

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147964.5 + phase: 0
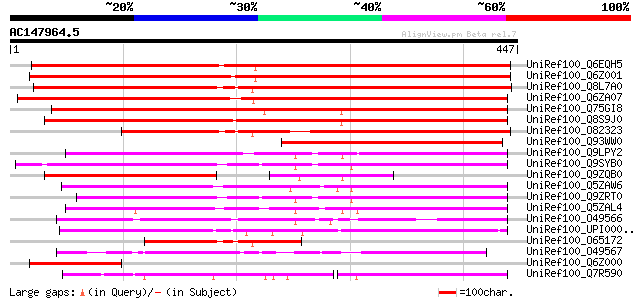
(447 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q6EQH5 Membrane protein-like [Oryza sativa] 576 e-163
UniRef100_Q6Z001 Membrane protein-like [Oryza sativa] 563 e-159
UniRef100_Q8L7A0 Hypothetical protein At2g25740 [Arabidopsis tha... 550 e-155
UniRef100_Q6ZA07 Hypothetical protein P0434E03.23 [Oryza sativa] 530 e-149
UniRef100_Q75GI8 Expressed protein [Oryza sativa] 421 e-116
UniRef100_Q8S9J0 At2g36630/F1O11.26 [Arabidopsis thaliana] 419 e-116
UniRef100_O82323 Hypothetical protein At2g25740 [Arabidopsis tha... 405 e-112
UniRef100_Q93WW0 Hypothetical protein [Musa acuminata] 247 6e-64
UniRef100_Q9LPY2 T23J18.20 [Arabidopsis thaliana] 226 8e-58
UniRef100_Q9SYB0 Hypothetical protein T13M11.10 [Arabidopsis tha... 220 6e-56
UniRef100_Q9ZQB0 Hypothetical protein At2g36630 [Arabidopsis tha... 214 4e-54
UniRef100_Q5ZAW6 Membrane protein-like [Oryza sativa] 213 1e-53
UniRef100_Q9ZRT0 Hypothetical protein [Arabidopsis thaliana] 213 1e-53
UniRef100_Q5ZAL4 Membrane protein-like [Oryza sativa] 206 1e-51
UniRef100_O49566 Hypothetical protein F7J7.190 [Arabidopsis thal... 199 1e-49
UniRef100_UPI000049A489 UPI000049A489 UniRef100 entry 187 4e-46
UniRef100_O65172 Hypothetical protein [Mesembryanthemum crystall... 169 1e-40
UniRef100_O49567 Hypothetical protein F7J7.200 [Arabidopsis thal... 145 2e-33
UniRef100_Q6Z000 Hypothetical protein P0473F05.27-3 [Oryza sativa] 119 1e-25
UniRef100_Q7R590 GLP_587_84274_82712 [Giardia lamblia ATCC 50803] 73 2e-11
>UniRef100_Q6EQH5 Membrane protein-like [Oryza sativa]
Length = 481
Score = 576 bits (1484), Expect = e-163
Identities = 276/426 (64%), Positives = 348/426 (80%), Gaps = 7/426 (1%)
Query: 20 VVMKKTSFFWYSGESSYERVWPEMKFGWRIVVGSIVGFFGAALGSVGGVGGGGIFIPMLT 79
++ K +F W + +SY VWPEM+ GW+IV+GS++GFFGAA GSVGGVGGGGIF+PMLT
Sbjct: 57 ILRKVANFLWQTDGNSYHHVWPEMELGWQIVLGSLIGFFGAAFGSVGGVGGGGIFVPMLT 116
Query: 80 LIIGFDPKSSTALSKCMITGAAGSTVYYNLRLRHPTLDMPLIDYDLALLFQPMLMLGISI 139
LIIGFDPKSSTA+SKCMI GAA STVYYNL+L+HPTLDMP+IDYDLALL QPMLMLGISI
Sbjct: 117 LIIGFDPKSSTAISKCMIMGAAVSTVYYNLKLKHPTLDMPVIDYDLALLIQPMLMLGISI 176
Query: 140 GVAFNVMFADWMVTILLIILFIGTSTKALVKGIDTWKKETIMKKEAFEEAAQMLESGSTP 199
GV FNV+F DW+VT+LLIILF+GTSTKA +KG++TWKKETI+K+EA A ++ ++ P
Sbjct: 177 GVIFNVLFPDWLVTVLLIILFLGTSTKAFLKGVETWKKETIIKREA---AKRLEQTSEEP 233
Query: 200 DYASEEDYKSLPADLQ---DEEVPLLDNIHWKELSVLMYVWVAFLIVQILKTYSKTCSIE 256
+YA AD + DE L+ NI+WKE +L +VW+AFL++Q+ K Y+ TCS
Sbjct: 234 EYAPLPTGPGAVADAKRPSDEAASLMKNIYWKEFGLLAFVWMAFLVLQVTKNYTATCSSW 293
Query: 257 YWLLNSLQVPIAISVTLFEAICLCKGTRVIASRGKE-ITWKFHKICLYCFCGIIAGMVSG 315
YW+LN LQ+P+++ VT++EA+ L G RV++S+G E T KFH++C+YCF GI AG+V G
Sbjct: 294 YWILNLLQIPVSVGVTMYEALGLMSGKRVLSSKGNEQTTLKFHQLCIYCFFGITAGVVGG 353
Query: 316 LLGLGGGFILGPLFLELGIPPQVASATSTFAMVFSSSMSVVQYYHLDRFPIPYASYLVLV 375
LLGLGGGFI+GPLFLELGIPPQV+SAT+TFAM+FSSSMSVV+YY L+RFP+PYA Y V+V
Sbjct: 354 LLGLGGGFIMGPLFLELGIPPQVSSATATFAMMFSSSMSVVEYYLLNRFPVPYALYFVIV 413
Query: 376 ATIAALTGQHVVRKIIAIFGRASIIVFILAFTIFVSAISLGGVGIGNMVEKMENEEYMGF 435
A IAA+ GQHVVR++I GRAS+I+FILAF IFVSAISLGGVGI NM+ ++ EYMGF
Sbjct: 414 AFIAAIIGQHVVRRLINWLGRASLIIFILAFMIFVSAISLGGVGISNMIHRINQHEYMGF 473
Query: 436 DNLCHH 441
+NLC +
Sbjct: 474 ENLCKY 479
>UniRef100_Q6Z001 Membrane protein-like [Oryza sativa]
Length = 465
Score = 563 bits (1451), Expect = e-159
Identities = 270/428 (63%), Positives = 346/428 (80%), Gaps = 7/428 (1%)
Query: 18 EIVVMKKTSFFWYSGESSYERVWPEMKFGWRIVVGSIVGFFGAALGSVGGVGGGGIFIPM 77
E+ M+K F +SGE+SY VWP M+FGW+IV+G ++GFFGAA GSVGGVGGGGIF+PM
Sbjct: 39 EVGYMRKVVNFLWSGEASYHHVWPPMEFGWKIVLGILIGFFGAAFGSVGGVGGGGIFVPM 98
Query: 78 LTLIIGFDPKSSTALSKCMITGAAGSTVYYNLRLRHPTLDMPLIDYDLALLFQPMLMLGI 137
LTLIIGFD KSSTA+SKCMI GAA STVYYNL+L+HPTLDMP+IDYDLALL QPMLMLGI
Sbjct: 99 LTLIIGFDAKSSTAISKCMIMGAAVSTVYYNLKLKHPTLDMPVIDYDLALLIQPMLMLGI 158
Query: 138 SIGVAFNVMFADWMVTILLIILFIGTSTKALVKGIDTWKKETIMKKEAFEEAAQMLESGS 197
SIGV FNV+F DW++T+LLIILF+GTSTKA +KG++TWKKETI+K+EA + Q+ E
Sbjct: 159 SIGVLFNVIFPDWLITVLLIILFLGTSTKAFLKGVETWKKETILKREAAKRLEQIAEE-- 216
Query: 198 TPDYASEEDYKSLPADLQ---DEEVPLLDNIHWKELSVLMYVWVAFLIVQILKTYSKTCS 254
P+Y+ + A+ + DE L N++WKE +L +VW+AFL++Q+ K Y TCS
Sbjct: 217 -PEYSPLPTGPNATAESKAPSDEAASLWQNVYWKEFGLLAFVWIAFLVLQVTKNYMPTCS 275
Query: 255 IEYWLLNSLQVPIAISVTLFEAICLCKGTRVIASRGKEIT-WKFHKICLYCFCGIIAGMV 313
YW+LN LQ+P+++ VT++E + L +G RVI+S+G E T KFH++ +YCF G++AG+V
Sbjct: 276 TWYWVLNFLQIPVSVGVTMYEGLGLMQGRRVISSKGDEQTNLKFHQLLVYCFFGMMAGVV 335
Query: 314 SGLLGLGGGFILGPLFLELGIPPQVASATSTFAMVFSSSMSVVQYYHLDRFPIPYASYLV 373
GLLGLGGGFI+GPLFLELGIPPQV+SAT+TFAM+FSSSMSVV+YY LDRFP+PYA +
Sbjct: 336 GGLLGLGGGFIMGPLFLELGIPPQVSSATATFAMMFSSSMSVVEYYLLDRFPVPYALFFT 395
Query: 374 LVATIAALTGQHVVRKIIAIFGRASIIVFILAFTIFVSAISLGGVGIGNMVEKMENEEYM 433
+VA AA+ GQH+VRK+I GRAS+I+FIL+F IF+SAISLGGVGI NM+ K+ EYM
Sbjct: 396 VVAFFAAIIGQHIVRKLINWLGRASLIIFILSFMIFISAISLGGVGISNMIGKIARHEYM 455
Query: 434 GFDNLCHH 441
GFDN+C++
Sbjct: 456 GFDNICNY 463
>UniRef100_Q8L7A0 Hypothetical protein At2g25740 [Arabidopsis thaliana]
Length = 476
Score = 550 bits (1418), Expect = e-155
Identities = 271/429 (63%), Positives = 342/429 (79%), Gaps = 14/429 (3%)
Query: 22 MKKTSFFWYSGESSYERVWPEMKFGWRIVVGSIVGFFGAALGSVGGVGGGGIFIPMLTLI 81
+K +F W S + Y VWPE +F W+IV+G++VGFFGAA GSVGGVGGGGIF+PML+LI
Sbjct: 53 LKAINFLWESDQIGYRHVWPEFEFNWQIVLGTLVGFFGAAFGSVGGVGGGGIFVPMLSLI 112
Query: 82 IGFDPKSSTALSKCMITGAAGSTVYYNLRLRHPTLDMPLIDYDLALLFQPMLMLGISIGV 141
IGFDPKS+TA+SKCMI GA+ STVYYNLRLRHPTLDMP+IDYDLALL QPMLMLGISIGV
Sbjct: 113 IGFDPKSATAISKCMIMGASVSTVYYNLRLRHPTLDMPIIDYDLALLIQPMLMLGISIGV 172
Query: 142 AFNVMFADWMVTILLIILFIGTSTKALVKGIDTWKKETIMKKEAFEEAAQMLESGSTPDY 201
AFNV+F DW+VT+LLI+LF+GTSTKA +KG +TW KETI KK EAA+ LES
Sbjct: 173 AFNVIFPDWLVTVLLIVLFLGTSTKAFLKGSETWNKETIEKK----EAAKRLESNGVS-- 226
Query: 202 ASEEDYKSLPA-------DLQDEEVPLLDNIHWKELSVLMYVWVAFLIVQILKTYSKTCS 254
+E +Y LPA + + EEV +++N++WKEL +L++VW+ FL +QI K CS
Sbjct: 227 GTEVEYVPLPAAPSTNPGNKKKEEVSIIENVYWKELGLLVFVWIVFLALQISKQNLANCS 286
Query: 255 IEYWLLNSLQVPIAISVTLFEAICLCKGTRVIASRGK-EITWKFHKICLYCFCGIIAGMV 313
+ YW++N LQ+P+A+ V+ +EA+ L +G R+IAS+G+ + + ++ +YC GIIAG+V
Sbjct: 287 VAYWVINLLQIPVAVGVSGYEAVALYQGRRIIASKGQGDSNFTVGQLVMYCTFGIIAGIV 346
Query: 314 SGLLGLGGGFILGPLFLELGIPPQVASATSTFAMVFSSSMSVVQYYHLDRFPIPYASYLV 373
GLLGLGGGFI+GPLFLELG+PPQV+SAT+TFAM FSSSMSVV+YY L RFP+PYA YLV
Sbjct: 347 GGLLGLGGGFIMGPLFLELGVPPQVSSATATFAMTFSSSMSVVEYYLLKRFPVPYALYLV 406
Query: 374 LVATIAALTGQHVVRKIIAIFGRASIIVFILAFTIFVSAISLGGVGIGNMVEKMENEEYM 433
VATIAA GQHVVR++IA GRAS+I+FILA IF+SAISLGGVGI NM+ K++ EYM
Sbjct: 407 GVATIAAWVGQHVVRRLIAAIGRASLIIFILASMIFISAISLGGVGIVNMIGKIQRHEYM 466
Query: 434 GFDNLCHHG 442
GF+NLC +G
Sbjct: 467 GFENLCKYG 475
>UniRef100_Q6ZA07 Hypothetical protein P0434E03.23 [Oryza sativa]
Length = 469
Score = 530 bits (1364), Expect = e-149
Identities = 261/438 (59%), Positives = 335/438 (75%), Gaps = 15/438 (3%)
Query: 8 AERVLEDEKPEIVVMKKTSFFWYSG-ESSYERVWPEMKFGWRIVVGSIVGFFGAALGSVG 66
A V E+ + + K +F W SG E+SY VWP M+FGW+IV+GS VGF GAA GS+G
Sbjct: 37 AAAVAEEGEEASHLRKVANFLWRSGGENSYHHVWPPMEFGWQIVLGSFVGFIGAAFGSIG 96
Query: 67 GVGGGGIFIPMLTLIIGFDPKSSTALSKCMITGAAGSTVYYNLRLRHPTLDMPLIDYDLA 126
GVGGGG F+PMLTLIIGFD KSS A+SKCMI GAA STVY NL+ +HPTLDMP+IDYDLA
Sbjct: 97 GVGGGGFFMPMLTLIIGFDAKSSVAISKCMIMGAAVSTVYCNLKRKHPTLDMPVIDYDLA 156
Query: 127 LLFQPMLMLGISIGVAFNVMFADWMVTILLIILFIGTSTKALVKGIDTWKKETIMKKEAF 186
LL QPMLMLGISIGV FNV+F DW+VT+LLIILF+GTSTKA +KGI+TWKKETI+K+EA
Sbjct: 157 LLIQPMLMLGISIGVIFNVIFPDWLVTVLLIILFLGTSTKAFLKGIETWKKETIIKREAE 216
Query: 187 EEAAQMLESGSTPDYASEEDYKSLPAD----LQDEEVPLLDNIHWKELSVLMYVWVAFLI 242
+ + Q E E +Y+ +PA DE V +L N++WKE +L +VW+AFL
Sbjct: 217 KRSEQTSE---------ELEYRPVPASESKPPSDEAVSILHNVYWKEFGLLAFVWIAFLA 267
Query: 243 VQILKTYSKTCSIEYWLLNSLQVPIAISVTLFEAICLCKGTRVIASRGKEIT-WKFHKIC 301
+Q+ K Y TCS YW+LN LQ+P+++ VT++E + L +G RVI+S G E T KFH++
Sbjct: 268 LQVTKNYMPTCSTWYWVLNLLQIPVSVGVTMYEGLGLMQGRRVISSNGNEQTNLKFHQLL 327
Query: 302 LYCFCGIIAGMVSGLLGLGGGFILGPLFLELGIPPQVASATSTFAMVFSSSMSVVQYYHL 361
+YCF GI AG+V+GLLG+GGG ILGP+FL+LG+PPQVASAT+TF+M+FS+SMS V+YY L
Sbjct: 328 MYCFFGITAGIVAGLLGVGGGSILGPMFLDLGVPPQVASATATFSMMFSASMSAVEYYFL 387
Query: 362 DRFPIPYASYLVLVATIAALTGQHVVRKIIAIFGRASIIVFILAFTIFVSAISLGGVGIG 421
DRFP+PYA YL +VA +A+ GQ +VRK+I GRASII+F L+ IF+S I LGG+GI
Sbjct: 388 DRFPVPYALYLTVVAFFSAIVGQRMVRKVINWLGRASIIIFTLSIMIFLSTIPLGGIGIV 447
Query: 422 NMVEKMENEEYMGFDNLC 439
N + K+E EYMGF+++C
Sbjct: 448 NWIGKIERHEYMGFEDIC 465
>UniRef100_Q75GI8 Expressed protein [Oryza sativa]
Length = 475
Score = 421 bits (1082), Expect = e-116
Identities = 210/414 (50%), Positives = 288/414 (68%), Gaps = 12/414 (2%)
Query: 38 RVWPEMKFGWRIVVGSIVGFFGAALGSVGGVGGGGIFIPMLTLIIGFDPKSSTALSKCMI 97
RVWP++ F WR+VV ++VGF G+A G+VGGVGGGGIF+PML L++GFD KS+ ALSKCMI
Sbjct: 60 RVWPDLAFNWRVVVATVVGFLGSAFGTVGGVGGGGIFVPMLNLLVGFDTKSAAALSKCMI 119
Query: 98 TGAAGSTVYYNLRLRHPTLDMPLIDYDLALLFQPMLMLGISIGVAFNVMFADWMVTILLI 157
GA+ S+V+YNL++ HPT + P+IDY LALLFQPMLMLGI+IGV +V+F W++T+L+I
Sbjct: 120 MGASASSVWYNLQVSHPTKEAPVIDYKLALLFQPMLMLGITIGVELSVIFPYWLITVLII 179
Query: 158 ILFIGTSTKALVKGIDTWKKETIMKKEAFEEAAQMLESGSTPDYASEEDYKS-LPADLQD 216
ILFIGTS+++ KGI WK ET ++ E E + S + D + + L Q
Sbjct: 180 ILFIGTSSRSFYKGILMWKDETRIQMETREREEESKSSCAARDVVIDPSCEEPLLCQPQP 239
Query: 217 EEVPLLD----NIHWKELSVLMYVWVAFLIVQILKTYSKTCSIEYWLLNSLQVPIAISVT 272
+E L+ N+ WK + VLM VW +FL++QI K S++CS YW++N LQVP+A+SV
Sbjct: 240 KEKSALETFLFNLRWKNILVLMTVWSSFLVLQIFKNNSQSCSTFYWVINILQVPVALSVF 299
Query: 273 LFEAICLCKGTRVIASRGK-------EITWKFHKICLYCFCGIIAGMVSGLLGLGGGFIL 325
L+E + LC+ +R G I W ++ FCG++ G V GLLG GGGFIL
Sbjct: 300 LWEGVQLCRESRARRMDGNWECVCEASIEWSPAQLIFCAFCGLLGGTVGGLLGSGGGFIL 359
Query: 326 GPLFLELGIPPQVASATSTFAMVFSSSMSVVQYYHLDRFPIPYASYLVLVATIAALTGQH 385
GPL LELG PQVASAT+TF M+FSSS+SVV++Y L+RFPIP+A YL+ ++ +A GQ
Sbjct: 360 GPLLLELGCIPQVASATATFVMMFSSSLSVVEFYFLNRFPIPFAVYLICISILAGFWGQS 419
Query: 386 VVRKIIAIFGRASIIVFILAFTIFVSAISLGGVGIGNMVEKMENEEYMGFDNLC 439
+VRK++ + RAS+IVFIL+ IF SA+++G VG + + N EYMGF + C
Sbjct: 420 LVRKLVHVLKRASLIVFILSSVIFASALTMGVVGTQKSISMINNHEYMGFLDFC 473
>UniRef100_Q8S9J0 At2g36630/F1O11.26 [Arabidopsis thaliana]
Length = 459
Score = 419 bits (1078), Expect = e-116
Identities = 202/417 (48%), Positives = 297/417 (70%), Gaps = 10/417 (2%)
Query: 31 SGESSYERVWPEMKFGWRIVVGSIVGFFGAALGSVGGVGGGGIFIPMLTLIIGFDPKSST 90
S S+ E++WP++KF W++V+ +++ F G+A G+VGGVGGGGIF+PMLTLI+GFD KS+
Sbjct: 42 SSLSATEKIWPDLKFSWKLVLATVIAFLGSACGTVGGVGGGGIFVPMLTLILGFDTKSAA 101
Query: 91 ALSKCMITGAAGSTVYYNLRLRHPTLDMPLIDYDLALLFQPMLMLGISIGVAFNVMFADW 150
A+SKCMI GA+ S+V+YN+R+RHPT ++P++DYDLALLFQPML+LGI++GV+ +V+F W
Sbjct: 102 AISKCMIMGASASSVWYNVRVRHPTKEVPILDYDLALLFQPMLLLGITVGVSLSVVFPYW 161
Query: 151 MVTILLIILFIGTSTKALVKGIDTWKKETIMKKEAFEEAAQMLESGSTPDYASEEDYKSL 210
++T+L+IILF+GTS+++ KGI+ WK+ET++K E ++ A M+ S + + +Y+ L
Sbjct: 162 LITVLIIILFVGTSSRSFFKGIEMWKEETLLKNEMAQQRANMVNSRG--ELLIDTEYEPL 219
Query: 211 -PADLQDEEVPLLDNIHWKELSVLMYVWVAFLIVQILKTYSKTCSIEYWLLNSLQVPIAI 269
P + + E + N+ WK L +L+ VW+ FL++QI+K K CS YW+L +Q P+A+
Sbjct: 220 YPREEKSELEIIRSNLKWKGLLILVTVWLTFLLIQIVKNEIKVCSTIYWILFIVQFPVAL 279
Query: 270 SVTLFEAICLCKGTRVIASRGK-------EITWKFHKICLYCFCGIIAGMVSGLLGLGGG 322
+V FEA L + + G I W + CG+I G+V GLLG GGG
Sbjct: 280 AVFGFEASKLYTANKKRLNSGNTECICEATIEWTPLSLIFCGLCGLIGGIVGGLLGSGGG 339
Query: 323 FILGPLFLELGIPPQVASATSTFAMVFSSSMSVVQYYHLDRFPIPYASYLVLVATIAALT 382
F+LGPL LE+G+ PQVASAT+TF M+FSSS+SVV++Y L RFPIPYA YL+ V+ +A
Sbjct: 340 FVLGPLLLEIGVIPQVASATATFVMMFSSSLSVVEFYLLKRFPIPYAMYLISVSILAGFW 399
Query: 383 GQHVVRKIIAIFGRASIIVFILAFTIFVSAISLGGVGIGNMVEKMENEEYMGFDNLC 439
GQ +RK++AI RASIIVF+L+ I SA+++G +GI ++ + N E+MGF C
Sbjct: 400 GQSFIRKLVAILRRASIIVFVLSGVICASALTMGVIGIEKSIKMIHNHEFMGFLGFC 456
>UniRef100_O82323 Hypothetical protein At2g25740 [Arabidopsis thaliana]
Length = 902
Score = 405 bits (1042), Expect = e-112
Identities = 211/351 (60%), Positives = 267/351 (75%), Gaps = 31/351 (8%)
Query: 99 GAAGSTVYYNLRLRHPTLDMPLIDYDLALLFQPMLMLGISIGVAFNVMFADWMVTILLII 158
GA+ STVYYNLRLRHPTLDMP+IDYDLALL QPMLMLGISIGVAFNV+F DW+VT+LLI+
Sbjct: 2 GASVSTVYYNLRLRHPTLDMPIIDYDLALLIQPMLMLGISIGVAFNVIFPDWLVTVLLIV 61
Query: 159 LFIGTSTKALVKGIDTWKKETIMKKEAFEEAAQMLESGSTPDYASEEDYKSLPA------ 212
LF+GTSTKA +KG +TW KETI KKEA A+ LES +E +Y LPA
Sbjct: 62 LFLGTSTKAFLKGSETWNKETIEKKEA----AKRLESNGVS--GTEVEYVPLPAAPSTNP 115
Query: 213 -DLQDEEVPLLDNIHWKELSVLMYVWVAFLIVQILKTYSKTCSIEYWLLNSLQVPIAISV 271
+ + EEV +++N++WKEL +L++VW+ FL +QI K +P+A+ V
Sbjct: 116 GNKKKEEVSIIENVYWKELGLLVFVWIVFLALQISK-----------------IPVAVGV 158
Query: 272 TLFEAICLCKGTRVIASRGK-EITWKFHKICLYCFCGIIAGMVSGLLGLGGGFILGPLFL 330
+ +EA+ L +G R+IAS+G+ + + ++ +YC GIIAG+V GLLGLGGGFI+GPLFL
Sbjct: 159 SGYEAVALYQGRRIIASKGQGDSNFTVGQLVMYCTFGIIAGIVGGLLGLGGGFIMGPLFL 218
Query: 331 ELGIPPQVASATSTFAMVFSSSMSVVQYYHLDRFPIPYASYLVLVATIAALTGQHVVRKI 390
ELG+PPQV+SAT+TFAM FSSSMSVV+YY L RFP+PYA YLV VATIAA GQHVVR++
Sbjct: 219 ELGVPPQVSSATATFAMTFSSSMSVVEYYLLKRFPVPYALYLVGVATIAAWVGQHVVRRL 278
Query: 391 IAIFGRASIIVFILAFTIFVSAISLGGVGIGNMVEKMENEEYMGFDNLCHH 441
IA GRAS+I+FILA IF+SAISLGGVGI NM+ K++ EYMGF+NLC +
Sbjct: 279 IAAIGRASLIIFILASMIFISAISLGGVGIVNMIGKIQRHEYMGFENLCKY 329
>UniRef100_Q93WW0 Hypothetical protein [Musa acuminata]
Length = 238
Score = 247 bits (630), Expect = 6e-64
Identities = 122/197 (61%), Positives = 160/197 (80%), Gaps = 2/197 (1%)
Query: 240 FLIVQILK-TYSKTCSIEYWLLNSLQVPIAISVTLFEAICLCKGTRVIASRGKEIT-WKF 297
FLI+Q+LK Y+ TCS+ YW+LN LQVP+++ V+ +EA+ L +G R+I+S+G E T +
Sbjct: 1 FLILQVLKQNYTSTCSLWYWILNLLQVPVSLGVSGYEAVSLYRGKRIISSKGLEGTDFTV 60
Query: 298 HKICLYCFCGIIAGMVSGLLGLGGGFILGPLFLELGIPPQVASATSTFAMVFSSSMSVVQ 357
++ YC G++AG+V GLLGLGGGFILGP+FLELG+PPQV+SAT+TFAM FSSSMSVV+
Sbjct: 61 IQLVFYCLIGVLAGVVGGLLGLGGGFILGPVFLELGVPPQVSSATATFAMTFSSSMSVVE 120
Query: 358 YYHLDRFPIPYASYLVLVATIAALTGQHVVRKIIAIFGRASIIVFILAFTIFVSAISLGG 417
YY L RFPIPYA Y V VA +AA GQH+V+++I I GRAS+I+FILA TIF+SAISLGG
Sbjct: 121 YYLLKRFPIPYALYFVSVALVAAFVGQHLVKRLIEILGRASLIIFILASTIFISAISLGG 180
Query: 418 VGIGNMVEKMENEEYMG 434
VGI NMV+K+++ E G
Sbjct: 181 VGISNMVQKIQHHESWG 197
>UniRef100_Q9LPY2 T23J18.20 [Arabidopsis thaliana]
Length = 491
Score = 226 bits (577), Expect = 8e-58
Identities = 133/409 (32%), Positives = 218/409 (52%), Gaps = 33/409 (8%)
Query: 50 VVGSIVGFFGAALGSVGGVGGGGIFIPMLTLIIGFDPKSSTALSKCMITGAAGSTVYYNL 109
++ +++ FF A++ S GG+GGGG+F+ ++T+I G + K++++ S M+TG + + V NL
Sbjct: 97 IIAAVLSFFAASISSAGGIGGGGLFLSIMTIIAGLEMKTASSFSAFMVTGVSFANVGCNL 156
Query: 110 RLRHP-TLDMPLIDYDLALLFQPMLMLGISIGVAFNVMFADWMVTILLIILFIGTSTKAL 168
LR+P + D LID+DLAL QP L+LG+SIGV N MF +W+V L + ++ K
Sbjct: 157 FLRNPKSRDKTLIDFDLALTIQPCLLLGVSIGVICNRMFPNWLVLFLFAVFLAWSTMKTC 216
Query: 169 VKGIDTWKKETIMKKEAFEEAAQMLESGSTPDYASEEDYKSLPADLQDEEVPLLDNIHWK 228
KG+ W E+ K +E +P + E + D + W
Sbjct: 217 KKGVSYWNLESERAKIKSPRDVDGIEVARSPLLSEERE---------DVRQRGMIRFPWM 267
Query: 229 ELSVLMYVWVAFLIVQILKTYS--------KTCSIEYWLLNSLQVPIAISVTLFEAICLC 280
+L VL+ +W+ F + + + K C YW L+SLQ+P+ T+F +C+
Sbjct: 268 KLGVLVIIWLLFFSINLFRGNKYGQGIISIKPCGALYWFLSSLQIPL----TIFFTLCIY 323
Query: 281 KGTRVIASRGKE----------ITWKFHKICLYCFCGIIAGMVSGLLGLGGGFILGPLFL 330
V ++ + + +K+ L ++AG++ GL G+GGG ++ PL L
Sbjct: 324 FSDNVQSNHTSHSNQNSEQETGVGGRQNKLMLPVMA-LLAGVLGGLFGIGGGMLISPLLL 382
Query: 331 ELGIPPQVASATSTFAMVFSSSMSVVQYYHLDRFPIPYASYLVLVATIAALTGQHVVRKI 390
++GI P+V +AT +F ++FSSSMS +QY L A+ LV +A+L G VV+K+
Sbjct: 383 QIGIAPEVTAATCSFMVLFSSSMSAIQYLLLGMEHAGTAAIFALVCFVASLVGLMVVKKV 442
Query: 391 IAIFGRASIIVFILAFTIFVSAISLGGVGIGNMVEKMENEEYMGFDNLC 439
IA +GRASIIVF + + +S + + G N+ + YMGF C
Sbjct: 443 IAKYGRASIIVFAVGIVMALSTVLMTTHGAFNVWNDFVSGRYMGFKLPC 491
>UniRef100_Q9SYB0 Hypothetical protein T13M11.10 [Arabidopsis thaliana]
Length = 458
Score = 220 bits (561), Expect = 6e-56
Identities = 134/457 (29%), Positives = 242/457 (52%), Gaps = 40/457 (8%)
Query: 6 SLAERVLEDEKPEIVVMKKTSFFWYSGESSYERVWPEMKFGWRIVVGSIVGFFGAALGSV 65
S+AE+ P ++ KTS + + S + P ++ ++ ++ F +++ S
Sbjct: 19 SIAEQEPSILSPVDQLLNKTSSYL---DFSTKFNQPRIELTTSTIIAGLLSFLASSISSA 75
Query: 66 GGVGGGGIFIPMLTLIIGFDPKSSTALSKCMITGAAGSTVYYNLRLRHPTLD-MPLIDYD 124
GG+GGGG+++P++T++ G D K++++ S M+TG + + V NL +R+P LID+D
Sbjct: 76 GGIGGGGLYVPIMTIVAGLDLKTASSFSAFMVTGGSIANVGCNLFVRNPKSGGKTLIDFD 135
Query: 125 LALLFQPMLMLGISIGVAFNVMFADWMVTILLIILFIGTSTKALVKGIDTWKKETIMKKE 184
LALL +P ++LG+SIGV N++F +W++T L + ++ K G+ W+ E+ M K
Sbjct: 136 LALLLEPCMLLGVSIGVICNLVFPNWLITSLFAVFLAWSTLKTFGNGLYYWRLESEMVK- 194
Query: 185 AFEEAAQMLESGSTPDYASEEDYKSLPADL-QDEEVPLLDNIHWKELSVLMYVWVAFLIV 243
+ ES + E+ +SL L +D + P W +L VL+ +W+++ V
Sbjct: 195 -------IRESNRIEEDDEEDKIESLKLPLLEDYQRP--KRFPWIKLGVLVIIWLSYFAV 245
Query: 244 QILKTYS--------KTCSIEYWLLNSLQVPIAISVTLFEAICLCKGTRVIASRGKEITW 295
+L+ + C YWL++S Q+P+ TLF + +C V + + +
Sbjct: 246 YLLRGNKYGEGIISIEPCGNAYWLISSSQIPL----TLFFTLWICFSDNVQSQQQSDYHV 301
Query: 296 KFHKI-------------CLYCFCGIIAGMVSGLLGLGGGFILGPLFLELGIPPQVASAT 342
+ C++ ++AG++ G+ G+GGG ++ PL L++GI P+V +AT
Sbjct: 302 SVKDVEDLRSNDGARSNKCMFPVMALLAGVLGGVFGIGGGMLISPLLLQVGIAPEVTAAT 361
Query: 343 STFAMVFSSSMSVVQYYHLDRFPIPYASYLVLVATIAALTGQHVVRKIIAIFGRASIIVF 402
+F ++FSS+MS +QY L AS ++ +A+L G VV+K+I +GRASIIVF
Sbjct: 362 CSFMVLFSSTMSAIQYLLLGMEHTGTASIFAVICFVASLVGLKVVQKVITEYGRASIIVF 421
Query: 403 ILAFTIFVSAISLGGVGIGNMVEKMENEEYMGFDNLC 439
+ + +S + + G ++ + YMGF C
Sbjct: 422 SVGIVMALSIVLMTSYGALDVWNDYVSGRYMGFKLPC 458
>UniRef100_Q9ZQB0 Hypothetical protein At2g36630 [Arabidopsis thaliana]
Length = 288
Score = 214 bits (545), Expect = 4e-54
Identities = 89/152 (58%), Positives = 133/152 (86%)
Query: 31 SGESSYERVWPEMKFGWRIVVGSIVGFFGAALGSVGGVGGGGIFIPMLTLIIGFDPKSST 90
S S+ E++WP++KF W++V+ +++ F G+A G+VGGVGGGGIF+PMLTLI+GFD KS+
Sbjct: 42 SSLSATEKIWPDLKFSWKLVLATVIAFLGSACGTVGGVGGGGIFVPMLTLILGFDTKSAA 101
Query: 91 ALSKCMITGAAGSTVYYNLRLRHPTLDMPLIDYDLALLFQPMLMLGISIGVAFNVMFADW 150
A+SKCMI GA+ S+V+YN+R+RHPT ++P++DYDLALLFQPML+LGI++GV+ +V+F W
Sbjct: 102 AISKCMIMGASASSVWYNVRVRHPTKEVPILDYDLALLFQPMLLLGITVGVSLSVVFPYW 161
Query: 151 MVTILLIILFIGTSTKALVKGIDTWKKETIMK 182
++T+L+IILF+GTS+++ KGI+ WK+ET++K
Sbjct: 162 LITVLIIILFVGTSSRSFFKGIEMWKEETLLK 193
Score = 66.2 bits (160), Expect = 2e-09
Identities = 44/121 (36%), Positives = 64/121 (52%), Gaps = 12/121 (9%)
Query: 230 LSVLMYVWVAFLIVQILKTYSKTCS----IEYWLLNSL-QVPIAISVTLFEAICLCKGTR 284
LSV+ W+ +++ IL + + S IE W +L + P+A++V FEA L +
Sbjct: 154 LSVVFPYWLITVLIIILFVGTSSRSFFKGIEMWKEETLLKFPVALAVFGFEASKLYTANK 213
Query: 285 VIASRGKE-------ITWKFHKICLYCFCGIIAGMVSGLLGLGGGFILGPLFLELGIPPQ 337
+ G I W + CG+I G+V GLLG GGGF+LGPL LE+G+ PQ
Sbjct: 214 KRLNSGNTECICEATIEWTPLSLIFCGLCGLIGGIVGGLLGSGGGFVLGPLLLEIGVIPQ 273
Query: 338 V 338
V
Sbjct: 274 V 274
>UniRef100_Q5ZAW6 Membrane protein-like [Oryza sativa]
Length = 461
Score = 213 bits (541), Expect = 1e-53
Identities = 135/411 (32%), Positives = 208/411 (49%), Gaps = 27/411 (6%)
Query: 46 GWRIVVGSIVGFFGAALGSVGGVGGGGIFIPMLTLIIGFDPKSSTALSKCMITGAAGSTV 105
G V ++ F A++ S GGVGGG +F+P+L L+ G K +TA S M+TG A S V
Sbjct: 61 GLNTVAAWLLSFLAASVSSAGGVGGGSLFLPILNLVAGLSLKRATAYSSFMVTGGAASNV 120
Query: 106 YYNLRLRH-PTLDMPLIDYDLALLFQPMLMLGISIGVAFNVMFADWMVTILLIILFIGTS 164
YNL +IDYD+ALLFQP L+LG+SIGV NVMF +W++T L + +
Sbjct: 121 LYNLLCTGCGRRAAAVIDYDIALLFQPCLLLGVSIGVVCNVMFPEWLITALFALFLAFCT 180
Query: 165 TKALVKGIDTWKKETIMKKEAFEEAAQMLESGSTPDYASEEDYKSLPADLQDEEVPLLD- 223
TK L G+ W E+ A + + +T E D + D
Sbjct: 181 TKTLRAGLRIWSSES--------RGATLAVAAATAHGREEPLLLPHGTDAGNGGGARGDA 232
Query: 224 NIHWKELSVLMYVWVAFLIVQIL--------KTYSKTCSIEYWLLNSLQVPIAISVTLFE 275
WK++SVL+ VW+ F ++ + K C + YWL+ QVP A++ T +
Sbjct: 233 GFPWKDVSVLVMVWLCFFVLHVFIGDKHGKGMIRIKPCGVAYWLITLSQVPFAVAFTAY- 291
Query: 276 AICLCKGTRVIA-----SRGKEITWKFHKI--CLYCFCGIIAGMVSGLLGLGGGFILGPL 328
I K + + + + K + L+ + G +SGL G+GGG +L P+
Sbjct: 292 -IIYAKRKKQVLHNQEDGKANPESTKMDTLPTLLFPLAAFVTGALSGLFGIGGGLLLNPV 350
Query: 329 FLELGIPPQVASATSTFAMVFSSSMSVVQYYHLDRFPIPYASYLVLVATIAALTGQHVVR 388
L++GIPPQ A+ATS+F ++F +SMS+VQ+ L I AS + +A++ G V+
Sbjct: 351 LLQIGIPPQTAAATSSFMVLFCASMSMVQFILLGMQGIGEASVYAGICFVASVVGAVVIE 410
Query: 389 KIIAIFGRASIIVFILAFTIFVSAISLGGVGIGNMVEKMENEEYMGFDNLC 439
+ I GR S+IVF++ + VS + + G ++ + + YMGF C
Sbjct: 411 RAIRKSGRVSLIVFLVTGIMAVSTVIITFFGALDVWAQYTSGAYMGFKLPC 461
>UniRef100_Q9ZRT0 Hypothetical protein [Arabidopsis thaliana]
Length = 389
Score = 213 bits (541), Expect = 1e-53
Identities = 124/403 (30%), Positives = 219/403 (53%), Gaps = 37/403 (9%)
Query: 60 AALGSVGGVGGGGIFIPMLTLIIGFDPKSSTALSKCMITGAAGSTVYYNLRLRHPTLD-M 118
+++ S GG+GGGG+++P++T++ G D K++++ S M+TG + + V NL +R+P
Sbjct: 1 SSISSAGGIGGGGLYVPIMTIVAGLDLKTASSFSAFMVTGGSIANVGCNLFVRNPKSGGK 60
Query: 119 PLIDYDLALLFQPMLMLGISIGVAFNVMFADWMVTILLIILFIGTSTKALVKGIDTWKKE 178
LID+DLALL +P ++LG+SIGV N++F +W++T L + ++ K G+ W+ E
Sbjct: 61 TLIDFDLALLLEPCMLLGVSIGVICNLVFPNWLITSLFAVFLAWSTLKTFGNGLYYWRLE 120
Query: 179 TIMKKEAFEEAAQMLESGSTPDYASEEDYKSLPADL-QDEEVPLLDNIHWKELSVLMYVW 237
+ M K + ES + E+ +SL L +D + P W +L VL+ +W
Sbjct: 121 SEMVK--------IRESNRIEEDDEEDKIESLKLPLLEDYQRP--KRFPWIKLGVLVIIW 170
Query: 238 VAFLIVQILKTYS--------KTCSIEYWLLNSLQVPIAISVTLFEAICLCKGTRVIASR 289
+++ V +L+ + C YWL++S Q+P+ TLF + +C V + +
Sbjct: 171 LSYFAVYLLRGNKYGEGIISIEPCGNAYWLISSSQIPL----TLFFTLWICFSDNVQSQQ 226
Query: 290 GKEITWKFHKI-------------CLYCFCGIIAGMVSGLLGLGGGFILGPLFLELGIPP 336
+ + C++ ++AG++ G+ G+GGG ++ PL L++GI P
Sbjct: 227 QSDYHVSVKDVEDLRSNDGARSNKCMFPVMALLAGVLGGVFGIGGGMLISPLLLQVGIAP 286
Query: 337 QVASATSTFAMVFSSSMSVVQYYHLDRFPIPYASYLVLVATIAALTGQHVVRKIIAIFGR 396
+V +AT +F ++FSS+MS +QY L AS ++ +A+L G VV+K+I +GR
Sbjct: 287 EVTAATCSFMVLFSSTMSAIQYLLLGMEHTGTASIFAVICFVASLVGLKVVQKVITEYGR 346
Query: 397 ASIIVFILAFTIFVSAISLGGVGIGNMVEKMENEEYMGFDNLC 439
ASIIVF + + +S + + G ++ + YMGF C
Sbjct: 347 ASIIVFSVGIVMALSIVLMTSYGALDVWNDYVSGRYMGFKLPC 389
>UniRef100_Q5ZAL4 Membrane protein-like [Oryza sativa]
Length = 397
Score = 206 bits (523), Expect = 1e-51
Identities = 128/411 (31%), Positives = 210/411 (50%), Gaps = 40/411 (9%)
Query: 50 VVGSIVGFFGAALGSVGGVGGGGIFIPMLTLIIGFDPKSSTALSKCMITGAAGSTVYYNL 109
V+ I+ F AA S GGVGGG +++P+L ++ G K++TA S M+TG S V Y L
Sbjct: 6 VLACILSFLAAAFSSAGGVGGGSLYVPILNIVAGLSLKTATAFSTFMVTGGTLSNVLYTL 65
Query: 110 ---RLRHPTLDMPLIDYDLALLFQPMLMLGISIGVAFNVMFADWMVTILLIILFIGTSTK 166
R PLIDYD+A++ QP L+LG+S+GV NVMF +W++T L + + K
Sbjct: 66 IVLRGHEKGGHQPLIDYDIAVVSQPCLLLGVSVGVICNVMFPEWLITALFAVFLASATFK 125
Query: 167 ALVKGIDTWKKETIMKKEAFEEAAQMLESGST-PDYASEEDYKSLPADLQDEEVPLLDNI 225
G+ W+ ET A +MLE GS+ D A E D +
Sbjct: 126 TYGTGMKRWRAETAA-------ARRMLEGGSSLGDGAGEALLGQKDGDGHRRQCV----- 173
Query: 226 HWKELSVLMYVWVAFLIVQILKTYS--------KTCSIEYWLLNSLQVPIAISVTLFEAI 277
+L VL+ +W+ F ++ + + C + YWL+ Q+PIA++ T
Sbjct: 174 ---DLMVLVTIWLCFFVIHLFIGGEGAKGVFDIEPCGVTYWLITIAQIPIAVAFTA---- 226
Query: 278 CLCKGTRVIASRGKE-------ITWKFHKICLYCF--CGIIAGMVSGLLGLGGGFILGPL 328
C+ R ++ + + K + +Y F ++ G++SGL G+GGG +L P+
Sbjct: 227 CIVHQKRKSHAQNSQEFDQAISVKSKLESLPVYVFPVAALLTGVMSGLFGIGGGLLLNPV 286
Query: 329 FLELGIPPQVASATSTFAMVFSSSMSVVQYYHLDRFPIPYASYLVLVATIAALTGQHVVR 388
L++G+PP+ AS+T+ F ++F +SMS+VQ+ L I A + +A++ G V++
Sbjct: 287 LLQIGVPPKTASSTTMFMVLFCASMSMVQFIILGVDGIVTALVYAITCFVASIVGLVVIQ 346
Query: 389 KIIAIFGRASIIVFILAFTIFVSAISLGGVGIGNMVEKMENEEYMGFDNLC 439
I GR S+IVF++A + +S + + G + + + +YMGF C
Sbjct: 347 GTIRKSGRVSLIVFMVAAILALSVVVIACSGAVRVWVQYTSGQYMGFKMPC 397
>UniRef100_O49566 Hypothetical protein F7J7.190 [Arabidopsis thaliana]
Length = 431
Score = 199 bits (506), Expect = 1e-49
Identities = 127/423 (30%), Positives = 212/423 (50%), Gaps = 67/423 (15%)
Query: 42 EMKFGWRIVVGSIVGFFGAALGSVGGVGGGGIFIPMLTLIIGFDPKSSTALSKCMITGAA 101
E+K I++ ++ F A + S GG+GGGG+FIP++T++ G D K++++ S M+TG +
Sbjct: 51 ELKLSSAIIMAGVLCFLAALISSAGGIGGGGLFIPIMTIVAGVDLKTASSFSAFMVTGGS 110
Query: 102 GSTVYYNLRLRHPTLDMPLIDYDLALLFQPMLMLGISIGVAFNVMFADWMVTILLIILFI 161
+ V NL L+DYDLALL +P ++LG+SIGV N + +W++T+L +
Sbjct: 111 IANVISNL-----FGGKALLDYDLALLLEPCMLLGVSIGVICNRVLPEWLITVLFAVFLA 165
Query: 162 GTSTKALVKGIDTWKKETIMKKEAFEEAAQMLESGSTPDYASEEDYKSLPADLQDEEVPL 221
+ K G+ WK E+ + +E+ + + EE+ K+L A L + +
Sbjct: 166 WSILKTCRSGVKFWKLESEIARESGHGRPERGQG------QIEEETKNLKAPLLEAQATK 219
Query: 222 -LDNIHWKELSVLMYVWVAFLIVQILKTYS--------KTCSIEYWLLNSLQVPIAISVT 272
I W +L VL+ VW +F ++ +L+ K C +EYW+L SLQ+P+A+
Sbjct: 220 NKSKIPWTKLGVLVIVWASFFVIYLLRGNKDGKGIITIKPCGVEYWILLSLQIPLAL--- 276
Query: 273 LFEAICLCK----------------GTRVIASRGKEITWKFHKICLYCFCGIIAGMVSGL 316
+F + L + GTR+ K KF + +AG++ G+
Sbjct: 277 IFTKLALSRTESRQEQSPNDQKNQEGTRL----DKSTRLKFPAM------SFLAGLLGGI 326
Query: 317 LGLGGGFILGPLFLELGIPPQVASATSTFAMVFSSSMSVVQYYHLDRFPIPYASYLVLVA 376
G+GGG ++ PL L+ GIPPQ+ +AT++F + FS++MS VQY
Sbjct: 327 FGIGGGMLISPLLLQSGIPPQITAATTSFMVFFSATMSAVQY------------------ 368
Query: 377 TIAALTGQHVVRKIIAIFGRASIIVFILAFTIFVSAISLGGVGIGNMVEKMENEEYMGFD 436
+ + K +A FGRASIIVF + + +S + + G ++ + MGF
Sbjct: 369 LLLGMQNTDTAYKAVAQFGRASIIVFSVGTVMSLSTVLMTSFGALDVWTDYVAGKDMGFK 428
Query: 437 NLC 439
C
Sbjct: 429 LPC 431
>UniRef100_UPI000049A489 UPI000049A489 UniRef100 entry
Length = 460
Score = 187 bits (476), Expect = 4e-46
Identities = 133/446 (29%), Positives = 205/446 (45%), Gaps = 56/446 (12%)
Query: 45 FGWRIVVGSIVGFFGAALGSVGGVGGGGIFIPMLTLIIGFDPKSSTALSKCMITGAAGST 104
F W+++VGSI F A L + G+GGG ++ + LI+ DP + LSK G A
Sbjct: 7 FDWKLIVGSIGSLFFAVLCAGSGIGGGCFYLVIFVLILQMDPHQAIPLSKITTFGVACGG 66
Query: 105 VYYNLRLRHPTLDM-PLIDYDLALLFQPMLMLGISIGVAFNVMFADWMVTILLIILFIGT 163
RHP + PLI Y AL+ +P+ + G IGV FN++ W++ I+L++L T
Sbjct: 67 FLILWMKRHPNVRYKPLISYPTALMVEPLTIYGTMIGVIFNIISPSWLIIIVLVLLLGFT 126
Query: 164 STKALVKGIDTWKKETIMKKEAFEEAAQMLESGSTPDYASEEDY---------------- 207
S K K I WK E +K +A +++E+ S PD A ++
Sbjct: 127 SYKTFAKAIKQWKNEN--EKRDAAKATELVET-SKPDIADNDNDDMKPSENGSNAVIVDE 183
Query: 208 --------------KSLPADLQDEEVPLLDNIHWKEL------------SVLMYVWVAFL 241
K LP D E I K L +L+ VW
Sbjct: 184 KVQEEEEEGQGTGPKLLPQDESQEAQKEAKKIEEKTLLKREIIKAILSVGILIIVWAVMF 243
Query: 242 IVQILKTYSKTCSIE--------YWLLNSLQVPIAISVTLFEAICLCKGTRVIASRGKEI 293
+ ILK K SI YW+L ++ P+ ++VT+ I L R G E+
Sbjct: 244 FIVILKGGEKMDSIVGIECGTPWYWVLTAIGGPLMLAVTIVVGIFLWWRQRGEEVEG-EV 302
Query: 294 TWKFHKICLYCFCGIIAGMVSGLLGLGGGFILGPLFLELGIPPQVASATSTFAMVFSSSM 353
W + AG+ + LG+GGG ++GP+ LE+G+ PQVA+ATS F ++F++S
Sbjct: 303 QWTVKNCLIIPIGAFFAGVSAAYLGIGGGMVIGPILLEIGVLPQVATATSAFMIMFTASS 362
Query: 354 SVVQYYHLDRFPIPYASYLVLVATIAALTGQHVVRKIIAIFGRASIIVFILAFTIFVSAI 413
S +QY + I Y + + I A GQ KI+ R SII F L I +S +
Sbjct: 363 SSLQYIIDGKLDIFYGIWYFAIGFIGAAFGQFGFSKIVQKLNRQSIIGFFLGVLIVLSTL 422
Query: 414 SLGGVGIGNMVEKMENEEYMGFDNLC 439
++ + + +V ++N+ +GF +LC
Sbjct: 423 AMIAITVVQLVSDVKNDN-LGFKHLC 447
>UniRef100_O65172 Hypothetical protein [Mesembryanthemum crystallinum]
Length = 132
Score = 169 bits (428), Expect = 1e-40
Identities = 84/140 (60%), Positives = 109/140 (77%), Gaps = 10/140 (7%)
Query: 120 LIDYDLALLFQPMLMLGISIGVAFNVMFADWMVTILLIILFIGTSTKALVKGIDTWKKET 179
+IDYDLALLFQPMLMLGISIGVAFNV+FADWMVT+LLI+LF+GTSTKA ++GIDTWKKET
Sbjct: 1 IIDYDLALLFQPMLMLGISIGVAFNVIFADWMVTVLLIVLFVGTSTKAFMRGIDTWKKET 60
Query: 180 IMKKEAFEEAAQMLESGSTPDYASEEDYKSLPA--DLQDEEVPLLDNIHWKELSVLMYVW 237
+M+K EAA+ ES A +Y+ LPA + +D E P+L+N++WKE+ +L +VW
Sbjct: 61 LMQK----EAAKRAESNG----ADGVEYEPLPAGPEKEDREAPILENVYWKEVGLLCFVW 112
Query: 238 VAFLIVQILKTYSKTCSIEY 257
VAFL +I+ + TCS+ Y
Sbjct: 113 VAFLAFEIINENTATCSVAY 132
>UniRef100_O49567 Hypothetical protein F7J7.200 [Arabidopsis thaliana]
Length = 393
Score = 145 bits (366), Expect = 2e-33
Identities = 104/379 (27%), Positives = 182/379 (47%), Gaps = 59/379 (15%)
Query: 42 EMKFGWRIVVGSIVGFFGAALGSVGGVGGGGIFIPMLTLIIGFDPKSSTALSKCMITGAA 101
E+K +VV ++ F A + S G+ D K++++ S M+TG +
Sbjct: 55 ELKLSPALVVAGVLCFTAALISSASGI----------------DLKAASSFSAFMVTGGS 98
Query: 102 GSTVYYNLRLRHPTLDMPLIDYDLALLFQPMLMLGISIGVAFNVMFADWMVTILLIILFI 161
+ + N H LIDYDLALL +P ++LG+S+GV N +F +W++T L ++ +
Sbjct: 99 IANLINN----HFGCKK-LIDYDLALLLEPCMLLGVSVGVICNKVFPEWLITGLFVVFLM 153
Query: 162 GTSTKALVKGIDTWKKETIMKKEAFEEAAQMLESGSTPDYASEEDYKSLPADLQDEEVPL 221
+S + G +WK I++++ +++ E +K L ++ E
Sbjct: 154 WSSMETCENGHTSWKLSLILREKEDMRDSRLAEVKRRRTIIF---FKHLYLKIKKTETK- 209
Query: 222 LDNIHWKELSVLMYVWVAFLIVQILKTYSKTCSIEYWLLNSLQVPIAISVTLFEAICLCK 281
+ + L ++ K CS+EYW+L SLQ+P+A+ T+ A+ +
Sbjct: 210 -QSFLGRNLGIISI---------------KPCSVEYWILLSLQIPLALVFTIL-ALSRTE 252
Query: 282 GTRVIASRGKEITWKFHKICLYCFCGIIAGMVSGLLGLGGGFILGPLFLELGIPPQVASA 341
+ + +E AG++ G+ G+GGG I+ PL L GIPPQV +A
Sbjct: 253 SLQEQSISNQE-----------------AGLLGGIFGIGGGMIISPLLLRAGIPPQVTAA 295
Query: 342 TSTFAMVFSSSMSVVQYYHLDRFPIPYASYLVLVATIAALTGQHVVRKIIAIFGRASIIV 401
T++F + FS++MS VQY L A ++ A+ G +K++ F RASIIV
Sbjct: 296 TTSFMVFFSATMSGVQYLLLGMQNTEAAYVFSVICFFASTLGLVFAQKVVPHFRRASIIV 355
Query: 402 FILAFTIFVSAISLGGVGI 420
F++ ++++ I + GI
Sbjct: 356 FLVGTMMYLTTIVMASFGI 374
>UniRef100_Q6Z000 Hypothetical protein P0473F05.27-3 [Oryza sativa]
Length = 132
Score = 119 bits (299), Expect = 1e-25
Identities = 54/81 (66%), Positives = 68/81 (83%)
Query: 18 EIVVMKKTSFFWYSGESSYERVWPEMKFGWRIVVGSIVGFFGAALGSVGGVGGGGIFIPM 77
E+ M+K F +SGE+SY VWP M+FGW+IV+G ++GFFGAA GSVGGVGGGGIF+PM
Sbjct: 39 EVGYMRKVVNFLWSGEASYHHVWPPMEFGWKIVLGILIGFFGAAFGSVGGVGGGGIFVPM 98
Query: 78 LTLIIGFDPKSSTALSKCMIT 98
LTLIIGFD KSSTA+SK +++
Sbjct: 99 LTLIIGFDAKSSTAISKFIVS 119
>UniRef100_Q7R590 GLP_587_84274_82712 [Giardia lamblia ATCC 50803]
Length = 520
Score = 72.8 bits (177), Expect = 2e-11
Identities = 68/293 (23%), Positives = 120/293 (40%), Gaps = 56/293 (19%)
Query: 47 WRIVVGSIVGFFGAALGSVGGVGGGGIFIPMLTLIIGFDPKSSTALSKCMITGAAGSTVY 106
W V+ +IV A L S GG+GGG IF+ ML L G P + LSK MI G +
Sbjct: 13 WMEVIVAIVCALFAMLASAGGIGGGVIFVSMLQLF-GVSPHVAAPLSKAMIFGGSCVLTC 71
Query: 107 YNLRLRHPTLD--MPLIDYDLALLFQPMLMLGISIGVAFNVMFADWMVTILLIILFIGTS 164
N+ +H + P I +DL + +P + G IG NV+ +W++ +L + + T+
Sbjct: 72 MNI-FQHEDNEPTKPSIIWDLVFIIEPAAVSGALIGALINVVLPEWLLLVLEVAFLLYTT 130
Query: 165 TKALVKGIDTWKKE-------------------TIMKKEAFEEAAQMLESGST-PDYASE 204
K L + T KE +I ++ + + + +E ST +
Sbjct: 131 QKMLRSSLATLNKERIAAGKRLLCTRKSRAPALSIDERGSPHQPSTFIEDQSTRSGNTTS 190
Query: 205 EDYKSLPADLQDEEVPLLDN---------------------------IHWKELS---VLM 234
+ ++ + E PLLD H+K +S ++M
Sbjct: 191 NEIQAYSTECSTEIAPLLDRQELTVKAEPQVQATTKKGRGRLGSCGVAHFKSISAVRMIM 250
Query: 235 YVWVAFLIV--QILKTYSKTCSIEYWLLNSLQVPIAISVTLFEAICLCKGTRV 285
++ LI+ Q++ + CS YW+ + I+I + + + + R+
Sbjct: 251 FILSTVLIMTCQVISQEFERCSTGYWIAFGVCFSISIITIIIIILSIKRSLRI 303
Score = 67.8 bits (164), Expect = 6e-10
Identities = 42/153 (27%), Positives = 69/153 (44%), Gaps = 3/153 (1%)
Query: 290 GKEITWKFHKICLYC---FCGIIAGMVSGLLGLGGGFILGPLFLELGIPPQVASATSTFA 346
G + FH I Y G+ AG++ +LG+GGG + P+ + GI P+ A ST
Sbjct: 368 GTDSLGAFHSILFYVKLVLAGLFAGILGAMLGIGGGLLKNPILISFGIDPERARTASTVM 427
Query: 347 MVFSSSMSVVQYYHLDRFPIPYASYLVLVATIAALTGQHVVRKIIAIFGRASIIVFILAF 406
+ F+S S++ Y + YA L+L ++G ++ II F S I F++
Sbjct: 428 IAFTSMSSMISYVVIGGLHFEYAWPLMLTVGAFFVSGYYLSELIIRCFKTKSFIPFLITA 487
Query: 407 TIFVSAISLGGVGIGNMVEKMENEEYMGFDNLC 439
I V + I ++ + GF +LC
Sbjct: 488 LIVVCTCFIVANMIIVFIDIAKTGHLPGFTSLC 520
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.326 0.142 0.431
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 726,026,560
Number of Sequences: 2790947
Number of extensions: 30162967
Number of successful extensions: 135533
Number of sequences better than 10.0: 962
Number of HSP's better than 10.0 without gapping: 492
Number of HSP's successfully gapped in prelim test: 472
Number of HSP's that attempted gapping in prelim test: 132986
Number of HSP's gapped (non-prelim): 2630
length of query: 447
length of database: 848,049,833
effective HSP length: 131
effective length of query: 316
effective length of database: 482,435,776
effective search space: 152449705216
effective search space used: 152449705216
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 76 (33.9 bits)
Medicago: description of AC147964.5


