVLIEENE 243
++ER+L ++ E
Sbjct: 376 DVERLLKKQRE 386
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.315 0.133 0.360
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 702,554,314
Number of Sequences: 2790947
Number of extensions: 30216565
Number of successful extensions: 216808
Number of sequences better than 10.0: 1383
Number of HSP's better than 10.0 without gapping: 358
Number of HSP's successfully gapped in prelim test: 1088
Number of HSP's that attempted gapping in prelim test: 202281
Number of HSP's gapped (non-prelim): 7652
length of query: 440
length of database: 848,049,833
effective HSP length: 130
effective length of query: 310
effective length of database: 485,226,723
effective search space: 150420284130
effective search space used: 150420284130
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 76 (33.9 bits)
Medicago: description of AC148289.2


