

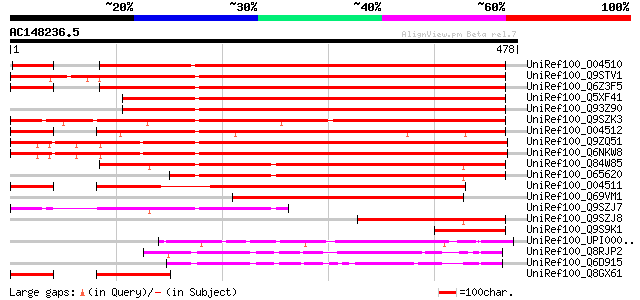
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148236.5 - phase: 0 /pseudo
(478 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_O04510 F21M12.27 protein [Arabidopsis thaliana] 561 e-158
UniRef100_Q9STV1 LG27/30-like gene [Arabidopsis thaliana] 551 e-155
UniRef100_Q6Z3F5 Putative MYST1 [Oryza sativa] 546 e-154
UniRef100_Q5XF41 At4g24430 [Arabidopsis thaliana] 498 e-139
UniRef100_Q93Z90 AT4g24430/T22A6_260 [Arabidopsis thaliana] 498 e-139
UniRef100_Q9SZK3 LG127/30 like gene [Arabidopsis thaliana] 489 e-137
UniRef100_O04512 F21M12.30 protein [Arabidopsis thaliana] 479 e-134
UniRef100_Q9ZQ51 Hypothetical protein At2g22620 [Arabidopsis tha... 475 e-132
UniRef100_Q6NKW8 Hypothetical protein At2g22620 [Arabidopsis tha... 473 e-132
UniRef100_Q84W85 Hypothetical protein At4g37950 [Arabidopsis tha... 455 e-126
UniRef100_O65620 MYST1 protein [Arabidopsis thaliana] 392 e-107
UniRef100_O04511 F21M12.28 protein [Arabidopsis thaliana] 367 e-100
UniRef100_Q69VM1 MYST1-like protein [Oryza sativa] 301 4e-80
UniRef100_Q9SZJ7 Hypothetical protein F20D10.70 [Arabidopsis tha... 211 4e-53
UniRef100_Q9SZJ8 Hypothetical protein F20D10.80 [Arabidopsis tha... 204 3e-51
UniRef100_Q9S9K1 T23K8.12 protein [Arabidopsis thaliana] 103 1e-20
UniRef100_UPI000042E7D6 UPI000042E7D6 UniRef100 entry 102 3e-20
UniRef100_Q8RJP2 Rhamnogalacturonate lyase precursor [Erwinia ch... 96 3e-18
UniRef100_Q6D915 Rhamnogalacturonate lyase [Erwinia carotovora] 89 4e-16
UniRef100_Q8GX61 Hypothetical protein At1g09910/F21M12_30 [Arabi... 87 8e-16
>UniRef100_O04510 F21M12.27 protein [Arabidopsis thaliana]
Length = 631
Score = 561 bits (1447), Expect = e-158
Identities = 258/383 (67%), Positives = 311/383 (80%), Gaps = 3/383 (0%)
Query: 85 WPAFDLDNTRVAFKLRKDKFNYMAIADNRQRFMPLPDDRLPPRGQVLAYPEAVRLVNPVE 144
WPA +LDN R+ FKL K KF+YMAI+D+RQR+MP+PDDR+PPRGQ LAYPEAV+L++P+E
Sbjct: 123 WPAVELDNMRLVFKLNKKKFHYMAISDDRQRYMPVPDDRVPPRGQPLAYPEAVQLLDPIE 182
Query: 145 PEFKGEVDDKYEYACESRSNQVHGWISIDSSSDSTGCWLITPSYEFRSAGPLKQYLGSHL 204
PEFKGEVDDKYEY+ ES+ +VHGWIS ++DS G W ITPS EFRSAGPLKQ+LGSH+
Sbjct: 183 PEFKGEVDDKYEYSMESKDIKVHGWIS---TNDSVGFWQITPSNEFRSAGPLKQFLGSHV 239
Query: 205 GPTMLSVFHSTHYSGADLIMKFGENEPWKKVFGPIFIYLNSNSDGFSPIKLWEDAKQQMV 264
GPT L+VFHSTHY GADLIM F E WKKVFGP+FIYLNS G P+ LW +AK Q
Sbjct: 240 GPTNLAVFHSTHYVGADLIMSFKNGEAWKKVFGPVFIYLNSFPKGVDPLLLWHEAKNQTK 299
Query: 265 NEVESWPYTFPASEDFLSSAQRGKFEGRLLVRDRYIRDAFVPVSGAYVGLAAPGDVGSWQ 324
E E WPY F AS+DF +S QRG GRLLVRDR+I +P +G+YVGLAAPGDVGSWQ
Sbjct: 300 IEEEKWPYNFTASDDFPASDQRGSVSGRLLVRDRFISSEDIPANGSYVGLAAPGDVGSWQ 359
Query: 325 REYKGYQFWTITDDKGYFSIINVLPGDYNLYSWVNGFIGDYQCNNIINITSGSEINVGEL 384
RE KGYQFW+ D+ G FSI NV G YNLY++ GFIGDY + + +I+ GS+I++G+L
Sbjct: 360 RECKGYQFWSKADENGSFSINNVRSGRYNLYAFAPGFIGDYHNDTVFDISPGSKISLGDL 419
Query: 385 VYEPPRHGPTLWEIGIPDRSAAEFYVPDPNPLYVNRLYVNHSDRFRQYGLWERYADLYPN 444
VYEPPR G TLWEIG+PDRSAAEFY+PDPNP +VN+LY+NHSD++RQYGLWERY++LYP+
Sbjct: 420 VYEPPRDGSTLWEIGVPDRSAAEFYIPDPNPSFVNKLYLNHSDKYRQYGLWERYSELYPD 479
Query: 445 EDLVYTVGISDYKKDWFFAQVTR 467
ED+VY V I DY K+WFF QVTR
Sbjct: 480 EDMVYNVDIDDYSKNWFFMQVTR 502
Score = 48.9 bits (115), Expect = 3e-04
Identities = 23/40 (57%), Positives = 30/40 (74%), Gaps = 1/40 (2%)
Query: 3 IDNDIVQVNISNPEGCVTGIQYNGLDNLLE-INNHESDRG 41
++N +Q+ +SNPEG VTGIQYNG+DN+L N E DRG
Sbjct: 1 MENRFLQLTLSNPEGFVTGIQYNGIDNVLAYYTNKEYDRG 40
>UniRef100_Q9STV1 LG27/30-like gene [Arabidopsis thaliana]
Length = 646
Score = 551 bits (1421), Expect = e-155
Identities = 284/509 (55%), Positives = 343/509 (66%), Gaps = 48/509 (9%)
Query: 1 VVIDNDIVQVNISNPEGCVTGIQYNGLDNLLEINNHE----------SDRG*KAGNDKYE 50
VV+ N V+V IS P+G VTGI Y G+DNLLE +N + SD G K E
Sbjct: 16 VVMGNGKVKVTISKPDGFVTGISYQGVDNLLETHNEDFNRGYWDLVWSDEGTPGTTGKSE 75
Query: 51 GDNGK*RTSRAIFH*NMEFIS*-------------------------RKACATQY**K-- 83
G TS + N E + RK Y
Sbjct: 76 RIKG---TSFEVVVENEELVEISFSRKWDSSLQDSIAPINVDKRFIMRKDVTGFYSYAIF 132
Query: 84 ----EWPAFDLDNTRVAFKLRKDKFNYMAIADNRQRFMPLPDDRLPPRGQVLAYPEAVRL 139
EWPAF+L TR+ +KLRKDKF YMAIADNRQR MPLP+DRL RG+ LAYPEAV L
Sbjct: 133 EHLAEWPAFNLPQTRIVYKLRKDKFKYMAIADNRQRKMPLPEDRLGKRGRPLAYPEAVLL 192
Query: 140 VNPVEPEFKGEVDDKYEYACESRSNQVHGWISIDSSSDSTGCWLITPSYEFRSAGPLKQY 199
V+PVE EFKGEVDDKYEY+ E++ +VHGWIS + GCW I PS EFRS G KQ
Sbjct: 193 VHPVEDEFKGEVDDKYEYSSENKDLKVHGWISHNLD---LGCWQIIPSNEFRSGGLSKQN 249
Query: 200 LGSHLGPTMLSVFHSTHYSGADLIMKFGENEPWKKVFGPIFIYLNSNSDGFS-PIKLWED 258
L SH+GP L++F S HY+G D++MK + WKKVFGP+F YLN D S P+ LW+D
Sbjct: 250 LTSHVGPISLAMFLSAHYAGEDMVMKVKAGDSWKKVFGPVFTYLNCLPDKTSDPLSLWQD 309
Query: 259 AKQQMVNEVESWPYTFPASEDFLSSAQRGKFEGRLLVRDRYIRDAFVPVSGAYVGLAAPG 318
AK QM+ EV+SWPY FPASEDF S +RG GRLLV D+++ D F+P +GA+VGLA PG
Sbjct: 310 AKNQMLTEVQSWPYDFPASEDFPVSDKRGCISGRLLVCDKFLSDDFLPANGAFVGLAPPG 369
Query: 319 DVGSWQREYKGYQFWTITDDKGYFSIINVLPGDYNLYSWVNGFIGDYQCNNIINITSGSE 378
+VGSWQ E KGYQFWT D GYF+I ++ G+YNL +V G+IGDYQ +INIT+G +
Sbjct: 370 EVGSWQLESKGYQFWTEADSDGYFAINDIREGEYNLNGYVTGWIGDYQYEQLINITAGCD 429
Query: 379 INVGELVYEPPRHGPTLWEIGIPDRSAAEFYVPDPNPLYVNRLYVNHSDRFRQYGLWERY 438
I+VG +VYEPPR GPT+WEIGIPDRSAAEF+VPDPNP Y+N+LY+ H DRFRQYGLWERY
Sbjct: 430 IDVGNIVYEPPRDGPTVWEIGIPDRSAAEFFVPDPNPKYINKLYIGHPDRFRQYGLWERY 489
Query: 439 ADLYPNEDLVYTVGISDYKKDWFFAQVTR 467
+LYP EDLV+T+G+SDYKKDWFFA VTR
Sbjct: 490 TELYPKEDLVFTIGVSDYKKDWFFAHVTR 518
>UniRef100_Q6Z3F5 Putative MYST1 [Oryza sativa]
Length = 645
Score = 546 bits (1406), Expect = e-154
Identities = 255/383 (66%), Positives = 301/383 (78%), Gaps = 3/383 (0%)
Query: 85 WPAFDLDNTRVAFKLRKDKFNYMAIADNRQRFMPLPDDRLPPRGQVLAYPEAVRLVNPVE 144
WP F L TRVAFKLRKDKF+YMA+AD+RQR MP+P+DR+PPRGQ LAYPEAV LV+P+
Sbjct: 145 WPGFSLGETRVAFKLRKDKFHYMALADDRQRIMPMPEDRVPPRGQQLAYPEAVLLVDPIN 204
Query: 145 PEFKGEVDDKYEYACESRSNQVHGWISIDSSSDSTGCWLITPSYEFRSAGPLKQYLGSHL 204
P+ +GEVDDKY+Y+CE + N VHGWIS D G W ITPS EFR+ GP+KQ L SH+
Sbjct: 205 PDLRGEVDDKYQYSCEDQYNNVHGWISFDPP---IGFWQITPSDEFRTGGPVKQNLTSHV 261
Query: 205 GPTMLSVFHSTHYSGADLIMKFGENEPWKKVFGPIFIYLNSNSDGFSPIKLWEDAKQQMV 264
GPTML++F S HY+G DL KF E WKKV GP+F+YLNS+ DG P LWEDAK QM+
Sbjct: 262 GPTMLAMFLSGHYAGDDLTPKFLTGEYWKKVHGPVFMYLNSSWDGSDPTLLWEDAKVQMM 321
Query: 265 NEVESWPYTFPASEDFLSSAQRGKFEGRLLVRDRYIRDAFVPVSGAYVGLAAPGDVGSWQ 324
E ESWPY F S+DF + QRG GRLLVRDRY+ DA + + AYVGLA PGDVGSWQ
Sbjct: 322 IEKESWPYCFALSDDFQKTEQRGCISGRLLVRDRYLDDADLYATSAYVGLALPGDVGSWQ 381
Query: 325 REYKGYQFWTITDDKGYFSIINVLPGDYNLYSWVNGFIGDYQCNNIINITSGSEINVGEL 384
RE KGYQFW +D G F I N++ GDYNLY+WV GFIGDY+ + + I+SG +I +G+L
Sbjct: 382 RECKGYQFWCRAEDDGSFCIRNIVAGDYNLYAWVPGFIGDYKLDAKLTISSGDDIYLGDL 441
Query: 385 VYEPPRHGPTLWEIGIPDRSAAEFYVPDPNPLYVNRLYVNHSDRFRQYGLWERYADLYPN 444
VYEPPR GPT+WEIGIPDRSA+EF+VPDPNP YVNRLY+NH DRFRQYGLWERYA+LYP+
Sbjct: 442 VYEPPRDGPTMWEIGIPDRSASEFFVPDPNPNYVNRLYINHPDRFRQYGLWERYAELYPD 501
Query: 445 EDLVYTVGISDYKKDWFFAQVTR 467
DLVYT+G SDY DWFFAQV R
Sbjct: 502 GDLVYTIGQSDYTTDWFFAQVNR 524
Score = 53.5 bits (127), Expect = 1e-05
Identities = 23/41 (56%), Positives = 32/41 (77%)
Query: 1 VVIDNDIVQVNISNPEGCVTGIQYNGLDNLLEINNHESDRG 41
V I N I ++ +SNP+G VTG++YNG+DNL+EI N E +RG
Sbjct: 25 VEIKNGIFELTLSNPDGIVTGVRYNGVDNLMEILNKEDNRG 65
>UniRef100_Q5XF41 At4g24430 [Arabidopsis thaliana]
Length = 487
Score = 498 bits (1282), Expect = e-139
Identities = 234/362 (64%), Positives = 282/362 (77%), Gaps = 4/362 (1%)
Query: 107 MAIADNRQRFMPLPDDRLPPRGQVLAYPEAVRLVNPVEPEFKGEVDDKYEYACESRSNQV 166
MAIADNRQR MPLP+DRL RG+ LAYPEAV LV+PVE EFKGEVDDKYEY+ E++ +V
Sbjct: 1 MAIADNRQRKMPLPEDRLGKRGRPLAYPEAVLLVHPVEDEFKGEVDDKYEYSSENKDLKV 60
Query: 167 HGWISIDSSSDSTGCWLITPSYEFRSAGPLKQYLGSHLGPTMLSVFHSTHYSGADLIMKF 226
HGWIS + GCW I PS EFRS G KQ L SH+GP L++F S HY+G D++MK
Sbjct: 61 HGWISHNLD---LGCWQIIPSNEFRSGGLSKQNLTSHVGPISLAMFLSAHYAGEDMVMKV 117
Query: 227 GENEPWKKVFGPIFIYLNSNSDGFS-PIKLWEDAKQQMVNEVESWPYTFPASEDFLSSAQ 285
+ WKKVFGP+F YLN D S P+ LW+DAK QM+ EV+SWPY FPASEDF S +
Sbjct: 118 KAGDSWKKVFGPVFTYLNCLPDKTSDPLSLWQDAKNQMLTEVQSWPYDFPASEDFPVSDK 177
Query: 286 RGKFEGRLLVRDRYIRDAFVPVSGAYVGLAAPGDVGSWQREYKGYQFWTITDDKGYFSII 345
RG GRLLV D+++ D F+P +GA+VGLA PG+VGSWQ E KGYQFWT D GYF+I
Sbjct: 178 RGCISGRLLVCDKFLSDDFLPANGAFVGLAPPGEVGSWQLESKGYQFWTEADSDGYFAIN 237
Query: 346 NVLPGDYNLYSWVNGFIGDYQCNNIINITSGSEINVGELVYEPPRHGPTLWEIGIPDRSA 405
++ G+YNL +V G+IGDYQ +INIT+G +I+VG +VYEPPR GPT+WEIGIPDRSA
Sbjct: 238 DIREGEYNLNGYVTGWIGDYQYEQLINITAGCDIDVGNIVYEPPRDGPTVWEIGIPDRSA 297
Query: 406 AEFYVPDPNPLYVNRLYVNHSDRFRQYGLWERYADLYPNEDLVYTVGISDYKKDWFFAQV 465
AEF+VPDPNP Y+N+LY+ H DRFRQYGLWERY +LYP EDLV+T+G+SDYKKDWFFA V
Sbjct: 298 AEFFVPDPNPKYINKLYIGHPDRFRQYGLWERYTELYPKEDLVFTIGVSDYKKDWFFAHV 357
Query: 466 TR 467
TR
Sbjct: 358 TR 359
>UniRef100_Q93Z90 AT4g24430/T22A6_260 [Arabidopsis thaliana]
Length = 487
Score = 498 bits (1282), Expect = e-139
Identities = 234/362 (64%), Positives = 282/362 (77%), Gaps = 4/362 (1%)
Query: 107 MAIADNRQRFMPLPDDRLPPRGQVLAYPEAVRLVNPVEPEFKGEVDDKYEYACESRSNQV 166
MAIADNRQR MPLP+DRL RG+ LAYPEAV LV+PVE EFKGEVDDKYEY+ E++ +V
Sbjct: 1 MAIADNRQRKMPLPEDRLGKRGRPLAYPEAVLLVHPVEDEFKGEVDDKYEYSIENKDLKV 60
Query: 167 HGWISIDSSSDSTGCWLITPSYEFRSAGPLKQYLGSHLGPTMLSVFHSTHYSGADLIMKF 226
HGWIS + GCW I PS EFRS G KQ L SH+GP L++F S HY+G D++MK
Sbjct: 61 HGWISHNLD---LGCWQIIPSNEFRSGGLSKQNLTSHVGPISLAMFLSAHYAGEDMVMKV 117
Query: 227 GENEPWKKVFGPIFIYLNSNSDGFS-PIKLWEDAKQQMVNEVESWPYTFPASEDFLSSAQ 285
+ WKKVFGP+F YLN D S P+ LW+DAK QM+ EV+SWPY FPASEDF S +
Sbjct: 118 KAGDSWKKVFGPVFTYLNCLPDKTSDPLSLWQDAKNQMLTEVQSWPYDFPASEDFPVSDK 177
Query: 286 RGKFEGRLLVRDRYIRDAFVPVSGAYVGLAAPGDVGSWQREYKGYQFWTITDDKGYFSII 345
RG GRLLV D+++ D F+P +GA+VGLA PG+VGSWQ E KGYQFWT D GYF+I
Sbjct: 178 RGCISGRLLVCDKFLSDDFLPANGAFVGLAPPGEVGSWQLESKGYQFWTEADSDGYFAIN 237
Query: 346 NVLPGDYNLYSWVNGFIGDYQCNNIINITSGSEINVGELVYEPPRHGPTLWEIGIPDRSA 405
++ G+YNL +V G+IGDYQ +INIT+G +I+VG +VYEPPR GPT+WEIGIPDRSA
Sbjct: 238 DIREGEYNLNGYVTGWIGDYQYEQLINITAGCDIDVGNIVYEPPRDGPTVWEIGIPDRSA 297
Query: 406 AEFYVPDPNPLYVNRLYVNHSDRFRQYGLWERYADLYPNEDLVYTVGISDYKKDWFFAQV 465
AEF+VPDPNP Y+N+LY+ H DRFRQYGLWERY +LYP EDLV+T+G+SDYKKDWFFA V
Sbjct: 298 AEFFVPDPNPKYINKLYIGHPDRFRQYGLWERYTELYPKEDLVFTIGVSDYKKDWFFAHV 357
Query: 466 TR 467
TR
Sbjct: 358 TR 359
>UniRef100_Q9SZK3 LG127/30 like gene [Arabidopsis thaliana]
Length = 649
Score = 489 bits (1259), Expect = e-137
Identities = 246/488 (50%), Positives = 323/488 (65%), Gaps = 33/488 (6%)
Query: 1 VVIDNDIVQVNISNPEGCVTGIQYNGLDNLLEINNHESDRG*KAGNDKY----------- 49
V++DN I++V+ SNP+G +TGI+Y G+DN+L + H DRG + N +
Sbjct: 54 VIVDNGIIRVSFSNPQGLITGIKYKGIDNVL--HPHLRDRGIEGTNFRIITQTQEQVEIS 111
Query: 50 ------EGDNGK*RTSRAIFH*NMEFIS*RKACATQY**KEWPAFDLDNTRVAFKLRKDK 103
+G R I N I EWP D+ R+AFKL ++
Sbjct: 112 FSRTWEDGHIPLNVDKRYIIRRNTSGIYMYGIFERL---PEWPELDMGLIRIAFKLNPER 168
Query: 104 FNYMAIADNRQRFMPLPDDRLPPRG--QVLAYPEAVRLVNPVEPEFKGEVDDKYEYACES 161
F+YMA+ADNRQR MP DDR RG + L Y EAV+L +P FK +VDDKY+Y CE
Sbjct: 169 FHYMAVADNRQREMPTEDDRDIKRGHAKALGYKEAVQLTHPHNSMFKNQVDDKYQYTCEI 228
Query: 162 RSNQVHGWISIDSSSDSTGCWLITPSYEFRSAGPLKQYLGSHLGPTMLSVFHSTHYSGAD 221
+ N+VHGWIS S G W+I+PS E+RS GP+KQ L SH+GPT ++ F S HY GAD
Sbjct: 229 KDNKVHGWISTKSR---VGFWIISPSGEYRSGGPIKQELTSHVGPTAITTFISGHYVGAD 285
Query: 222 LIMKFGENEPWKKVFGPIFIYLNSNSDGFSPIK--LWEDAKQQMVNEVESWPYTFPASED 279
+ + E WKKV GP+FIYLNS+S + + LWEDAKQQ EV++WPY F AS D
Sbjct: 286 MEAHYRPGEAWKKVLGPVFIYLNSDSTSNNKPQDLLWEDAKQQSEKEVKAWPYDFVASSD 345
Query: 280 FLSSAQRGKFEGRLLVRDRYIRDAFVPVSGAYVGLAAPGDVGSWQREYKGYQFWTITDDK 339
+LS +RG GRLLV DR++ P AYVGLA PG+ GSWQ KGYQFWT T++
Sbjct: 346 YLSRRERGSVTGRLLVNDRFL----TPGKSAYVGLAPPGEAGSWQTNTKGYQFWTKTNET 401
Query: 340 GYFSIINVLPGDYNLYSWVNGFIGDYQCNNIINITSGSEINVGELVYEPPRHGPTLWEIG 399
GYF+I NV PG YNLY WV GFIGD++ N++N+ +GS I++G +VY+PPR+GPTLWEIG
Sbjct: 402 GYFTIENVRPGTYNLYGWVPGFIGDFRYQNLVNVAAGSVISLGRVVYKPPRNGPTLWEIG 461
Query: 400 IPDRSAAEFYVPDPNPLYVNRLYVNHSDRFRQYGLWERYADLYPNEDLVYTVGISDYKKD 459
+PDR+A E+++P+P +N LY+NH+D+FRQYGLW+RY +LYP DLVYTVG+S+Y +D
Sbjct: 462 VPDRTAREYFIPEPYKDTMNPLYLNHTDKFRQYGLWQRYTELYPTHDLVYTVGVSNYSQD 521
Query: 460 WFFAQVTR 467
WF+AQVTR
Sbjct: 522 WFYAQVTR 529
>UniRef100_O04512 F21M12.30 protein [Arabidopsis thaliana]
Length = 728
Score = 479 bits (1233), Expect = e-134
Identities = 236/458 (51%), Positives = 294/458 (63%), Gaps = 76/458 (16%)
Query: 83 KEWPAFDLDNTRVAFKLRKDK--------------------------------------- 103
K+WP F+L TR+AFKLRKDK
Sbjct: 155 KDWPGFELGETRIAFKLRKDKKLEMLVIACPCEEYHRFQAKELCNMSDIYLLGLIVRKHK 214
Query: 104 --FNYMAIADNRQRFMPLPDDRLPPRGQVLAYPEAVRLVNPVEPEFKGEVDDKYEYACES 161
F+YMA+AD+R+R MP PDD R Q L Y EA L P +P +GEVDDKY+Y+CE+
Sbjct: 215 SVFHYMAVADDRKRIMPFPDDLCKGRCQTLDYQEASLLTAPCDPRLQGEVDDKYQYSCEN 274
Query: 162 RSNQVHGWISIDSSSDSTGCWLITPSYEFRSAGPLKQYLGSHLGPTMLSV--------FH 213
+ +VHGWIS D G W ITPS EFRS GPLKQ L SH+GPT L+V FH
Sbjct: 275 KDLRVHGWISFDPP---VGFWQITPSNEFRSGGPLKQNLTSHVGPTTLAVSVPVTYVVFH 331
Query: 214 STHYSGADLIMKFGENEPWKKVFGPIFIYLNSNSDGFSPIKLWEDAKQQMVNEVESWPYT 273
STHY+G ++ +F EPWKKV+GP+FIYLNS ++G P+ LW+DAK +M+ EVE WPY+
Sbjct: 332 STHYAGKTMMPRFEHGEPWKKVYGPVFIYLNSTANGDDPLCLWDDAKIKMMAEVERWPYS 391
Query: 274 FPASEDFLSSAQRGKFEGRLLVRDRYIRDAFVPVSGAYVGLAAPGDVGSWQREYKGYQFW 333
F AS+D+ S +RG GRLL+RDR+I + + GAYVGLA PGD GSWQ E KGYQFW
Sbjct: 392 FVASDDYPKSEERGTARGRLLIRDRFINNDLISARGAYVGLAPPGDSGSWQIECKGYQFW 451
Query: 334 TITDDKGYFSIINVLPGDYNLYSWVNGFIGDYQCNNIINI-----------TSGSEINVG 382
I D+ GYFSI NV PG+YNLY+WV FIGDY I+ + +G I +G
Sbjct: 452 AIADEAGYFSIGNVRPGEYNLYAWVPSFIGDYHNGTIVRVPAYQPNFDGQNNTGCMIEMG 511
Query: 383 ELVYEPPRHGPTLWEIGIPDRSAAEFYVPDPNPLYVNRLYVNHSDR-------------F 429
++VYEPPR GPTLWEIGIPDR A+EF++PDP+P VNR+ V+H DR F
Sbjct: 512 DIVYEPPRDGPTLWEIGIPDRKASEFFIPDPDPTLVNRVLVHHQDRFISYTFVNLTAWMF 571
Query: 430 RQYGLWERYADLYPNEDLVYTVGISDYKKDWFFAQVTR 467
RQYGLW++Y D+YPN+DLVYTVG+SDY++DWFFA V R
Sbjct: 572 RQYGLWKKYTDMYPNDDLVYTVGVSDYRRDWFFAHVPR 609
Score = 58.2 bits (139), Expect = 5e-07
Identities = 24/41 (58%), Positives = 34/41 (82%)
Query: 1 VVIDNDIVQVNISNPEGCVTGIQYNGLDNLLEINNHESDRG 41
VV+DN I+QV +S P G +TGI+YNG+DN+LE+ N E++RG
Sbjct: 20 VVMDNGILQVTLSKPGGIITGIEYNGIDNVLEVRNKETNRG 60
>UniRef100_Q9ZQ51 Hypothetical protein At2g22620 [Arabidopsis thaliana]
Length = 677
Score = 475 bits (1222), Expect = e-132
Identities = 257/505 (50%), Positives = 314/505 (61%), Gaps = 43/505 (8%)
Query: 1 VVIDNDIVQVNISNPEGCVTGIQY----NGLDNLLEINNH------ESDRG*KAGNDKYE 50
VV+DN IVQV SNPEG +TGI+Y N LD+ ++ + E ++ K DK E
Sbjct: 62 VVVDNGIVQVTFSNPEGLITGIKYHGIDNVLDDKIDDRGYWDVVWYEPEK--KQKTDKLE 119
Query: 51 GDNGK*RTSRA----IFH*NMEFIS*RKACATQY**KE---------------------W 85
G + T I IS R + K W
Sbjct: 120 GTKFEIITQNEEQIEISFTRTWTISRRGSLVPLNVDKRYIIRSGVSGLYMYGILERLEGW 179
Query: 86 PAFDLDNTRVAFKLRKDKFNYMAIADNRQRFMPLPDDRLPPRGQVLAYPEAVRLVNPVEP 145
P D+D R+ FKL KF++MAI+D+RQR MP DR + LAY EAV L NP P
Sbjct: 180 PDVDMDQIRIVFKLNPKKFDFMAISDDRQRSMPSMADR--ENSKSLAYKEAVLLTNPSNP 237
Query: 146 EFKGEVDDKYEYACESRSNQVHGWISIDSSSDSTGCWLITPSYEFRSAGPLKQYLGSHLG 205
FKGEVDDKY Y+ E + N VHGWIS D G W+ITPS EFR GP+KQ L SH G
Sbjct: 238 MFKGEVDDKYMYSMEDKDNNVHGWISSDPP---VGFWMITPSDEFRLGGPIKQDLTSHAG 294
Query: 206 PTMLSVFHSTHYSGADLIMKFGENEPWKKVFGPIFIYLNSNSDGFSPIKLWEDAKQQMVN 265
P LS+F STHY+G ++ M + EPWKKVFGP+ YLNS S S ++LW DAK+QM
Sbjct: 295 PITLSMFTSTHYAGKEMRMDYRNGEPWKKVFGPVLAYLNSVSPKDSTLRLWRDAKRQMAA 354
Query: 266 EVESWPYTFPASEDFLSSAQRGKFEGRLLVRDRYIRDAFVPVSGAYVGLAAPGDVGSWQR 325
EV+SWPY F SED+ QRG EG+ L++D Y+ + A+VGLA G+ GSWQ
Sbjct: 355 EVKSWPYDFITSEDYPLRHQRGTLEGQFLIKDSYVSRLKIYGKFAFVGLAPIGEAGSWQT 414
Query: 326 EYKGYQFWTITDDKGYFSIINVLPGDYNLYSWVNGFIGDYQCNNIINITSGSEINVGELV 385
E KGYQFWT D +G F I NV G+Y+LY+W +GFIGDY+ I IT GSE+NVG LV
Sbjct: 415 ESKGYQFWTKADRRGRFIIENVRAGNYSLYAWGSGFIGDYKYEQNITITPGSEMNVGPLV 474
Query: 386 YEPPRHGPTLWEIGIPDRSAAEFYVPDPNPLYVNRLYVNH-SDRFRQYGLWERYADLYPN 444
YEPPR+GPTLWEIG+PDR+A EFY+PDP P +N+LYVN DRFRQYGLW+RYADLYP
Sbjct: 475 YEPPRNGPTLWEIGVPDRTAGEFYIPDPYPTLMNKLYVNPLQDRFRQYGLWDRYADLYPQ 534
Query: 445 EDLVYTVGISDYKKDWFFAQVTRFV 469
DLVYT+G+SDY+ DWFFA V R V
Sbjct: 535 NDLVYTIGVSDYRSDWFFAHVARNV 559
>UniRef100_Q6NKW8 Hypothetical protein At2g22620 [Arabidopsis thaliana]
Length = 677
Score = 473 bits (1216), Expect = e-132
Identities = 256/505 (50%), Positives = 314/505 (61%), Gaps = 43/505 (8%)
Query: 1 VVIDNDIVQVNISNPEGCVTGIQY----NGLDNLLEINNH------ESDRG*KAGNDKYE 50
VV+DN IVQV SNPEG +TGI+Y N LD+ ++ + E ++ K DK E
Sbjct: 62 VVVDNGIVQVTFSNPEGLITGIKYHGIDNVLDDKIDDRGYWDVVWYEPEK--KQKTDKLE 119
Query: 51 GDNGK*RTSRA----IFH*NMEFIS*RKACATQY**KE---------------------W 85
G + T I IS R + K W
Sbjct: 120 GTKFEIITQNEEQIEISFTRTWTISRRGSLVPLNVDKRYIIRSGVSGLYMYGILERLEGW 179
Query: 86 PAFDLDNTRVAFKLRKDKFNYMAIADNRQRFMPLPDDRLPPRGQVLAYPEAVRLVNPVEP 145
P D+D R+ FKL KF++MAI+D+R+R MP DR + LAY EAV L NP P
Sbjct: 180 PDVDMDPIRIVFKLNPKKFDFMAISDDRRRSMPSMADR--ENSKSLAYKEAVLLTNPSNP 237
Query: 146 EFKGEVDDKYEYACESRSNQVHGWISIDSSSDSTGCWLITPSYEFRSAGPLKQYLGSHLG 205
FKGEVDDKY Y+ E + N VHGWIS D G W+ITPS EFR GP+KQ L SH G
Sbjct: 238 MFKGEVDDKYMYSMEDKDNNVHGWISSDPP---VGFWMITPSDEFRLGGPIKQDLTSHAG 294
Query: 206 PTMLSVFHSTHYSGADLIMKFGENEPWKKVFGPIFIYLNSNSDGFSPIKLWEDAKQQMVN 265
P LS+F STHY+G ++ M + EPWKKVFGP+ YLNS S S ++LW DAK+QM
Sbjct: 295 PITLSMFTSTHYAGKEMRMDYRNGEPWKKVFGPVLAYLNSVSPKDSTLRLWRDAKRQMAA 354
Query: 266 EVESWPYTFPASEDFLSSAQRGKFEGRLLVRDRYIRDAFVPVSGAYVGLAAPGDVGSWQR 325
EV+SWPY F SED+ QRG EG+ L++D Y+ + A+VGLA G+ GSWQ
Sbjct: 355 EVKSWPYDFITSEDYPLRHQRGTLEGQFLIKDSYVSRLKIYGKFAFVGLAPIGEAGSWQT 414
Query: 326 EYKGYQFWTITDDKGYFSIINVLPGDYNLYSWVNGFIGDYQCNNIINITSGSEINVGELV 385
E KGYQFWT D +G F I NV G+Y+LY+W +GFIGDY+ I IT GSE+NVG LV
Sbjct: 415 ESKGYQFWTKADRRGRFIIENVRAGNYSLYAWGSGFIGDYKYEQNITITPGSEMNVGPLV 474
Query: 386 YEPPRHGPTLWEIGIPDRSAAEFYVPDPNPLYVNRLYVNH-SDRFRQYGLWERYADLYPN 444
YEPPR+GPTLWEIG+PDR+A EFY+PDP P +N+LYVN DRFRQYGLW+RYADLYP
Sbjct: 475 YEPPRNGPTLWEIGVPDRTAGEFYIPDPYPTLMNKLYVNPLQDRFRQYGLWDRYADLYPQ 534
Query: 445 EDLVYTVGISDYKKDWFFAQVTRFV 469
DLVYT+G+SDY+ DWFFA V R V
Sbjct: 535 NDLVYTIGVSDYRSDWFFAHVARNV 559
>UniRef100_Q84W85 Hypothetical protein At4g37950 [Arabidopsis thaliana]
Length = 678
Score = 455 bits (1170), Expect = e-126
Identities = 221/389 (56%), Positives = 277/389 (70%), Gaps = 12/389 (3%)
Query: 85 WPAFDLDNTRVAFKLRKDKFNYMAIADNRQRFMPLPDDRLPPRGQV--LAYPEAVRLVNP 142
WP D+D R+ FKL KF++MA++DNRQ+ MP DR +G+ LAY EAV L+NP
Sbjct: 176 WPDVDMDQIRIVFKLNTTKFDFMAVSDNRQKIMPFDTDRDITKGRASPLAYKEAVHLINP 235
Query: 143 VEPEFKGEVDDKYEYACESRSNQVHGWISIDSSSDSTGCWLITPSYEFRSAGPLKQYLGS 202
KG+VDDKY Y+ E++ N+VHGWIS D G W+ITPS EF + GP+KQ L S
Sbjct: 236 QNHMLKGQVDDKYMYSVENKDNKVHGWISSDQR---IGFWMITPSDEFHACGPIKQDLTS 292
Query: 203 HLGPTMLSVFHSTHYSGADLIMKFGENEPWKKVFGPIFIYLNSNSDGFSPIKLWEDAKQQ 262
H+GPT LS+F S HY+G D+ + EPWKKVFGP+F+YLNS S S LW DAK+Q
Sbjct: 293 HVGPTTLSMFTSVHYAGKDMNTNYKSKEPWKKVFGPVFVYLNSAS---SRNLLWTDAKRQ 349
Query: 263 MVNEVESWPYTFPASEDFLSSAQRGKFEGRLLVRDRYIRDA-FVPVSGAYVGLAAPGDVG 321
MV+EV+SWPY F S D+ QRG +G+L V DRYI++ ++ A+VGLA PG+ G
Sbjct: 350 MVSEVQSWPYDFVKSVDYPLHHQRGTVKGQLFVIDRYIKNVTYLFGQFAFVGLALPGEAG 409
Query: 322 SWQREYKGYQFWTITDDKGYFSIINVLPGDYNLYSWVNGFIGDYQCNNIINITSGSEINV 381
SWQ E KGYQFWT D G F+I NV PG Y+LY+WV+GFIGDY+ I IT G EI+V
Sbjct: 410 SWQTENKGYQFWTRADKMGMFTIANVRPGTYSLYAWVSGFIGDYKYVRDITITPGREIDV 469
Query: 382 GELVYEPPRHGPTLWEIGIPDRSAAEFYVPDPNPLYVNRLYVNHS---DRFRQYGLWERY 438
G +VY PPR+GPTLWEIG PDR+AAEFY+PDP+P +LY+N+S DRFRQYGLW+RY
Sbjct: 470 GHIVYVPPRNGPTLWEIGQPDRTAAEFYIPDPDPTLFTKLYLNYSNPQDRFRQYGLWDRY 529
Query: 439 ADLYPNEDLVYTVGISDYKKDWFFAQVTR 467
+ LYP DLV+T G+SDYKKDWF+A V R
Sbjct: 530 SVLYPRNDLVFTAGVSDYKKDWFYAHVNR 558
Score = 43.5 bits (101), Expect = 0.013
Identities = 20/41 (48%), Positives = 30/41 (72%), Gaps = 2/41 (4%)
Query: 1 VVIDNDIVQVNISNPEGCVTGIQYNGLDNLLEINNHESDRG 41
VV+DN I+ V S+P+G +T I+YNGL+N+L N+ +RG
Sbjct: 61 VVVDNGIIDVTFSSPQGLITRIKYNGLNNVL--NDQIENRG 99
>UniRef100_O65620 MYST1 protein [Arabidopsis thaliana]
Length = 435
Score = 392 bits (1006), Expect = e-107
Identities = 189/321 (58%), Positives = 234/321 (72%), Gaps = 10/321 (3%)
Query: 151 VDDKYEYACESRSNQVHGWISIDSSSDSTGCWLITPSYEFRSAGPLKQYLGSHLGPTMLS 210
VDDKY Y+ E++ N+VHGWIS D G W+ITPS EF + GP+KQ L SH+GPT LS
Sbjct: 1 VDDKYMYSVENKDNKVHGWISSDQR---IGFWMITPSDEFHACGPIKQDLTSHVGPTTLS 57
Query: 211 VFHSTHYSGADLIMKFGENEPWKKVFGPIFIYLNSNSDGFSPIKLWEDAKQQMVNEVESW 270
+F S HY+G D+ + EPWKKVFGP+F+YLNS S S LW DAK+QMV+EV+SW
Sbjct: 58 MFTSVHYAGKDMNTNYKSKEPWKKVFGPVFVYLNSAS---SRNLLWTDAKRQMVSEVQSW 114
Query: 271 PYTFPASEDFLSSAQRGKFEGRLLVRDRYIRDA-FVPVSGAYVGLAAPGDVGSWQREYKG 329
PY F S D+ QRG +G+L V DRYI++ ++ A+VGLA PG+ GSWQ E KG
Sbjct: 115 PYDFVKSVDYPLHHQRGTVKGQLFVIDRYIKNVTYLFGQFAFVGLALPGEAGSWQTENKG 174
Query: 330 YQFWTITDDKGYFSIINVLPGDYNLYSWVNGFIGDYQCNNIINITSGSEINVGELVYEPP 389
YQFWT D G F+I NV PG Y+LY+WV+GFIGDY+ I IT G EI+VG +VY PP
Sbjct: 175 YQFWTRADKMGMFTIANVRPGTYSLYAWVSGFIGDYKYVRDITITPGREIDVGHIVYVPP 234
Query: 390 RHGPTLWEIGIPDRSAAEFYVPDPNPLYVNRLYVNHS---DRFRQYGLWERYADLYPNED 446
R+GPTLWEIG PDR+AAEFY+PDP+P +LY+N+S DRFRQYGLW+RY+ LYP D
Sbjct: 235 RNGPTLWEIGQPDRTAAEFYIPDPDPTLFTKLYLNYSNPQDRFRQYGLWDRYSVLYPRND 294
Query: 447 LVYTVGISDYKKDWFFAQVTR 467
LV+T G+SDYKKDWF+A V R
Sbjct: 295 LVFTAGVSDYKKDWFYAHVNR 315
>UniRef100_O04511 F21M12.28 protein [Arabidopsis thaliana]
Length = 447
Score = 367 bits (941), Expect = e-100
Identities = 185/348 (53%), Positives = 225/348 (64%), Gaps = 46/348 (13%)
Query: 83 KEWPAFDLDNTRVAFKLRKDKFNYMAIADNRQRFMPLPDDRLPPRGQVLAYPEAVRLVNP 142
KEWPAF L TR+AFKLRK+KF+YMA+ D+RQRFMPLPDDRLP RGQ LAYPEAV LVNP
Sbjct: 145 KEWPAFSLAETRIAFKLRKEKFHYMAVTDDRQRFMPLPDDRLPDRGQALAYPEAVLLVNP 204
Query: 143 VEPEFKGEVDDKYEYACESRSNQVHGWISIDSSSDSTGCWLITPSYEFRSAGPLKQYLGS 202
+ E + G +K
Sbjct: 205 L---------------------------------------------ESQFKGEVKFIHFF 219
Query: 203 HLGPTMLSVFHSTHYSGADLIMKFGENEPWKKVFGPIFIYLNSNSDGFS-PIKLWEDAKQ 261
L + S S DL+ KF E E WKKVFGP+F+YLNS++D + P+ LW+DAK
Sbjct: 220 FLFKIIREKLFSCVRSWEDLVPKFSEGEAWKKVFGPVFVYLNSSTDDDNDPLWLWQDAKS 279
Query: 262 QMVNEVESWPYTFPASEDFLSSAQRGKFEGRLLVRDRYIRDAFVPVSGAYVGLAAPGDVG 321
QM E ESWPY+FPAS+D++ + QRG GRLLV+DRY+ F+ + YVGLA PG G
Sbjct: 280 QMNVEAESWPYSFPASDDYVKTEQRGNVVGRLLVQDRYVDKDFIAANRGYVGLAVPGAAG 339
Query: 322 SWQREYKGYQFWTITDDKGYFSIINVLPGDYNLYSWVNGFIGDYQCNNIINITSGSEINV 381
SWQRE K YQFWT TD++G+F I + PG YNLY+W+ GFIGDY+ +++I ITSG I V
Sbjct: 340 SWQRECKEYQFWTRTDEEGFFYISGIRPGQYNLYAWIPGFIGDYKYDDVITITSGCYIYV 399
Query: 382 GELVYEPPRHGPTLWEIGIPDRSAAEFYVPDPNPLYVNRLYVNHSDRF 429
+LVY+PPR+G TLWEIG PDRSAAEFYVPDPNP Y+N LY NH DRF
Sbjct: 400 EDLVYQPPRNGATLWEIGFPDRSAAEFYVPDPNPKYINNLYQNHPDRF 447
Score = 57.4 bits (137), Expect = 9e-07
Identities = 25/41 (60%), Positives = 33/41 (79%)
Query: 1 VVIDNDIVQVNISNPEGCVTGIQYNGLDNLLEINNHESDRG 41
VV+DN I +V +S P+G VTGI+YNG+DNLLE+ N E +RG
Sbjct: 15 VVMDNGIARVTLSKPDGIVTGIEYNGIDNLLEVLNEEVNRG 55
>UniRef100_Q69VM1 MYST1-like protein [Oryza sativa]
Length = 220
Score = 301 bits (770), Expect = 4e-80
Identities = 141/218 (64%), Positives = 168/218 (76%)
Query: 211 VFHSTHYSGADLIMKFGENEPWKKVFGPIFIYLNSNSDGFSPIKLWEDAKQQMVNEVESW 270
+F S HY+G DL KF E WKKV GP+F+YLNS+ DG +P LW+DAK QM+ E ESW
Sbjct: 1 MFLSGHYAGDDLTPKFMTGEYWKKVHGPVFMYLNSSWDGSNPTLLWKDAKVQMMIEKESW 60
Query: 271 PYTFPASEDFLSSAQRGKFEGRLLVRDRYIRDAFVPVSGAYVGLAAPGDVGSWQREYKGY 330
PY F S+DF + QRG+ GRLLVRDRY+ DA + + AYVGLA PGDVGSWQRE KGY
Sbjct: 61 PYYFALSDDFQKTEQRGRISGRLLVRDRYLHDADLYGTSAYVGLALPGDVGSWQRECKGY 120
Query: 331 QFWTITDDKGYFSIINVLPGDYNLYSWVNGFIGDYQCNNIINITSGSEINVGELVYEPPR 390
QFW D G FSI N++ GDYNLY+WV GFIGDY+ + + I+SG +I +G+LVYEPPR
Sbjct: 121 QFWCRAHDGGSFSIRNIVAGDYNLYAWVPGFIGDYKLDAKLTISSGDDIYLGDLVYEPPR 180
Query: 391 HGPTLWEIGIPDRSAAEFYVPDPNPLYVNRLYVNHSDR 428
GPT+WEIGIPDRSA+EFYVPDPNP YVNRLY+NH DR
Sbjct: 181 DGPTMWEIGIPDRSASEFYVPDPNPNYVNRLYINHPDR 218
>UniRef100_Q9SZJ7 Hypothetical protein F20D10.70 [Arabidopsis thaliana]
Length = 296
Score = 211 bits (537), Expect = 4e-53
Identities = 115/265 (43%), Positives = 152/265 (56%), Gaps = 51/265 (19%)
Query: 1 VVIDNDIVQVNISNPEGCVTGIQYNGLDNLLEINNHESDRG*KAGNDKYEGDNGK*RTSR 60
VV+DN I+ V S+P+G +T I+YNGL+N+L N+ +RG
Sbjct: 69 VVVDNGIIDVTFSSPQGLITRIKYNGLNNVL--NDQIENRG------------------- 107
Query: 61 AIFH*NMEFIS*RKACATQY**KEWPAFDLDNTRVAFKLRKDKFNYMAIADNRQRFMPLP 120
WP D+D R+ FKL KF++MA++DNRQ+ MP
Sbjct: 108 ----------------------YSWPDVDMDQIRIVFKLNTTKFDFMAVSDNRQKIMPFD 145
Query: 121 DDRLPPRGQV--LAYPEAVRLVNPVEPEFKGEVDDKYEYACESRSNQVHGWISIDSSSDS 178
DR +G+ LAY EAV L+NP KG+VDDKY Y+ E++ N+VHGWIS D
Sbjct: 146 TDRDITKGRASPLAYKEAVHLINPQNHMLKGQVDDKYMYSVENKDNKVHGWISSDQR--- 202
Query: 179 TGCWLITPSYEFRSAGPLKQYLGSHLGPTMLSVFHSTHYSGADLIMKFGENEPWKKVFGP 238
G W+ITPS EF + GP+KQ L SH+GPT LS+F S HY+G D+ + EPWKKVFGP
Sbjct: 203 IGFWMITPSDEFHACGPIKQDLTSHVGPTTLSMFTSVHYAGKDMNTNYKSKEPWKKVFGP 262
Query: 239 IFIYLNSNSDGFSPIKLWEDAKQQM 263
+F+YLNS S S LW DAK+Q+
Sbjct: 263 VFVYLNSAS---SRNLLWTDAKRQV 284
>UniRef100_Q9SZJ8 Hypothetical protein F20D10.80 [Arabidopsis thaliana]
Length = 289
Score = 204 bits (520), Expect = 3e-51
Identities = 92/142 (64%), Positives = 111/142 (77%), Gaps = 3/142 (2%)
Query: 329 GYQFWTITDDKGYFSIINVLPGDYNLYSWVNGFIGDYQCNNIINITSGSEINVGELVYEP 388
GYQFWT D G F+I NV PG Y+LY+WV+GFIGDY+ I IT G EI+VG +VY P
Sbjct: 28 GYQFWTRADKMGMFTIANVRPGTYSLYAWVSGFIGDYKYVRDITITPGREIDVGHIVYVP 87
Query: 389 PRHGPTLWEIGIPDRSAAEFYVPDPNPLYVNRLYVNHS---DRFRQYGLWERYADLYPNE 445
PR+GPTLWEIG PDR+AAEFY+PDP+P +LY+N+S DRFRQYGLW+RY+ LYP
Sbjct: 88 PRNGPTLWEIGQPDRTAAEFYIPDPDPTLFTKLYLNYSNPQDRFRQYGLWDRYSVLYPRN 147
Query: 446 DLVYTVGISDYKKDWFFAQVTR 467
DLV+T G+SDYKKDWF+A V R
Sbjct: 148 DLVFTAGVSDYKKDWFYAHVNR 169
>UniRef100_Q9S9K1 T23K8.12 protein [Arabidopsis thaliana]
Length = 248
Score = 103 bits (256), Expect = 1e-20
Identities = 40/67 (59%), Positives = 56/67 (82%)
Query: 401 PDRSAAEFYVPDPNPLYVNRLYVNHSDRFRQYGLWERYADLYPNEDLVYTVGISDYKKDW 460
P + A E++VP+P +N LY+NH+D+FRQYGLW+RY +LYPN DL+YT+G+S+Y KDW
Sbjct: 62 PHQRAREYFVPEPYKNTMNPLYLNHTDKFRQYGLWQRYTELYPNHDLIYTIGVSNYSKDW 121
Query: 461 FFAQVTR 467
F++QVTR
Sbjct: 122 FYSQVTR 128
Score = 42.0 bits (97), Expect = 0.039
Identities = 15/31 (48%), Positives = 26/31 (83%)
Query: 1 VVIDNDIVQVNISNPEGCVTGIQYNGLDNLL 31
V++DN I+ V+ S+P+G +TGI+Y G++N+L
Sbjct: 30 VIVDNGIISVSFSSPQGLITGIKYKGVNNVL 60
>UniRef100_UPI000042E7D6 UPI000042E7D6 UniRef100 entry
Length = 678
Score = 102 bits (253), Expect = 3e-20
Identities = 85/343 (24%), Positives = 156/343 (44%), Gaps = 35/343 (10%)
Query: 141 NPVEPEFKGEVDDKYEYACESRSNQVHGWISIDSSSDST--GCWLITPSYEFRSAGPLKQ 198
NP E + KYE+A + HG+ + ++S+ + G W + + E + GPL+
Sbjct: 222 NPYRTE-SSDYFTKYEWADLYGDHFAHGFYADGTASNGSTLGFWTVFNNLEGYTGGPLRS 280
Query: 199 YLGSHLGPTMLSVFHSTHYSGADLIMKFGENEPWKKVFGPIFIYLNSNSDGFSPIKLWED 258
L + T + ++++++ GA + + ++FGP FIYLN + D + L++D
Sbjct: 281 DL---VVDTNIYNYYASNHRGASTM---NITSGFDRIFGPTFIYLNKDGDLHN---LYDD 331
Query: 259 AKQQMVNEVESWPYTFPAS--EDFLSSAQRGKFEGRLLVRDRYIRDAFVPVSGAYVGLAA 316
AK + Y A +++S+ RG F+ ++ + + S + LA
Sbjct: 332 AKSYANTSFAADFYDDVADLIPGYVNSSGRGDFKAQISLPEG--------ASNPKIILAQ 383
Query: 317 PGDVGSWQREYKGYQFWTITDDKGYFSIINVLPGDYNLYSWVNGFIGDYQCNNIINITSG 376
G +Y Q+WT G +I V G Y + + G G ++ + ++
Sbjct: 384 SGVDPQDNVDYTAKQYWTNVSSDGRVTIPRVQAGTYRVTLYAEGIFGQFEQDEVVVSAGD 443
Query: 377 SEINVGELVYEPPRHGPTLWEIGIPDRSAAEF---YVPDPNPLYVNRLYVNHSDRFRQY- 432
+ + +E HG LW +G+PD++A EF + DP+ + H + +R+Y
Sbjct: 444 GDGAEFRINWEAECHGIELWRLGVPDKTAGEFLHGFAKDPD-------HTLHQEEYREYW 496
Query: 433 GLWERYADLYPNEDLVYTVGISDYKKDWFFAQVTRFVLPFLLP 475
G W+ D +P + + YT+G SD KDW + +R+ F P
Sbjct: 497 GAWDFPTD-FP-DGVNYTIGTSDPSKDWNYVHYSRYGGSFTRP 537
>UniRef100_Q8RJP2 Rhamnogalacturonate lyase precursor [Erwinia chrysanthemi]
Length = 578
Score = 95.5 bits (236), Expect = 3e-18
Identities = 87/341 (25%), Positives = 142/341 (41%), Gaps = 52/341 (15%)
Query: 127 RGQVLAYPEAVRLVNPVEPEFK---GEVDDKYEYACESRSNQVHGWISIDSSSDSTGCWL 183
RG L Y E +L + ++ G V KY++A R ++ G + + G W+
Sbjct: 175 RGTPLLYDELEQLPKVQDETWRLPDGSVYSKYDFAGYQRESRYWGVMG-----NGYGAWM 229
Query: 184 ITPSYEFRSAGPLKQYLGSHLGPTMLSVFHSTHYSGADLIMKFGENEPWKKVFGPIFIYL 243
+ S E+ S LKQ L H +L+ +H+ D++ + G ++K++GP +Y+
Sbjct: 230 VPASGEYYSGDALKQELLVHQDAIILNYLTGSHFGTPDMVAQPG----FEKLYGPWLLYI 285
Query: 244 NSNSDGFSPIKLWEDAKQQMVNEVESWPYTFPASEDFLSSAQRGKFEGRLLVRDRYIRDA 303
N +D +L D ++ +E SWPY + +D QR GRL
Sbjct: 286 NQGNDR----ELVADVSRRAEHERASWPYRW--LDDARYPRQRATVSGRLRTE------- 332
Query: 304 FVPVSGAYVGLAAPGDVGSWQREYKGYQFWTITDDKGYFSIINVLPGDYNLYSWVNGFIG 363
+ + ++ + GY F T+ G FS+ NV PG+Y L ++ +G
Sbjct: 333 -----APHATVVLNSSAENFDIQTTGYLFSARTNRDGRFSLSNVPPGEYRLSAYADGGTQ 387
Query: 364 DYQCNNIINITSGSEINVGELVYEPPRHGPTLWEIGIPDRSAAEFYVPDPNPLYVNRLYV 423
G + +G++ P P W IG DR A EF
Sbjct: 388 IGLLAQQTVRVEGKKTRLGQIDARQP--APLAWAIGQADRRADEF--------------- 430
Query: 424 NHSDRFRQYGLWERYADLYPNEDLVYTVGISDYKKDWFFAQ 464
D+ RQY R+ P DL + +G S +KDW++AQ
Sbjct: 431 RFGDKPRQY----RWQTEVP-ADLTFEIGKSRERKDWYYAQ 466
>UniRef100_Q6D915 Rhamnogalacturonate lyase [Erwinia carotovora]
Length = 576
Score = 88.6 bits (218), Expect = 4e-16
Identities = 81/317 (25%), Positives = 138/317 (42%), Gaps = 51/317 (16%)
Query: 149 GEVDDKYEYACESRSNQVHGWISIDSSSDSTGCWLITPSYEFRSAGPLKQYLGSHLGPTM 208
G + KY++A R+ G + G WLI + E+ S LKQ L H +
Sbjct: 201 GSIYSKYDFAGYMRTAPFWGVFG-----NGVGAWLIHGNREYFSGDALKQDLLVHQDAII 255
Query: 209 LSVFHSTHYSGADLIMKFGENEPWKKVFGPIFIYLNSNSDGFSPIKLWEDAKQQMVNEVE 268
L+ +H+ D+ G WKK +GP +Y+N ++ DA++Q + E
Sbjct: 256 LNYMTGSHFGTPDMKAPPG----WKKFYGPWLLYINEGDTE----QVLADAQRQALTETV 307
Query: 269 SWPYTFPASEDFLSSAQRGKFEGRLLVRDRYIRDAFVPVSGAYVGLAAPGDVGSWQREYK 328
SWPY + +D + R + GR+ + + V +S + L P DV + +
Sbjct: 308 SWPYKWV--DDSRYARDRTQVSGRVASQ----QPVTVVLSSS---LDEPFDV-----QTR 353
Query: 329 GYQFWTITDDKGYFSIINVLPGDYNLYSWVN-GFIGDYQCNNIINITSGSEINVGELVYE 387
GY + TD +G F+I +V PG+Y+L + N G ++++ ++ +
Sbjct: 354 GYSYQATTDSQGNFAIPHVRPGNYHLSVYANSGTQPGILAEQALSVSGDKQVLP---MIT 410
Query: 388 PPRHGPTLWEIGIPDRSAAEFYVPDPNPLYVNRLYVNHSDRFRQYGLWERYADLYPNEDL 447
P+ +W IG +R A+EF RF R+ P +L
Sbjct: 411 LPKAESVVWAIGQANRQASEF-------------------RFGNEVRNTRWQHEVP-ANL 450
Query: 448 VYTVGISDYKKDWFFAQ 464
+ +G SDY++DW++AQ
Sbjct: 451 TFDIGRSDYQRDWYYAQ 467
>UniRef100_Q8GX61 Hypothetical protein At1g09910/F21M12_30 [Arabidopsis thaliana]
Length = 243
Score = 87.4 bits (215), Expect = 8e-16
Identities = 40/69 (57%), Positives = 49/69 (70%)
Query: 83 KEWPAFDLDNTRVAFKLRKDKFNYMAIADNRQRFMPLPDDRLPPRGQVLAYPEAVRLVNP 142
K+WP F+L TR+AFKLRKDKF+YMA+AD+R+R MP PDD R Q L Y EA L P
Sbjct: 175 KDWPGFELGETRIAFKLRKDKFHYMAVADDRKRIMPFPDDLCKGRCQTLDYQEASLLTAP 234
Query: 143 VEPEFKGEV 151
+P +GEV
Sbjct: 235 CDPRLQGEV 243
Score = 55.1 bits (131), Expect = 4e-06
Identities = 23/41 (56%), Positives = 33/41 (80%)
Query: 1 VVIDNDIVQVNISNPEGCVTGIQYNGLDNLLEINNHESDRG 41
VV+DN I+QV +S P G +TGI+YNG+D +LE+ N E++RG
Sbjct: 58 VVMDNGILQVTLSKPGGIITGIEYNGIDYVLEVRNKETNRG 98
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.326 0.144 0.466
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 884,219,410
Number of Sequences: 2790947
Number of extensions: 40730794
Number of successful extensions: 96356
Number of sequences better than 10.0: 34
Number of HSP's better than 10.0 without gapping: 19
Number of HSP's successfully gapped in prelim test: 15
Number of HSP's that attempted gapping in prelim test: 96261
Number of HSP's gapped (non-prelim): 54
length of query: 478
length of database: 848,049,833
effective HSP length: 131
effective length of query: 347
effective length of database: 482,435,776
effective search space: 167405214272
effective search space used: 167405214272
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 77 (34.3 bits)
Medicago: description of AC148236.5


