

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148289.14 - phase: 0
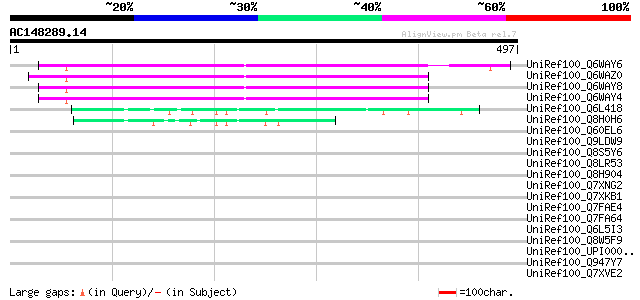
(497 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q6WAY6 Hypothetical protein [Pisum sativum] 278 3e-73
UniRef100_Q6WAZ0 Hypothetical protein [Pisum sativum] 275 3e-72
UniRef100_Q6WAY8 Hypothetical protein [Pisum sativum] 269 1e-70
UniRef100_Q6WAY4 Hypothetical protein [Pisum sativum] 265 3e-69
UniRef100_Q6L418 Hypothetical protein PGEC989P08.9 [Solanum demi... 72 5e-11
UniRef100_Q8H0H6 Hypothetical protein B27 [Nicotiana tabacum] 61 6e-08
UniRef100_Q60EL6 Putative polyprotein [Oryza sativa] 43 0.024
UniRef100_Q9LDW9 EST C28952(C62945) corresponds to a region of t... 42 0.041
UniRef100_Q8S5Y6 Putative transposase related protein [Oryza sat... 42 0.041
UniRef100_Q8LR53 Putative mutator-like transposase [Oryza sativa] 42 0.041
UniRef100_Q8H904 Putative transposase [Oryza sativa] 42 0.041
UniRef100_Q7XNG2 OSJNBa0096F01.11 protein [Oryza sativa] 42 0.041
UniRef100_Q7XKB1 OSJNBa0064G10.20 protein [Oryza sativa] 42 0.041
UniRef100_Q7FAE4 OSJNBa0052P16.6 protein [Oryza sativa] 42 0.041
UniRef100_Q7FA64 OSJNBa0011F23.13 protein [Oryza sativa] 42 0.041
UniRef100_Q6L5I3 Putative mutator-like transposase [Oryza sativa] 42 0.041
UniRef100_Q8W5F9 Hypothetical protein OSJNBb0008A05.4 [Oryza sat... 42 0.041
UniRef100_UPI000042E5B2 UPI000042E5B2 UniRef100 entry 42 0.053
UniRef100_Q947Y7 Putative mutator-like transposase [Oryza sativa] 42 0.053
UniRef100_Q7XVE2 OSJNBa0083D01.20 protein [Oryza sativa] 42 0.053
>UniRef100_Q6WAY6 Hypothetical protein [Pisum sativum]
Length = 562
Score = 278 bits (711), Expect = 3e-73
Identities = 167/469 (35%), Positives = 249/469 (52%), Gaps = 27/469 (5%)
Query: 29 RRNTRKFTFRKLDLENLKKLAFEVTS--LENFRDRHGNLLGVLWTNIEKGCLETLVQFYD 86
+R T ++F + L +L +L+ +TS + F D+HG++L +L ++ L+TL+QFYD
Sbjct: 24 KRKTCSYSFHREPLTSLIELSNLMTSGNQKGFVDQHGDILTLLKMVVDPVPLQTLLQFYD 83
Query: 87 PAYHCFTFPDYQLMPTLEEYSHLIGLPVLDKVPFTGLEPFPKAATIANALHLKTSLIKEK 146
P HCFTF DYQL PTLEEYS L+ +PV +VPF + +A ALHL + +
Sbjct: 84 PELHCFTFQDYQLAPTLEEYSILLSVPVRYQVPFLDVPKEVDFGVVARALHLGIKEVSDS 143
Query: 147 LTLKGNLPSLPTKFLYQQASEFSKTNNVEAFYSILALLIYGLVLFPNIDNYVDIHAIQIF 206
G++ LP KFL + A E ++ + EAF + LA++IYG+VLFP++ N+VD A+ IF
Sbjct: 144 WKYSGDVVGLPLKFLLRVAREEAEKGSWEAFRAQLAVMIYGIVLFPSMPNFVDYAAVSIF 203
Query: 207 LTKNPVPTLLADIYHSIHDRTQVGRGAILGCAPLLYKWFTSHLPQTHSFQANPENLSWPK 266
+ NPVPTLLAD Y++IH R G GAI C PLL +WF S LP + F W +
Sbjct: 204 IGGNPVPTLLADTYYAIHSRHGKG-GAIRCCLPLLLRWFLSLLPVSGPFVDAQSTHKWTQ 262
Query: 267 RIMSLTPSDITWYRATCNFTNIIVSCGEYSNVPLLGTQGGISYNPILAKRQFGCPMEAKP 326
R+MSLT DI W + N+I+SCG + NVPL+GT+G I+YNP+L+ RQ G M+ +P
Sbjct: 263 RVMSLTSYDIRWQSYRMDVRNVIMSCGGFRNVPLVGTRGCINYNPVLSLRQLGFVMKGRP 322
Query: 327 DNIYLQGEFYFNHEDPSNKRGRFVQAWHAIRTLNRSQLARRLDSLQGSYTQWVVNRASDL 386
+ YF + + + +AW +I + S L ++ YT WV R L
Sbjct: 323 LEAEVAESVYFEKQSDPARLEQIGRAWKSIGVKDGSVLGKKFVVAMPDYTDWVKERVETL 382
Query: 387 VLPYHLPRYLSSTTPAPALPLAPATVEECQEKLARSECVSATWKRKYDEAMLKVETISGK 446
+LPY L P+ PA Y +A+++ + GK
Sbjct: 383 LLPYDRMEPLQEQPPSILAESVPA--------------------EHYKQALMENRRLRGK 422
Query: 447 IEQQEHEVHKLRRQIVKKNVQIRA----QSTRLSQFISAGERWEFFKDA 491
+ + E++K + + Q+R ++RL + E E DA
Sbjct: 423 EQDTQLELYKAKADKLNLAHQLRGVREEDASRLRSKKRSYEEMESMLDA 471
>UniRef100_Q6WAZ0 Hypothetical protein [Pisum sativum]
Length = 562
Score = 275 bits (702), Expect = 3e-72
Identities = 154/394 (39%), Positives = 225/394 (57%), Gaps = 3/394 (0%)
Query: 19 LVFVNIIGMERRNTRKFTFRKLDLENLKKLAFEVTS--LENFRDRHGNLLGVLWTNIEKG 76
L+ + I+ +R T ++F + L +L +L+ +TS + F D++G++L +L ++
Sbjct: 14 LIEMAIVPELKRKTCSYSFHREPLTSLIELSNLMTSGNQKGFVDQYGDILTLLKMVVDPV 73
Query: 77 CLETLVQFYDPAYHCFTFPDYQLMPTLEEYSHLIGLPVLDKVPFTGLEPFPKAATIANAL 136
L+TL+QFYDP HCFTF DYQL PTLEEYS L+ +PV +VPF + +A AL
Sbjct: 74 PLQTLLQFYDPELHCFTFQDYQLAPTLEEYSILLSVPVRYQVPFLDVPKEVDFRVVARAL 133
Query: 137 HLKTSLIKEKLTLKGNLPSLPTKFLYQQASEFSKTNNVEAFYSILALLIYGLVLFPNIDN 196
HL + + G++ LP KFL + A E ++ + EAF++ LA++IYG+VLFP++ N
Sbjct: 134 HLGIKEVSDNWKSSGDVVGLPLKFLLRVAREEAEKGSWEAFHAQLAVMIYGIVLFPSMPN 193
Query: 197 YVDIHAIQIFLTKNPVPTLLADIYHSIHDRTQVGRGAILGCAPLLYKWFTSHLPQTHSFQ 256
+VD A+ IF+ NPVPTLLAD Y++IH R G GAI C PLL +WF S LP + F
Sbjct: 194 FVDYAAVSIFIGGNPVPTLLADTYYAIHSRHGKG-GAIRCCLPLLLRWFLSLLPVSGPFV 252
Query: 257 ANPENLSWPKRIMSLTPSDITWYRATCNFTNIIVSCGEYSNVPLLGTQGGISYNPILAKR 316
W +R+MSLT DI W + N+I+SCG + NVPL+GT+G I+YNP+L+ R
Sbjct: 253 DAQSTHKWTQRVMSLTSYDIRWQSYRMDVRNVIMSCGGFRNVPLVGTRGCINYNPVLSLR 312
Query: 317 QFGCPMEAKPDNIYLQGEFYFNHEDPSNKRGRFVQAWHAIRTLNRSQLARRLDSLQGSYT 376
Q G M+ +P + YF + + + +AW +I + S L ++ YT
Sbjct: 313 QLGFVMKGRPLEAEIAESVYFEKQSDPARLEQIGRAWKSIGVKDGSVLGKKFVVAMPDYT 372
Query: 377 QWVVNRASDLVLPYHLPRYLSSTTPAPALPLAPA 410
WV R L+LPY L P PA
Sbjct: 373 DWVKERVETLLLPYDRMEPLQEQPPLILAESVPA 406
>UniRef100_Q6WAY8 Hypothetical protein [Pisum sativum]
Length = 562
Score = 269 bits (688), Expect = 1e-70
Identities = 151/384 (39%), Positives = 218/384 (56%), Gaps = 3/384 (0%)
Query: 29 RRNTRKFTFRKLDLENLKKLAFEVTS--LENFRDRHGNLLGVLWTNIEKGCLETLVQFYD 86
+R T ++F + L +L +L+ +TS + F D++G++L +L ++ L+TL+QFYD
Sbjct: 24 KRKTCSYSFHREPLTSLIELSNLMTSGNQKGFVDQYGDILTLLKMVVDPVPLQTLLQFYD 83
Query: 87 PAYHCFTFPDYQLMPTLEEYSHLIGLPVLDKVPFTGLEPFPKAATIANALHLKTSLIKEK 146
P HCFTF DYQL PTLEEYS L+ +PV +VPF + +A ALHL + +
Sbjct: 84 PELHCFTFQDYQLAPTLEEYSILLSVPVRYQVPFLDVPKEVDFRVVARALHLGIKEVSDS 143
Query: 147 LTLKGNLPSLPTKFLYQQASEFSKTNNVEAFYSILALLIYGLVLFPNIDNYVDIHAIQIF 206
G++ LP KFL + A E ++ + EAF++ LA++IYG+VLFP++ N+VD A+ IF
Sbjct: 144 WKSSGDVVGLPLKFLLRVAREEAEKGSWEAFHAQLAVMIYGIVLFPSMPNFVDYAAVSIF 203
Query: 207 LTKNPVPTLLADIYHSIHDRTQVGRGAILGCAPLLYKWFTSHLPQTHSFQANPENLSWPK 266
+ NPVPTLLAD Y++IH R G GAI C PLL +WF S LP + F W +
Sbjct: 204 IGGNPVPTLLADTYYAIHSRHGKG-GAIRCCLPLLLRWFLSLLPVSGPFVDAQSTHKWTQ 262
Query: 267 RIMSLTPSDITWYRATCNFTNIIVSCGEYSNVPLLGTQGGISYNPILAKRQFGCPMEAKP 326
R+MSLT DI W + N+I+SCG + NVPL+GT+G I+YNP+L+ RQ G M+ +P
Sbjct: 263 RVMSLTSYDIRWQSYRMDVRNVIMSCGGFRNVPLIGTRGCINYNPVLSLRQLGFVMKGRP 322
Query: 327 DNIYLQGEFYFNHEDPSNKRGRFVQAWHAIRTLNRSQLARRLDSLQGSYTQWVVNRASDL 386
+ F + + +AW +I + S L ++ YT WV R L
Sbjct: 323 LEAEVAESVCFEKRSDPARLEQIGRAWKSIGMKDGSVLGKKFAVAMPDYTDWVKERVETL 382
Query: 387 VLPYHLPRYLSSTTPAPALPLAPA 410
+LPY L P PA
Sbjct: 383 LLPYDRMEPLQEQPPLILAESVPA 406
>UniRef100_Q6WAY4 Hypothetical protein [Pisum sativum]
Length = 546
Score = 265 bits (676), Expect = 3e-69
Identities = 148/384 (38%), Positives = 213/384 (54%), Gaps = 3/384 (0%)
Query: 29 RRNTRKFTFRKLDLENLKKLAFEVTS--LENFRDRHGNLLGVLWTNIEKGCLETLVQFYD 86
+R T ++F + L +L +L + S L+ F + G++L +L T ++ L+TL+QFYD
Sbjct: 8 KRKTCSYSFYREPLTSLIELGSLMPSDQLKGFVGQFGDILTLLKTVVDPVPLQTLLQFYD 67
Query: 87 PAYHCFTFPDYQLMPTLEEYSHLIGLPVLDKVPFTGLEPFPKAATIANALHLKTSLIKEK 146
P CFTF DYQL PTLEEYS L+ +P+ +VPF + IA AL + + +
Sbjct: 68 PELRCFTFQDYQLAPTLEEYSILMSIPIQHQVPFLDVPKEVDFRVIARALRMSVKEVCDN 127
Query: 147 LTLKGNLPSLPTKFLYQQASEFSKTNNVEAFYSILALLIYGLVLFPNIDNYVDIHAIQIF 206
G + +P KFL + A E ++ N E F++ LA +IYG+VLFP++ N++D AI IF
Sbjct: 128 WKPSGEVVGMPLKFLLRVAREEAEKGNWEVFHAQLAAMIYGIVLFPSMPNFIDHAAISIF 187
Query: 207 LTKNPVPTLLADIYHSIHDRTQVGRGAILGCAPLLYKWFTSHLPQTHSFQANPENLSWPK 266
+ NPVPTLLAD Y++IH R G GAI C PLL +WF S LP + F L W +
Sbjct: 188 IGGNPVPTLLADTYYAIHSRHGKG-GAIRCCLPLLLRWFMSLLPASGPFMDTQSTLKWTQ 246
Query: 267 RIMSLTPSDITWYRATCNFTNIIVSCGEYSNVPLLGTQGGISYNPILAKRQFGCPMEAKP 326
R+MSLT DI W + ++++SCGE+ NVPL+GT+G I+YNP+L+ RQ G M +P
Sbjct: 247 RVMSLTSYDIRWQSYRMDVKDVVMSCGEFRNVPLVGTKGCINYNPVLSLRQLGFIMSRRP 306
Query: 327 DNIYLQGEFYFNHEDPSNKRGRFVQAWHAIRTLNRSQLARRLDSLQGSYTQWVVNRASDL 386
+ YF + + +AW +I + S L ++ YT WV R L
Sbjct: 307 LEAEIVESVYFEKRTDPVRLEQIGRAWKSIGVKDGSVLGKKFAIAMPDYTDWVKKRVETL 366
Query: 387 VLPYHLPRYLSSTTPAPALPLAPA 410
+LPY L P PA
Sbjct: 367 LLPYDRMEPLQEQPPLILADSVPA 390
>UniRef100_Q6L418 Hypothetical protein PGEC989P08.9 [Solanum demissum]
Length = 607
Score = 71.6 bits (174), Expect = 5e-11
Identities = 106/471 (22%), Positives = 174/471 (36%), Gaps = 84/471 (17%)
Query: 61 RHGNLLGVLWTNIEKGCLETLVQFYDPAYHCFTFPDYQLMPTLEEYSHLIGLPVLDKVPF 120
R G L +L+ + +E LV F+DP + F F D++L PTLEE IG K
Sbjct: 47 RLGFLRSLLYVEPRRDLIEALVHFWDPIKNVFHFSDFELTPTLEEIGAFIGR---GKNLH 103
Query: 121 TGLEPFPKAATIANALHLKTSLIKEKLTLKGNLPS--LPTKFLYQQASEFSKTNNVEAF- 177
G PK L L L + + G L + +P +FLY++ + + + E +
Sbjct: 104 EGEPMIPKHINGRRFLEL---LHINENEIGGCLDNGWVPLEFLYKR---YGRKDGFELYG 157
Query: 178 ----------------YSILALLIYGLVLFPNIDNYVDIH--AIQIFLTKNP----VPTL 215
Y + + G+++F ++I+ A+ K P VP +
Sbjct: 158 KKLHNNGCRLTWETHSYDAITVAFLGIMVFLKRGGKININLAAVITAFEKKPNITLVPMI 217
Query: 216 LADIYHSIHDRTQVGRGAILGCAPLLYKWFTSHLP-----------------------QT 252
LADIY ++ + G GC LL W H+ +
Sbjct: 218 LADIYRAL-TICKNGGDYFEGCNMLLQLWMIEHIRHHPYVVDFKVECNDYIGGHEERIKD 276
Query: 253 HSFQANPENLSWPKRIMSLTPSDITWYRATCNFTNIIVSCGEYSNVPLLGTQGGISYNPI 312
HSF E +W K + +LT I W +I S + L+G +G Y P+
Sbjct: 277 HSFPKGIE--AWKKYLNNLTADKIVWNYHWFPSAEVIYMSTFRSFIVLMGLRGVQPYMPL 334
Query: 313 LAKRQFGCPMEAKPDNIYLQGEFYFNHEDPSNKRGRFVQAWHAIRTLNRSQLA--RRLDS 370
RQ G P+ + + F+ E P K F + W + R
Sbjct: 335 RVMRQLGRRQFLPPNEDIREFMYEFHPEIPLRKSEIF-KIWGGCMLSSPHDRVEDRTKGE 393
Query: 371 LQGSYTQWVVNRASDLVLP-----------------YHLPRYLSSTTPAPALPLAPATVE 413
+ +Y +WV ++ S VLP R + L ++
Sbjct: 394 VDQAYLEWVHDQPSPKVLPEGSVKGPVDREAEIEVRIKQARLEVERSYRSTLDCLSNDLK 453
Query: 414 ECQEKLARSECVSATWKRKYDEAMLKVE----TISGKIEQQEHEVHKLRRQ 460
+E+LA+ + + RK+ +L ++ + +EQQE E K + Q
Sbjct: 454 NAKEELAQRDAIFEVRVRKHRSTILTLQEDLGIVISAMEQQEEEYTKEKVQ 504
>UniRef100_Q8H0H6 Hypothetical protein B27 [Nicotiana tabacum]
Length = 464
Score = 61.2 bits (147), Expect = 6e-08
Identities = 75/305 (24%), Positives = 115/305 (37%), Gaps = 60/305 (19%)
Query: 63 GNLLGVLWTNIEKGCLETLVQFYDPAYHCFTFPDYQLMPTLEEYSHLIGLPVLDKVPFTG 122
G L ++ +E LV F+DP ++ F F D++L PTLEE G +
Sbjct: 39 GALTDIMKIKPRDDLIEALVTFWDPVHNVFCFSDFELTPTLEEIDGYSGF---GRDLRNQ 95
Query: 123 LEPFPKAATIANALHLK--TSLIKEKLTLKGNLPSLPTKFLYQQASEFSKTNNVE----- 175
FP+A ++ L + I++ +KG FLY S F + N E
Sbjct: 96 ELIFPRALSVHRFFDLLNISKQIRKTNVVKG---CCSFYFLY---SRFGQPNGFEMHEKG 149
Query: 176 --------------AFYSILALLIYGLVLFPNIDNYVDIH---AIQIFLTKNP---VPTL 215
F I+A L G+++FPN + +D +Q+ TK P +
Sbjct: 150 LNNKQNKDTWHIHRCFAFIMAFL--GIMVFPNRERTIDTRIARVVQVLTTKEHHTLAPII 207
Query: 216 LADIYHSIHDRTQVGRGAILGCAPLLYKWFTSHL---PQTHSFQANPENL---------- 262
L+DIY ++ + G GC LL W HL P+ S+ + +N
Sbjct: 208 LSDIYRAL-TLCKSGAKFFEGCNILLQMWLIEHLRHHPKFMSYGPSKDNFIDSYEERVKE 266
Query: 263 --------SWPKRIMSLTPSDITWYRATCNFTNIIVSCGEYSNVPLLGTQGGISYNPILA 314
+W + SL I W +I S++ LLG + Y P
Sbjct: 267 YNSPEGVETWISHLRSLNACQIEWTLGWLPLREVIHMSALKSHLLLLGLRSVQPYMPHRV 326
Query: 315 KRQFG 319
RQ G
Sbjct: 327 LRQLG 331
>UniRef100_Q60EL6 Putative polyprotein [Oryza sativa]
Length = 1542
Score = 42.7 bits (99), Expect = 0.024
Identities = 51/196 (26%), Positives = 87/196 (44%), Gaps = 29/196 (14%)
Query: 74 EKGCLETLVQFYDPAYHCFTFPDYQLMPTLEEYSHLIGLPVLDKV--PFTGLEPFPK--A 129
++ L LV + P H F P ++ PTL++ S+L+GLP+ P G+ + +
Sbjct: 842 DRSLLAALVDRWRPETHTFHLPCGEMAPTLQDVSYLLGLPLAGAPVGPVDGVFGWKEDIT 901
Query: 130 ATIANALHL----KTSLIKEKLTL---KGNLPSLPTKFLYQQASEFSKTNNVEAFYSILA 182
A +HL T+ + T+ K L L+ A ++S ++EA+
Sbjct: 902 ARFEQVMHLPHLGPTTTLPPYSTVGPSKAWLLQFTADLLHPDADDYSVRRSLEAY----L 957
Query: 183 LLIYGLVLFPNIDNY-VD---IHAIQIFLTKNP--VP------TLLADIYHSIHDR-TQV 229
L ++G V+F + + VD +H + P VP +LA Y ++ + T+
Sbjct: 958 LWLFGWVMFTSTHGHAVDFRLVHYARSIADAQPQDVPQWSWGSAVLAATYRALCEACTKT 1017
Query: 230 GRGAIL-GCAPLLYKW 244
GAI+ GC LL W
Sbjct: 1018 DAGAIIAGCPMLLQLW 1033
>UniRef100_Q9LDW9 EST C28952(C62945) corresponds to a region of the predicted gene
[Oryza sativa]
Length = 1591
Score = 42.0 bits (97), Expect = 0.041
Identities = 49/196 (25%), Positives = 84/196 (42%), Gaps = 29/196 (14%)
Query: 74 EKGCLETLVQFYDPAYHCFTFPDYQLMPTLEEYSHLIGLPVLDKV--PFTGLEPFPKAAT 131
++ L LV + P H F P ++ PTL++ S+L+GLP+ P G+ + + T
Sbjct: 945 DRSLLAALVDRWRPETHTFHLPCGEMAPTLQDVSYLLGLPLAGAAVGPVDGVFGWKEDIT 1004
Query: 132 IANALHLKTSLIKEKLTL---------KGNLPSLPTKFLYQQASEFSKTNNVEAFYSILA 182
++ + TL K L L+ A ++S ++EA+
Sbjct: 1005 ARFEQVMRLPHLGPTTTLPPYSTVGPSKAWLLQFTADLLHPDADDYSVRRSLEAY----L 1060
Query: 183 LLIYGLVLFPNIDNY-VD---IHAIQIFLTKNP--VP------TLLADIYHSIHDR-TQV 229
L ++G V+F + + VD +H + P VP +LA Y ++ + T+
Sbjct: 1061 LWLFGWVMFTSTHGHAVDFRLVHYARSIADAQPQDVPQWSWGSAVLAATYRALCEACTKT 1120
Query: 230 GRGAIL-GCAPLLYKW 244
GAI+ GC LL W
Sbjct: 1121 DAGAIIAGCPMLLQLW 1136
>UniRef100_Q8S5Y6 Putative transposase related protein [Oryza sativa]
Length = 1557
Score = 42.0 bits (97), Expect = 0.041
Identities = 49/196 (25%), Positives = 84/196 (42%), Gaps = 29/196 (14%)
Query: 74 EKGCLETLVQFYDPAYHCFTFPDYQLMPTLEEYSHLIGLPVLDKV--PFTGLEPFPKAAT 131
++ L LV + P H F P ++ PTL++ S+L+GLP+ P G+ + + T
Sbjct: 929 DRSLLAALVDRWRPETHTFHLPCGEMAPTLQDVSYLLGLPLAGAAVGPVDGVFGWKEDIT 988
Query: 132 IANALHLKTSLIKEKLTL---------KGNLPSLPTKFLYQQASEFSKTNNVEAFYSILA 182
++ + TL K L L+ A ++S ++EA+
Sbjct: 989 ARFEQVMRLPHLGPTTTLPPYSTVGPSKAWLLQFTADLLHPDADDYSVRRSLEAY----L 1044
Query: 183 LLIYGLVLFPNIDNY-VD---IHAIQIFLTKNP--VP------TLLADIYHSIHDR-TQV 229
L ++G V+F + + VD +H + P VP +LA Y ++ + T+
Sbjct: 1045 LWLFGWVMFTSTHGHAVDFRLVHYARSIADAQPQDVPQWSWGSAVLAATYRALCEACTKT 1104
Query: 230 GRGAIL-GCAPLLYKW 244
GAI+ GC LL W
Sbjct: 1105 DAGAIIAGCPMLLQLW 1120
>UniRef100_Q8LR53 Putative mutator-like transposase [Oryza sativa]
Length = 1605
Score = 42.0 bits (97), Expect = 0.041
Identities = 49/196 (25%), Positives = 84/196 (42%), Gaps = 29/196 (14%)
Query: 74 EKGCLETLVQFYDPAYHCFTFPDYQLMPTLEEYSHLIGLPVLDKV--PFTGLEPFPKAAT 131
++ L LV + P H F P ++ PTL++ S+L+GLP+ P G+ + + T
Sbjct: 956 DRSLLAALVDRWRPETHTFHLPCGEMAPTLQDVSYLLGLPLAGAAVGPVDGVFGWKEDIT 1015
Query: 132 IANALHLKTSLIKEKLTL---------KGNLPSLPTKFLYQQASEFSKTNNVEAFYSILA 182
++ + TL K L L+ A ++S ++EA+
Sbjct: 1016 ARFEQVMRLPHLGPTTTLPPYSTVGPSKAWLLQFTADLLHPDADDYSVRRSLEAY----L 1071
Query: 183 LLIYGLVLFPNIDNY-VD---IHAIQIFLTKNP--VP------TLLADIYHSIHDR-TQV 229
L ++G V+F + + VD +H + P VP +LA Y ++ + T+
Sbjct: 1072 LWLFGWVMFTSTHGHAVDFRLVHYARSIADAQPQDVPQWSWGSAVLAATYRALCEACTKT 1131
Query: 230 GRGAIL-GCAPLLYKW 244
GAI+ GC LL W
Sbjct: 1132 DAGAIIAGCPMLLQLW 1147
>UniRef100_Q8H904 Putative transposase [Oryza sativa]
Length = 1597
Score = 42.0 bits (97), Expect = 0.041
Identities = 49/196 (25%), Positives = 84/196 (42%), Gaps = 29/196 (14%)
Query: 74 EKGCLETLVQFYDPAYHCFTFPDYQLMPTLEEYSHLIGLPVLDKV--PFTGLEPFPKAAT 131
++ L LV + P H F P ++ PTL++ S+L+GLP+ P G+ + + T
Sbjct: 956 DRSLLAALVDRWRPETHTFHLPCGEMAPTLQDVSYLLGLPLAGAAVGPVDGVFGWKEDIT 1015
Query: 132 IANALHLKTSLIKEKLTL---------KGNLPSLPTKFLYQQASEFSKTNNVEAFYSILA 182
++ + TL K L L+ A ++S ++EA+
Sbjct: 1016 ARFEQVMRLPHLGPTTTLPPYSTVGPSKAWLLQFTADLLHPDADDYSVRRSLEAY----L 1071
Query: 183 LLIYGLVLFPNIDNY-VD---IHAIQIFLTKNP--VP------TLLADIYHSIHDR-TQV 229
L ++G V+F + + VD +H + P VP +LA Y ++ + T+
Sbjct: 1072 LWLFGWVMFTSTHGHAVDFRLVHYARSIADAQPQDVPQWSWGSAVLAATYRALCEACTKT 1131
Query: 230 GRGAIL-GCAPLLYKW 244
GAI+ GC LL W
Sbjct: 1132 DAGAIIAGCPMLLQLW 1147
>UniRef100_Q7XNG2 OSJNBa0096F01.11 protein [Oryza sativa]
Length = 1365
Score = 42.0 bits (97), Expect = 0.041
Identities = 49/196 (25%), Positives = 84/196 (42%), Gaps = 29/196 (14%)
Query: 74 EKGCLETLVQFYDPAYHCFTFPDYQLMPTLEEYSHLIGLPVLDKV--PFTGLEPFPKAAT 131
++ L LV + P H F P ++ PTL++ S+L+GLP+ P G+ + + T
Sbjct: 805 DRSLLAALVDHWRPETHTFHLPCGEMAPTLQDVSYLLGLPLAGAPVGPVDGVFGWKEDIT 864
Query: 132 IANALHLKTSLIKEKLTL---------KGNLPSLPTKFLYQQASEFSKTNNVEAFYSILA 182
++ + TL K L L+ A ++ ++EA+
Sbjct: 865 ARFEQVMRLPHLGPANTLPPYSTVGPSKAWLLQFTADLLHPDADDYLVRRSLEAY----L 920
Query: 183 LLIYGLVLFPNIDNY-VD---IHAIQIFLTKNP--VP------TLLADIYHSIHDR-TQV 229
L ++G V+F + + VD +H + P VP +LA YH++ + T+
Sbjct: 921 LWLFGWVMFTSTHGHAVDFRLVHYARSIADAQPQDVPQWSWGSAVLAATYHALCEACTKT 980
Query: 230 GRGAIL-GCAPLLYKW 244
GAI+ GC LL W
Sbjct: 981 DAGAIIAGCPMLLQLW 996
>UniRef100_Q7XKB1 OSJNBa0064G10.20 protein [Oryza sativa]
Length = 1602
Score = 42.0 bits (97), Expect = 0.041
Identities = 49/191 (25%), Positives = 82/191 (42%), Gaps = 41/191 (21%)
Query: 74 EKGCLETLVQFYDPAYHCFTFPDYQLMPTLEEYSHLIGLPVLDKV------PFTGLEPFP 127
++ L LV + P H F P ++ PTL++ S+L+GLP+ P T L P+
Sbjct: 924 DRSLLAALVDRWRPETHTFHLPCGEMAPTLQDVSYLLGLPLAGAPVGPVDGPTTTLPPY- 982
Query: 128 KAATIANALHLKTSLIKEKLTLKGNLPSLPTKFLYQQASEFSKTNNVEAFYSILALLIYG 187
+T+ + K L L+ A ++S ++EA+ L ++G
Sbjct: 983 --STVGPS--------------KAWLLQFTADLLHPDADDYSVRRSLEAY----LLWLFG 1022
Query: 188 LVLFPNIDNY-VD---IHAIQIFLTKNP--VP------TLLADIYHSIHDR-TQVGRGAI 234
V+F + + VD +H + P VP +LA Y ++ + T+ GAI
Sbjct: 1023 WVMFTSTHGHAVDFRLVHYARSIADAQPQDVPQWSWGSAVLAATYRALCEACTKTDAGAI 1082
Query: 235 L-GCAPLLYKW 244
+ GC LL W
Sbjct: 1083 IAGCPMLLQLW 1093
>UniRef100_Q7FAE4 OSJNBa0052P16.6 protein [Oryza sativa]
Length = 1489
Score = 42.0 bits (97), Expect = 0.041
Identities = 49/196 (25%), Positives = 84/196 (42%), Gaps = 29/196 (14%)
Query: 74 EKGCLETLVQFYDPAYHCFTFPDYQLMPTLEEYSHLIGLPVLDKV--PFTGLEPFPKAAT 131
++ L LV + P H F P ++ PTL++ S+L+GLP+ P G+ + + T
Sbjct: 805 DRSLLAALVDHWRPETHTFHLPCGEMAPTLQDVSYLLGLPLAGAPVGPVDGVFGWKEDIT 864
Query: 132 IANALHLKTSLIKEKLTL---------KGNLPSLPTKFLYQQASEFSKTNNVEAFYSILA 182
++ + TL K L L+ A ++ ++EA+
Sbjct: 865 ARFEQVMRLPHLGPANTLPPYSTVGPSKAWLLQFTADLLHPDADDYLVRRSLEAY----L 920
Query: 183 LLIYGLVLFPNIDNY-VD---IHAIQIFLTKNP--VP------TLLADIYHSIHDR-TQV 229
L ++G V+F + + VD +H + P VP +LA YH++ + T+
Sbjct: 921 LWLFGWVMFTSTHGHAVDFRLVHYARSIADAQPQDVPQWSWGSAVLAATYHALCEACTKT 980
Query: 230 GRGAIL-GCAPLLYKW 244
GAI+ GC LL W
Sbjct: 981 DAGAIIAGCPMLLQLW 996
>UniRef100_Q7FA64 OSJNBa0011F23.13 protein [Oryza sativa]
Length = 1396
Score = 42.0 bits (97), Expect = 0.041
Identities = 49/196 (25%), Positives = 84/196 (42%), Gaps = 29/196 (14%)
Query: 74 EKGCLETLVQFYDPAYHCFTFPDYQLMPTLEEYSHLIGLPVLDKV--PFTGLEPFPKAAT 131
++ L LV + P H F P ++ PTL++ S+L+GLP+ P G+ + + T
Sbjct: 805 DRSLLAALVDRWRPETHTFHLPCGEMAPTLQDVSYLLGLPLAGAAVGPVDGVFGWKEDIT 864
Query: 132 IANALHLKTSLIKEKLTL---------KGNLPSLPTKFLYQQASEFSKTNNVEAFYSILA 182
++ + TL K L L+ A ++S ++EA+
Sbjct: 865 ARFEQVMRLPHLGPTTTLPPYSTVGPSKAWLLQFTADLLHPDADDYSVRRSLEAY----L 920
Query: 183 LLIYGLVLFPNIDNY-VD---IHAIQIFLTKNP--VP------TLLADIYHSIHDR-TQV 229
L ++G V+F + + VD +H + P VP +LA Y ++ + T+
Sbjct: 921 LWLFGWVMFTSTHGHAVDFRLVHYARSIADAQPQDVPQWSWGSAVLAATYRALCEACTKT 980
Query: 230 GRGAIL-GCAPLLYKW 244
GAI+ GC LL W
Sbjct: 981 DAGAIIAGCPMLLQLW 996
>UniRef100_Q6L5I3 Putative mutator-like transposase [Oryza sativa]
Length = 1684
Score = 42.0 bits (97), Expect = 0.041
Identities = 49/196 (25%), Positives = 84/196 (42%), Gaps = 29/196 (14%)
Query: 74 EKGCLETLVQFYDPAYHCFTFPDYQLMPTLEEYSHLIGLPVLDKV--PFTGLEPFPKAAT 131
++ L LV + P H F P ++ PTL++ S+L+GLP+ P G+ + + T
Sbjct: 968 DRSLLAALVDRWRPETHTFHLPCGEMAPTLQDVSYLLGLPLAGAAVGPVDGVFGWKEDIT 1027
Query: 132 IANALHLKTSLIKEKLTL---------KGNLPSLPTKFLYQQASEFSKTNNVEAFYSILA 182
++ + TL K L L+ A ++S ++EA+
Sbjct: 1028 ARFEQVMRLPHLGPTTTLPPYSTVGPSKAWLLQFTADLLHPDADDYSVRRSLEAY----L 1083
Query: 183 LLIYGLVLFPNIDNY-VD---IHAIQIFLTKNP--VP------TLLADIYHSIHDR-TQV 229
L ++G V+F + + VD +H + P VP +LA Y ++ + T+
Sbjct: 1084 LWLFGWVMFTSTHGHAVDFRLVHYARSIADAQPQDVPQWSWGSAVLAATYRALCEACTKT 1143
Query: 230 GRGAIL-GCAPLLYKW 244
GAI+ GC LL W
Sbjct: 1144 DAGAIIAGCPMLLQLW 1159
>UniRef100_Q8W5F9 Hypothetical protein OSJNBb0008A05.4 [Oryza sativa]
Length = 831
Score = 42.0 bits (97), Expect = 0.041
Identities = 49/196 (25%), Positives = 84/196 (42%), Gaps = 29/196 (14%)
Query: 74 EKGCLETLVQFYDPAYHCFTFPDYQLMPTLEEYSHLIGLPVLDKV--PFTGLEPFPKAAT 131
++ L LV + P H F P ++ PTL++ S+L+GLP+ P G+ + + T
Sbjct: 236 DRSLLAALVDRWRPETHTFHLPCGEMAPTLQDVSYLLGLPLAGAAVGPVDGVFGWKEDIT 295
Query: 132 IANALHLKTSLIKEKLTL---------KGNLPSLPTKFLYQQASEFSKTNNVEAFYSILA 182
++ + TL K L L+ A ++S ++EA+
Sbjct: 296 ARFEQVMRLPHLGPTTTLPPYSTVGPSKAWLLQFTADLLHPDADDYSVRRSLEAY----L 351
Query: 183 LLIYGLVLFPNIDNY-VD---IHAIQIFLTKNP--VP------TLLADIYHSIHDR-TQV 229
L ++G V+F + + VD +H + P VP +LA Y ++ + T+
Sbjct: 352 LWLFGWVMFTSTHGHAVDFRLVHYARSIADAQPQDVPQWSWGSAVLAATYRALCEACTKT 411
Query: 230 GRGAIL-GCAPLLYKW 244
GAI+ GC LL W
Sbjct: 412 DAGAIIAGCPMLLQLW 427
>UniRef100_UPI000042E5B2 UPI000042E5B2 UniRef100 entry
Length = 1948
Score = 41.6 bits (96), Expect = 0.053
Identities = 17/67 (25%), Positives = 40/67 (59%)
Query: 411 TVEECQEKLARSECVSATWKRKYDEAMLKVETISGKIEQQEHEVHKLRRQIVKKNVQIRA 470
T++ Q +L + E + ++ K +EA KV + GK+E++ E+ + + +I K ++ +
Sbjct: 1075 TIDSLQIQLKKDEDYKSRYESKIEEAREKVTLLKGKVEKKSQEIDQYKSEIKKLKAELAS 1134
Query: 471 QSTRLSQ 477
+T+++Q
Sbjct: 1135 SNTKITQ 1141
>UniRef100_Q947Y7 Putative mutator-like transposase [Oryza sativa]
Length = 2421
Score = 41.6 bits (96), Expect = 0.053
Identities = 49/196 (25%), Positives = 84/196 (42%), Gaps = 29/196 (14%)
Query: 74 EKGCLETLVQFYDPAYHCFTFPDYQLMPTLEEYSHLIGLPVLDKV--PFTGLEPFPKAAT 131
++ L LV + P H F P ++ PTL++ S+L+GLP+ P G+ + + T
Sbjct: 862 DRSLLAALVDRWRPETHTFHLPCGEMAPTLQDVSYLLGLPLAGAPVGPIDGVFGWKEDIT 921
Query: 132 IANALHLKTSLIKEKLTL---------KGNLPSLPTKFLYQQASEFSKTNNVEAFYSILA 182
++ + TL K L L+ A ++S ++EA+
Sbjct: 922 ARFEQVMRLPHLGPTTTLPPYSTVGPSKAWLLQFTADLLHPDADDYSVRRSLEAY----L 977
Query: 183 LLIYGLVLFPNIDNY-VD---IHAIQIFLTKNP--VP------TLLADIYHSIHDR-TQV 229
L ++G V+F + + VD +H + P VP +LA Y ++ + T+
Sbjct: 978 LWLFGWVMFTSTHGHAVDFRLVHYARSIADAQPQDVPQWSWGSAVLAATYRALCEACTKT 1037
Query: 230 GRGAIL-GCAPLLYKW 244
GAI+ GC LL W
Sbjct: 1038 DAGAIIAGCPMLLQLW 1053
>UniRef100_Q7XVE2 OSJNBa0083D01.20 protein [Oryza sativa]
Length = 1590
Score = 41.6 bits (96), Expect = 0.053
Identities = 48/196 (24%), Positives = 84/196 (42%), Gaps = 29/196 (14%)
Query: 74 EKGCLETLVQFYDPAYHCFTFPDYQLMPTLEEYSHLIGLPVLDKV--PFTGLEPFPKAAT 131
++ L L+ + P H F P ++ PTL++ S+L+GLP+ P G+ + + T
Sbjct: 956 DRSLLAALIDRWRPETHTFHLPCGEMAPTLQDVSYLLGLPLAGAAVGPVDGVFGWKEDIT 1015
Query: 132 IANALHLKTSLIKEKLTL---------KGNLPSLPTKFLYQQASEFSKTNNVEAFYSILA 182
++ + TL K L L+ A ++S ++EA+
Sbjct: 1016 ARFEQVMRLPHLGPTTTLPPYSTVGPSKAWLLQFTADLLHPDADDYSVRRSLEAY----L 1071
Query: 183 LLIYGLVLFPNIDNY-VD---IHAIQIFLTKNP--VP------TLLADIYHSIHDR-TQV 229
L ++G V+F + + VD +H + P VP +LA Y ++ + T+
Sbjct: 1072 LWLFGWVMFTSTHGHAVDFRLVHYARSIADAQPQDVPQWSWGSAVLAATYRALCEACTKT 1131
Query: 230 GRGAIL-GCAPLLYKW 244
GAI+ GC LL W
Sbjct: 1132 DAGAIIAGCPMLLQLW 1147
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.322 0.137 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 853,981,226
Number of Sequences: 2790947
Number of extensions: 37114408
Number of successful extensions: 90380
Number of sequences better than 10.0: 93
Number of HSP's better than 10.0 without gapping: 80
Number of HSP's successfully gapped in prelim test: 14
Number of HSP's that attempted gapping in prelim test: 90280
Number of HSP's gapped (non-prelim): 108
length of query: 497
length of database: 848,049,833
effective HSP length: 132
effective length of query: 365
effective length of database: 479,644,829
effective search space: 175070362585
effective search space used: 175070362585
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 77 (34.3 bits)
Medicago: description of AC148289.14


