

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147875.5 - phase: 0
(459 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
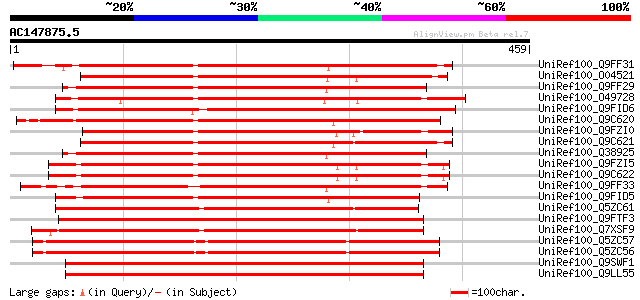
UniRef100_Q9FF31 Receptor serine/threonine kinase [Arabidopsis t... 384 e-105
UniRef100_O04521 F20P5.3 protein [Arabidopsis thaliana] 380 e-104
UniRef100_Q9FF29 Receptor serine/threonine kinase [Arabidopsis t... 378 e-103
UniRef100_O49728 Receptor serine/threonine kinase-like protein [... 378 e-103
UniRef100_Q9FID6 Receptor protein kinase-like protein [Arabidops... 377 e-103
UniRef100_Q9C620 Receptor serine/threonine kinase PR5K, putative... 375 e-102
UniRef100_Q9FZI0 F1O19.6 protein [Arabidopsis thaliana] 375 e-102
UniRef100_Q9C621 Receptor serine/threonine kinase PR5K, putative... 374 e-102
UniRef100_Q38925 Receptor serine/threonine kinase PR5K [Arabidop... 374 e-102
UniRef100_Q9FZI5 Hypothetical protein F1O19.1 [Arabidopsis thali... 373 e-102
UniRef100_Q9C622 Receptor serine/threonine kinase PR5K, putative... 373 e-102
UniRef100_Q9FF33 Receptor serine/threonine kinase-like protein [... 370 e-101
UniRef100_Q9FID5 Receptor protein kinase-like protein [Arabidops... 361 3e-98
UniRef100_Q5ZC61 Putative receptor serine/threonine kinase PR5K ... 359 8e-98
UniRef100_Q9FTF3 Putative receptor serine/threonine kinase PR5K ... 357 5e-97
UniRef100_Q7XSF9 OSJNBb0051N19.4 protein [Oryza sativa] 355 1e-96
UniRef100_Q5ZC57 Putative receptor serine/threonine kinase PR5K ... 355 1e-96
UniRef100_Q5ZC56 Putative receptor serine/threonine kinase PR5K ... 355 1e-96
UniRef100_Q9SWF1 Receptor kinase [Oryza sativa] 353 4e-96
UniRef100_Q9LL55 Receptor-like kinase [Oryza sativa] 353 4e-96
>UniRef100_Q9FF31 Receptor serine/threonine kinase [Arabidopsis thaliana]
Length = 638
Score = 384 bits (987), Expect = e-105
Identities = 198/394 (50%), Positives = 269/394 (68%), Gaps = 31/394 (7%)
Query: 4 INSGASVAGFGVTMFFIIFISCYFKKGIRRPQMTIFRKRRKHV---DNNVEVFMQSYNLS 60
I G G T+ + + +F+K R+ H+ DNN++ +Q
Sbjct: 263 IGIGLGCGFLGATLITVCLLCFFFQK----------RRTSHHLRPRDNNLKGLVQ----- 307
Query: 61 IARRYSYAEVKRITNSFRDKLGHGGYGVVYKASLTDGRQVAVKVINESKGNGEEFINEVA 120
++YSYAEV++IT F LG GG+G VY +L DGR+VAVK++ + K NGE+FINEVA
Sbjct: 308 -LKQYSYAEVRKITKLFSHTLGKGGFGTVYGGNLCDGRKVAVKILKDFKSNGEDFINEVA 366
Query: 121 SISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIYKSGFPDAVCDCDSNTLFQIA 180
S+S+TSH+NIVSLLG+CYE +KRA++YEF+ GSLD+F+ + + D +TL++IA
Sbjct: 367 SMSQTSHVNIVSLLGFCYEGSKRAIVYEFLENGSLDQFLSEK----KSLNLDVSTLYRIA 422
Query: 181 IGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQMNDSIVSIPGTR 240
+G+ARGL+YLH GC +RI+H DIKPQNILLD+ FCPK+SDFGLAK+C+ +SI+S+ R
Sbjct: 423 LGVARGLDYLHHGCKTRIVHFDIKPQNILLDDTFCPKVSDFGLAKLCEKRESILSLLDAR 482
Query: 241 GTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGRKN--YQTGGSCTSEMYFPDWIY 298
GTIGY+APEVFS +G VS+KSDVYSYGML+LEMIG + +T S +S YFPDWIY
Sbjct: 483 GTIGYIAPEVFSGMYGRVSHKSDVYSYGMLVLEMIGAKNKEIEETAASNSSSAYFPDWIY 542
Query: 299 KDLEQGNDLLN-SLTISEEENDMVKKITMVSLWCIQTNPLDRPPMNKVIEMLQGPLSSVT 357
K+LE G D IS E+ ++ KK+T+V LWCIQ +PL+RPPMN+++EM++G L +
Sbjct: 543 KNLENGEDTWKFGDEISREDKEVAKKMTLVGLWCIQPSPLNRPPMNRIVEMMEGSLDVLE 602
Query: 358 FPPKPVLFSPKRPPLQLSNMSSSDWQETNSITTE 391
PPKP + P QLS+ S E NSI TE
Sbjct: 603 VPPKPSIHYSAEPLPQLSSFS-----EENSIYTE 631
>UniRef100_O04521 F20P5.3 protein [Arabidopsis thaliana]
Length = 676
Score = 380 bits (975), Expect = e-104
Identities = 192/331 (58%), Positives = 242/331 (73%), Gaps = 14/331 (4%)
Query: 63 RRYSYAEVKRITNSFRDKLGHGGYGVVYKASLTDG-RQVAVKVINESKGNGEEFINEVAS 121
+R+SY +VK++T SF + LG GG+G VYK L DG R VAVK++ ES +GE+FINE+AS
Sbjct: 324 KRFSYVQVKKMTKSFENVLGKGGFGTVYKGKLPDGSRDVAVKILKESNEDGEDFINEIAS 383
Query: 122 ISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIYKSGFPDAVCDCDSNTLFQIAI 181
+SRTSH NIVSLLG+CYE K+A+IYE MP GSLDKFI K+ + TL+ IA+
Sbjct: 384 MSRTSHANIVSLLGFCYEGRKKAIIYELMPNGSLDKFISKN----MSAKMEWKTLYNIAV 439
Query: 182 GIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQMNDSIVSIPGTRG 241
G++ GLEYLH C SRI+H DIKPQNIL+D + CPKISDFGLAK+C+ N+SI+S+ RG
Sbjct: 440 GVSHGLEYLHSHCVSRIVHFDIKPQNILIDGDLCPKISDFGLAKLCKNNESIISMLHARG 499
Query: 242 TIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGRK--NYQTGGSCTSEMYFPDWIYK 299
TIGY+APEVFS+ FGGVS+KSDVYSYGM++LEMIG R Q GS + MYFPDWIYK
Sbjct: 500 TIGYIAPEVFSQNFGGVSHKSDVYSYGMVVLEMIGARNIGRAQNAGSSNTSMYFPDWIYK 559
Query: 300 DLEQGN--DLLNSLTISEEENDMVKKITMVSLWCIQTNPLDRPPMNKVIEMLQGPLSSVT 357
DLE+G L EE+ +VKK+ +V LWCIQTNP DRPPM+KV+EML+G L ++
Sbjct: 560 DLEKGEIMSFLADQITEEEDEKIVKKMVLVGLWCIQTNPYDRPPMSKVVEMLEGSLEALQ 619
Query: 358 FPPKPVLFSPK-RPPLQLSNMSSSDWQETNS 387
PPKP+L P P+ + D QET+S
Sbjct: 620 IPPKPLLCLPAITAPITV----DEDIQETSS 646
>UniRef100_Q9FF29 Receptor serine/threonine kinase [Arabidopsis thaliana]
Length = 665
Score = 378 bits (970), Expect = e-103
Identities = 188/326 (57%), Positives = 242/326 (73%), Gaps = 14/326 (4%)
Query: 47 DNNVEVFMQSYNLSIARRYSYAEVKRITNSFRDKLGHGGYGVVYKASLTD-GRQVAVKVI 105
D NVE +++ +RYSY VK++TNSF LG GG+G VYK L D GR VAVK++
Sbjct: 309 DQNVEA------VAMLKRYSYTRVKKMTNSFAHVLGKGGFGTVYKGKLADSGRDVAVKIL 362
Query: 106 NESKGNGEEFINEVASISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIYKSGFP 165
S+GNGEEFINEVAS+SRTSH+NIVSLLG+CYE NKRA+IYEFMP GSLDK+I +
Sbjct: 363 KVSEGNGEEFINEVASMSRTSHVNIVSLLGFCYEKNKRAIIYEFMPNGSLDKYISAN--- 419
Query: 166 DAVCDCDSNTLFQIAIGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAK 225
+ L+ +A+GI+RGLEYLH C +RI+H DIKPQNIL+DEN CPKISDFGLAK
Sbjct: 420 -MSTKMEWERLYDVAVGISRGLEYLHNRCVTRIVHFDIKPQNILMDENLCPKISDFGLAK 478
Query: 226 ICQMNDSIVSIPGTRGTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGR--KNYQT 283
+C+ +SI+S+ RGT GY+APE+FS+ FG VS+KSDVYSYGM++LEMIG + + +
Sbjct: 479 LCKNKESIISMLHMRGTFGYIAPEMFSKNFGAVSHKSDVYSYGMVVLEMIGAKNIEKVEY 538
Query: 284 GGSCTSEMYFPDWIYKDLEQGN-DLLNSLTISEEENDMVKKITMVSLWCIQTNPLDRPPM 342
GS MYFP+W+YKD E+G + +I++EE + KK+ +V+LWCIQ NP DRPPM
Sbjct: 539 SGSNNGSMYFPEWVYKDFEKGEITRIFGDSITDEEEKIAKKLVLVALWCIQMNPSDRPPM 598
Query: 343 NKVIEMLQGPLSSVTFPPKPVLFSPK 368
KVIEML+G L ++ PP P+LFSP+
Sbjct: 599 IKVIEMLEGNLEALQVPPNPLLFSPE 624
>UniRef100_O49728 Receptor serine/threonine kinase-like protein [Arabidopsis
thaliana]
Length = 687
Score = 378 bits (970), Expect = e-103
Identities = 195/372 (52%), Positives = 266/372 (71%), Gaps = 23/372 (6%)
Query: 41 KRRKHV-DNNVEVFMQSYNLSIARRYSYAEVKRITNSFRDKLGHGGYGVVYKASLTD--G 97
KR+ + D N+E + + +RYS+ +VK++TNSF +G GG+G VYK L D G
Sbjct: 324 KRKSELNDENIEAVV------MLKRYSFEKVKKMTNSFDHVIGKGGFGTVYKGKLPDASG 377
Query: 98 RQVAVKVINESKGNGEEFINEVASISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDK 157
R +A+K++ ESKGNGEEFINE+ S+SR SH+NIVSL G+CYE ++RA+IYEFMP GSLDK
Sbjct: 378 RDIALKILKESKGNGEEFINELVSMSRASHVNIVSLFGFCYEGSQRAIIYEFMPNGSLDK 437
Query: 158 FIYKSGFPDAVCDCDSNTLFQIAIGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPK 217
FI ++ + TL+ IA+G+ARGLEYLH C S+I+H DIKPQNIL+DE+ CPK
Sbjct: 438 FISEN----MSTKIEWKTLYNIAVGVARGLEYLHNSCVSKIVHFDIKPQNILIDEDLCPK 493
Query: 218 ISDFGLAKICQMNDSIVSIPGTRGTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGG 277
ISDFGLAK+C+ +SI+S+ RGT+GY+APE+FS+ +GGVS+KSDVYSYGM++LEMIG
Sbjct: 494 ISDFGLAKLCKKKESIISMLDARGTVGYIAPEMFSKNYGGVSHKSDVYSYGMVVLEMIGA 553
Query: 278 --RKNYQTGGSCTSEMYFPDWIYKDLEQGND--LLNSLTISEEEND-MVKKITMVSLWCI 332
R+ +T + S MYFPDW+Y+DLE+ LL I EEE + +VK++T+V LWCI
Sbjct: 554 TKREEVETSATDKSSMYFPDWVYEDLERKETMRLLEDHIIEEEEEEKIVKRMTLVGLWCI 613
Query: 333 QTNPLDRPPMNKVIEMLQGP-LSSVTFPPKPVLFSPKRPPLQLSNMSSSDWQETNSITTE 391
QTNP DRPPM KV+EML+G L ++ PPKP+L + +S D Q+T+ ++T+
Sbjct: 614 QTNPSDRPPMRKVVEMLEGSRLEALQVPPKPLL----NLHVVTDWETSEDSQQTSRLSTQ 669
Query: 392 TELEEEPIEGYQ 403
+ LE + YQ
Sbjct: 670 SLLERKTRSNYQ 681
>UniRef100_Q9FID6 Receptor protein kinase-like protein [Arabidopsis thaliana]
Length = 813
Score = 377 bits (968), Expect = e-103
Identities = 190/357 (53%), Positives = 257/357 (71%), Gaps = 13/357 (3%)
Query: 41 KRRKHVDNNVEVFMQSYNLSIARRYSYAEVKRITNSFRDKLGHGGYGVVYKASLTDGRQV 100
KR+K+ N + + + ++Y YAE+K+IT SF +G GG+G VY+ +L++GR V
Sbjct: 466 KRKKNKKENSVIMFKL----LLKQYIYAELKKITKSFSHTVGKGGFGTVYRGNLSNGRTV 521
Query: 101 AVKVINESKGNGEEFINEVASISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIY 160
AVKV+ + KGNG++FINEV S+S+TSH+NIVSLLG+CYE +KRA+I EF+ GSLD+FI
Sbjct: 522 AVKVLKDLKGNGDDFINEVTSMSQTSHVNIVSLLGFCYEGSKRAIISEFLEHGSLDQFIS 581
Query: 161 --KSGFPDAVCDCDSNTLFQIAIGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKI 218
KS P+ TL+ IA+GIARGLEYLH GC +RI+H DIKPQNILLD+NFCPK+
Sbjct: 582 RNKSLTPNVT------TLYGIALGIARGLEYLHYGCKTRIVHFDIKPQNILLDDNFCPKV 635
Query: 219 SDFGLAKICQMNDSIVSIPGTRGTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGR 278
+DFGLAK+C+ +SI+S+ TRGTIGY+APEV SR +GG+S+KSDVYSYGML+L+MIG R
Sbjct: 636 ADFGLAKLCEKRESILSLIDTRGTIGYIAPEVVSRMYGGISHKSDVYSYGMLVLDMIGAR 695
Query: 279 KNYQTGGSCTSEMYFPDWIYKDLEQGNDL-LNSLTISEEENDMVKKITMVSLWCIQTNPL 337
+T S YFPDWIYKDLE G+ + I+EE+N +VKK+ +VSLWCI+ P
Sbjct: 696 NKVETTTCNGSTAYFPDWIYKDLENGDQTWIIGDEINEEDNKIVKKMILVSLWCIRPCPS 755
Query: 338 DRPPMNKVIEMLQGPLSSVTFPPKPVLFSPKRPPLQLSNMSSSDWQETNSITTETEL 394
DRPPMNKV+EM++G L ++ PPKP L+ S++S E + T ++ +
Sbjct: 756 DRPPMNKVVEMIEGSLDALELPPKPSRHISTELVLESSSLSDGQEAEKQTQTLDSTI 812
>UniRef100_Q9C620 Receptor serine/threonine kinase PR5K, putative [Arabidopsis
thaliana]
Length = 655
Score = 375 bits (964), Expect = e-102
Identities = 199/378 (52%), Positives = 263/378 (68%), Gaps = 11/378 (2%)
Query: 7 GASVAGFGVTMFFIIFISCYFKKGIRRPQMTIFRKRRKHVDNNVEVFMQSYNLSIARRYS 66
G A F + M I+ ++C IRR + T+ R + D++ + +++ L + YS
Sbjct: 273 GIGAASFAM-MGVILVVTC-LNCLIRRQRKTLNDPRMRTSDDSRQQNLKA--LIPLKHYS 328
Query: 67 YAEVKRITNSFRDKLGHGGYGVVYKASLTDGRQVAVKVINESKGNGEEFINEVASISRTS 126
YA+V IT SF + +G GG+G VY+ +L DGR VAVKV+ ES+GNGE+FINEVAS+S+TS
Sbjct: 329 YAQVTSITKSFAEVIGKGGFGTVYRGTLYDGRSVAVKVLKESQGNGEDFINEVASMSQTS 388
Query: 127 HMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIYKSGFPDAVCDCDSNTLFQIAIGIARG 186
H+NIV+LLG+C E KRA+IYEFM GSLDKFI D L+ IA+G+ARG
Sbjct: 389 HVNIVTLLGFCSEGYKRAIIYEFMENGSLDKFISSK----KSSTMDWRELYGIALGVARG 444
Query: 187 LEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQMNDSIVSIPGTRGTIGYM 246
LEYLH GC +RI+H DIKPQN+LLD+N PK+SDFGLAK+C+ +SI+S+ TRGTIGY+
Sbjct: 445 LEYLHHGCRTRIVHFDIKPQNVLLDDNLSPKVSDFGLAKLCERKESILSLMDTRGTIGYI 504
Query: 247 APEVFSRAFGGVSYKSDVYSYGMLILEMIGGRKNYQTGG--SCTSEMYFPDWIYKDLEQG 304
APEVFSR +G VS+KSDVYSYGML+L++IG R T S TS MYFP+WIY+DLE+
Sbjct: 505 APEVFSRVYGRVSHKSDVYSYGMLVLDIIGARNKTSTEDTTSSTSSMYFPEWIYRDLEKA 564
Query: 305 -NDLLNSLTISEEENDMVKKITMVSLWCIQTNPLDRPPMNKVIEMLQGPLSSVTFPPKPV 363
N IS EE+++ KK+T+V LWCIQ PLDRP MN+V+EM++G L ++ PP+PV
Sbjct: 565 HNGKSIETAISNEEDEIAKKMTLVGLWCIQPWPLDRPAMNRVVEMMEGNLDALEVPPRPV 624
Query: 364 LFSPKRPPLQLSNMSSSD 381
L LQ S+ S D
Sbjct: 625 LQQIPTATLQESSTFSED 642
>UniRef100_Q9FZI0 F1O19.6 protein [Arabidopsis thaliana]
Length = 1111
Score = 375 bits (962), Expect = e-102
Identities = 184/334 (55%), Positives = 251/334 (75%), Gaps = 18/334 (5%)
Query: 65 YSYAEVKRITNSFRDKLGHGGYGVVYKASLTDGRQVAVKVINESKGNGEEFINEVASISR 124
Y+YA+VKRIT SF + +G GG+G+VYK +L+DGR VAVKV+ ++KGNGE+FINEVA++SR
Sbjct: 788 YTYAQVKRITKSFAEVVGRGGFGIVYKGTLSDGRVVAVKVLKDTKGNGEDFINEVATMSR 847
Query: 125 TSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIYKSGFPDAVCDCDSNTLFQIAIGIA 184
TSH+NIVSLLG+C E +KRA+IYEF+ GSLDKFI + D L++IA+G+A
Sbjct: 848 TSHLNIVSLLGFCSEGSKRAIIYEFLENGSLDKFILGK----TSVNMDWTALYRIALGVA 903
Query: 185 RGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQMNDSIVSIPGTRGTIG 244
GLEYLH C +RI+H DIKPQN+LLD++FCPK+SDFGLAK+C+ +SI+S+ TRGTIG
Sbjct: 904 HGLEYLHHSCKTRIVHFDIKPQNVLLDDSFCPKVSDFGLAKLCEKKESILSMLDTRGTIG 963
Query: 245 YMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGRKNYQTGGSC---TSEMYFPDWIYKDL 301
Y+APE+ SR +G VS+KSDVYSYGML+LE+IG R + +C TS MYFP+W+Y+DL
Sbjct: 964 YIAPEMISRVYGNVSHKSDVYSYGMLVLEIIGARNKEKANQACASNTSSMYFPEWVYRDL 1023
Query: 302 E---QGNDLLNSLTISEEENDMVKKITMVSLWCIQTNPLDRPPMNKVIEMLQGPLSSVTF 358
E G + + I+ EE+++ KK+T+V LWCIQ +P+DRP MN+V+EM++G L ++
Sbjct: 1024 ESCKSGRHIEDG--INSEEDELAKKMTLVGLWCIQPSPVDRPAMNRVVEMMEGSLEALEV 1081
Query: 359 PPKPVLFSPKRPPLQLSNM-SSSDWQETNSITTE 391
PP+PVL + +SN+ SS E S+ TE
Sbjct: 1082 PPRPVL-----QQIPISNLHESSILSEDVSVYTE 1110
>UniRef100_Q9C621 Receptor serine/threonine kinase PR5K, putative [Arabidopsis
thaliana]
Length = 609
Score = 374 bits (960), Expect = e-102
Identities = 191/332 (57%), Positives = 247/332 (73%), Gaps = 13/332 (3%)
Query: 63 RRYSYAEVKRITNSFRDKLGHGGYGVVYKASLTDGRQVAVKVINESKGN-GEEFINEVAS 121
++YSY +VKRITNSF + +G GG+G+VY+ +L+DGR VAVKV+ + KGN GE+FINEVAS
Sbjct: 287 KQYSYEQVKRITNSFAEVVGRGGFGIVYRGTLSDGRMVAVKVLKDLKGNNGEDFINEVAS 346
Query: 122 ISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIYKSGFPDAVCDCDSNTLFQIAI 181
+S+TSH+NIV+LLG+C E KRA+IYEFM GSLDKFI D L+ IA+
Sbjct: 347 MSQTSHVNIVTLLGFCSEGYKRAIIYEFMENGSLDKFISSK----KSSTMDWRELYGIAL 402
Query: 182 GIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQMNDSIVSIPGTRG 241
G+ARGLEYLH GC +RI+H DIKPQN+LLD+N PK+SDFGLAK+C+ +SI+S+ TRG
Sbjct: 403 GVARGLEYLHHGCRTRIVHFDIKPQNVLLDDNLSPKVSDFGLAKLCERKESILSLMDTRG 462
Query: 242 TIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGRKNYQTGG--SCTSEMYFPDWIYK 299
TIGY+APEVFSR +G VS+KSDVYSYGML+L++IG R T S TS MYFP+WIYK
Sbjct: 463 TIGYIAPEVFSRVYGSVSHKSDVYSYGMLVLDIIGARNKTSTEDTTSSTSSMYFPEWIYK 522
Query: 300 DLEQGNDLLNSLTISEEENDMVKKITMVSLWCIQTNPLDRPPMNKVIEMLQGPLSSVTFP 359
DLE+G D + EE+++ KK+T+V LWCIQ PLDRP MN+V+EM++G L ++ P
Sbjct: 523 DLEKG-DNGRLIVNRSEEDEIAKKMTLVGLWCIQPWPLDRPAMNRVVEMMEGNLDALEVP 581
Query: 360 PKPVLFSPKRPPLQLSNMSSSDWQETNSITTE 391
P+PVL P L S +S E NSI++E
Sbjct: 582 PRPVLQCSVVPHLDSSWIS-----EENSISSE 608
>UniRef100_Q38925 Receptor serine/threonine kinase PR5K [Arabidopsis thaliana]
Length = 665
Score = 374 bits (960), Expect = e-102
Identities = 187/326 (57%), Positives = 239/326 (72%), Gaps = 14/326 (4%)
Query: 47 DNNVEVFMQSYNLSIARRYSYAEVKRITNSFRDKLGHGGYGVVYKASLTD-GRQVAVKVI 105
D NVE +++ +RYSY VK++TNSF LG GG+G VYK L D GR VAVK++
Sbjct: 309 DQNVEA------VAMLKRYSYTRVKKMTNSFAHVLGKGGFGTVYKGKLADSGRDVAVKIL 362
Query: 106 NESKGNGEEFINEVASISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIYKSGFP 165
S+GNGEEFINEVAS+SRTSH+NIVSLLG+CYE NKRA+IYEFMP GSLDK+I +
Sbjct: 363 KVSEGNGEEFINEVASMSRTSHVNIVSLLGFCYEKNKRAIIYEFMPNGSLDKYISAN--- 419
Query: 166 DAVCDCDSNTLFQIAIGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAK 225
+ L+ +A+GI+RGLEYLH C +RI+H DIKPQNIL+DEN CPKISDFGLAK
Sbjct: 420 -MSTKMEWERLYDVAVGISRGLEYLHNRCVTRIVHFDIKPQNILMDENLCPKISDFGLAK 478
Query: 226 ICQMNDSIVSIPGTRGTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGR--KNYQT 283
+C+ +SI+S+ RGT GY+APE+FS+ FG VS+KSDVYSYGM++LEMIG + + +
Sbjct: 479 LCKNKESIISMLHMRGTFGYIAPEMFSKNFGAVSHKSDVYSYGMVVLEMIGAKNIEKVEY 538
Query: 284 GGSCTSEMYFPDWIYKDLEQGN-DLLNSLTISEEENDMVKKITMVSLWCIQTNPLDRPPM 342
S MYFP+W+YKD E+G + +I+EEE KK+ +V+LWCIQ NP DRPPM
Sbjct: 539 SESNNGSMYFPEWVYKDFEKGEITRIFGNSITEEEEKFAKKLVLVALWCIQMNPSDRPPM 598
Query: 343 NKVIEMLQGPLSSVTFPPKPVLFSPK 368
KV EML+G L ++ PP P+LFSP+
Sbjct: 599 IKVTEMLEGNLEALQVPPNPLLFSPE 624
>UniRef100_Q9FZI5 Hypothetical protein F1O19.1 [Arabidopsis thaliana]
Length = 727
Score = 373 bits (958), Expect = e-102
Identities = 191/363 (52%), Positives = 261/363 (71%), Gaps = 20/363 (5%)
Query: 35 QMTIFRKRRKHVDNNVEVFMQSYNLSIARRYSYAEVKRITNSFRDKLGHGGYGVVYKASL 94
++ IFR R+ D E L + Y+YA+VKR+T SF + +G GG+G+VY+ +L
Sbjct: 362 KVRIFRNRKTSDDRRQEKLKALIPL---KHYTYAQVKRMTKSFAEVVGRGGFGIVYRGTL 418
Query: 95 TDGRQVAVKVINESKGNG-EEFINEVASISRTSHMNIVSLLGYCYEANKRALIYEFMPKG 153
DGR VAVKV+ ESKGN E+FINEV+S+S+TSH+NIVSLLG+C E ++RA+IYEF+ G
Sbjct: 419 CDGRMVAVKVLKESKGNNSEDFINEVSSMSQTSHVNIVSLLGFCSEGSRRAIIYEFLENG 478
Query: 154 SLDKFIYKSGFPDAVCDCDSNTLFQIAIGIARGLEYLHQGCSSRILHLDIKPQNILLDEN 213
SLDKFI + D L+ IA+G+ARGLEYLH GC +RI+H DIKPQN+LLD+N
Sbjct: 479 SLDKFISEK----TSVILDLTALYGIALGVARGLEYLHYGCKTRIVHFDIKPQNVLLDDN 534
Query: 214 FCPKISDFGLAKICQMNDSIVSIPGTRGTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILE 273
PK+SDFGLAK+C+ +S++S+ TRGTIGY+APE+ SR +G VS+KSDVYSYGML+ E
Sbjct: 535 LSPKVSDFGLAKLCEKKESVMSLMDTRGTIGYIAPEMISRVYGSVSHKSDVYSYGMLVFE 594
Query: 274 MIGGRKNYQTGGSCT--SEMYFPDWIYKDLEQGN--DLLN-SLTISEEENDMVKKITMVS 328
MIG RK + G + S MYFP+WIYKDLE+ + DL + + IS EE ++ KK+T+V
Sbjct: 595 MIGARKKERFGQNSANGSSMYFPEWIYKDLEKADNGDLEHIEIGISSEEEEIAKKMTLVG 654
Query: 329 LWCIQTNPLDRPPMNKVIEMLQGPLSSVTFPPKPVLFSPKRPPLQLSNMSSSDW--QETN 386
LWCIQ++P DRPPMNKV+EM++G L ++ PP+PVL + + + S W +E++
Sbjct: 655 LWCIQSSPSDRPPMNKVVEMMEGSLDALEVPPRPVL-----QQIHVGPLLESSWITEESS 709
Query: 387 SIT 389
SI+
Sbjct: 710 SIS 712
>UniRef100_Q9C622 Receptor serine/threonine kinase PR5K, putative [Arabidopsis
thaliana]
Length = 876
Score = 373 bits (958), Expect = e-102
Identities = 191/363 (52%), Positives = 261/363 (71%), Gaps = 20/363 (5%)
Query: 35 QMTIFRKRRKHVDNNVEVFMQSYNLSIARRYSYAEVKRITNSFRDKLGHGGYGVVYKASL 94
++ IFR R+ D E L + Y+YA+VKR+T SF + +G GG+G+VY+ +L
Sbjct: 511 KVRIFRNRKTSDDRRQEKLKALIPL---KHYTYAQVKRMTKSFAEVVGRGGFGIVYRGTL 567
Query: 95 TDGRQVAVKVINESKGNG-EEFINEVASISRTSHMNIVSLLGYCYEANKRALIYEFMPKG 153
DGR VAVKV+ ESKGN E+FINEV+S+S+TSH+NIVSLLG+C E ++RA+IYEF+ G
Sbjct: 568 CDGRMVAVKVLKESKGNNSEDFINEVSSMSQTSHVNIVSLLGFCSEGSRRAIIYEFLENG 627
Query: 154 SLDKFIYKSGFPDAVCDCDSNTLFQIAIGIARGLEYLHQGCSSRILHLDIKPQNILLDEN 213
SLDKFI + D L+ IA+G+ARGLEYLH GC +RI+H DIKPQN+LLD+N
Sbjct: 628 SLDKFISEK----TSVILDLTALYGIALGVARGLEYLHYGCKTRIVHFDIKPQNVLLDDN 683
Query: 214 FCPKISDFGLAKICQMNDSIVSIPGTRGTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILE 273
PK+SDFGLAK+C+ +S++S+ TRGTIGY+APE+ SR +G VS+KSDVYSYGML+ E
Sbjct: 684 LSPKVSDFGLAKLCEKKESVMSLMDTRGTIGYIAPEMISRVYGSVSHKSDVYSYGMLVFE 743
Query: 274 MIGGRKNYQTGGSCT--SEMYFPDWIYKDLEQGN--DLLN-SLTISEEENDMVKKITMVS 328
MIG RK + G + S MYFP+WIYKDLE+ + DL + + IS EE ++ KK+T+V
Sbjct: 744 MIGARKKERFGQNSANGSSMYFPEWIYKDLEKADNGDLEHIEIGISSEEEEIAKKMTLVG 803
Query: 329 LWCIQTNPLDRPPMNKVIEMLQGPLSSVTFPPKPVLFSPKRPPLQLSNMSSSDW--QETN 386
LWCIQ++P DRPPMNKV+EM++G L ++ PP+PVL + + + S W +E++
Sbjct: 804 LWCIQSSPSDRPPMNKVVEMMEGSLDALEVPPRPVL-----QQIHVGPLLESSWITEESS 858
Query: 387 SIT 389
SI+
Sbjct: 859 SIS 861
>UniRef100_Q9FF33 Receptor serine/threonine kinase-like protein [Arabidopsis
thaliana]
Length = 611
Score = 370 bits (949), Expect = e-101
Identities = 195/381 (51%), Positives = 263/381 (68%), Gaps = 32/381 (8%)
Query: 10 VAGFGVTMFFIIFISCYFKKGIRRPQMTIFRKRRKHVDNNVEVFMQSYNLSIARRYSYAE 69
V GF V + ++ I+ F RR ++ RK+ N+E + S RRYSY +
Sbjct: 254 VVGFHVFIIVVMIIAFLFW---RRKKVNDLRKQ------NLEALVTS------RRYSYRQ 298
Query: 70 VKRITNSFRDKLGHGGYGVVYKASLTDGRQVAVKVINESKGNGEEFINEVASISRTSHMN 129
+K+IT SF + +G GG+G VYK +L DGR+VAVK++ +S GN E+FINEVASIS+TSH+N
Sbjct: 299 IKKITKSFTEVVGRGGFGTVYKGNLRDGRKVAVKILKDSNGNCEDFINEVASISQTSHVN 358
Query: 130 IVSLLGYCYEANKRALIYEFMPKGSLDKFIYKSGFPDAVCDCDSNTLFQIAIGIARGLEY 189
IVSLLG+C+E +KRA++YEF+ GSLD+ + D +TL+ IA+G+ARG+EY
Sbjct: 359 IVSLLGFCFEKSKRAIVYEFLENGSLDQS----------SNLDVSTLYGIALGVARGIEY 408
Query: 190 LHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQMNDSIVSIPGTRGTIGYMAPE 249
LH GC RI+H DIKPQN+LLDEN PK++DFGLAK+C+ +SI+S+ TRGTIGY+APE
Sbjct: 409 LHFGCKKRIVHFDIKPQNVLLDENLKPKVADFGLAKLCEKQESILSLLDTRGTIGYIAPE 468
Query: 250 VFSRAFGGVSYKSDVYSYGMLILEMIGGR--KNYQTGGSCTSEMYFPDWIYKDLEQGNDL 307
+FSR +G VS+KSDVYSYGML+LEM G R + Q S S YFPDWI+KDLE G+ +
Sbjct: 469 LFSRVYGNVSHKSDVYSYGMLVLEMTGARNKERVQNADSNNSSAYFPDWIFKDLENGDYV 528
Query: 308 -LNSLTISEEENDMVKKITMVSLWCIQTNPLDRPPMNKVIEMLQGPLSSVTFPPKPVLFS 366
L + ++ EE D+ KK+ +V LWCIQ P DRP MNKV+ M++G L S+ PPKP+L
Sbjct: 529 KLLADGLTREEEDIAKKMILVGLWCIQFRPSDRPSMNKVVGMMEGNLDSLDPPPKPLL-- 586
Query: 367 PKRPPLQLSNMSSSDWQETNS 387
P+Q +N SS E +S
Sbjct: 587 --HMPMQNNNAESSQPSEEDS 605
>UniRef100_Q9FID5 Receptor protein kinase-like protein [Arabidopsis thaliana]
Length = 806
Score = 361 bits (926), Expect = 3e-98
Identities = 177/325 (54%), Positives = 240/325 (73%), Gaps = 12/325 (3%)
Query: 41 KRRKHVDNNVEVFMQSYNLSIARRYSYAEVKRITNSFRDKLGHGGYGVVYKASLTDGRQV 100
KR+ + V +F + N+ Y+YAE+K+IT SF +G GG+G VY +L++GR+V
Sbjct: 469 KRKNRKEERVVMFKKLLNM-----YTYAELKKITKSFSYIIGKGGFGTVYGGNLSNGRKV 523
Query: 101 AVKVINESKGNGEEFINEVASISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIY 160
AVKV+ + KG+ E+FINEVAS+S+TSH+NIVSLLG+C+E +KRA++YEF+ GSLD+F+
Sbjct: 524 AVKVLKDLKGSAEDFINEVASMSQTSHVNIVSLLGFCFEGSKRAIVYEFLENGSLDQFMS 583
Query: 161 KSGFPDAVCDCDSNTLFQIAIGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISD 220
++ D TL+ IA+GIARGLEYLH GC +RI+H DIKPQNILLD N CPK+SD
Sbjct: 584 RN----KSLTQDVTTLYGIALGIARGLEYLHYGCKTRIVHFDIKPQNILLDGNLCPKVSD 639
Query: 221 FGLAKICQMNDSIVSIPGTRGTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGRKN 280
FGLAK+C+ +S++S+ TRGTIGY+APEVFSR +G VS+KSDVYS+GML+++MIG R
Sbjct: 640 FGLAKLCEKRESVLSLMDTRGTIGYIAPEVFSRMYGRVSHKSDVYSFGMLVIDMIGARSK 699
Query: 281 --YQTGGSCTSEMYFPDWIYKDLEQGNDL-LNSLTISEEENDMVKKITMVSLWCIQTNPL 337
+T S S YFPDWIYKDLE G + I++EE ++ KK+ +V LWCIQ P
Sbjct: 700 EIVETVDSAASSTYFPDWIYKDLEDGEQTWIFGDEITKEEKEIAKKMIVVGLWCIQPCPS 759
Query: 338 DRPPMNKVIEMLQGPLSSVTFPPKP 362
DRP MN+V+EM++G L ++ PPKP
Sbjct: 760 DRPSMNRVVEMMEGSLDALEIPPKP 784
>UniRef100_Q5ZC61 Putative receptor serine/threonine kinase PR5K [Oryza sativa]
Length = 598
Score = 359 bits (922), Expect = 8e-98
Identities = 174/321 (54%), Positives = 232/321 (72%), Gaps = 4/321 (1%)
Query: 41 KRRKHVDNNVEVFMQSYNLSIARRYSYAEVKRITNSFRDKLGHGGYGVVYKASLTDGRQV 100
K R ++N++ + SY +RY Y+EV +IT+ +KLG GGYGVV+K L DGR V
Sbjct: 275 KTRSRNESNIQKLIVSYGSLAPKRYKYSEVAKITSFLSNKLGEGGYGVVFKGKLQDGRLV 334
Query: 101 AVKVINESKGNGEEFINEVASISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIY 160
AVK +++SKGNGEEF+NEV SI RTSH+NIVSL G+C E +KRALIY++MP SLD +IY
Sbjct: 335 AVKFLHDSKGNGEEFVNEVMSIGRTSHVNIVSLFGFCLEGSKRALIYDYMPNSSLDNYIY 394
Query: 161 KSGFPDAVCDCDSNTLFQIAIGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISD 220
+ + L+ IAIGIARGLEYLH GC++RI+H DIKPQNILLD++FCPKI+D
Sbjct: 395 SEKPKETL---GWEKLYDIAIGIARGLEYLHHGCNTRIVHFDIKPQNILLDQDFCPKIAD 451
Query: 221 FGLAKICQMNDSIVSIPGTRGTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGRKN 280
FGLAK+C +S +S+ G RGTIG++APEV R+FG VS KSDVYSYGM++LEMIGGRKN
Sbjct: 452 FGLAKLCCTKESKLSMTGARGTIGFIAPEVLYRSFGVVSIKSDVYSYGMMLLEMIGGRKN 511
Query: 281 YQTGGSCTSEMYFPDWIYKDLEQGNDLLNSLTISEEENDMVKKITMVSLWCIQTNPLDRP 340
++ +SE YFPDWIY QG D L + ++ E ++ KK+T++ LWC+Q P+ RP
Sbjct: 512 VKSMVQNSSEKYFPDWIYDHFYQG-DGLQACEVTSEVEEIAKKMTLIGLWCVQVLPMHRP 570
Query: 341 PMNKVIEMLQGPLSSVTFPPK 361
+ +V++M + L + PPK
Sbjct: 571 TITQVLDMFEKALDELDMPPK 591
>UniRef100_Q9FTF3 Putative receptor serine/threonine kinase PR5K [Oryza sativa]
Length = 621
Score = 357 bits (915), Expect = 5e-97
Identities = 169/323 (52%), Positives = 232/323 (71%)
Query: 44 KHVDNNVEVFMQSYNLSIARRYSYAEVKRITNSFRDKLGHGGYGVVYKASLTDGRQVAVK 103
K + VE+F+++Y S RY++++VK+IT F++KLGHGG+G VYK L +G VAVK
Sbjct: 293 KEIHLKVEMFLKTYGTSKPMRYTFSDVKKITRRFKNKLGHGGFGSVYKGELPNGVPVAVK 352
Query: 104 VINESKGNGEEFINEVASISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIYKSG 163
++ S G GEEFINEVA+I R H NIV LLG+C E +RALIYEFMP SL+K+I+ +G
Sbjct: 353 MLENSLGEGEEFINEVATIGRIHHANIVRLLGFCSEGTRRALIYEFMPNESLEKYIFSNG 412
Query: 164 FPDAVCDCDSNTLFQIAIGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGL 223
+ + IA+GIARG+EYLHQGC+ RILH DIKP NILLD +F PKISDFGL
Sbjct: 413 SNISREFLVPKKMLDIALGIARGMEYLHQGCNQRILHFDIKPHNILLDYSFSPKISDFGL 472
Query: 224 AKICQMNDSIVSIPGTRGTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGRKNYQT 283
AK+C + SIV++ RGT+GY+APE++SR+FG +SYKSDVYS+GML+LEM+ GR+N
Sbjct: 473 AKLCARDQSIVTLTAARGTMGYIAPELYSRSFGAISYKSDVYSFGMLVLEMVSGRRNTDP 532
Query: 284 GGSCTSEMYFPDWIYKDLEQGNDLLNSLTISEEENDMVKKITMVSLWCIQTNPLDRPPMN 343
+E YFP+WIY+ + G +L+ ++ ++ E + V+++ +V+LWCIQ NP +RP M
Sbjct: 533 TVENQNEFYFPEWIYERVINGQELVLNMETTQGEKETVRQLAIVALWCIQWNPTNRPSMT 592
Query: 344 KVIEMLQGPLSSVTFPPKPVLFS 366
KV+ ML G L + PPKP + S
Sbjct: 593 KVVNMLTGRLQKLQVPPKPFISS 615
>UniRef100_Q7XSF9 OSJNBb0051N19.4 protein [Oryza sativa]
Length = 356
Score = 355 bits (912), Expect = 1e-96
Identities = 176/350 (50%), Positives = 239/350 (68%), Gaps = 8/350 (2%)
Query: 20 IIFISCYFKKGIRRP---QMTIFRKRRKHVDNNVEVFMQSYNLSIARRYSYAEVKRITNS 76
++ ISC+ IR+ Q+ I K + N+E + +Y +RY Y+++K +T S
Sbjct: 9 LLVISCFIVVKIRKSGKFQLRIIGKN-SNPKENIEELLDNYGSLAPKRYKYSQLKDMTGS 67
Query: 77 FRDKLGHGGYGVVYKASLTDGRQVAVKVINESKGNGEEFINEVASISRTSHMNIVSLLGY 136
F +KLG GGYG+VYK + DG VAVK +++ NGEEF+NEV SI RTSH+N+V+L+G+
Sbjct: 68 FSEKLGEGGYGMVYKGTSPDGHSVAVKFLHDLTRNGEEFVNEVISIRRTSHVNVVTLVGF 127
Query: 137 CYEANKRALIYEFMPKGSLDKFIYKSGFPDAVCDCDSNTLFQIAIGIARGLEYLHQGCSS 196
C E +KRALIYE+MP GSL+KFIY + + L+ IA+GIARGLEYLH+GC++
Sbjct: 128 CLEGSKRALIYEYMPNGSLEKFIYAENSKTTL---GWDKLYDIAVGIARGLEYLHRGCNT 184
Query: 197 RILHLDIKPQNILLDENFCPKISDFGLAKICQMNDSIVSIPGTRGTIGYMAPEVFSRAFG 256
RI+H DIKP NILLD +F PKI+DFGLAK+C +S +S+ G RGTIG++APEVFSR FG
Sbjct: 185 RIIHFDIKPHNILLDHDFVPKIADFGLAKLCNPKESYLSMAGMRGTIGFIAPEVFSRRFG 244
Query: 257 GVSYKSDVYSYGMLILEMIGGRKNYQTGGSCTSEMYFPDWIYKDLEQGNDLLNSLTISEE 316
VS KSDVYSYGM++LEM+GGRKN + SEMYFPDWIY+ L L+S + E
Sbjct: 245 VVSTKSDVYSYGMMLLEMVGGRKNLKASVDNPSEMYFPDWIYRCLADVGS-LHSFDMEHE 303
Query: 317 ENDMVKKITMVSLWCIQTNPLDRPPMNKVIEMLQGPLSSVTFPPKPVLFS 366
++ +K+ + LWCIQ +P RP M+KV+EM + + PPK +S
Sbjct: 304 TEEIARKMASIGLWCIQVSPSSRPTMSKVLEMFERSADELEIPPKHCFYS 353
>UniRef100_Q5ZC57 Putative receptor serine/threonine kinase PR5K [Oryza sativa]
Length = 674
Score = 355 bits (911), Expect = 1e-96
Identities = 180/360 (50%), Positives = 245/360 (68%), Gaps = 5/360 (1%)
Query: 21 IFISCYFKKGIRRPQMTIFRKRRKHVDNNVEVFMQSYNLSIARRYSYAEVKRITNSFRDK 80
I++ + KG ++ + ++ K + N+E + SY RY Y+EV +IT+ K
Sbjct: 282 IYVLIWHGKG-KQLRYFLYTKTSSTSERNIEALIISYGSIAPTRYKYSEVTKITSFLNYK 340
Query: 81 LGHGGYGVVYKASLTDGRQVAVKVINESKGNGEEFINEVASISRTSHMNIVSLLGYCYEA 140
LG GGYGVV+K L DGR VAVK +++SKGNGEEF+NEV SI RTSH+NIVSL G+C E
Sbjct: 341 LGEGGYGVVFKGRLQDGRLVAVKFLHDSKGNGEEFVNEVMSIGRTSHINIVSLFGFCLEG 400
Query: 141 NKRALIYEFMPKGSLDKFIYKSGFPDAVCDCDSNTLFQIAIGIARGLEYLHQGCSSRILH 200
+KRAL+YE+MP GSLD +IY P + + L+ IAIGIARGLEYLH C++RI+H
Sbjct: 401 SKRALLYEYMPNGSLDDYIYSEN-PKEILGWEK--LYGIAIGIARGLEYLHHSCNTRIIH 457
Query: 201 LDIKPQNILLDENFCPKISDFGLAKICQMNDSIVSIPGTRGTIGYMAPEVFSRAFGGVSY 260
DIKPQNILLD++FCPKI+DFGLAK+C+ +S +S+ G RGTIG++APEV R+FG VS
Sbjct: 458 FDIKPQNILLDQDFCPKIADFGLAKLCRTKESKLSMTGARGTIGFIAPEVIYRSFGIVST 517
Query: 261 KSDVYSYGMLILEMIGGRKNYQTGGSCTSEMYFPDWIYKDLEQGNDLLNSLTISEEENDM 320
KSDVYSYGM++LEM+GGRKN ++ +SE YFPDWIY D +D L + ++ E +
Sbjct: 518 KSDVYSYGMMLLEMVGGRKNAKSMVENSSEKYFPDWIY-DHFALDDGLQACEVTSEVEQI 576
Query: 321 VKKITMVSLWCIQTNPLDRPPMNKVIEMLQGPLSSVTFPPKPVLFSPKRPPLQLSNMSSS 380
KK+T++ LWC+Q P+ RP + +V++M + L + PPK P Q N S+
Sbjct: 577 AKKMTLIGLWCVQVLPMHRPTITQVLDMFERSLDELEMPPKQNFSELLEHPAQEINTEST 636
>UniRef100_Q5ZC56 Putative receptor serine/threonine kinase PR5K [Oryza sativa]
Length = 419
Score = 355 bits (911), Expect = 1e-96
Identities = 180/360 (50%), Positives = 245/360 (68%), Gaps = 5/360 (1%)
Query: 21 IFISCYFKKGIRRPQMTIFRKRRKHVDNNVEVFMQSYNLSIARRYSYAEVKRITNSFRDK 80
I++ + KG ++ + ++ K + N+E + SY RY Y+EV +IT+ K
Sbjct: 27 IYVLIWHGKG-KQLRYFLYTKTSSTSERNIEALIISYGSIAPTRYKYSEVTKITSFLNYK 85
Query: 81 LGHGGYGVVYKASLTDGRQVAVKVINESKGNGEEFINEVASISRTSHMNIVSLLGYCYEA 140
LG GGYGVV+K L DGR VAVK +++SKGNGEEF+NEV SI RTSH+NIVSL G+C E
Sbjct: 86 LGEGGYGVVFKGRLQDGRLVAVKFLHDSKGNGEEFVNEVMSIGRTSHINIVSLFGFCLEG 145
Query: 141 NKRALIYEFMPKGSLDKFIYKSGFPDAVCDCDSNTLFQIAIGIARGLEYLHQGCSSRILH 200
+KRAL+YE+MP GSLD +IY P + + L+ IAIGIARGLEYLH C++RI+H
Sbjct: 146 SKRALLYEYMPNGSLDDYIYSEN-PKEILGWEK--LYGIAIGIARGLEYLHHSCNTRIIH 202
Query: 201 LDIKPQNILLDENFCPKISDFGLAKICQMNDSIVSIPGTRGTIGYMAPEVFSRAFGGVSY 260
DIKPQNILLD++FCPKI+DFGLAK+C+ +S +S+ G RGTIG++APEV R+FG VS
Sbjct: 203 FDIKPQNILLDQDFCPKIADFGLAKLCRTKESKLSMTGARGTIGFIAPEVIYRSFGIVST 262
Query: 261 KSDVYSYGMLILEMIGGRKNYQTGGSCTSEMYFPDWIYKDLEQGNDLLNSLTISEEENDM 320
KSDVYSYGM++LEM+GGRKN ++ +SE YFPDWIY D +D L + ++ E +
Sbjct: 263 KSDVYSYGMMLLEMVGGRKNAKSMVENSSEKYFPDWIY-DHFALDDGLQACEVTSEVEQI 321
Query: 321 VKKITMVSLWCIQTNPLDRPPMNKVIEMLQGPLSSVTFPPKPVLFSPKRPPLQLSNMSSS 380
KK+T++ LWC+Q P+ RP + +V++M + L + PPK P Q N S+
Sbjct: 322 AKKMTLIGLWCVQVLPMHRPTITQVLDMFERSLDELEMPPKQNFSELLEHPAQEINTEST 381
>UniRef100_Q9SWF1 Receptor kinase [Oryza sativa]
Length = 657
Score = 353 bits (907), Expect = 4e-96
Identities = 167/317 (52%), Positives = 228/317 (71%)
Query: 50 VEVFMQSYNLSIARRYSYAEVKRITNSFRDKLGHGGYGVVYKASLTDGRQVAVKVINESK 109
VE+F+++Y S RY+++EVK+I F+DKLGHG +G VYK L +G VAVK++ S
Sbjct: 335 VEMFLKTYGTSKPTRYTFSEVKKIARRFKDKLGHGAFGTVYKGELPNGVPVAVKMLENSV 394
Query: 110 GNGEEFINEVASISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIYKSGFPDAVC 169
G G+EFINEVA+I R H NIV LLG+C E +RALIYE MP SL+K+I+ G +
Sbjct: 395 GEGQEFINEVATIGRIHHANIVRLLGFCSEGTRRALIYELMPNESLEKYIFPHGSNISRE 454
Query: 170 DCDSNTLFQIAIGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQM 229
+ + IA+GIARG+EYLHQGC+ RILH DIKP NILLD +F PKISDFGLAK+C
Sbjct: 455 LLVPDKMLDIALGIARGMEYLHQGCNQRILHFDIKPHNILLDYSFNPKISDFGLAKLCAR 514
Query: 230 NDSIVSIPGTRGTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGRKNYQTGGSCTS 289
+ SIV++ RGT+GY+APE++SR FG +SYKSDVYS+GML+LEM+ GR+N +
Sbjct: 515 DQSIVTLTAARGTMGYIAPELYSRNFGAISYKSDVYSFGMLVLEMVSGRRNTDPPAENQN 574
Query: 290 EMYFPDWIYKDLEQGNDLLNSLTISEEENDMVKKITMVSLWCIQTNPLDRPPMNKVIEML 349
E YFP+W+++ + G DL+ ++ ++ E +MV+++ +V+LWCIQ NP DRP M KV+ ML
Sbjct: 575 EFYFPEWVFERVMNGQDLVLTMETTQGEKEMVRQLAIVALWCIQWNPKDRPSMTKVVNML 634
Query: 350 QGPLSSVTFPPKPVLFS 366
G L ++ PPKP + S
Sbjct: 635 TGRLQNLQVPPKPFISS 651
>UniRef100_Q9LL55 Receptor-like kinase [Oryza sativa]
Length = 657
Score = 353 bits (907), Expect = 4e-96
Identities = 167/317 (52%), Positives = 228/317 (71%)
Query: 50 VEVFMQSYNLSIARRYSYAEVKRITNSFRDKLGHGGYGVVYKASLTDGRQVAVKVINESK 109
VE+F+++Y S RY+++EVK+I F+DKLGHG +G VYK L +G VAVK++ S
Sbjct: 335 VEMFLKTYGTSKPTRYTFSEVKKIARRFKDKLGHGAFGTVYKGELPNGVPVAVKMLENSV 394
Query: 110 GNGEEFINEVASISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIYKSGFPDAVC 169
G G+EFINEVA+I R H NIV LLG+C E +RALIYE MP SL+K+I+ G +
Sbjct: 395 GEGQEFINEVATIGRIHHANIVRLLGFCSEGTRRALIYELMPNESLEKYIFPHGSNISRE 454
Query: 170 DCDSNTLFQIAIGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQM 229
+ + IA+GIARG+EYLHQGC+ RILH DIKP NILLD +F PKISDFGLAK+C
Sbjct: 455 LLVPDKMLDIALGIARGMEYLHQGCNQRILHFDIKPHNILLDYSFNPKISDFGLAKLCAR 514
Query: 230 NDSIVSIPGTRGTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGRKNYQTGGSCTS 289
+ SIV++ RGT+GY+APE++SR FG +SYKSDVYS+GML+LEM+ GR+N +
Sbjct: 515 DQSIVTLTAARGTMGYIAPELYSRNFGAISYKSDVYSFGMLVLEMVSGRRNTDPPAENQN 574
Query: 290 EMYFPDWIYKDLEQGNDLLNSLTISEEENDMVKKITMVSLWCIQTNPLDRPPMNKVIEML 349
E YFP+W+++ + G DL+ ++ ++ E +MV+++ +V+LWCIQ NP DRP M KV+ ML
Sbjct: 575 EFYFPEWVFERVMNGQDLVLTMETTQGEKEMVRQLAIVALWCIQWNPKDRPSMTKVVNML 634
Query: 350 QGPLSSVTFPPKPVLFS 366
G L ++ PPKP + S
Sbjct: 635 TGRLQNLQVPPKPFISS 651
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.320 0.138 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 727,050,896
Number of Sequences: 2790947
Number of extensions: 30623672
Number of successful extensions: 116213
Number of sequences better than 10.0: 18272
Number of HSP's better than 10.0 without gapping: 7496
Number of HSP's successfully gapped in prelim test: 10778
Number of HSP's that attempted gapping in prelim test: 82833
Number of HSP's gapped (non-prelim): 20439
length of query: 459
length of database: 848,049,833
effective HSP length: 131
effective length of query: 328
effective length of database: 482,435,776
effective search space: 158238934528
effective search space used: 158238934528
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 76 (33.9 bits)
Medicago: description of AC147875.5


