

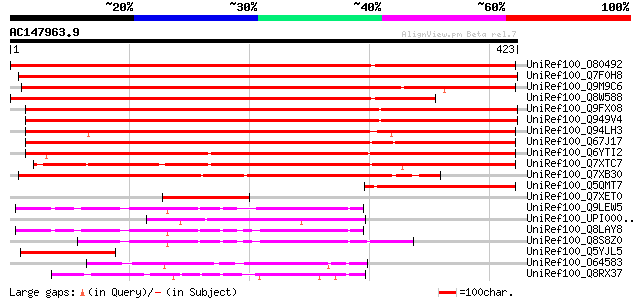
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147963.9 - phase: 0
(423 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_O80492 T12M4.15 protein [Arabidopsis thaliana] 635 0.0
UniRef100_Q7F0H8 Protein phosphatase 2C-like [Oryza sativa] 574 e-162
UniRef100_Q9M9C6 Hypothetical protein T2E12.9 [Arabidopsis thali... 569 e-161
UniRef100_Q8W588 At1g09160/T12M4_13 [Arabidopsis thaliana] 560 e-158
UniRef100_Q9FX08 T3F24.2 protein [Arabidopsis thaliana] 478 e-133
UniRef100_Q949V4 Hypothetical protein At1g47380 [Arabidopsis tha... 477 e-133
UniRef100_Q94LH3 Hypothetical protein OSJNBb0004M10.5 [Oryza sat... 444 e-123
UniRef100_Q67J17 Putative calmodulin-binding protein phosphatase... 437 e-121
UniRef100_Q6YTI2 Putative calmodulin-binding protein phosphatase... 434 e-120
UniRef100_Q7XTC7 OSJNBa0064H22.20 protein [Oryza sativa] 408 e-112
UniRef100_Q7XB30 Calmodulin-binding protein phosphatase [Physcom... 370 e-101
UniRef100_Q5QMT7 Phosphatase-like protein [Oryza sativa] 122 1e-26
UniRef100_Q7XET0 Putative transposase [Oryza sativa] 117 6e-25
UniRef100_Q9LEW5 Protein phosphatase 2C-like protein [Arabidopsi... 116 1e-24
UniRef100_UPI0000291C80 UPI0000291C80 UniRef100 entry 114 7e-24
UniRef100_Q8LAY8 Protein phosphatase 2C-like protein [Arabidopsi... 112 2e-23
UniRef100_Q8S8Z0 Protein phosphatase 2C [Mesembryanthemum crysta... 112 2e-23
UniRef100_Q5YJL5 Protein phosphatase 2C-like protein [Hyacinthus... 112 2e-23
UniRef100_O64583 Hypothetical protein At2g34740 [Arabidopsis tha... 109 1e-22
UniRef100_Q8RX37 Hypothetical protein At1g07160 [Arabidopsis tha... 109 2e-22
>UniRef100_O80492 T12M4.15 protein [Arabidopsis thaliana]
Length = 428
Score = 635 bits (1639), Expect = 0.0
Identities = 317/422 (75%), Positives = 359/422 (84%), Gaps = 2/422 (0%)
Query: 1 MSKAELSRMKQPLVPLATLIGRELRNGKTEKPFVKYGQAGLAKKGEDYFLIKTDCHRVPG 60
MS ++ SR + LVPLATLIGRELR+ K EKPFVKYGQA LAKKGEDYFLIKTDC RVPG
Sbjct: 1 MSVSKASRTQHSLVPLATLIGRELRSEKVEKPFVKYGQAALAKKGEDYFLIKTDCERVPG 60
Query: 61 DSSTSFSVFAILDGHNGISAAIFAKENIINNVMSAIPQGVSREEWLQALPRALVVAFVKT 120
D S++FSVF I DGHNG SAAI+ KE+++ NV+SAIPQG SR+EWLQALPRALV FVKT
Sbjct: 61 DPSSAFSVFGIFDGHNGNSAAIYTKEHLLENVVSAIPQGASRDEWLQALPRALVAGFVKT 120
Query: 121 DMEFQKKGETSGTTATFVIIDGWTVTVASVGDSRCILDTQGGVVSLLTVDHRLEENVEER 180
D+EFQ+KGETSGTT TFVIIDGWT+TVASVGDSRCILDTQGGVVSLLTVDHRLEENVEER
Sbjct: 121 DIEFQQKGETSGTTVTFVIIDGWTITVASVGDSRCILDTQGGVVSLLTVDHRLEENVEER 180
Query: 181 ERVTASGGEVGRLNVFGGNEVGPLRCWPGGLCLSRSIGDTDVGEYIVPIPHVKQVKLSNA 240
ER+TASGGEVGRLNVFGGNEVGPLRCWPGGLCLSRSIGDTDVGE+IVPIPHVKQVKL +A
Sbjct: 181 ERITASGGEVGRLNVFGGNEVGPLRCWPGGLCLSRSIGDTDVGEFIVPIPHVKQVKLPDA 240
Query: 241 GGRLIIASDGIWDTLSSDMAAKSCRGVPAELAAKLVVKEALRSRGLKDDTTCLVVDIIPS 300
GGRLIIASDGIWD LSSD+AAK+CRG+ A+LAAKLVVKEALR++GLKDDTTC+VVDI+PS
Sbjct: 241 GGRLIIASDGIWDILSSDVAAKACRGLSADLAAKLVVKEALRTKGLKDDTTCVVVDIVPS 300
Query: 301 DYPVLPMPATPRKKHNVLSSLLFGKKSQNLANKGTNKLSAVGVVEELFEEGSAMLTERLG 360
+ L + P KK N +S L K + NK NKLSAVGVVEELFEEGSA+L +RLG
Sbjct: 301 GH--LSLAPAPMKKQNPFTSFLSRKNHMDTNNKNGNKLSAVGVVEELFEEGSAVLADRLG 358
Query: 361 NNVPSDTNSGIHRCAVCLADQPSGDGLSVNNDHFITPVSKPWEGPFLCTNCQKKKDAMEG 420
++ S+T +G+ +CAVC D+ + LS N I+ SK WEGPFLCT C+KKKDAMEG
Sbjct: 359 KDLLSNTETGLLKCAVCQIDESPSEDLSSNGGSIISSASKRWEGPFLCTICKKKKDAMEG 418
Query: 421 KR 422
KR
Sbjct: 419 KR 420
>UniRef100_Q7F0H8 Protein phosphatase 2C-like [Oryza sativa]
Length = 431
Score = 574 bits (1479), Expect = e-162
Identities = 280/417 (67%), Positives = 334/417 (79%), Gaps = 1/417 (0%)
Query: 8 RMKQPLVPLATLIGRELRNGKTEKPFVKYGQAGLAKKGEDYFLIKTDCHRVPGDSSTSFS 67
R P +PLATLIGRELR G +E+P ++YG AG AK+GEDYFL+K DC RVPGD+ST+FS
Sbjct: 8 RRLPPALPLATLIGRELRAGGSERPSLRYGHAGFAKRGEDYFLVKPDCLRVPGDTSTAFS 67
Query: 68 VFAILDGHNGISAAIFAKENIINNVMSAIPQGVSREEWLQALPRALVVAFVKTDMEFQKK 127
VFA+ DGHNG+SAA+++KE+++ +VMSA+P + R++WLQALPRALV FVK D++FQ+K
Sbjct: 68 VFAVFDGHNGVSAAVYSKEHLLEHVMSALPPDIGRDDWLQALPRALVAGFVKADIDFQRK 127
Query: 128 GETSGTTATFVIIDGWTVTVASVGDSRCILDTQGGVVSLLTVDHRLEENVEERERVTASG 187
GE SGTTAT V++DG+TVTVASVGDSRCILDTQGG V LLTVDHRLEEN EERERVTASG
Sbjct: 128 GEVSGTTATLVVVDGFTVTVASVGDSRCILDTQGGEVQLLTVDHRLEENAEERERVTASG 187
Query: 188 GEVGRLNVFGGNEVGPLRCWPGGLCLSRSIGDTDVGEYIVPIPHVKQVKLSNAGGRLIIA 247
GEVGRLN+FGG EVGPLRCWPGGLCLSRSIGD DVGE+IVPIPHVKQVKLSN GGRLIIA
Sbjct: 188 GEVGRLNLFGGQEVGPLRCWPGGLCLSRSIGDMDVGEFIVPIPHVKQVKLSNIGGRLIIA 247
Query: 248 SDGIWDTLSSDMAAKSCRGVPAELAAKLVVKEALRSRGLKDDTTCLVVDIIPSDYPVLPM 307
SDGIWD L S+ AAK+CRG+PAELAAKLVVK+AL+ GLKDDTTC+VVDIIPSDY +
Sbjct: 248 SDGIWDALPSEAAAKACRGLPAELAAKLVVKQALKKSGLKDDTTCVVVDIIPSDYRLTSP 307
Query: 308 PATPRKKHNVLSSLLFGKKSQNLANKGTNKLSAVGVVEELFEEGSAMLTERLGNNVPSDT 367
+P++ + SLLFG++S + K K ++ G VEELFEEGSAML ERLG N+
Sbjct: 308 QLSPKRNQSKFKSLLFGRRSHSSIGKLGGKSASFGSVEELFEEGSAMLEERLGRNLSLKA 367
Query: 368 NSGIHRCAVCLADQ-PSGDGLSVNNDHFITPVSKPWEGPFLCTNCQKKKDAMEGKRS 423
S RCA+C DQ P ++ + + PW GP+LC C+KKKDAMEGKRS
Sbjct: 368 TSAPLRCAICQVDQEPFESMMTEKGGSYCSSPCAPWGGPYLCLECRKKKDAMEGKRS 424
>UniRef100_Q9M9C6 Hypothetical protein T2E12.9 [Arabidopsis thaliana]
Length = 436
Score = 569 bits (1467), Expect = e-161
Identities = 284/415 (68%), Positives = 336/415 (80%), Gaps = 4/415 (0%)
Query: 11 QPLVPLATLIGRELRNGKTEKPFVKYGQAGLAKKGEDYFLIKTDCHRVPGDSSTSFSVFA 70
+ LVPLA LI RE + K EKP V++GQA ++KGEDY LIKTD RVP +SST+FSVFA
Sbjct: 16 EKLVPLAALISRETKAAKMEKPIVRFGQAAQSRKGEDYVLIKTDSLRVPSNSSTAFSVFA 75
Query: 71 ILDGHNGISAAIFAKENIINNVMSAIPQGVSREEWLQALPRALVVAFVKTDMEFQKKGET 130
+ DGHNG +AA++ +EN++N+V+SA+P G+SR+EWL ALPRALV FVKTD EFQ +GET
Sbjct: 76 VFDGHNGKAAAVYTRENLLNHVISALPSGLSRDEWLHALPRALVSGFVKTDKEFQSRGET 135
Query: 131 SGTTATFVIIDGWTVTVASVGDSRCILDTQGGVVSLLTVDHRLEENVEERERVTASGGEV 190
SGTTATFVI+DGWTVTVA VGDSRCILDT+GG VS LTVDHRLE+N EERERVTASGGEV
Sbjct: 136 SGTTATFVIVDGWTVTVACVGDSRCILDTKGGSVSNLTVDHRLEDNTEERERVTASGGEV 195
Query: 191 GRLNVFGGNEVGPLRCWPGGLCLSRSIGDTDVGEYIVPIPHVKQVKLSNAGGRLIIASDG 250
GRL++ GG E+GPLRCWPGGLCLSRSIGD DVGE+IVP+P VKQVKLSN GGRLIIASDG
Sbjct: 196 GRLSIVGGVEIGPLRCWPGGLCLSRSIGDMDVGEFIVPVPFVKQVKLSNLGGRLIIASDG 255
Query: 251 IWDTLSSDMAAKSCRGVPAELAAKLVVKEALRSRGLKDDTTCLVVDIIPSDYPVLPMPAT 310
IWD LSS++AAK+CRG+ AELAA+ VVKEALR RGLKDDTTC+VVDIIP + P P+
Sbjct: 256 IWDALSSEVAAKTCRGLSAELAARQVVKEALRRRGLKDDTTCIVVDIIPPENFQEPPPSP 315
Query: 311 PRKKHNVLSSLLFGKKSQNLANKGTNKLSAVGVVEELFEEGSAMLTERLGN---NVPSDT 367
P+K +N SLLF KKS N +NK + KLS VG+VEELFEEGSAML ERLG+ + S T
Sbjct: 316 PKKHNNFFKSLLFRKKS-NSSNKLSKKLSTVGIVEELFEEGSAMLAERLGSGDCSKESTT 374
Query: 368 NSGIHRCAVCLADQPSGDGLSVNNDHFITPVSKPWEGPFLCTNCQKKKDAMEGKR 422
GI CA+C D +G+SV+ + KPW+GPFLCT+C+ KKDAMEGKR
Sbjct: 375 GGGIFTCAICQLDLAPSEGISVHAGSIFSTSLKPWQGPFLCTDCRDKKDAMEGKR 429
>UniRef100_Q8W588 At1g09160/T12M4_13 [Arabidopsis thaliana]
Length = 355
Score = 560 bits (1443), Expect = e-158
Identities = 281/355 (79%), Positives = 312/355 (87%), Gaps = 2/355 (0%)
Query: 1 MSKAELSRMKQPLVPLATLIGRELRNGKTEKPFVKYGQAGLAKKGEDYFLIKTDCHRVPG 60
MS ++ SR + LVPLATLIGRELR+ K EKPFVKYGQA LAKKGEDYFLIKTDC RVPG
Sbjct: 1 MSVSKASRTQHSLVPLATLIGRELRSEKVEKPFVKYGQAALAKKGEDYFLIKTDCERVPG 60
Query: 61 DSSTSFSVFAILDGHNGISAAIFAKENIINNVMSAIPQGVSREEWLQALPRALVVAFVKT 120
D S++FSVF I DGHNG SAAI+ KE+++ NV+SAIPQG SR+EWLQALPRALV FVKT
Sbjct: 61 DPSSAFSVFGIFDGHNGNSAAIYTKEHLLENVVSAIPQGASRDEWLQALPRALVAGFVKT 120
Query: 121 DMEFQKKGETSGTTATFVIIDGWTVTVASVGDSRCILDTQGGVVSLLTVDHRLEENVEER 180
D+EFQ+KGETSGTT TFVIIDGWT+TVASVGDSRCILDTQGGVVSLLTVDHRLEENVEER
Sbjct: 121 DIEFQQKGETSGTTVTFVIIDGWTITVASVGDSRCILDTQGGVVSLLTVDHRLEENVEER 180
Query: 181 ERVTASGGEVGRLNVFGGNEVGPLRCWPGGLCLSRSIGDTDVGEYIVPIPHVKQVKLSNA 240
ER+TASGGEVGRLNVFGGNEVGPLRCWPGGLCLSRSIGDTDVGE+IVPIPHVKQVKL +A
Sbjct: 181 ERITASGGEVGRLNVFGGNEVGPLRCWPGGLCLSRSIGDTDVGEFIVPIPHVKQVKLPDA 240
Query: 241 GGRLIIASDGIWDTLSSDMAAKSCRGVPAELAAKLVVKEALRSRGLKDDTTCLVVDIIPS 300
GGRLIIASDGIWD LSSD+AAK+CRG+ A+LAAKLVVKEALR++GLKDDTTC+VVDI+PS
Sbjct: 241 GGRLIIASDGIWDILSSDVAAKACRGLSADLAAKLVVKEALRTKGLKDDTTCVVVDIVPS 300
Query: 301 DYPVLPMPATPRKKHNVLSSLLFGKKSQNLANKGTNKLSAVGVVEELFEEGSAML 355
+ L + P KK N +S L K + NK NKLSAVGVVEELFEEGSA+L
Sbjct: 301 GH--LSLAPAPMKKQNPFTSFLSRKNHMDTNNKNGNKLSAVGVVEELFEEGSAVL 353
>UniRef100_Q9FX08 T3F24.2 protein [Arabidopsis thaliana]
Length = 428
Score = 478 bits (1231), Expect = e-133
Identities = 232/410 (56%), Positives = 304/410 (73%), Gaps = 1/410 (0%)
Query: 14 VPLATLIGRELRNGKTEKPFVKYGQAGLAKKGEDYFLIKTDCHRVPGDSSTSFSVFAILD 73
VPL+ L+ RE N K + P + +GQ +KKGED+ L+KT+C RV GD T+FSVF + D
Sbjct: 10 VPLSVLLKRESANEKIDNPELIHGQHNQSKKGEDFTLVKTECQRVMGDGVTTFSVFGLFD 69
Query: 74 GHNGISAAIFAKENIINNVMSAIPQGVSREEWLQALPRALVVAFVKTDMEFQKKGETSGT 133
GHNG +AAI+ KEN++NNV++AIP ++R+EW+ ALPRALV FVKTD +FQ++ TSGT
Sbjct: 70 GHNGSAAAIYTKENLLNNVLAAIPSDLNRDEWVAALPRALVAGFVKTDKDFQERARTSGT 129
Query: 134 TATFVIIDGWTVTVASVGDSRCILDTQGGVVSLLTVDHRLEENVEERERVTASGGEVGRL 193
T TFVI++GW V+VASVGDSRCIL+ G V L+ DHRLE N EER+RVTASGGEVGRL
Sbjct: 130 TVTFVIVEGWVVSVASVGDSRCILEPAEGGVYYLSADHRLEINEEERDRVTASGGEVGRL 189
Query: 194 NVFGGNEVGPLRCWPGGLCLSRSIGDTDVGEYIVPIPHVKQVKLSNAGGRLIIASDGIWD 253
N GG E+GPLRCWPGGLCLSRSIGD DVGEYIVP+P+VKQVKLS+AGGRLII+SDG+WD
Sbjct: 190 NTGGGTEIGPLRCWPGGLCLSRSIGDLDVGEYIVPVPYVKQVKLSSAGGRLIISSDGVWD 249
Query: 254 TLSSDMAAKSCRGVPAELAAKLVVKEALRSRGLKDDTTCLVVDIIPSDYPVLPMPATPRK 313
+S++ A CRG+P E +A+ +VKEA+ +G++DDTTC+VVDI+P + P +P P+K
Sbjct: 250 AISAEEALDCCRGLPPESSAEHIVKEAVGKKGIRDDTTCIVVDILPLEKPAASVP-PPKK 308
Query: 314 KHNVLSSLLFGKKSQNLANKGTNKLSAVGVVEELFEEGSAMLTERLGNNVPSDTNSGIHR 373
+ + +F +K+ + ++ + + VVEELFEEGSAML+ERL P +
Sbjct: 309 QGKGMLKSMFKRKTSDSSSNIEKEYAEPDVVEELFEEGSAMLSERLDTKYPLCNMFKLFM 368
Query: 374 CAVCLADQPSGDGLSVNNDHFITPVSKPWEGPFLCTNCQKKKDAMEGKRS 423
CAVC + G+G+S++ +PW+GPFLC +CQ KKDAMEGKRS
Sbjct: 369 CAVCQVEVKPGEGVSIHAGSDNCRKLRPWDGPFLCASCQDKKDAMEGKRS 418
>UniRef100_Q949V4 Hypothetical protein At1g47380 [Arabidopsis thaliana]
Length = 428
Score = 477 bits (1227), Expect = e-133
Identities = 231/410 (56%), Positives = 304/410 (73%), Gaps = 1/410 (0%)
Query: 14 VPLATLIGRELRNGKTEKPFVKYGQAGLAKKGEDYFLIKTDCHRVPGDSSTSFSVFAILD 73
VPL+ L+ RE N K + P + +GQ +KKGED+ L+KT+C RV GD T+FSVF + D
Sbjct: 10 VPLSVLLKRESANEKIDNPELIHGQHNQSKKGEDFTLVKTECQRVMGDGVTTFSVFGLFD 69
Query: 74 GHNGISAAIFAKENIINNVMSAIPQGVSREEWLQALPRALVVAFVKTDMEFQKKGETSGT 133
GHNG +AAI+ KEN++NNV++AIP ++R+EW+ ALPRALV FVKTD +F+++ TSGT
Sbjct: 70 GHNGSAAAIYTKENLLNNVLAAIPSDLNRDEWVAALPRALVAGFVKTDKDFRERARTSGT 129
Query: 134 TATFVIIDGWTVTVASVGDSRCILDTQGGVVSLLTVDHRLEENVEERERVTASGGEVGRL 193
T TFVI++GW V+VASVGDSRCIL+ G V L+ DHRLE N EER+RVTASGGEVGRL
Sbjct: 130 TVTFVIVEGWVVSVASVGDSRCILEPAEGGVYYLSADHRLEINEEERDRVTASGGEVGRL 189
Query: 194 NVFGGNEVGPLRCWPGGLCLSRSIGDTDVGEYIVPIPHVKQVKLSNAGGRLIIASDGIWD 253
N GG E+GPLRCWPGGLCLSRSIGD DVGEYIVP+P+VKQVKLS+AGGRLII+SDG+WD
Sbjct: 190 NTGGGTEIGPLRCWPGGLCLSRSIGDLDVGEYIVPVPYVKQVKLSSAGGRLIISSDGVWD 249
Query: 254 TLSSDMAAKSCRGVPAELAAKLVVKEALRSRGLKDDTTCLVVDIIPSDYPVLPMPATPRK 313
+S++ A CRG+P E +A+ +VKEA+ +G++DDTTC+VVDI+P + P +P P+K
Sbjct: 250 AISAEEALDCCRGLPPESSAEHIVKEAVGKKGIRDDTTCIVVDILPLEKPAASVP-PPKK 308
Query: 314 KHNVLSSLLFGKKSQNLANKGTNKLSAVGVVEELFEEGSAMLTERLGNNVPSDTNSGIHR 373
+ + +F +K+ + ++ + + VVEELFEEGSAML+ERL P +
Sbjct: 309 QGKGMLKSMFKRKTSDSSSNIEKEYAEPDVVEELFEEGSAMLSERLDTKYPLCNMFKLFM 368
Query: 374 CAVCLADQPSGDGLSVNNDHFITPVSKPWEGPFLCTNCQKKKDAMEGKRS 423
CAVC + G+G+S++ +PW+GPFLC +CQ KKDAMEGKRS
Sbjct: 369 CAVCQVEVKPGEGVSIHAGSDNCRKLRPWDGPFLCASCQDKKDAMEGKRS 418
>UniRef100_Q94LH3 Hypothetical protein OSJNBb0004M10.5 [Oryza sativa]
Length = 439
Score = 444 bits (1141), Expect = e-123
Identities = 231/424 (54%), Positives = 289/424 (67%), Gaps = 20/424 (4%)
Query: 14 VPLATLIGRELRNGKTEKPFVKYGQAGLAKKGEDYFLIKTDCHRVPGDSST--------- 64
VPLA L+ REL N K E+P + +G+A +KKGED+ + C R PG +
Sbjct: 12 VPLAVLLKRELCNQKVERPDMLFGEASKSKKGEDFTFLLPKCSRRPGQAQADGEDAGGAG 71
Query: 65 ---SFSVFAILDGHNGISAAIFAKENIINNVMSAIPQGVSREEWLQALPRALVVAFVKTD 121
+ SVFAI DGHNG +AAI+ +EN++NNV++AIP ++ EEW ALPRALV FVKTD
Sbjct: 72 DDDTISVFAIFDGHNGSAAAIYTRENLLNNVLAAIPPNLTSEEWTTALPRALVAGFVKTD 131
Query: 122 MEFQKKGETSGTTATFVIIDGWTVTVASVGDSRCILDTQGGVVSLLTVDHRLEENVEERE 181
EFQ K SGTT TFVIIDGW VTVASVGDSRCIL++ G V L+ DHRL+ N EE E
Sbjct: 132 KEFQTKAARSGTTVTFVIIDGWVVTVASVGDSRCILESAEGSVYFLSADHRLDTNEEEVE 191
Query: 182 RVTASGGEVGRLNVFGGNEVGPLRCWPGGLCLSRSIGDTDVGEYIVPIPHVKQVKLSNAG 241
RVTASGG+VGR+N+ GG +GPLRCWPGGLCLSRSIGD DVGE+IVP+PHVKQVKLSNAG
Sbjct: 192 RVTASGGDVGRINIAGGAGIGPLRCWPGGLCLSRSIGDIDVGEFIVPVPHVKQVKLSNAG 251
Query: 242 GRLIIASDGIWDTLSSDMAAKSCRGVPAELAAKLVVKEALRSRGLKDDTTCLVVDIIPSD 301
GRL+IASDG+WD L A RG+PAE AA +VKE++ S+GL+DDTTC+VVDI+P +
Sbjct: 252 GRLVIASDGVWDALRFQEALNYTRGLPAEAAASRIVKESVSSKGLRDDTTCIVVDILPPE 311
Query: 302 YPVLPMPATPRKKHNV--LSSLLFGKKSQNLANKGTNK-LSAVGVVEELFEEGSAMLTER 358
+ P KKH + +L + S L ++ VVEE++EEGSAML +R
Sbjct: 312 -----KLSPPLKKHGKGGIKALFRRRPSDELTEDQMDRGCLEPDVVEEIYEEGSAMLAQR 366
Query: 359 LGNNVPSDTNSGIHRCAVCLADQPSGDGLSVNNDHFITPVSKPWEGPFLCTNCQKKKDAM 418
L N P+ +H CAVC G+G+SV+ PW GPFLC++CQ KK+AM
Sbjct: 367 LKINYPTGNMFKLHDCAVCQLVMKPGEGISVHGSIPRNSRVDPWGGPFLCSSCQLKKEAM 426
Query: 419 EGKR 422
EGK+
Sbjct: 427 EGKQ 430
>UniRef100_Q67J17 Putative calmodulin-binding protein phosphatase [Oryza sativa]
Length = 422
Score = 437 bits (1124), Expect = e-121
Identities = 219/410 (53%), Positives = 295/410 (71%), Gaps = 3/410 (0%)
Query: 14 VPLATLIGRELRNGKTEKPFVKYGQAGLAKKGEDYFLIKTDC-HRVPGDSSTSFSVFAIL 72
VPL L+ RE+ + + E+P V G+A ++KGED+ L+ + RV GD STSFSVFA+
Sbjct: 10 VPLGVLLRREVTSERMERPDVLCGEAARSRKGEDFTLLLAEAGERVAGDPSTSFSVFALF 69
Query: 73 DGHNGISAAIFAKENIINNVMSAIPQGVSREEWLQALPRALVVAFVKTDMEFQKKGETSG 132
DGHNG AA++AK+N++NN++ AIP G+SR+EWL LPRALV AFVKTD +FQ ETSG
Sbjct: 70 DGHNGSGAAMYAKKNLLNNLLRAIPSGLSRDEWLAVLPRALVAAFVKTDKDFQAVAETSG 129
Query: 133 TTATFVIIDGWTVTVASVGDSRCILDTQGGVVSLLTVDHRLEENVEERERVTASGGEVGR 192
TT TFV+ID W VTVASVGDSRCIL++ G + L+ DHR + N +E +RVTA G +VG+
Sbjct: 130 TTVTFVVIDEWVVTVASVGDSRCILESADGSLYHLSADHRFDSNQDEVQRVTACGSKVGK 189
Query: 193 LNVFGGNEVGPLRCWPGGLCLSRSIGDTDVGEYIVPIPHVKQVKLSNAGGRLIIASDGIW 252
LN+ GG EVGPLRCWPGGLCLSRSIGD DVGE I+P+PHVKQVKLSNAGGR+IIASDG+W
Sbjct: 190 LNLVGGPEVGPLRCWPGGLCLSRSIGDMDVGECIIPVPHVKQVKLSNAGGRIIIASDGVW 249
Query: 253 DTLSSDMAAKSCRGVPAELAAKLVVKEALRSRGLKDDTTCLVVDIIPSDYPVLPMPATPR 312
D L+ +MA + RG P+++AA +V EA+ RGL+DDTTC+VVDI+P + + P P T R
Sbjct: 250 DDLTFEMALECSRGFPSDIAANRIVNEAIHPRGLRDDTTCIVVDILPPE-KLAPSPPTKR 308
Query: 313 KKHNVLSSLLFGKKSQNLANKGTNKLSAVGVVEELFEEGSAMLTERLGNNVPSDTNSGIH 372
+ V ++ +F +K +++ + + VEE+F++GSAML++RL +
Sbjct: 309 QGKIVFNN-MFRRKHTDVSFILDREYAEPDEVEEIFDDGSAMLSKRLAAGYALQSMFEPF 367
Query: 373 RCAVCLADQPSGDGLSVNNDHFITPVSKPWEGPFLCTNCQKKKDAMEGKR 422
CAVC +G G+SV+++ + W+GPFLC +C +KKDA+EGKR
Sbjct: 368 SCAVCQVQLKAGQGISVHSNPLQHEKLQGWQGPFLCQSCNEKKDAIEGKR 417
>UniRef100_Q6YTI2 Putative calmodulin-binding protein phosphatase [Oryza sativa]
Length = 442
Score = 434 bits (1115), Expect = e-120
Identities = 218/412 (52%), Positives = 296/412 (70%), Gaps = 5/412 (1%)
Query: 14 VPLATLIGRELRNGKT--EKPFVKYGQAGLAKKGEDYFLIKTDCHRVPGDSSTSFSVFAI 71
VPLA L+ RE+ + KT E+P ++ G AKKGEDY +K DC R+PG S+SFS F +
Sbjct: 19 VPLAVLLRREVVSEKTAAERPELQVGLFSQAKKGEDYTFLKPDCERLPGVPSSSFSAFGL 78
Query: 72 LDGHNGISAAIFAKENIINNVMSAIPQGVSREEWLQALPRALVVAFVKTDMEFQKKGETS 131
DGHNG AAI+ KEN+++N+++AIP ++RE+WL ALPRA+V AFVKTD +FQ K +S
Sbjct: 79 FDGHNGNGAAIYTKENLLSNILTAIPADLNREDWLAALPRAMVAAFVKTDKDFQTKARSS 138
Query: 132 GTTATFVIIDGWTVTVASVGDSRCILDTQGGVVSLLTVDHRLEENVEERERVTASGGEVG 191
GTT TFVIIDG +TVASVGDSRC+L+ +G + L + DHR + + EE +RVT SGG+VG
Sbjct: 139 GTTVTFVIIDGLFITVASVGDSRCVLEAEGSIYHL-SADHRFDASKEEVDRVTESGGDVG 197
Query: 192 RLNVFGGNEVGPLRCWPGGLCLSRSIGDTDVGEYIVPIPHVKQVKLSNAGGRLIIASDGI 251
RLNV GG E+GPLRCWPGGLCLSRSIGD DVG++IVP+P+VKQVKLS AGGRLII+SDG+
Sbjct: 198 RLNVVGGAEIGPLRCWPGGLCLSRSIGDQDVGQFIVPVPYVKQVKLSTAGGRLIISSDGV 257
Query: 252 WDTLSSDMAAKSCRGVPAELAAKLVVKEALRSRGLKDDTTCLVVDIIPSDYPVLPMPATP 311
WD L++++A R +P E AA+ +VKEA++ +GL+DDTTC+VVDI+P D L MP T
Sbjct: 258 WDVLTAEVAFNCSRTLPPEAAAEQIVKEAVQQKGLRDDTTCIVVDILP-DKANLTMPHTK 316
Query: 312 RKKHNVLSSLLFGKKSQNLANKGTNK-LSAVGVVEELFEEGSAMLTERLGNNVPSDTNSG 370
++ + +F KK+ + ++ T++ +VEE+FE+G A L++RL + P
Sbjct: 317 KQPGMGVFKNMFRKKTPSDSSSHTDREYMDPDIVEEIFEDGCAFLSKRLDSEYPVRNMFK 376
Query: 371 IHRCAVCLADQPSGDGLSVNNDHFITPVSKPWEGPFLCTNCQKKKDAMEGKR 422
+ CA+C + G+SV+ D + W+GPFLC CQ+KK+AMEGKR
Sbjct: 377 LFICAICQVELKPSQGISVHEDSSQPGNLRRWDGPFLCQGCQEKKEAMEGKR 428
>UniRef100_Q7XTC7 OSJNBa0064H22.20 protein [Oryza sativa]
Length = 457
Score = 408 bits (1049), Expect = e-112
Identities = 211/404 (52%), Positives = 278/404 (68%), Gaps = 13/404 (3%)
Query: 21 GRELRNGKTEKPFVKYGQAGLAKKGEDYFLIKTDCHRVPGDSSTSFSVFAILDGHNGISA 80
GRE + +P V GQA AKKGED+ L+K C R+P + FS FA+ DGHNG A
Sbjct: 51 GRE----RERRPSVAAGQACRAKKGEDFALLKPACERLPAGGAP-FSAFALFDGHNGSGA 105
Query: 81 AIFAKENIINNVMSAIPQGVSREEWLQALPRALVVAFVKTDMEFQKKGETSGTTATFVII 140
A++AKENI++NVM +P +S +EWL ALPRALV FVKTD +FQ T GTT TFVII
Sbjct: 106 AVYAKENILSNVMCCVPADLSGDEWLAALPRALVAGFVKTDKDFQ----TRGTTVTFVII 161
Query: 141 DGWTVTVASVGDSRCILDTQGGVVSLLTVDHRLEENVEERERVTASGGEVGRLNVFGGNE 200
DG+ VTVASVGDSRC+L+ +G + L + DHR + + EE RVT GGEVGRLNV GG E
Sbjct: 162 DGYVVTVASVGDSRCVLEAEGTIYHL-SADHRFDASEEEVGRVTECGGEVGRLNVVGGAE 220
Query: 201 VGPLRCWPGGLCLSRSIGDTDVGEYIVPIPHVKQVKLSNAGGRLIIASDGIWDTLSSDMA 260
+GPLRCWPGGLCLSRSIGD DVGE+I+P+P+VKQ+KLS+AGGR+II+SDG+WD L+ D A
Sbjct: 221 IGPLRCWPGGLCLSRSIGDQDVGEFIIPVPYVKQIKLSSAGGRIIISSDGVWDALTVDTA 280
Query: 261 AKSCRGVPAELAAKLVVKEALRSRGLKDDTTCLVVDIIPSDYPVLPMPATPRKKHNVLSS 320
RG+P E AA +VKEA+ S+GL+DDTTC+V+DIIP + + P +K L
Sbjct: 281 FSCARGLPPEAAADQIVKEAIASKGLRDDTTCIVIDIIPPE-KISPTVQPAKKAGKGLFK 339
Query: 321 LLFGKK--SQNLANKGTNKLSAVGVVEELFEEGSAMLTERLGNNVPSDTNSGIHRCAVCL 378
+F KK S + + ++ + +VEE+FE+G L+ RL + P + CA+C
Sbjct: 340 NIFYKKATSDSPCHADKDQCTQPDLVEEVFEDGCPSLSRRLDSEYPVRNMFKLFICAICQ 399
Query: 379 ADQPSGDGLSVNNDHFITPVSKPWEGPFLCTNCQKKKDAMEGKR 422
+ SG G+S++ + +PW+GPFLC +CQ+KK+AMEGKR
Sbjct: 400 VELESGQGISIHEGLSKSGKLRPWDGPFLCHSCQEKKEAMEGKR 443
>UniRef100_Q7XB30 Calmodulin-binding protein phosphatase [Physcomitrella patens]
Length = 373
Score = 370 bits (950), Expect = e-101
Identities = 191/354 (53%), Positives = 253/354 (70%), Gaps = 12/354 (3%)
Query: 8 RMKQPLVPLATLIGRELRNG-KTEKPFVKYGQAGLAKKGEDYFLIKTDCHRVPGDSSTSF 66
R LV L++L + + K KP V YGQA AKK ED+ ++KT+C R+PGD S+++
Sbjct: 26 RRAHHLVTLSSLFNKGTQPTIKANKPSVSYGQALQAKKEEDFVIVKTNCQRIPGDGSSTY 85
Query: 67 SVFAILDGHNGISAAIFAKENIINNVMSAIPQGVSREEWLQALPRALVVAFVKTDMEFQK 126
FA+LDGHNG +A F KEN++NNV++A+P G+SR+EWL ALPRA+V FVKTD +++
Sbjct: 86 DAFAVLDGHNGSAAGAFTKENLLNNVVNAVPSGLSRDEWLAALPRAMVAGFVKTDKDWRS 145
Query: 127 KGETSGTTATFVIIDGWTVTVASVGDSRCILDTQGGVVSLLTVDHRLEENVEERERVTAS 186
KG SGTT T VI+DG+ VT A VGDSRC+LD Q G+V+ LT+DHR + N EE ERV AS
Sbjct: 146 KGIFSGTTVTLVIVDGYIVTCACVGDSRCVLDAQ-GIVTALTIDHRFDTNEEECERVVAS 204
Query: 187 GGEVGRLNVFGGNEVGPLRCWPGGLCLSRSIGDTDVGEYIVPIPHVKQVKLSNAGGRLII 246
GG+V RL+ + G ++GPLR WPGG+CLSR+IGD D GE+IVP+PHVKQ+KLS GGRLII
Sbjct: 205 GGQVARLHPY-GCDIGPLRSWPGGICLSRTIGDVDAGEHIVPVPHVKQIKLSPGGGRLII 263
Query: 247 ASDGIWDTLSSDMAAKSCRGVP-AELAAKLVVKEALRSRGLKDDTTCLVVDIIPSDYPVL 305
ASDG+WD L+S+ AAK CRG E+AAK +VKE ++ +GL+DDTTCLVVDI+P
Sbjct: 264 ASDGVWDALTSEKAAKVCRGEKHPEVAAKNIVKEVIKVQGLRDDTTCLVVDIVPPGRASR 323
Query: 306 PMPATPRKKHNVLSSLLFGKKSQNLANKGTNKLSAVGVVEELFEEGSAMLTERL 359
RK+ V+ F +++++ G+ +EELFEE SA L+ERL
Sbjct: 324 SFAPAVRKQMMVMR---FLRRTRSKKGSGSG-----APMEELFEESSATLSERL 369
>UniRef100_Q5QMT7 Phosphatase-like protein [Oryza sativa]
Length = 133
Score = 122 bits (307), Expect = 1e-26
Identities = 67/128 (52%), Positives = 88/128 (68%), Gaps = 4/128 (3%)
Query: 297 IIPSDYPVLPMPATPRKKHNVLSSLLFGKKSQNLANKGTNKLSAVGVVEELFEEGSAMLT 356
+IP D + +P KK N L SL+F KK+++ NK T +LSA G+VEELFEEGSAML+
Sbjct: 1 MIPPDQTI--RHPSPPKKINKLKSLIFRKKTKDHPNKLTKQLSAAGMVEELFEEGSAMLS 58
Query: 357 ERLGNNVPS-DTNSGIHRCAVCLADQPSGDGLSVN-NDHFITPVSKPWEGPFLCTNCQKK 414
ERLGN+ T+S + CA+C D +G+SV+ F + SKPWEGPFLC++C+ K
Sbjct: 59 ERLGNDSSGRRTSSSLFTCAICQVDLEPSEGISVHAGSIFSSSSSKPWEGPFLCSDCRDK 118
Query: 415 KDAMEGKR 422
KDAMEGKR
Sbjct: 119 KDAMEGKR 126
>UniRef100_Q7XET0 Putative transposase [Oryza sativa]
Length = 205
Score = 117 bits (293), Expect = 6e-25
Identities = 58/73 (79%), Positives = 65/73 (88%)
Query: 128 GETSGTTATFVIIDGWTVTVASVGDSRCILDTQGGVVSLLTVDHRLEENVEERERVTASG 187
GE SGTTAT V+I+G+TVTV SVGDSRCILDTQGG V LLTVD+ LE+N EERERV+ASG
Sbjct: 5 GEVSGTTATLVVINGFTVTVESVGDSRCILDTQGGEVQLLTVDNCLEKNAEERERVSASG 64
Query: 188 GEVGRLNVFGGNE 200
GEVGRLN+FGG E
Sbjct: 65 GEVGRLNLFGGQE 77
>UniRef100_Q9LEW5 Protein phosphatase 2C-like protein [Arabidopsis thaliana]
Length = 348
Score = 116 bits (290), Expect = 1e-24
Identities = 90/296 (30%), Positives = 150/296 (50%), Gaps = 38/296 (12%)
Query: 6 LSRMKQPLVPLATLIGREL-RNGKTEKPFVKYGQAGLAKKGEDYFLIKTDCHRVPGDSST 64
LS QP A G L +NGK + AG ED+F + D G +
Sbjct: 8 LSYSNQPQTVEAPASGGGLSQNGKFSYGYAS--SAGKRSSMEDFFETRID-----GINGE 60
Query: 65 SFSVFAILDGHNGISAAIFAKENIINNVMSAIPQGVSREEWLQALPRALVVAFVKTDMEF 124
+F + DGH G AA + K ++ +N+ ++ +++ A+ A+ TD E
Sbjct: 61 IVGLFGVFDGHGGARAAEYVKRHLFSNL-------ITHPKFISDTKSAITDAYNHTDSEL 113
Query: 125 QKKGET----SGTTATFVIIDGWTVTVASVGDSRCILDTQGGVVSLLTVDHRLEENVEER 180
K + +G+TA+ I+ G + VA+VGDSR ++ ++GG ++ DH+ +++ +ER
Sbjct: 114 LKSENSHNRDAGSTASTAILVGDRLVVANVGDSRAVI-SRGGKAIAVSRDHKPDQS-DER 171
Query: 181 ERVTASGGEVGRLNVFGGNEVGPLRCWPGGLCLSRSIGDTDVGEYIVPIPHVKQVKLSNA 240
ER+ +GG V W G L +SR+ GD + +Y+V P +++ K+ +
Sbjct: 172 ERIENAGGFV---------------MWAGVLAVSRAFGDRLLKQYVVADPEIQEEKIDDT 216
Query: 241 GGRLIIASDGIWDTLSSDMAAKSCRGV-PAELAAKLVVKEALRSRGLKDDTTCLVV 295
LI+ASDG+WD S++ A + V E +AK +V EA++ RG D+ TC+VV
Sbjct: 217 LEFLILASDGLWDVFSNEAAVAMVKEVEDPEDSAKKLVGEAIK-RGSADNITCVVV 271
>UniRef100_UPI0000291C80 UPI0000291C80 UniRef100 entry
Length = 253
Score = 114 bits (284), Expect = 7e-24
Identities = 71/200 (35%), Positives = 106/200 (52%), Gaps = 18/200 (9%)
Query: 115 VAFVKTDMEFQKKGETSGTTATFVIID--GWTVTVASVGDSRCILDTQGGVVSLLTVDHR 172
+AF + E ++ +GTTAT +IID A+VGDS ++ +++T HR
Sbjct: 3 IAFDRCHTEIRENSPKAGTTATVLIIDLINSYCLCANVGDSWALVIDHSDS-AIITTSHR 61
Query: 173 LEENVEERERVTASGGEVGRLNVFGGNEVGPLRCWPGGLCLSRSIGDTDVGEYIVPIPHV 232
L +N EE RV A+G + R G +G LR PGGL LSRS+GD D GE ++ P +
Sbjct: 62 LGDNEEECTRVVAAGSRLARAADISGAPIGALRMVPGGLALSRSLGDADCGEALLSEPAI 121
Query: 233 KQVKLSNAGG--------------RLIIASDGIWDTLSSDMAAKSCRGVP-AELAAKLVV 277
+ V++ AGG L++ASDG+WD++ RG E A +V
Sbjct: 122 EVVEMYCAGGGGEGGGNGGGGGGLTLVVASDGVWDSMRVPEVVSLARGTSNPEAIASRIV 181
Query: 278 KEALRSRGLKDDTTCLVVDI 297
K A+ RGL+DDT+ +V+ +
Sbjct: 182 KGAIDKRGLRDDTSVIVIQL 201
>UniRef100_Q8LAY8 Protein phosphatase 2C-like protein [Arabidopsis thaliana]
Length = 354
Score = 112 bits (281), Expect = 2e-23
Identities = 92/297 (30%), Positives = 154/297 (50%), Gaps = 34/297 (11%)
Query: 6 LSRMKQPLVPLATLIGREL-RNGKTEKPFVKYGQAGLAKKGEDYFLIKTDCHRVPGDSST 64
LS QP A G L +NGK + AG ED+F + D G +
Sbjct: 8 LSYSNQPQTVEAPASGGGLSQNGKFSYGYAS--SAGKRSSMEDFFETRID-----GINGE 60
Query: 65 SFSVFAILDGHNGISAAIFAKENIINNVMSAIPQGVSREEWLQALPRALVVAFVKTDMEF 124
+F + DGH G AA + K ++ +N+ ++ +++ A+ A+ TD E
Sbjct: 61 IVGLFGVFDGHGGARAAEYVKRHLFSNL-------ITHPKFISDTKSAITDAYNHTDSEL 113
Query: 125 QKKGET----SGTTATFVIIDGWTVTVASVGDSRCILDTQGGVVSLLTVDHRLEENVEER 180
K + +G+TA+ I+ G + VA+VGDSR ++ ++GG ++ DH+ +++ +ER
Sbjct: 114 LKSENSHNRDAGSTASTAILVGDRLVVANVGDSRAVI-SRGGKAIAVSRDHKPDQS-DER 171
Query: 181 ERVTASGGEVGRLNVFGGN-EVGPLRCWPGGLCLSRSIGDTDVGEYIVPIPHVKQVKLSN 239
ER+ +GG V ++ G VG G L +SR+ GD + +Y+V P +++ K+ +
Sbjct: 172 ERIENAGGFV----MWAGTWRVG------GVLAVSRAFGDRLLKQYVVADPEIQEEKIDD 221
Query: 240 AGGRLIIASDGIWDTLSSDMAAKSCRGV-PAELAAKLVVKEALRSRGLKDDTTCLVV 295
LI+ASDG+WD S++ A + V E +AK +V EA++ RG D+ TC+VV
Sbjct: 222 TLEFLILASDGLWDVFSNEAAVAMVKEVEDPEDSAKKLVGEAIK-RGSADNITCVVV 277
>UniRef100_Q8S8Z0 Protein phosphatase 2C [Mesembryanthemum crystallinum]
Length = 319
Score = 112 bits (281), Expect = 2e-23
Identities = 84/287 (29%), Positives = 150/287 (51%), Gaps = 28/287 (9%)
Query: 57 RVPGDSSTSFSVFAILDGHNGISAAIFAKENIINNVMSAIPQGVSREEWLQALPRALVVA 116
R+ G +F + DGH G AA + K+N+ +N+ + +++ A+ A
Sbjct: 53 RIDGVEGEVVGLFGVFDGHGGARAAEYVKQNLFSNL-------IKHPKFISDTKSAIAEA 105
Query: 117 FVKTDMEFQKKGET----SGTTATFVIIDGWTVTVASVGDSRCILDTQGGVVSLLTVDHR 172
+ TD EF K T +G+TA+ I+ G + VA+VGDSR ++ +GG ++ DH+
Sbjct: 106 YTHTDSEFLKSENTQNRDAGSTASTAILVGDRLLVANVGDSRAVI-CRGGEAIAVSRDHK 164
Query: 173 LEENVEERERVTASGGEVGRLNVFGGN-EVGPLRCWPGGLCLSRSIGDTDVGEYIVPIPH 231
+++ +ER+R+ +GG V ++ G VG G L +SR+ GD + +Y+V P
Sbjct: 165 PDQS-DERQRIEDAGGFV----MWAGTWRVG------GVLAVSRAFGDKLLKQYVVADPE 213
Query: 232 VKQVKLSNAGGRLIIASDGIWDTLSSDMAAKSCRGV-PAELAAKLVVKEALRSRGLKDDT 290
+++ + ++ LI+ASDG+WD ++++ A + + E AAK +++EA + RG D+
Sbjct: 214 IQEEVVDSSLEFLILASDGLWDVVTNEEAVTMVKPIQDTEEAAKKLMQEAYQ-RGSADNI 272
Query: 291 TCLVVDIIPSDYPVLPMPATPRKKHNVLSSLLFGKKSQNLANKGTNK 337
TC+VV + D P+ H SS + G + +N+ K
Sbjct: 273 TCVVVRFL--DNPIDSSSNRISGNHGDTSSRVLGGSHTDSSNRFLGK 317
>UniRef100_Q5YJL5 Protein phosphatase 2C-like protein [Hyacinthus orientalis]
Length = 118
Score = 112 bits (280), Expect = 2e-23
Identities = 51/79 (64%), Positives = 67/79 (84%)
Query: 10 KQPLVPLATLIGRELRNGKTEKPFVKYGQAGLAKKGEDYFLIKTDCHRVPGDSSTSFSVF 69
+ PLVPLATLIGRELR ++E+P +++G AGLAK+GED FLIK DC R+ G+ ST+FSV+
Sbjct: 12 RPPLVPLATLIGRELRGERSERPSLRHGHAGLAKRGEDLFLIKPDCLRIHGNPSTAFSVY 71
Query: 70 AILDGHNGISAAIFAKENI 88
AI DGHNG+SAA+F KE++
Sbjct: 72 AIFDGHNGVSAAVFTKEHL 90
>UniRef100_O64583 Hypothetical protein At2g34740 [Arabidopsis thaliana]
Length = 239
Score = 109 bits (273), Expect = 1e-22
Identities = 79/239 (33%), Positives = 133/239 (55%), Gaps = 24/239 (10%)
Query: 65 SFSVFAILDGHNGISAAIFAKENIINNVMSAIPQGVSREEWLQALPRALVVAFVKTDMEF 124
+ ++AI DGH+G A + + ++ +N++S + ++ + +A+ A+ TD
Sbjct: 16 NLGLYAIFDGHSGSDVADYLQNHLFDNILS-------QPDFWRNPKKAIKRAYKSTDDYI 68
Query: 125 QKK--GETSGTTA-TFVIIDGWTVTVASVGDSRCILDTQGGVVSLLTVDHRLEENVEERE 181
+ G G+TA T ++IDG + VA+VGDSR IL + VV +TVDH E +ER+
Sbjct: 69 LQNVVGPRGGSTAVTAIVIDGKKIVVANVGDSRAILCRESDVVKQITVDH---EPDKERD 125
Query: 182 RVTASGGEVGRLNVFGGNEVGPLRCWPGGLCLSRSIGDTDVGEYIVPIPHVKQVKLSNAG 241
V + GG F + G + G L ++R+ GD + E+I IP+++ ++ +
Sbjct: 126 LVKSKGG-------FVSQKPGNVPRVDGQLAMTRAFGDGGLKEHISVIPNIEIAEIHDDT 178
Query: 242 GRLIIASDGIWDTLSSDMAAKSC--RGVPAELAAKLVVKEALRSRGLKDDTTCLVVDII 298
LI+ASDG+W +S+D RG AE AAK+++ +AL +RG KDD +C+VV +
Sbjct: 179 KFLILASDGLWKVMSNDEVWDQIKKRG-NAEEAAKMLIDKAL-ARGSKDDISCVVVSFL 235
>UniRef100_Q8RX37 Hypothetical protein At1g07160 [Arabidopsis thaliana]
Length = 380
Score = 109 bits (272), Expect = 2e-22
Identities = 85/274 (31%), Positives = 138/274 (50%), Gaps = 35/274 (12%)
Query: 36 YGQAGLAKKGEDYFLIKTDCHRVPGDSSTSFSVFAILDGHNGISAAIFAKENIINNVMSA 95
Y + G + ED F T+ P + +F + DGH G +AA FA +N+ +N++
Sbjct: 126 YCKRGKREAMEDRFSAITNLQGDPKQA-----IFGVYDGHGGPTAAEFAAKNLCSNILGE 180
Query: 96 IPQGVSREEWLQALPRALVVAFVKTDMEFQKKGETSGTTA--TFVIIDGWTVTVASVGDS 153
I G + + +A+ R ++ TD EF K+ G + T +I DG + VA+ GD
Sbjct: 181 IVGGRNESKIEEAVKRG----YLATDSEFLKEKNVKGGSCCVTALISDG-NLVVANAGDC 235
Query: 154 RCILDTQGGVVSLLTVDHRLEENVEERERVTASGGEVGRLNVFGGNEVGPLRCW--PGGL 211
R +L GG LT DHR + +ER R+ +SGG V N W G L
Sbjct: 236 RAVLSV-GGFAEALTSDHRPSRD-DERNRIESSGGYVDTFN----------SVWRIQGSL 283
Query: 212 CLSRSIGDTDVGEYIVPIPHVKQVKLSNAGGRLIIASDGIWDTLSS----DMAAKSCRGV 267
+SR IGD + ++I+ P + ++++ LI+ASDG+WD +S+ D+A C+G
Sbjct: 284 AVSRGIGDAHLKQWIISEPEINILRINPQHEFLILASDGLWDKVSNQEAVDIARPFCKGT 343
Query: 268 PAE----LAAKLVVKEALRSRGLKDDTTCLVVDI 297
+ LA K +V ++ SRG DD + +++ +
Sbjct: 344 DQKRKPLLACKKLVDLSV-SRGSLDDISVMLIQL 376
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.316 0.135 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 708,433,135
Number of Sequences: 2790947
Number of extensions: 30476332
Number of successful extensions: 71324
Number of sequences better than 10.0: 682
Number of HSP's better than 10.0 without gapping: 256
Number of HSP's successfully gapped in prelim test: 426
Number of HSP's that attempted gapping in prelim test: 70071
Number of HSP's gapped (non-prelim): 846
length of query: 423
length of database: 848,049,833
effective HSP length: 130
effective length of query: 293
effective length of database: 485,226,723
effective search space: 142171429839
effective search space used: 142171429839
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 76 (33.9 bits)
Medicago: description of AC147963.9


