

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147960.7 + phase: 0
(248 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
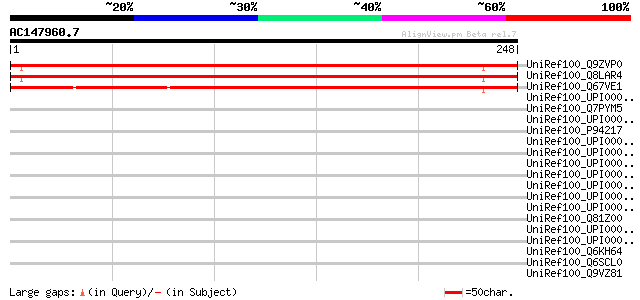
UniRef100_Q9ZVP0 T7A14.7 protein [Arabidopsis thaliana] 288 1e-76
UniRef100_Q8LAR4 Hypothetical protein [Arabidopsis thaliana] 286 4e-76
UniRef100_Q67VE1 Hypothetical protein OSJNBa0052G07.29 [Oryza sa... 229 6e-59
UniRef100_UPI00002437D1 UPI00002437D1 UniRef100 entry 40 0.072
UniRef100_Q7PYM5 ENSANGP00000007849 [Anopheles gambiae str. PEST] 40 0.072
UniRef100_UPI000033637F UPI000033637F UniRef100 entry 39 0.16
UniRef100_P94217 S-layer protein EA1 precursor [Bacillus anthracis] 38 0.21
UniRef100_UPI000028E1FA UPI000028E1FA UniRef100 entry 37 0.61
UniRef100_UPI000033B07E UPI000033B07E UniRef100 entry 36 1.0
UniRef100_UPI0000313722 UPI0000313722 UniRef100 entry 35 1.4
UniRef100_UPI00002EB26D UPI00002EB26D UniRef100 entry 35 1.4
UniRef100_UPI00002E3F1B UPI00002E3F1B UniRef100 entry 35 1.4
UniRef100_UPI000029235A UPI000029235A UniRef100 entry 35 1.4
UniRef100_UPI000028919F UPI000028919F UniRef100 entry 35 1.4
UniRef100_Q81Z00 Prophage LambdaBa04, tape measure protein, puta... 35 1.4
UniRef100_UPI0000268DDE UPI0000268DDE UniRef100 entry 34 3.9
UniRef100_UPI000027927E UPI000027927E UniRef100 entry 33 5.1
UniRef100_Q6KH64 Pyruvate dehydrogenase E3 component dihydrolipo... 33 5.1
UniRef100_Q6SCL0 S-layer protein [Bacillus thuringiensis] 33 5.1
UniRef100_Q9VZ81 CG13708-PA [Drosophila melanogaster] 33 5.1
>UniRef100_Q9ZVP0 T7A14.7 protein [Arabidopsis thaliana]
Length = 253
Score = 288 bits (736), Expect = 1e-76
Identities = 146/252 (57%), Positives = 184/252 (72%), Gaps = 4/252 (1%)
Query: 1 MSFL-AGRLAGKEAAYFFQESKQAVTKLAQKNNPISKTNVVDQRHVVQDNADVLPEVLRH 59
MSFL AGRLAGKEAAYFFQESK AV +LA+K+ K + DVLPE+LRH
Sbjct: 1 MSFLGAGRLAGKEAAYFFQESKHAVNRLAEKSPATGKKLPSSPPDPPEIQPDVLPEILRH 60
Query: 60 SLPSKLFRDETASSSSFSASKWVLQSDPKLRSSVSPDAINPLRAFVSLPQVTFGARRWEL 119
SLPSK++ SS SKW L+SDP S+SPD +NPLR +VSLPQVTFG RRW+L
Sbjct: 61 SLPSKIYGRPPDPSSLSQFSKWALESDPNATVSISPDVLNPLRGYVSLPQVTFGRRRWDL 120
Query: 120 PEAKHGVSASTANELRQDRYDVNVNPEKLKAASEGLANLGKAFAIATAVVFGGAAMVIGM 179
PE+++ V ASTANELR+DRY VNPEKLKAA EGL ++GKAFA AT ++FG A +V G
Sbjct: 121 PESENSVLASTANELRRDRYGTPVNPEKLKAAGEGLQHIGKAFAAATIIIFGSATLVFGT 180
Query: 180 VASKLELHNMGDLKTKGKDVVEPQLENIKNYFVPMKVWAENMSRKWHLERE---DVKQKA 236
ASKL++ N D++TKGKD+ +P+LE++K P++ WAENMS+KWH+E E +K+K
Sbjct: 181 AASKLDMRNADDIRTKGKDLFQPKLESMKEQVEPLRTWAENMSKKWHIENEGGNTIKEKP 240
Query: 237 IVKDLSKILGSK 248
I+K+LSKILG K
Sbjct: 241 ILKELSKILGPK 252
>UniRef100_Q8LAR4 Hypothetical protein [Arabidopsis thaliana]
Length = 253
Score = 286 bits (731), Expect = 4e-76
Identities = 145/252 (57%), Positives = 183/252 (72%), Gaps = 4/252 (1%)
Query: 1 MSFL-AGRLAGKEAAYFFQESKQAVTKLAQKNNPISKTNVVDQRHVVQDNADVLPEVLRH 59
MSFL AGRL GKEAAYFFQESK AV +LA+K+ K + DVLPE+LRH
Sbjct: 1 MSFLGAGRLPGKEAAYFFQESKHAVNRLAEKSPATGKKLPSSPPDPPEIQPDVLPEILRH 60
Query: 60 SLPSKLFRDETASSSSFSASKWVLQSDPKLRSSVSPDAINPLRAFVSLPQVTFGARRWEL 119
SLPSK++ SS SKW L+SDP S+SPD +NPLR +VSLPQVTFG RRW+L
Sbjct: 61 SLPSKIYGRPPDPSSLSQFSKWALESDPNATVSISPDVLNPLRGYVSLPQVTFGRRRWDL 120
Query: 120 PEAKHGVSASTANELRQDRYDVNVNPEKLKAASEGLANLGKAFAIATAVVFGGAAMVIGM 179
PE+++ V ASTANELR+DRY VNPEKLKAA EGL ++GKAFA AT ++FG A +V G
Sbjct: 121 PESENSVLASTANELRRDRYGTPVNPEKLKAAGEGLQHIGKAFAAATIIIFGSATLVFGT 180
Query: 180 VASKLELHNMGDLKTKGKDVVEPQLENIKNYFVPMKVWAENMSRKWHLERE---DVKQKA 236
ASKL++ N D++TKGKD+ +P+LE++K P++ WAENMS+KWH+E E +K+K
Sbjct: 181 AASKLDMRNADDIRTKGKDLFQPKLESMKEQVEPLRTWAENMSKKWHIENEGGNTIKEKP 240
Query: 237 IVKDLSKILGSK 248
I+K+LSKILG K
Sbjct: 241 ILKELSKILGPK 252
>UniRef100_Q67VE1 Hypothetical protein OSJNBa0052G07.29 [Oryza sativa]
Length = 251
Score = 229 bits (583), Expect = 6e-59
Identities = 120/249 (48%), Positives = 165/249 (66%), Gaps = 3/249 (1%)
Query: 1 MSFLAGRLAGKEAAYFFQESKQAVTKLAQKNNPISKTNVVDQRHVVQDNADVLPEVLRHS 60
MSFLAGRLA KE AYF QESK A +LA+K P S + DVLPE+LRH+
Sbjct: 1 MSFLAGRLAAKEGAYFLQESKHAAGRLAEKL-PASAPAPAPAPGSTSPSPDVLPEILRHA 59
Query: 61 LPSKLFRDETASSSSFSASKWVLQSDPKLRSSVSPDAINPLRAFVSLPQVTFGARRWELP 120
+P K S S S S+W + + +SPDA+NPLR++VSLPQ TFG +RW+LP
Sbjct: 60 VPIKATPPPGEPSLSAS-SRWAVPRGGAEAAGLSPDALNPLRSYVSLPQATFGPKRWQLP 118
Query: 121 EAKHGVSASTANELRQDRYDVNVNPEKLKAASEGLANLGKAFAIATAVVFGGAAMVIGMV 180
+ S+STANE R+DR+ ++PEKLKA G + +GKAF AT +VFGG+ V+
Sbjct: 119 NEQPNYSSSTANERRRDRHPPPMDPEKLKAVIAGYSQIGKAFIAATILVFGGSTAVLLYT 178
Query: 181 ASKLELHNMGDLKTKGKDVVEPQLENIKNYFVPMKVWAENMSRKWHLERE-DVKQKAIVK 239
A KL+LH++ D++TKG+D V+P+ + IK P++ WAE MSRKWH E + D K+K+I++
Sbjct: 179 ADKLQLHSVDDVRTKGRDAVQPRADMIKEQIAPLRSWAEEMSRKWHFEGDKDAKEKSIIR 238
Query: 240 DLSKILGSK 248
+LS+ LGS+
Sbjct: 239 ELSRALGSR 247
>UniRef100_UPI00002437D1 UPI00002437D1 UniRef100 entry
Length = 829
Score = 39.7 bits (91), Expect = 0.072
Identities = 18/46 (39%), Positives = 30/46 (65%)
Query: 53 LPEVLRHSLPSKLFRDETASSSSFSASKWVLQSDPKLRSSVSPDAI 98
LPE L L +L +ETA +S +A +W++Q+DP L++ V +A+
Sbjct: 710 LPEGLLQPLLQRLRLEETAHASKMTAKEWLMQADPSLKNIVGKEAL 755
>UniRef100_Q7PYM5 ENSANGP00000007849 [Anopheles gambiae str. PEST]
Length = 1082
Score = 39.7 bits (91), Expect = 0.072
Identities = 18/46 (39%), Positives = 30/46 (65%)
Query: 53 LPEVLRHSLPSKLFRDETASSSSFSASKWVLQSDPKLRSSVSPDAI 98
LPE L L +L +ETA +S +A +W++Q+DP L++ V +A+
Sbjct: 963 LPEGLLQPLLQRLRLEETAHASKMTAKEWLMQADPSLKNIVGKEAL 1008
>UniRef100_UPI000033637F UPI000033637F UniRef100 entry
Length = 98
Score = 38.5 bits (88), Expect = 0.16
Identities = 26/73 (35%), Positives = 36/73 (48%), Gaps = 3/73 (4%)
Query: 165 ATAVVFGGAAMVIGMVASKLELHNMGDLKTKGKDVVEPQLENIKNYFVPMKVWAENMSRK 224
A V+ G A M IG + MG G V +P + IKNYFVP + A+ ++ K
Sbjct: 25 AAQVMSGQAQMSIGGGVESMSRVPMGS--DGGAMVADPSVA-IKNYFVPQGISADMIASK 81
Query: 225 WHLEREDVKQKAI 237
W R+DV A+
Sbjct: 82 WGFTRDDVDAYAV 94
>UniRef100_P94217 S-layer protein EA1 precursor [Bacillus anthracis]
Length = 862
Score = 38.1 bits (87), Expect = 0.21
Identities = 37/126 (29%), Positives = 54/126 (42%), Gaps = 8/126 (6%)
Query: 103 AFVSLPQVTFGARRWELPEAKHGVSASTANELRQDRYD---VNVNPEKLKAASEGLANLG 159
AF + V+ + + P+ K ++ ST E + +Y V +PE L EG
Sbjct: 616 AFKNFELVSKVGQYGQSPDTKLDLNVSTTVEYQLSKYTSDRVYSDPENL----EGYEVES 671
Query: 160 KAFAIATAVVFGGAAMVIGMVASKLELHNMGDLKTKGKDVVEPQLENIKNYFVPMK-VWA 218
K A+A A + G +V G K+++H + T GK VE E I V K V
Sbjct: 672 KNLAVADAKIVGNKVVVTGKTPGKVDIHLTKNGATAGKATVEIVQETIAIKSVNFKPVQT 731
Query: 219 ENMSRK 224
EN K
Sbjct: 732 ENFVEK 737
>UniRef100_UPI000028E1FA UPI000028E1FA UniRef100 entry
Length = 239
Score = 36.6 bits (83), Expect = 0.61
Identities = 35/119 (29%), Positives = 54/119 (44%), Gaps = 8/119 (6%)
Query: 120 PEAKHGVSASTANELRQDRYDVNVNPEKL-KAASEGLANLGKAFAIATAVVFGGAAMVIG 178
P + G + L D YD +V+ ++ + + GL A A V+ G A M IG
Sbjct: 60 PVGEQGACIARTAVLNAD-YDQSVSGVQINRFCASGLEATNMA---AAQVMSGQAQMSIG 115
Query: 179 MVASKLELHNMGDLKTKGKDVVEPQLENIKNYFVPMKVWAENMSRKWHLEREDVKQKAI 237
+ MG G V +P + IKNYFVP + A+ ++ K+ R+DV A+
Sbjct: 116 GGVESMSRVPMGS--DGGAMVADPSVA-IKNYFVPQGISADLIATKYGFSRDDVDSYAV 171
>UniRef100_UPI000033B07E UPI000033B07E UniRef100 entry
Length = 169
Score = 35.8 bits (81), Expect = 1.0
Identities = 24/67 (35%), Positives = 33/67 (48%), Gaps = 3/67 (4%)
Query: 165 ATAVVFGGAAMVIGMVASKLELHNMGDLKTKGKDVVEPQLENIKNYFVPMKVWAENMSRK 224
A V+ G A M IG + MG G V +P + IKNYFVP + A+ ++ K
Sbjct: 102 AAQVMSGQAQMSIGGGVESMSRVPMGS--DGGAMVADPSVA-IKNYFVPQGISADMIASK 158
Query: 225 WHLERED 231
W R+D
Sbjct: 159 WGFTRDD 165
>UniRef100_UPI0000313722 UPI0000313722 UniRef100 entry
Length = 894
Score = 35.4 bits (80), Expect = 1.4
Identities = 20/62 (32%), Positives = 37/62 (59%), Gaps = 5/62 (8%)
Query: 179 MVASKLELHNMGDLKTK--GKDVVEPQLENIKNYFVPMKVWAENMSRKWHLEREDVKQKA 236
+++ +L+ ++GDL+ DV+E L NI +P+K W + S ++ REDV+QK
Sbjct: 686 IISDELDSVSLGDLEPHEWNTDVLESSLSNIFGLQIPIKQWIASES---NMTREDVEQKI 742
Query: 237 IV 238
++
Sbjct: 743 LM 744
>UniRef100_UPI00002EB26D UPI00002EB26D UniRef100 entry
Length = 226
Score = 35.4 bits (80), Expect = 1.4
Identities = 34/119 (28%), Positives = 54/119 (44%), Gaps = 8/119 (6%)
Query: 120 PEAKHGVSASTANELRQDRYDVNVNPEKL-KAASEGLANLGKAFAIATAVVFGGAAMVIG 178
P + G + L D YD +V+ ++ + + GL A A V+ G + M IG
Sbjct: 60 PVGEQGACIARTAVLNAD-YDQSVSGVQINRFCASGLEATNMA---AAQVMSGQSQMSIG 115
Query: 179 MVASKLELHNMGDLKTKGKDVVEPQLENIKNYFVPMKVWAENMSRKWHLEREDVKQKAI 237
+ MG G V +P + IKNYFVP + A+ ++ K+ R+DV A+
Sbjct: 116 GGVESMSRVPMGS--DGGAMVADPSVA-IKNYFVPQGISADLIATKYGFSRDDVDSYAV 171
>UniRef100_UPI00002E3F1B UPI00002E3F1B UniRef100 entry
Length = 228
Score = 35.4 bits (80), Expect = 1.4
Identities = 34/119 (28%), Positives = 54/119 (44%), Gaps = 8/119 (6%)
Query: 120 PEAKHGVSASTANELRQDRYDVNVNPEKL-KAASEGLANLGKAFAIATAVVFGGAAMVIG 178
P + G + L D YD +V+ ++ + + GL A A V+ G + M IG
Sbjct: 60 PVGEQGACIARTAVLNAD-YDQSVSGVQINRFCASGLEATNMA---AAQVMSGQSQMSIG 115
Query: 179 MVASKLELHNMGDLKTKGKDVVEPQLENIKNYFVPMKVWAENMSRKWHLEREDVKQKAI 237
+ MG G V +P + IKNYFVP + A+ ++ K+ R+DV A+
Sbjct: 116 GGVESMSRVPMGS--DGGAMVADPSVA-IKNYFVPQGISADLIATKYGFSRDDVDSYAV 171
>UniRef100_UPI000029235A UPI000029235A UniRef100 entry
Length = 185
Score = 35.4 bits (80), Expect = 1.4
Identities = 34/119 (28%), Positives = 54/119 (44%), Gaps = 8/119 (6%)
Query: 120 PEAKHGVSASTANELRQDRYDVNVNPEKL-KAASEGLANLGKAFAIATAVVFGGAAMVIG 178
P + G + L D YD +V+ ++ + + GL A A V+ G + M IG
Sbjct: 60 PVGEQGACIARTAVLNAD-YDQSVSGVQINRFCASGLEATNMA---AAQVMSGQSQMSIG 115
Query: 179 MVASKLELHNMGDLKTKGKDVVEPQLENIKNYFVPMKVWAENMSRKWHLEREDVKQKAI 237
+ MG G V +P + IKNYFVP + A+ ++ K+ R+DV A+
Sbjct: 116 GGVESMSRVPMGS--DGGAMVADPSVA-IKNYFVPQGISADLIATKYGFSRDDVDSYAV 171
>UniRef100_UPI000028919F UPI000028919F UniRef100 entry
Length = 402
Score = 35.4 bits (80), Expect = 1.4
Identities = 34/119 (28%), Positives = 54/119 (44%), Gaps = 8/119 (6%)
Query: 120 PEAKHGVSASTANELRQDRYDVNVNPEKL-KAASEGLANLGKAFAIATAVVFGGAAMVIG 178
P + G + L D YD +V+ ++ + + GL A A V+ G + M IG
Sbjct: 60 PVGEQGACIARTAVLNAD-YDQSVSGVQINRFCASGLEATNMA---AAQVMSGQSQMSIG 115
Query: 179 MVASKLELHNMGDLKTKGKDVVEPQLENIKNYFVPMKVWAENMSRKWHLEREDVKQKAI 237
+ MG G V +P + IKNYFVP + A+ ++ K+ R+DV A+
Sbjct: 116 GGVESMSRVPMGS--DGGAMVADPSVA-IKNYFVPQGISADLIATKYGFSRDDVDSYAV 171
>UniRef100_Q81Z00 Prophage LambdaBa04, tape measure protein, putative [Bacillus
anthracis]
Length = 1311
Score = 35.4 bits (80), Expect = 1.4
Identities = 32/129 (24%), Positives = 50/129 (37%), Gaps = 8/129 (6%)
Query: 127 SASTANELRQDRYDVNVNPEKLKAASEGLANLGKAF--AIATAVVFGGAAMVIGMVASKL 184
S N ++ R D+N ++ + ASE + LG + V GG VIG
Sbjct: 197 SVDDGNSIQNIRNDLNQLSQEAEEASESVKGLGVELENVLGGIVAGGGIQEVIGQALDMS 256
Query: 185 ELHN----MGDLKTKGKDVVEPQLENIKNYFVPMKVWAENMSRKWHLERE--DVKQKAIV 238
EL D+ K VE + + Y + E + R+W L ++ D I+
Sbjct: 257 ELKTKIDITFDVPESSKRSVEDAIRTVVAYGGDAEEALEGVRRQWSLNKDASDAANTEII 316
Query: 239 KDLSKILGS 247
K + I S
Sbjct: 317 KGAANIASS 325
>UniRef100_UPI0000268DDE UPI0000268DDE UniRef100 entry
Length = 162
Score = 33.9 bits (76), Expect = 3.9
Identities = 14/56 (25%), Positives = 34/56 (60%)
Query: 189 MGDLKTKGKDVVEPQLENIKNYFVPMKVWAENMSRKWHLEREDVKQKAIVKDLSKI 244
+ DLK K D ++ +++N + +++W E SRK ++ ++++KA+ ++ K+
Sbjct: 86 LSDLKEKRSDRLKKEIKNNASIINLVQLWKEEESRKKLIKLAEIRKKAVSNEVEKL 141
>UniRef100_UPI000027927E UPI000027927E UniRef100 entry
Length = 264
Score = 33.5 bits (75), Expect = 5.1
Identities = 24/84 (28%), Positives = 39/84 (45%), Gaps = 9/84 (10%)
Query: 130 TANELRQDRYDVNVNPEKLKAASEGLANLGKAFAIATAVVFGGAAMVIGMVASKLELHNM 189
T NE+ + N ++K A N KAF I TA+ G ++ V S+ + +
Sbjct: 184 TKNEINLGYVAITENANEIKTA----INERKAFVIRTAISVGFVILIFSFVLSRYFIKPI 239
Query: 190 GDL-----KTKGKDVVEPQLENIK 208
+L + K K V+P +EN+K
Sbjct: 240 QNLVGYTIRIKEKSQVKPGIENLK 263
>UniRef100_Q6KH64 Pyruvate dehydrogenase E3 component dihydrolipoamide dehydrogenase
[Mycoplasma mobile]
Length = 600
Score = 33.5 bits (75), Expect = 5.1
Identities = 42/173 (24%), Positives = 72/173 (41%), Gaps = 18/173 (10%)
Query: 68 DETASSSSFSASKWVLQSDPKLRSSVSPDAINPLRAFVSLPQVTFGARRWELPEAKHGVS 127
D+ +SS S ++++ ++ + + VS ++FG + P + +
Sbjct: 76 DDGSSSQSIEVKPAEIKAEAPKKAGGGASVVGEVA--VSDEVMSFGKKTSSTPTSSTSIQ 133
Query: 128 ASTANELRQDRYDVNV---NPEKLKAASEGLANLGKAFAIATAVVFGGAAMVIGMVASKL 184
++ N D+YDV V P AA E N GK I +GG + +G + +K
Sbjct: 134 PTSFNGKITDKYDVIVLGSGPGGYLAAEEAGKN-GKKTLIIEKEYWGGVCLNVGCIPTKA 192
Query: 185 ELHNMGDLKTKGKDVVEPQLENIKNYFVPMKVWAENMSRKWHLEREDVKQKAI 237
L K +V E QL + +Y + + V M+ K ER KQK +
Sbjct: 193 LL--------KSTEVFE-QLSHASDYGLDIDVSKLKMNWKKMQER---KQKVV 233
>UniRef100_Q6SCL0 S-layer protein [Bacillus thuringiensis]
Length = 862
Score = 33.5 bits (75), Expect = 5.1
Identities = 32/109 (29%), Positives = 46/109 (41%), Gaps = 8/109 (7%)
Query: 120 PEAKHGVSASTANELRQDRYD---VNVNPEKLKAASEGLANLGKAFAIATAVVFGGAAMV 176
P+ K ++ S E + +Y V +PE L EG K A+A A + G +V
Sbjct: 633 PDTKLDLNVSNKVEYQLSKYTSDRVYSDPENL----EGYVVESKNPAVAEAKIVGNKVVV 688
Query: 177 IGMVASKLELHNMGDLKTKGKDVVEPQLENIKNYFVPMK-VWAENMSRK 224
G K+++H + T GK +E E I V K V EN K
Sbjct: 689 TGKTPGKVDIHLTKNGATAGKATIEIVQETIAIESVNFKPVQTENFVEK 737
>UniRef100_Q9VZ81 CG13708-PA [Drosophila melanogaster]
Length = 1301
Score = 33.5 bits (75), Expect = 5.1
Identities = 18/47 (38%), Positives = 29/47 (61%), Gaps = 1/47 (2%)
Query: 53 LPEVLRHSLPSKLFRDETASSSSFSASKWVLQSDPK-LRSSVSPDAI 98
+PEV+ L ++L DET ++S S +W+L+ D K LR V +A+
Sbjct: 1162 MPEVMLQPLLARLRLDETCTASKLSPKEWLLRPDNKSLRLVVGKEAL 1208
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.314 0.129 0.360
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 385,598,861
Number of Sequences: 2790947
Number of extensions: 14659527
Number of successful extensions: 35224
Number of sequences better than 10.0: 29
Number of HSP's better than 10.0 without gapping: 8
Number of HSP's successfully gapped in prelim test: 21
Number of HSP's that attempted gapping in prelim test: 35207
Number of HSP's gapped (non-prelim): 30
length of query: 248
length of database: 848,049,833
effective HSP length: 124
effective length of query: 124
effective length of database: 501,972,405
effective search space: 62244578220
effective search space used: 62244578220
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 73 (32.7 bits)
Medicago: description of AC147960.7


