

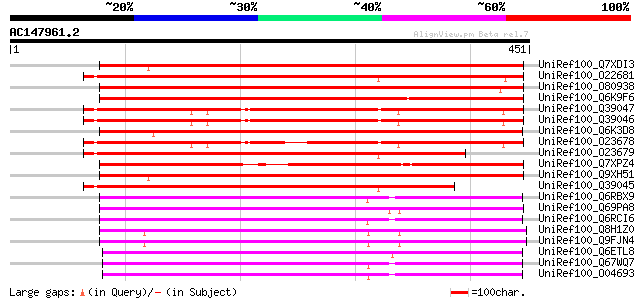
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147961.2 - phase: 0 /pseudo
(451 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q7XDI3 Putative CER1 [Oryza sativa] 439 e-122
UniRef100_O22681 Arabidopsis thaliana gl1 homolog [Arabidopsis t... 419 e-116
UniRef100_O80938 CER1-like protein [Arabidopsis thaliana] 403 e-111
UniRef100_Q6K9F6 Putative CER1 protein [Oryza sativa] 396 e-109
UniRef100_Q39047 CER1-like protein [Arabidopsis thaliana] 391 e-107
UniRef100_Q39046 CER1-like protein [Arabidopsis thaliana] 384 e-105
UniRef100_Q6K3D8 Putative CER1 [Oryza sativa] 371 e-101
UniRef100_O23678 CER1-like protein [Arabidopsis thaliana] 366 e-100
UniRef100_O23679 CER1 protein [Arabidopsis thaliana] 346 7e-94
UniRef100_Q7XPZ4 OSJNBa0004N05.14 protein [Oryza sativa] 340 4e-92
UniRef100_Q9XH51 CER1 [Oryza sativa] 340 6e-92
UniRef100_Q39045 Possible aldehyde decarbonylase [Arabidopsis th... 334 3e-90
UniRef100_Q6RBX9 Glossy1 protein [Zea mays] 210 6e-53
UniRef100_Q69PA8 Putative Gl1 protein [Oryza sativa] 210 6e-53
UniRef100_Q6RCI6 Gl1 protein [Zea mays] 209 1e-52
UniRef100_Q8H1Z0 Cuticle protein [Arabidopsis thaliana] 204 3e-51
UniRef100_Q9FJN4 Lipid transfer protein; glossy1 homolog [Arabid... 204 3e-51
UniRef100_Q6ETL8 Putative glossy1 protein [Oryza sativa] 201 4e-50
UniRef100_Q67WQ7 Putative Gl1 [Oryza sativa] 196 9e-49
UniRef100_O04693 Glossy1 homolog [Oryza sativa] 195 3e-48
>UniRef100_Q7XDI3 Putative CER1 [Oryza sativa]
Length = 621
Score = 439 bits (1130), Expect = e-122
Identities = 208/372 (55%), Positives = 266/372 (70%), Gaps = 4/372 (1%)
Query: 79 FHSLHHTKFRRNYSLFMPMYDYIYGTVDKSTDVIYETSLMR--PKESPDVVHLTHLTTFN 136
FHSLHHT+FR NYSLFMP YDYIY T+DKS+D +YE SL +E+ DVVHLTHLTT +
Sbjct: 247 FHSLHHTQFRTNYSLFMPFYDYIYNTMDKSSDTLYENSLKNNEEEEAVDVVHLTHLTTLH 306
Query: 137 SIYQLRLGFASLASNPQTSKWYLHLMWPFTMFSMLMTWICGRAFVLESNSFKNLKLQCWL 196
SIY +R GFA AS P S+WY+ +MWP + SM++TW G +F +E N K +++Q W
Sbjct: 307 SIYHMRPGFAEFASRPYVSRWYMRMMWPLSWLSMVLTWTYGSSFTVERNVMKKIRMQSWA 366
Query: 197 IPRFKRQYFSKWQSKTFNNLIEEAIVEAELNGAKVISLGLFNKNHQLNERHEHYIGRLPQ 256
IPR+ Y W+ + N+LIE+A+ EA+ NGAKV+SLGL N+ H LN+ E Y+ + P+
Sbjct: 367 IPRYSFHYGLDWEKEAINDLIEKAVCEADKNGAKVVSLGLLNQAHTLNKSGEQYLLKYPK 426
Query: 257 LKIKVVDGSSLAAATVLNNIPKGTNQVLLRGKFNKVAFVIANALCKKNVQVVVLYKDELK 316
L ++VDG+SLAAA V+N+IP+GT+QV+L G +KVA +A ALCKKN++V + K +
Sbjct: 427 LGARIVDGTSLAAAVVVNSIPQGTDQVILAGNVSKVARAVAQALCKKNIKVTMTNKQDYH 486
Query: 317 ELEQRI-NTSKGNLALSPFNTPKIWLVGDEWDEYEQMEAPKGSLFIPFSHFPPKKMRKD- 374
L+ I T NL+ S T K+WL+GD D EQ A KG+LFIP+S FPPK +RKD
Sbjct: 487 LLKPEIPETVADNLSFSKTGTAKVWLIGDGLDSAEQFRAQKGTLFIPYSQFPPKMVRKDS 546
Query: 375 CFYHYTPAMITPTTFMNSHSCENWLPRRVMSAWRIAGIIHALEGWNVHECGDTILSTEKV 434
C Y TPAM P T N HSCENWLPRRVMSAWRIAGI+HALEGWN HECGD +L +KV
Sbjct: 547 CSYSTTPAMAVPKTLQNVHSCENWLPRRVMSAWRIAGILHALEGWNEHECGDKVLDMDKV 606
Query: 435 WEASIRHGFQPL 446
W A+I HGF P+
Sbjct: 607 WSAAIMHGFCPV 618
>UniRef100_O22681 Arabidopsis thaliana gl1 homolog [Arabidopsis thaliana]
Length = 625
Score = 419 bits (1077), Expect = e-116
Identities = 203/388 (52%), Positives = 263/388 (67%), Gaps = 8/388 (2%)
Query: 65 FSPSFFNCH*ACHLFHSLHHTKFRRNYSLFMPMYDYIYGTVDKSTDVIYETSLMRPKESP 124
F P F C+ +HSLHHT+FR NYSLFMP+YDYIYGT+D+STD +YE +L R +
Sbjct: 235 FPPLKFLCY--TPSYHSLHHTQFRTNYSLFMPLYDYIYGTMDESTDTLYEKTLERGDDRV 292
Query: 125 DVVHLTHLTTFNSIYQLRLGFASLASNPQTSKWYLHLMWPFTMFSMLMTWICGRAFVLES 184
DVVHLTHLTT SIY LR+G S AS P +W++ L+WPFT SM+ T R FV E
Sbjct: 293 DVVHLTHLTTPESIYHLRIGLPSFASYPFAYRWFMRLLWPFTSLSMIFTLFYARLFVAER 352
Query: 185 NSFKNLKLQCWLIPRFKRQYFSKWQSKTFNNLIEEAIVEAELNGAKVISLGLFNKNHQLN 244
NSF L LQ W+IPR+ QY KW+ + NN+IE+AI+EA+ G KV+SLGL N+ +LN
Sbjct: 353 NSFNKLNLQSWVIPRYNLQYLLKWRKEAINNMIEKAILEADKKGVKVLSLGLMNQGEELN 412
Query: 245 ERHEHYIGRLPQLKIKVVDGSSLAAATVLNNIPKGTNQVLLRGKFNKVAFVIANALCKKN 304
E YI P +K+++VDGS LAAA V+N++PK T V++ G KVA+ IA+ALC++
Sbjct: 413 RNGEVYIHNHPDMKVRLVDGSRLAAAVVINSVPKATTSVVMTGNLTKVAYTIASALCQRG 472
Query: 305 VQVVVLYKDELKELE----QRINTSKGNLALSPFNTPKIWLVGDEWDEYEQMEAPKGSLF 360
VQV L DE +++ Q L ++ K+WLVG+ EQ +A KG+LF
Sbjct: 473 VQVSTLRLDEYEKIRSCVPQECRDHLVYLTSEALSSNKVWLVGEGTTREEQEKATKGTLF 532
Query: 361 IPFSHFPPKKMRKDCFYHYTPAMITPTTFMNSHSCENWLPRRVMSAWRIAGIIHALEGWN 420
IPFS FP K++R DC YH TPA+I P + +N HSCENWLPR+ MSA R+AGI+HALEGW
Sbjct: 533 IPFSQFPLKQLRSDCIYHTTPALIVPKSLVNVHSCENWLPRKAMSATRVAGILHALEGWE 592
Query: 421 VHECGDTIL--STEKVWEASIRHGFQPL 446
HECG ++L +KVWEA + HGFQPL
Sbjct: 593 THECGTSLLLSDLDKVWEACLSHGFQPL 620
>UniRef100_O80938 CER1-like protein [Arabidopsis thaliana]
Length = 635
Score = 403 bits (1036), Expect = e-111
Identities = 202/373 (54%), Positives = 263/373 (70%), Gaps = 5/373 (1%)
Query: 79 FHSLHHTKFRRNYSLFMPMYDYIYGTVDKSTDVIYETSLMRPKESPDVVHLTHLTTFNSI 138
FHSLHHT+FR NYSLFMPMYDYIYGT D+ +D +YETSL + +E PD +HLTHLT+ +SI
Sbjct: 256 FHSLHHTQFRTNYSLFMPMYDYIYGTTDECSDSLYETSLEKEEEKPDAIHLTHLTSLDSI 315
Query: 139 YQLRLGFASLASNPQTSKWYLHLMWPFTMF-SMLMTWICGRAFVLESNSFKNLKLQCWLI 197
Y LRLGFASL+S+P +S+ YL LM PF + S ++ + FV+E N F++L L L+
Sbjct: 316 YHLRLGFASLSSHPLSSRCYLFLMKPFALILSFILRSFSFQTFVVERNRFRDLTLHSHLL 375
Query: 198 PRFKRQYFSKWQSKTFNNLIEEAIVEAELNGAKVISLGLFNKNHQLNERHEHYIGRLPQL 257
P+F Y S Q + N +IE AI+EA+ G KV+SLGL N+ +LN E Y+ R P+L
Sbjct: 376 PKFSSHYMSHQQKECINKMIEAAILEADKKGVKVMSLGLLNQGEELNGYGEMYVRRHPKL 435
Query: 258 KIKVVDGSSLAAATVLNNIPKGTNQVLLRGKFNKVAFVIANALCKKNVQVVVLYKDELKE 317
KI++VDG SLAA VL++IP GT +VL RG+ KVA I +LC+ ++V+VL K+E
Sbjct: 436 KIRIVDGGSLAAEVVLHSIPVGTKEVLFRGQITKVARAIVFSLCQNAIKVMVLRKEEHSM 495
Query: 318 LEQRINTS-KGNLALSPFNTPKIWLVGDEWDEYEQMEAPKGSLFIPFSHFPPKKMRKDCF 376
L + ++ K NL L+ P IWLVGD EQ A G+LF+PFS FPPK +RKDCF
Sbjct: 496 LAEFLDDKCKENLVLTTNYYPMIWLVGDGLSTKEQKMAKDGTLFLPFSQFPPKTLRKDCF 555
Query: 377 YHYTPAMITPTTFMNSHSCENWLPRRVMSAWRIAGIIHALEGWNVHECG---DTILSTEK 433
YH TPAMI P + N SCENWL RRVMSAWR+ GI+HALEGW HECG ++I++ +
Sbjct: 556 YHTTPAMIIPHSAQNIDSCENWLGRRVMSAWRVGGIVHALEGWKEHECGLDDNSIINPPR 615
Query: 434 VWEASIRHGFQPL 446
VWEA++R+GFQPL
Sbjct: 616 VWEAALRNGFQPL 628
>UniRef100_Q6K9F6 Putative CER1 protein [Oryza sativa]
Length = 619
Score = 396 bits (1018), Expect = e-109
Identities = 194/371 (52%), Positives = 257/371 (68%), Gaps = 4/371 (1%)
Query: 79 FHSLHHTKFRRNYSLFMPMYDYIYGTVDKSTDVIYETSLMRPKESPDVVHLTHLTTFNSI 138
FHSLHHT+FR NYSLFMP YDYIY T+DKS+D +YE+SL +E+PD+VHLTH+T S
Sbjct: 247 FHSLHHTQFRTNYSLFMPFYDYIYNTMDKSSDELYESSLKGTEETPDLVHLTHMTNLQSA 306
Query: 139 YQLRLGFASLASNPQT-SKWYLHLMWPFTMFSMLMTWICGR-AFVLESNSFKNLKLQCWL 196
Y LR+G AS+AS P + S WY+ +WP SM++ WI G AFV+E +K+Q W
Sbjct: 307 YHLRIGIASIASKPYSDSAWYMWTLWPLAWLSMVLAWIYGSSAFVVERIKLNKMKMQTWA 366
Query: 197 IPRFKRQYFSKWQSKTFNNLIEEAIVEAELNGAKVISLGLFNKNHQLNERHEHYIGRLPQ 256
+PR+ QY W+ + N+LIE+AI++A++ G KVISLGL N+ QLN E + + P+
Sbjct: 367 LPRYNFQYGLTWEREPINDLIEKAILDADMKGVKVISLGLLNQAKQLNGNGELFRQKYPK 426
Query: 257 LKIKVVDGSSLAAATVLNNIPKGTNQVLLRGKFNKVAFVIANALCKKNVQVVVLYKDELK 316
L ++++DGS LA A VL +IP +V LR +K+A IA ALC + VQV++ K+
Sbjct: 427 LGVRIIDGSGLATAVVLKSIPSDAKKVFLRTGTSKIARAIAIALCDRGVQVIMNEKEVYH 486
Query: 317 ELEQRINTSKGN-LALSPFNTPKIWLVGDEWDEYEQMEAPKGSLFIPFSHFPPKKMRKDC 375
L+ +I ++ + L LS N P++W+V + D+ EQ APKG++FIP S FP KK+RKDC
Sbjct: 487 MLKSQIPENRASYLKLSSDNVPQLWIVHNI-DDNEQKMAPKGTIFIPISQFPLKKLRKDC 545
Query: 376 FYHYTPAMITPTTFMNSHSCENWLPRRVMSAWRIAGIIHALEGWNVHECGDTILSTEKVW 435
Y TPAM P N HSCENWLPRRVMSAW IAGI+HALEGWN+HECGD ++ EK W
Sbjct: 546 TYMSTPAMRIPEEMKNIHSCENWLPRRVMSAWHIAGILHALEGWNMHECGDEMMDIEKSW 605
Query: 436 EASIRHGFQPL 446
A+IRHGF PL
Sbjct: 606 SAAIRHGFLPL 616
>UniRef100_Q39047 CER1-like protein [Arabidopsis thaliana]
Length = 623
Score = 391 bits (1005), Expect = e-107
Identities = 207/392 (52%), Positives = 267/392 (67%), Gaps = 18/392 (4%)
Query: 65 FSPSFFNCH*ACHLFHSLHHTKFRRNYSLFMPMYDYIYGTVDKSTDVIYETSLMRPKESP 124
F P F C+ FHSLHHT+FR NYSLFMP+YD+IYGT D TD +YE SL +ESP
Sbjct: 235 FPPLKFLCY--TPSFHSLHHTQFRTNYSLFMPIYDFIYGTTDNLTDSLYERSLEIEEESP 292
Query: 125 DVVHLTHLTTFNSIYQLRLGFASLASNPQTSK--WYLH-LMWPFTMFSM--LMTWICGRA 179
DV+HLTHLTT NSIYQ+RLGF SL+S P S+ WYL MWPFT+ L + I R
Sbjct: 293 DVIHLTHLTTHNSIYQMRLGFPSLSSCPLWSRPPWYLTCFMWPFTLLCSFALTSAIPLRT 352
Query: 180 FVLESNSFKNLKLQCWLIPRFKRQYFSKWQSKTFNNLIEEAIVEAELNGAKVISLGLFNK 239
FV E N ++L + L+P+F F + ++ N +IEEAI+EA+ G KV+SLGL N
Sbjct: 353 FVFERNRLRDLTVHSHLLPKFS---FHR-HHESINTIIEEAILEADEKGVKVMSLGLMNN 408
Query: 240 NHQLNERHEHYIGRLPQLKIKVVDGSSLAAATVLNNIPKGTNQVLLRGKFNKVAFVIANA 299
+LN E Y+ + P+LKI++VDGSS+AA V+NNIPK +++ RG KVA + A
Sbjct: 409 REELNGSGEMYVQKYPKLKIRLVDGSSMAATVVINNIPKEATEIVFRGNLTKVASAVVFA 468
Query: 300 LCKKNVQVVVLYKDELKELEQRINTSKGNLALSPFNT---PKIWLVGDEWDEYEQMEAPK 356
LC+K V+VVVL ++E +L + + NL LS N+ PK+WLVGD + EQM+A +
Sbjct: 469 LCQKGVKVVVLREEEHSKLIK--SGVDKNLVLSTSNSYYSPKVWLVGDGIENEEQMKAKE 526
Query: 357 GSLFIPFSHFPPKKMRKDCFYHYTPAMITPTTFMNSHSCENWLPRRVMSAWRIAGIIHAL 416
G+LF+PFSHFPP K+RKDCFY TPAM P + N SCENWL RRVMSAW+I GI+HAL
Sbjct: 527 GTLFVPFSHFPPNKLRKDCFYQSTPAMRVPKSAQNIDSCENWLGRRVMSAWKIGGIVHAL 586
Query: 417 EGWNVHECGDT--ILSTEKVWEASIRHGFQPL 446
EGW H+CG+T +L +WEA++RH FQPL
Sbjct: 587 EGWEEHDCGNTCNVLRLHAIWEAALRHDFQPL 618
>UniRef100_Q39046 CER1-like protein [Arabidopsis thaliana]
Length = 622
Score = 384 bits (986), Expect = e-105
Identities = 205/392 (52%), Positives = 265/392 (67%), Gaps = 18/392 (4%)
Query: 65 FSPSFFNCH*ACHLFHSLHHTKFRRNYSLFMPMYDYIYGTVDKSTDVIYETSLMRPKESP 124
F P F C+ FHSLHHT+FR NYSLFMP+YD+IYGT D TD +YE SL +ESP
Sbjct: 234 FPPLKFLCY--TPSFHSLHHTQFRTNYSLFMPIYDFIYGTTDNLTDSLYERSLEIEEESP 291
Query: 125 DVVHLTHLTTFNSIYQLRLGFASLASNPQTSK--WYLH-LMWPFTMFSM--LMTWICGRA 179
DV+HLTHLTT NSIYQ+RLGF SL+S P S+ WYL M PFT+ L + I R
Sbjct: 292 DVIHLTHLTTHNSIYQMRLGFPSLSSCPLWSRPPWYLTCFMXPFTLLCSFALTSAIPLRT 351
Query: 180 FVLESNSFKNLKLQCWLIPRFKRQYFSKWQSKTFNNLIEEAIVEAELNGAKVISLGLFNK 239
FV E N ++L + L+P+F F + ++ N +IEEAI+EA+ G KV+SLGL N
Sbjct: 352 FVFERNRLRDLTVHSHLLPKFS---FHR-HHESINTIIEEAILEADEKGVKVMSLGLMNN 407
Query: 240 NHQLNERHEHYIGRLPQLKIKVVDGSSLAAATVLNNIPKGTNQVLLRGKFNKVAFVIANA 299
+LN E Y+ + P+LKI++VDGSS+AA V+NNIPK +++ RG KVA + A
Sbjct: 408 REELNGSGEMYVQKYPKLKIRLVDGSSMAATVVINNIPKEATEIVFRGNLTKVASAVVFA 467
Query: 300 LCKKNVQVVVLYKDELKELEQRINTSKGNLALSPFNT---PKIWLVGDEWDEYEQMEAPK 356
LC+K V+VVVL ++E +L + + NL LS N+ PK+WLVGD + EQM+ +
Sbjct: 468 LCQKGVKVVVLREEEHXKLIK--SGVDKNLVLSTSNSYYSPKVWLVGDGIENEEQMKPKE 525
Query: 357 GSLFIPFSHFPPKKMRKDCFYHYTPAMITPTTFMNSHSCENWLPRRVMSAWRIAGIIHAL 416
G+LF+PFSHFPP K+RKDCFY TPAM P + N SCENWL RRVMSAW+I GI+HAL
Sbjct: 526 GTLFVPFSHFPPNKLRKDCFYQSTPAMRVPKSAQNIDSCENWLGRRVMSAWKIGGIVHAL 585
Query: 417 EGWNVHECGDT--ILSTEKVWEASIRHGFQPL 446
EGW H+CG+T +L +WEA++RH FQPL
Sbjct: 586 EGWEEHDCGNTCNVLRLHAIWEAALRHDFQPL 617
>UniRef100_Q6K3D8 Putative CER1 [Oryza sativa]
Length = 635
Score = 371 bits (953), Expect = e-101
Identities = 184/371 (49%), Positives = 249/371 (66%), Gaps = 4/371 (1%)
Query: 79 FHSLHHTKFRRNYSLFMPMYDYIYGTVDKSTDVIYETSLMRPKES---PDVVHLTHLTTF 135
FHSLHHT+FR NYSLFMP+YDYIYGT DKS+D +YE +L E+ PDVVHLTHLTT
Sbjct: 248 FHSLHHTQFRTNYSLFMPVYDYIYGTTDKSSDELYERTLQGRDEAAWRPDVVHLTHLTTP 307
Query: 136 NSIYQLRLGFASLASNPQTSKWYLHLMWPFTMFSMLMTWICGRAFVLESNSFKNLKLQCW 195
S++ RLGFA++ASNP + HL+ + + + + F E+N L ++ W
Sbjct: 308 ESVFHNRLGFAAVASNPLGAAASGHLLRAASAVASPLLSLFASTFRSEANRLDKLNIETW 367
Query: 196 LIPRFKRQYFSKWQSKTFNNLIEEAIVEAELNGAKVISLGLFNKNHQLNERHEHYIGRLP 255
+IPRF Y SK + LIE+A+ +AE +GA+V++LGL N+ + LN E Y+ R P
Sbjct: 368 VIPRFTSHYTSKSDGYKVSRLIEKAVSDAEASGARVLTLGLLNQGYDLNRNGELYVVRKP 427
Query: 256 QLKIKVVDGSSLAAATVLNNIPKGTNQVLLRGKFNKVAFVIANALCKKNVQVVVLYKDEL 315
LK K+VDG+SLA A VLN IP+GT VLL G NK++ V+ +LCK+ +QV ++ K+
Sbjct: 428 SLKTKIVDGTSLAVAAVLNMIPQGTKDVLLLGNANKISLVLTLSLCKREIQVRMVNKELY 487
Query: 316 KELEQRINTS-KGNLALSPFNTPKIWLVGDEWDEYEQMEAPKGSLFIPFSHFPPKKMRKD 374
+ L+Q++ + +L LS + K+WLVGD + EQM+A KGS F+P+S FPP K R D
Sbjct: 488 ECLKQQLQPEMQEHLVLSCSYSSKVWLVGDGVTDEEQMKAQKGSHFVPYSQFPPNKARND 547
Query: 375 CFYHYTPAMITPTTFMNSHSCENWLPRRVMSAWRIAGIIHALEGWNVHECGDTILSTEKV 434
C YH TPA++ P +F N H CENWLPRRVMSAWR AGI+HALE W+ HECG + +K
Sbjct: 548 CVYHCTPALLVPESFENLHVCENWLPRRVMSAWRAAGIVHALEKWDGHECGGRVTGVQKA 607
Query: 435 WEASIRHGFQP 445
W A++ GF+P
Sbjct: 608 WSAALARGFRP 618
>UniRef100_O23678 CER1-like protein [Arabidopsis thaliana]
Length = 604
Score = 366 bits (939), Expect = e-100
Identities = 200/392 (51%), Positives = 256/392 (65%), Gaps = 37/392 (9%)
Query: 65 FSPSFFNCH*ACHLFHSLHHTKFRRNYSLFMPMYDYIYGTVDKSTDVIYETSLMRPKESP 124
F P F C+ FHSLHHT+FR NYSLFMP+YD+IYGT D TD +YE SL +ESP
Sbjct: 235 FPPLKFLCY--TPSFHSLHHTQFRTNYSLFMPIYDFIYGTTDNLTDSLYERSLEIEEESP 292
Query: 125 DVVHLTHLTTFNSIYQLRLGFASLASNPQTSK--WYLH-LMWPFTMFSM--LMTWICGRA 179
DV+HLTHLTT NSIYQ+RLGF SL+S P S+ WYL MWPFT+ L + I R
Sbjct: 293 DVIHLTHLTTHNSIYQMRLGFPSLSSCPLWSRPPWYLTCFMWPFTLLCSFALTSAIPLRT 352
Query: 180 FVLESNSFKNLKLQCWLIPRFKRQYFSKWQSKTFNNLIEEAIVEAELNGAKVISLGLFNK 239
FV E N ++L + L+P+F F + ++ N +IEEAI+EA+ G KV+SLGL N
Sbjct: 353 FVFERNRLRDLTVHSHLLPKFS---FHR-HHESINTIIEEAILEADEKGVKVMSLGLMNN 408
Query: 240 NHQLNERHEHYIGRLPQLKIKVVDGSSLAAATVLNNIPKGTNQVLLRGKFNKVAFVIANA 299
I++VDGSS+AA V+NNIPK +++ RG KVA + A
Sbjct: 409 -------------------IRLVDGSSMAATVVINNIPKEATEIVFRGNLTKVASAVVFA 449
Query: 300 LCKKNVQVVVLYKDELKELEQRINTSKGNLALSPFNT---PKIWLVGDEWDEYEQMEAPK 356
LC+K V+VVVL ++E +L + + NL LS N+ PK+WLVGD + EQM+A +
Sbjct: 450 LCQKGVKVVVLREEEHSKLIK--SGVDKNLVLSTSNSYYSPKVWLVGDGIENEEQMKAKE 507
Query: 357 GSLFIPFSHFPPKKMRKDCFYHYTPAMITPTTFMNSHSCENWLPRRVMSAWRIAGIIHAL 416
G+LF+PFSHFPP K+RKDCFY TPAM P + N SCENWL RRVMSAW+I GI+HAL
Sbjct: 508 GTLFVPFSHFPPNKLRKDCFYQSTPAMRVPKSAQNIDSCENWLGRRVMSAWKIGGIVHAL 567
Query: 417 EGWNVHECGDT--ILSTEKVWEASIRHGFQPL 446
EGW H+CG+T +L +WEA++RH FQPL
Sbjct: 568 EGWEEHDCGNTCNVLRLHAIWEAALRHDFQPL 599
>UniRef100_O23679 CER1 protein [Arabidopsis thaliana]
Length = 580
Score = 346 bits (888), Expect = 7e-94
Identities = 170/336 (50%), Positives = 224/336 (66%), Gaps = 6/336 (1%)
Query: 65 FSPSFFNCH*ACHLFHSLHHTKFRRNYSLFMPMYDYIYGTVDKSTDVIYETSLMRPKESP 124
F P F C+ +HSLHHT+FR NYSLFMP+YDYIYGT+D+STD +YE +L R +
Sbjct: 235 FPPLKFLCY--TPSYHSLHHTQFRTNYSLFMPLYDYIYGTMDESTDTLYEKTLERGDDIV 292
Query: 125 DVVHLTHLTTFNSIYQLRLGFASLASNPQTSKWYLHLMWPFTMFSMLMTWICGRAFVLES 184
DVVHLTHLTT SIY LR+G AS AS P +W++ L+WPFT SM+ T R FV E
Sbjct: 293 DVVHLTHLTTPESIYHLRIGLASFASYPFAYRWFMRLLWPFTSLSMIFTLFYARLFVAER 352
Query: 185 NSFKNLKLQCWLIPRFKRQYFSKWQSKTFNNLIEEAIVEAELNGAKVISLGLFNKNHQLN 244
NSF L LQ W+IPR+ QY KW+ + NN+IE+AI+EA+ G KV+SLGL N+ +LN
Sbjct: 353 NSFNKLNLQSWVIPRYNLQYLLKWRKEAINNMIEKAILEADKKGVKVLSLGLMNQGEELN 412
Query: 245 ERHEHYIGRLPQLKIKVVDGSSLAAATVLNNIPKGTNQVLLRGKFNKVAFVIANALCKKN 304
E YI P +K+++VDGS LAAA V+N++PK T V++ G KVA+ IA+ALC++
Sbjct: 413 RNGEVYIHNHPDMKVRLVDGSRLAAAVVINSVPKATTSVVMTGNLTKVAYTIASALCQRG 472
Query: 305 VQVVVLYKDELKELE----QRINTSKGNLALSPFNTPKIWLVGDEWDEYEQMEAPKGSLF 360
VQV L DE +++ Q L ++ K+WLVG+ EQ +A KG+LF
Sbjct: 473 VQVSTLRLDEYEKIRSCVPQECRDHLVYLTSEALSSNKVWLVGEGTTREEQEKATKGTLF 532
Query: 361 IPFSHFPPKKMRKDCFYHYTPAMITPTTFMNSHSCE 396
IPFS FP K++R+DC YH TPA+I P + +N HSCE
Sbjct: 533 IPFSQFPLKQLRRDCIYHTTPALIVPKSLVNVHSCE 568
>UniRef100_Q7XPZ4 OSJNBa0004N05.14 protein [Oryza sativa]
Length = 597
Score = 340 bits (873), Expect = 4e-92
Identities = 176/371 (47%), Positives = 233/371 (62%), Gaps = 35/371 (9%)
Query: 79 FHSLHHTKFRRNYSLFMPMYDYIYGTVDKSTDVIYETSLMRPKESPDVVHLTHLTTFNSI 138
FHSLHHT+FR NYSLFMP YDYIY T+D S+D +YE SL +E+PD+VHLTH+T+ S
Sbjct: 247 FHSLHHTQFRTNYSLFMPFYDYIYNTMDSSSDELYERSLKGTEETPDIVHLTHMTSLKST 306
Query: 139 YQLRLGFASLASNP-QTSKWYLHLMWPFTMFSMLMTWICGR-AFVLESNSFKNLKLQCWL 196
Y LR+G S++S P S WY+ ++WP SM++ WI G AFV+E K +Q W
Sbjct: 307 YHLRIGITSISSKPCNDSVWYMWMLWPVAWLSMVLAWIYGSSAFVVERLKLKKFSMQVWA 366
Query: 197 IPRFKRQYFSKWQSKTFNNLIEEAIVEAELNGAKVISLGLFNKNHQLNERHEHYIGRLPQ 256
+PR+ Q +++ + E QLN E + + P+
Sbjct: 367 LPRYNFQ------------VMDSSAAE------------------QLNGSGELFAKKYPR 396
Query: 257 LKIKVVDGSSLAAATVLNNIPKGTNQVLLRGKFNKVAFVIANALCKKNVQVVVLYKDELK 316
L+++++DGS LA A VLN+IP GT QV L G +KV A ALC++ VQV++ + E
Sbjct: 397 LRVRLIDGSGLATAVVLNSIPFGTKQVFLCGSNSKVTRATAIALCQRGVQVILNQEKEYG 456
Query: 317 ELEQRINTSKG-NLALSPFNTPKIWLVGDEWDEYEQMEAPKGSLFIPFSHFPPKKMRKDC 375
L+ R+ S+ L S TP+IW +GD D+ Q APKG++FIP S FP KK RKDC
Sbjct: 457 MLKSRVPESRAIYLKFSNDETPQIW-IGDSIDD-AQGRAPKGTIFIPTSQFPLKKARKDC 514
Query: 376 FYHYTPAMITPTTFMNSHSCENWLPRRVMSAWRIAGIIHALEGWNVHECGDTILSTEKVW 435
Y PAM P T N H+CENWLPRRVMSAWRIAGI+HALEGW +HECGD +++ EK W
Sbjct: 515 TYLSNPAMKIPETMQNVHTCENWLPRRVMSAWRIAGILHALEGWEMHECGDDMMTIEKTW 574
Query: 436 EASIRHGFQPL 446
A+I+HGF+PL
Sbjct: 575 SAAIKHGFKPL 585
>UniRef100_Q9XH51 CER1 [Oryza sativa]
Length = 621
Score = 340 bits (871), Expect = 6e-92
Identities = 173/372 (46%), Positives = 230/372 (61%), Gaps = 4/372 (1%)
Query: 79 FHSLHHTKFRRNYSLFMPMYDYIYGTVDKSTDVIYETSLMR--PKESPDVVHLTHLTTFN 136
FHSLHHT+FR NYSLFMP YDYIY T+DKS+D +YE SL +E+ DVVHLTHLTT +
Sbjct: 247 FHSLHHTQFRTNYSLFMPFYDYIYNTMDKSSDTLYENSLKNNDEEEAVDVVHLTHLTTLH 306
Query: 137 SIYQLRLGFASLASNPQTSKWYLHLMWPFTMFSMLMTWICGRAFVLESNSFKNLKLQCWL 196
SIY +R GFA AS P S+WY+ +MWP + SM++TW G +F +E N ++
Sbjct: 307 SIYHMRPGFAEFASRPYVSRWYMRMMWPLSWLSMVLTWTYGSSFTVERNVMRDQDAVMGH 366
Query: 197 IPRFKRQYFSKWQSKTFNNLIEEAIVEAELNGAKVISLGLFNKNHQLNERHEHYIGRLPQ 256
Y W+ + N+LIE+A+ EA+ NGAKV+SLGL N+ H LN+ E Y+ + P+
Sbjct: 367 YQDTSFHYGLDWEKEAINDLIEKAVCEADKNGAKVVSLGLLNQAHTLNKSGEQYLLKYPK 426
Query: 257 LKIKVVDGSSLAAATVLNNIPKGTNQVLLRGKFNKVAFVIANALCKKNVQVVVLYKDELK 316
L ++VDG+SLAAA V+N+IP+GT+QV+L G +KVA +A ALCKKN++V + K +
Sbjct: 427 LGARIVDGTSLAAAVVVNSIPQGTDQVILAGNVSKVARAVAQALCKKNIKVTMTNKQDYH 486
Query: 317 ELEQRI-NTSKGNLALSPFNTPKIWLVGDEWDEYEQMEAPKGSLFIPFSHFPPKKMRKD- 374
L+ I T NL+ S T K+WL+GD D EQ A KG+LFIP+S FPPK +RKD
Sbjct: 487 LLKPEIPETVADNLSFSKTGTAKVWLIGDGLDSAEQFRAQKGTLFIPYSQFPPKMVRKDS 546
Query: 375 CFYHYTPAMITPTTFMNSHSCENWLPRRVMSAWRIAGIIHALEGWNVHECGDTILSTEKV 434
C Y TPA+ + G + GWN HECGD +L KV
Sbjct: 547 CSYSTTPAIGCTKNAAECAFMRELAAKEGYGRMANGGNSSCVGGWNEHECGDKVLGMAKV 606
Query: 435 WEASIRHGFQPL 446
W +I HG P+
Sbjct: 607 WTDTIEHGLCPV 618
>UniRef100_Q39045 Possible aldehyde decarbonylase [Arabidopsis thaliana]
Length = 567
Score = 334 bits (856), Expect = 3e-90
Identities = 165/326 (50%), Positives = 216/326 (65%), Gaps = 6/326 (1%)
Query: 65 FSPSFFNCH*ACHLFHSLHHTKFRRNYSLFMPMYDYIYGTVDKSTDVIYETSLMRPKESP 124
F P F C+ +HSLHHT+FR NYSLFMP+YDYIYGT+D+STD +YE +L R +
Sbjct: 235 FPPLKFLCY--TPSYHSLHHTQFRTNYSLFMPLYDYIYGTMDESTDTLYEKTLERGDDRV 292
Query: 125 DVVHLTHLTTFNSIYQLRLGFASLASNPQTSKWYLHLMWPFTMFSMLMTWICGRAFVLES 184
DVVHLTHLTT SIY LR+G AS AS P +W++ L+WPFT SM+ T R FV E
Sbjct: 293 DVVHLTHLTTPESIYHLRIGLASFASYPFAYRWFMRLLWPFTSLSMIFTLFYARLFVAER 352
Query: 185 NSFKNLKLQCWLIPRFKRQYFSKWQSKTFNNLIEEAIVEAELNGAKVISLGLFNKNHQLN 244
NSF L LQ W+IPR+ QY KW+ + NN+IE+AI+EA+ G KV+SLGL N+ +LN
Sbjct: 353 NSFNKLNLQSWVIPRYNLQYLLKWRKEAINNMIEKAILEADKKGVKVLSLGLMNQGEELN 412
Query: 245 ERHEHYIGRLPQLKIKVVDGSSLAAATVLNNIPKGTNQVLLRGKFNKVAFVIANALCKKN 304
E YI P +K+++VDGS LAAA V+N++PK T V++ G KVA+ IA+ALC++
Sbjct: 413 RNGEVYIHNHPDMKVRLVDGSRLAAAVVINSVPKATTSVVMTGNLTKVAYTIASALCQRG 472
Query: 305 VQVVVLYKDELKELE----QRINTSKGNLALSPFNTPKIWLVGDEWDEYEQMEAPKGSLF 360
VQV L DE +++ Q L ++ K+WLVG+ EQ +A KG+LF
Sbjct: 473 VQVSTLRLDEYEKIRSCVPQECRDHLVYLTSEALSSNKVWLVGEGTTREEQEKATKGTLF 532
Query: 361 IPFSHFPPKKMRKDCFYHYTPAMITP 386
IPFS FP K++R DC YH TPA+I P
Sbjct: 533 IPFSQFPLKQLRSDCIYHTTPALIVP 558
>UniRef100_Q6RBX9 Glossy1 protein [Zea mays]
Length = 621
Score = 210 bits (535), Expect = 6e-53
Identities = 127/379 (33%), Positives = 190/379 (49%), Gaps = 16/379 (4%)
Query: 79 FHSLHHTKFRRNYSLFMPMYDYIYGTVDKSTDVIYETSLMRPKESPDVVHLTHLTTFNSI 138
+H++HHTK N+ LFMP++D + GT+D+ + + E PD V L H+
Sbjct: 246 YHAIHHTKKEANFCLFMPLFDLLGGTIDRRSWDMQRKMSAGVDEVPDFVFLAHVVDVMQS 305
Query: 139 YQLRLGFASLASNPQTSKWYLHLMWPFTMFSMLMTWICGRAFVLESNSFKNLKLQCWLIP 198
+ + AS P + + +L MWPF ML W+ + FV+ + + Q W +P
Sbjct: 306 LHVPFVMRTFASTPFSVQLFLLPMWPFAFLVMLAMWVWSKTFVISCYNLRGRLHQIWAVP 365
Query: 199 RFKRQYFSKWQSKTFNNLIEEAIVEAELNGAKVISLGLFNKNHQLNERHEHYIGRLPQLK 258
R+ QYF + N IE AI+ A+ G KV+SL NKN LN ++ + P L+
Sbjct: 366 RYGFQYFLPFAKDGINRQIELAILRADKMGVKVLSLAALNKNEALNGGGTLFVNKHPDLR 425
Query: 259 IKVVDGSSLAAATVLNNIPKGTNQVLLRGKFNKVAFVIANALCKKNVQVVV--------- 309
++VV G++L AA +LN IPKGT +V L G +K+ IA LCKK V+V++
Sbjct: 426 VRVVHGNTLTAAVILNEIPKGTAEVFLTGATSKLGRAIALYLCKKRVRVMMMTLSTERFQ 485
Query: 310 -LYKDELKELEQ-RINTSKGNLALSPFNTPKIWLVGDEWDEYEQMEAPKGSLFIPFSHFP 367
+ K+ E +Q + +K A + W+VG EQ AP G+ F F P
Sbjct: 486 KIQKEAPAEFQQYLVQVTKYRSA----QHCRTWIVGKWLSPREQRWAPPGTHFHQFVVPP 541
Query: 368 PKKMRKDCFYHYTPAMITPTTFMNSHSCENWLPRRVMSAWRIAGIIHALEGWNVHECGD- 426
R+DC Y AM P +CE L R V+ A G++H LEG+ HE G
Sbjct: 542 IIGFRRDCTYGKLAAMRLPKDVRGLGACEYSLERGVVHACHAGGVVHFLEGYTHHEVGAI 601
Query: 427 TILSTEKVWEASIRHGFQP 445
+ + VWEA+++HG +P
Sbjct: 602 DVDRIDVVWEAALKHGLRP 620
>UniRef100_Q69PA8 Putative Gl1 protein [Oryza sativa]
Length = 619
Score = 210 bits (535), Expect = 6e-53
Identities = 127/376 (33%), Positives = 194/376 (50%), Gaps = 8/376 (2%)
Query: 79 FHSLHHTKFRRNYSLFMPMYDYIYGTVDKSTDVIYETSLMRPKESPDVVHLTHLTTFNSI 138
+H++HHTK N+ LFMP++D I GT+D + + + + E P+ V L H+
Sbjct: 244 YHTIHHTKEDANFCLFMPLFDLIGGTLDAQSWEMQKKTSAGVDEVPEFVFLAHVVDVMQS 303
Query: 139 YQLRLGFASLASNPQTSKWYLHLMWPFTMFSMLMTWICGRAFVLESNSFKNLKLQCWLIP 198
+ + AS P + + +L MWPF MLM W + FV+ + Q W +P
Sbjct: 304 LHVPFVLRTFASTPFSVQPFLLPMWPFAFLVMLMMWAWSKTFVISCYRLRGRLHQMWAVP 363
Query: 199 RFKRQYFSKWQSKTFNNLIEEAIVEAELNGAKVISLGLFNKNHQLNERHEHYIGRLPQLK 258
R+ YF + NN IE AI+ A+ GAKV+SL NKN LN ++ + P L+
Sbjct: 364 RYGFHYFLPFAKDGINNQIELAILRADKMGAKVVSLAALNKNEALNGGGTLFVNKHPGLR 423
Query: 259 IKVVDGSSLAAATVLNNIPKGTNQVLLRGKFNKVAFVIANALCKKNVQVVVL-YKDELKE 317
++VV G++L AA +LN IP+GT +V + G +K+ IA LC+K V+V+++ E +
Sbjct: 424 VRVVHGNTLTAAVILNEIPQGTTEVFMTGATSKLGRAIALYLCRKKVRVMMMTLSTERFQ 483
Query: 318 LEQRINTSKGN---LALSPFNTP---KIWLVGDEWDEYEQMEAPKGSLFIPFSHFPPKKM 371
QR T + + ++ + + K W+VG EQ AP G+ F F P
Sbjct: 484 KIQREATPEHQQYLVQVTKYRSAQHCKTWIVGKWLSPREQRWAPPGTHFHQFVVPPIIGF 543
Query: 372 RKDCFYHYTPAMITPTTFMNSHSCENWLPRRVMSAWRIAGIIHALEGWNVHECGD-TILS 430
R+DC Y AM P +CE L R V+ A G++H LEG+ HE G +
Sbjct: 544 RRDCTYGKLAAMRLPKDVQGLGACEYSLERGVVHACHAGGVVHFLEGYTHHEVGAIDVDR 603
Query: 431 TEKVWEASIRHGFQPL 446
+ VWEA++RHG +P+
Sbjct: 604 IDVVWEAALRHGLRPV 619
>UniRef100_Q6RCI6 Gl1 protein [Zea mays]
Length = 621
Score = 209 bits (532), Expect = 1e-52
Identities = 126/379 (33%), Positives = 190/379 (49%), Gaps = 16/379 (4%)
Query: 79 FHSLHHTKFRRNYSLFMPMYDYIYGTVDKSTDVIYETSLMRPKESPDVVHLTHLTTFNSI 138
+H++HHTK N+ LFMP++D + GT+D+ + + E PD V L H+
Sbjct: 246 YHAIHHTKKEANFCLFMPLFDLLGGTIDRRSWDMQRKMSAGVDEVPDFVFLAHVVDVMQS 305
Query: 139 YQLRLGFASLASNPQTSKWYLHLMWPFTMFSMLMTWICGRAFVLESNSFKNLKLQCWLIP 198
+ + AS P + + +L MWPF ML W+ + FV+ + + Q W +P
Sbjct: 306 LHVPFVMRTFASTPFSVQLFLLPMWPFAFLVMLAMWVWSKTFVISCYNLRGRLHQIWAVP 365
Query: 199 RFKRQYFSKWQSKTFNNLIEEAIVEAELNGAKVISLGLFNKNHQLNERHEHYIGRLPQLK 258
R+ QYF + N IE AI+ A+ G KV+SL NKN LN ++ + P L+
Sbjct: 366 RYGFQYFLPFAKDGINRQIELAILRADKMGVKVLSLAALNKNEALNGGGTLFVNKHPDLR 425
Query: 259 IKVVDGSSLAAATVLNNIPKGTNQVLLRGKFNKVAFVIANALCKKNVQVVV--------- 309
++VV G++L AA +LN IPKGT +V L G +K+ IA LCKK V+V++
Sbjct: 426 VRVVHGNTLTAAVILNEIPKGTAEVFLTGATSKLGRAIALYLCKKRVRVMMMTLSTERFQ 485
Query: 310 -LYKDELKELEQ-RINTSKGNLALSPFNTPKIWLVGDEWDEYEQMEAPKGSLFIPFSHFP 367
+ K+ E +Q + +K A + W+VG EQ AP G+ F F P
Sbjct: 486 KIQKEAPAEFQQYLVQVTKYRSA----QHCRTWIVGKWLSPREQRWAPPGTHFHQFVVPP 541
Query: 368 PKKMRKDCFYHYTPAMITPTTFMNSHSCENWLPRRVMSAWRIAGIIHALEGWNVHECGD- 426
R+DC Y AM P +CE L R ++ A G++H LEG+ HE G
Sbjct: 542 IIGFRRDCTYGKLAAMRLPKDVRGLGACEYSLERGLVHACHAGGVVHFLEGYTHHEVGAI 601
Query: 427 TILSTEKVWEASIRHGFQP 445
+ + VWEA+++HG +P
Sbjct: 602 DVDRIDVVWEAALKHGLRP 620
>UniRef100_Q8H1Z0 Cuticle protein [Arabidopsis thaliana]
Length = 632
Score = 204 bits (520), Expect = 3e-51
Identities = 126/382 (32%), Positives = 194/382 (49%), Gaps = 11/382 (2%)
Query: 79 FHSLHHTKFRRNYSLFMPMYDYIYGTVDKSTDVIYET---SLMRPKESPDVVHLTHLTTF 135
+HSLHH + N+ LFMP++D + T + ++ + + S K P+ V L H
Sbjct: 249 YHSLHHQEMGTNFCLFMPLFDVLGDTQNPNSWELQKKIRLSAGERKRVPEFVFLAHGVDV 308
Query: 136 NSIYQLRLGFASLASNPQTSKWYLHLMWPFTMFSMLMTWICGRAFVLESNSFKNLKLQCW 195
S F S AS P T++ +L MWPFT ML W + F+ + +N Q W
Sbjct: 309 MSAMHAPFVFRSFASMPYTTRIFLLPMWPFTFCVMLGMWAWSKTFLFSFYTLRNNLCQTW 368
Query: 196 LIPRFKRQYFSKWQSKTFNNLIEEAIVEAELNGAKVISLGLFNKNHQLNERHEHYIGRLP 255
+PRF QYF + +K N+ IE AI+ A+ G KVISL NKN LN ++ + P
Sbjct: 369 GVPRFGFQYFLPFATKGINDQIEAAILRADKIGVKVISLAALNKNEALNGGGTLFVNKHP 428
Query: 256 QLKIKVVDGSSLAAATVLNNIPKGTNQVLLRGKFNKVAFVIANALCKKNVQVVVL----Y 311
L+++VV G++L AA +L IPK N+V L G +K+ IA LC++ V+V++L
Sbjct: 429 DLRVRVVHGNTLTAAVILYEIPKDVNEVFLTGATSKLGRAIALYLCRRGVRVLMLTLSME 488
Query: 312 KDELKELEQRINTSKGNLALSPFNTP---KIWLVGDEWDEYEQMEAPKGSLFIPFSHFPP 368
+ + + E + + ++ +N K W+VG EQ AP G+ F F P
Sbjct: 489 RFQKIQKEAPVEFQNNLVQVTKYNAAQHCKTWIVGKWLTPREQSWAPAGTHFHQFVVPPI 548
Query: 369 KKMRKDCFYHYTPAMITPTTFMNSHSCENWLPRRVMSAWRIAGIIHALEGWNVHECGD-T 427
K R++C Y AM P +CE + R V+ A G++H LEGW HE G
Sbjct: 549 LKFRRNCTYGDLAAMKLPKDVEGLGTCEYTMERGVVHACHAGGVVHMLEGWKHHEVGAID 608
Query: 428 ILSTEKVWEASIRHGFQPLKNI 449
+ + VWEA++++G + ++
Sbjct: 609 VDRIDLVWEAAMKYGLSAVSSL 630
>UniRef100_Q9FJN4 Lipid transfer protein; glossy1 homolog [Arabidopsis thaliana]
Length = 566
Score = 204 bits (520), Expect = 3e-51
Identities = 126/382 (32%), Positives = 194/382 (49%), Gaps = 11/382 (2%)
Query: 79 FHSLHHTKFRRNYSLFMPMYDYIYGTVDKSTDVIYET---SLMRPKESPDVVHLTHLTTF 135
+HSLHH + N+ LFMP++D + T + ++ + + S K P+ V L H
Sbjct: 183 YHSLHHQEMGTNFCLFMPLFDVLGDTQNPNSWELQKKIRLSAGERKRVPEFVFLAHGVDV 242
Query: 136 NSIYQLRLGFASLASNPQTSKWYLHLMWPFTMFSMLMTWICGRAFVLESNSFKNLKLQCW 195
S F S AS P T++ +L MWPFT ML W + F+ + +N Q W
Sbjct: 243 MSAMHAPFVFRSFASMPYTTRIFLLPMWPFTFCVMLGMWAWSKTFLFSFYTLRNNLCQTW 302
Query: 196 LIPRFKRQYFSKWQSKTFNNLIEEAIVEAELNGAKVISLGLFNKNHQLNERHEHYIGRLP 255
+PRF QYF + +K N+ IE AI+ A+ G KVISL NKN LN ++ + P
Sbjct: 303 GVPRFGFQYFLPFATKGINDQIEAAILRADKIGVKVISLAALNKNEALNGGGTLFVNKHP 362
Query: 256 QLKIKVVDGSSLAAATVLNNIPKGTNQVLLRGKFNKVAFVIANALCKKNVQVVVL----Y 311
L+++VV G++L AA +L IPK N+V L G +K+ IA LC++ V+V++L
Sbjct: 363 DLRVRVVHGNTLTAAVILYEIPKDVNEVFLTGATSKLGRAIALYLCRRGVRVLMLTLSME 422
Query: 312 KDELKELEQRINTSKGNLALSPFNTP---KIWLVGDEWDEYEQMEAPKGSLFIPFSHFPP 368
+ + + E + + ++ +N K W+VG EQ AP G+ F F P
Sbjct: 423 RFQKIQKEAPVEFQNNLVQVTKYNAAQHCKTWIVGKWLTPREQSWAPAGTHFHQFVVPPI 482
Query: 369 KKMRKDCFYHYTPAMITPTTFMNSHSCENWLPRRVMSAWRIAGIIHALEGWNVHECGD-T 427
K R++C Y AM P +CE + R V+ A G++H LEGW HE G
Sbjct: 483 LKFRRNCTYGDLAAMKLPKDVEGLGTCEYTMERGVVHACHAGGVVHMLEGWKHHEVGAID 542
Query: 428 ILSTEKVWEASIRHGFQPLKNI 449
+ + VWEA++++G + ++
Sbjct: 543 VDRIDLVWEAAMKYGLSAVSSL 564
>UniRef100_Q6ETL8 Putative glossy1 protein [Oryza sativa]
Length = 628
Score = 201 bits (511), Expect = 4e-50
Identities = 125/374 (33%), Positives = 186/374 (49%), Gaps = 9/374 (2%)
Query: 81 SLHHTKFRRNYSLFMPMYDYIYGTVD-KSTDVIYETSLMRPKESPDVVHLTHLTTFNSIY 139
SLHH + N+ LFMP++D + GT++ KS ++ E L + ++PD V L H+ +
Sbjct: 254 SLHHREKDSNFCLFMPIFDLLGGTLNHKSWELQKEVYLGKNDQAPDFVFLAHVVDIMASM 313
Query: 140 QLRLGFASLASNPQTSKWYLHLMWPFTMFSMLMTWICGRAFVLESNSFKNLKLQCWLIPR 199
+ S +S P + + L WP ML+ W C + F++ S + Q W +PR
Sbjct: 314 HVPFVLRSCSSTPFANHFVLLPFWPVAFGFMLLMWCCSKTFLVSSYRLRGNLHQMWTVPR 373
Query: 200 FKRQYFSKWQSKTFNNLIEEAIVEAELNGAKVISLGLFNKNHQLNERHEHYIGRLPQLKI 259
+ QYF K N IE AI+ A+ G KV+SL NKN LN ++ + P+L++
Sbjct: 374 YGFQYFIPAAKKGINEQIELAILRADRMGVKVLSLAALNKNEALNGGGTLFVNKHPELRV 433
Query: 260 KVVDGSSLAAATVLNNIPKGTNQVLLRGKFNKVAFVIANALCKKNVQVVVLYKDELKELE 319
+VV G++L AA +LN IP V L G +K+ IA LC+K ++V++L + L+
Sbjct: 434 RVVHGNTLTAAVILNEIPSNVKDVFLTGATSKLGRAIALYLCRKKIRVLMLTLSSERFLK 493
Query: 320 -QRINTSKGNLAL------SPFNTPKIWLVGDEWDEYEQMEAPKGSLFIPFSHFPPKKMR 372
QR ++ L P K WLVG EQ AP G+ F F P R
Sbjct: 494 IQREAPAEFQQYLVQVTKYQPAQNCKTWLVGKWLSPREQRWAPAGTHFHQFVVPPIIGFR 553
Query: 373 KDCFYHYTPAMITPTTFMNSHSCENWLPRRVMSAWRIAGIIHALEGWNVHECGD-TILST 431
+DC Y AM P CE + R V+ A G++H LEGW HE G +
Sbjct: 554 RDCTYGKLAAMRLPKDVQGLGYCEYTMERGVVHACHAGGVVHFLEGWEHHEVGAIDVDRI 613
Query: 432 EKVWEASIRHGFQP 445
+ VW+A+++HG P
Sbjct: 614 DVVWKAALKHGLTP 627
>UniRef100_Q67WQ7 Putative Gl1 [Oryza sativa]
Length = 627
Score = 196 bits (499), Expect = 9e-49
Identities = 124/378 (32%), Positives = 184/378 (47%), Gaps = 17/378 (4%)
Query: 81 SLHHTKFRRNYSLFMPMYDYIYGTVD-KSTDVIYETSLMRPKESPDVVHLTHLTTFNSIY 139
SLHH + N+ LFMP++D + GT++ KS + E L + PD V L H+ S
Sbjct: 253 SLHHREKDSNFCLFMPLFDALGGTLNPKSWQLQKEVDLGKNHRVPDFVFLVHVVDVVSSM 312
Query: 140 QLRLGFASLASNPQTSKWYLHLMWPFTMFSMLMTWICGRAFVLESNSFKNLKLQCWLIPR 199
+ F + +S P + L +WP ML+ W C + F + + Q W +PR
Sbjct: 313 HVPFAFRACSSLPFATHLVLLPLWPIAFGFMLLQWFCSKTFTVSFYKLRGFLHQTWSVPR 372
Query: 200 FKRQYFSKWQSKTFNNLIEEAIVEAELNGAKVISLGLFNKNHQLNERHEHYIGRLPQLKI 259
+ QYF K N +IE AI+ A+ G KV+SL NKN LN ++ + P L++
Sbjct: 373 YGFQYFIPSAKKGINEMIELAILRADKMGVKVLSLAALNKNEALNGGGTLFVRKHPDLRV 432
Query: 260 KVVDGSSLAAATVLNNIPKGTNQVLLRGKFNKVAFVIANALCKKNVQVVVL--------- 310
+VV G++L AA +LN IP +V L G +K+ IA LC+K ++V++L
Sbjct: 433 RVVHGNTLTAAVILNEIPGDVAEVFLTGATSKLGRAIALYLCRKKIRVLMLTLSTERFMN 492
Query: 311 -YKDELKELEQ-RINTSKGNLALSPFNTPKIWLVGDEWDEYEQMEAPKGSLFIPFSHFPP 368
++ E +Q + +K A K W+VG EQ AP G+ F F P
Sbjct: 493 IQREAPAEFQQYLVQVTKYQAA----QNCKTWIVGKWLSPREQRWAPAGTHFHQFVVPPI 548
Query: 369 KKMRKDCFYHYTPAMITPTTFMNSHSCENWLPRRVMSAWRIAGIIHALEGWNVHECGD-T 427
R+DC Y AM P +CE + R V+ A G++H LEGW+ HE G
Sbjct: 549 IGFRRDCTYGKLAAMRLPEDVEGLGTCEYTMGRGVVHACHAGGVVHFLEGWDHHEVGAID 608
Query: 428 ILSTEKVWEASIRHGFQP 445
+ + VW A++RHG P
Sbjct: 609 VDRIDAVWNAALRHGLTP 626
>UniRef100_O04693 Glossy1 homolog [Oryza sativa]
Length = 555
Score = 195 bits (495), Expect = 3e-48
Identities = 123/378 (32%), Positives = 183/378 (47%), Gaps = 17/378 (4%)
Query: 81 SLHHTKFRRNYSLFMPMYDYIYGTVD-KSTDVIYETSLMRPKESPDVVHLTHLTTFNSIY 139
SLHH + N+ LFMP++D + GT++ KS + E L + PD V L H+ S
Sbjct: 181 SLHHREKDSNFCLFMPLFDALGGTLNPKSWQLQKEVDLGKNHRVPDFVFLVHVVDVVSSM 240
Query: 140 QLRLGFASLASNPQTSKWYLHLMWPFTMFSMLMTWICGRAFVLESNSFKNLKLQCWLIPR 199
+ F + +S P + L +WP ML+ W C + F + + Q W +PR
Sbjct: 241 HVPFAFRACSSLPFATHLVLLPLWPIAFGFMLLQWFCSKTFTVSFYKLRGFLHQTWSVPR 300
Query: 200 FKRQYFSKWQSKTFNNLIEEAIVEAELNGAKVISLGLFNKNHQLNERHEHYIGRLPQLKI 259
+ QYF K N +IE AI+ A+ G KV+SL NKN LN ++ + P L++
Sbjct: 301 YGFQYFIPSAKKGINEMIELAILRADKMGVKVLSLAALNKNEALNGGGTLFVRKHPDLRV 360
Query: 260 KVVDGSSLAAATVLNNIPKGTNQVLLRGKFNKVAFVIANALCKKNVQVVVL--------- 310
+VV G++L AA +LN IP +V L G +K+ IA C+K ++V++L
Sbjct: 361 RVVHGNTLTAAVILNEIPGDVAEVFLTGATSKLGRAIALYFCRKKIRVLMLTLSTERFMN 420
Query: 311 -YKDELKELEQ-RINTSKGNLALSPFNTPKIWLVGDEWDEYEQMEAPKGSLFIPFSHFPP 368
++ E +Q + +K A K W+VG EQ AP G+ F F P
Sbjct: 421 IQREAPAEFQQYLVQVTKYQAA----QNCKTWIVGKWLSPREQRWAPAGTHFHQFVVPPI 476
Query: 369 KKMRKDCFYHYTPAMITPTTFMNSHSCENWLPRRVMSAWRIAGIIHALEGWNVHECGD-T 427
R+DC Y AM P +CE + R V+ A G++H LEGW+ HE G
Sbjct: 477 IGFRRDCTYGKLAAMRLPEDVEGLGTCEYTMGRGVVHACHAGGVVHFLEGWDHHEVGAID 536
Query: 428 ILSTEKVWEASIRHGFQP 445
+ + VW A++RHG P
Sbjct: 537 VDRIDAVWNAALRHGLTP 554
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.328 0.139 0.455
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 779,775,865
Number of Sequences: 2790947
Number of extensions: 32836229
Number of successful extensions: 86238
Number of sequences better than 10.0: 47
Number of HSP's better than 10.0 without gapping: 27
Number of HSP's successfully gapped in prelim test: 20
Number of HSP's that attempted gapping in prelim test: 86138
Number of HSP's gapped (non-prelim): 50
length of query: 451
length of database: 848,049,833
effective HSP length: 131
effective length of query: 320
effective length of database: 482,435,776
effective search space: 154379448320
effective search space used: 154379448320
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.8 bits)
S2: 76 (33.9 bits)
Medicago: description of AC147961.2


