

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147875.4 + phase: 0
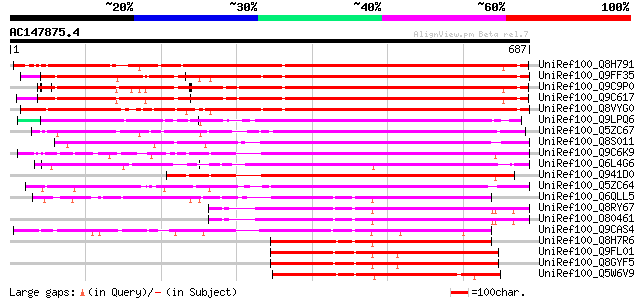
(687 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8H791 Hypothetical protein [Arabidopsis thaliana] 550 e-155
UniRef100_Q9FF35 Similarity to protein kinase [Arabidopsis thali... 544 e-153
UniRef100_Q9C9P0 Protein kinase, putative [Arabidopsis thaliana] 542 e-152
UniRef100_Q9C617 Wall-associated kinase, putative [Arabidopsis t... 542 e-152
UniRef100_Q8VYG0 Hypothetical protein At5g38210 [Arabidopsis tha... 533 e-150
UniRef100_Q9LPQ6 F15H18.11 [Arabidopsis thaliana] 497 e-139
UniRef100_Q5ZC67 Serine/threonine-specific protein kinase-like [... 466 e-130
UniRef100_Q8S011 P0034C09.1 protein [Oryza sativa] 459 e-127
UniRef100_Q9C6K9 Wall-associated kinase, putative [Arabidopsis t... 443 e-123
UniRef100_Q6L4G6 Putative receptor-like protein kinase [Oryza sa... 442 e-122
UniRef100_Q941D0 At1g25390/F2J7_14 [Arabidopsis thaliana] 412 e-113
UniRef100_Q5ZC64 Putative receptor protein kinase CRINKLY4 [Oryz... 410 e-113
UniRef100_Q6QLL5 WAK-like kinase [Lycopersicon esculentum] 297 8e-79
UniRef100_Q8RY67 At2g23450/F26B6.10 [Arabidopsis thaliana] 288 3e-76
UniRef100_O80461 Hypothetical protein At2g23450 [Arabidopsis tha... 288 3e-76
UniRef100_Q9CAS4 Hypothetical protein T17F3.6 [Arabidopsis thali... 284 5e-75
UniRef100_Q8H7R6 Hypothetical protein OSJNBa0081P02.15 [Oryza sa... 281 3e-74
UniRef100_Q9FL01 Similarity to protein kinase [Arabidopsis thali... 265 3e-69
UniRef100_Q8GYF5 Hypothetical protein At5g66790/MUD21_3 [Arabido... 265 3e-69
UniRef100_Q5W6V9 Hypothetical protein OSJNBb0059K16.4 [Oryza sat... 256 2e-66
>UniRef100_Q8H791 Hypothetical protein [Arabidopsis thaliana]
Length = 663
Score = 550 bits (1416), Expect = e-155
Identities = 334/692 (48%), Positives = 424/692 (61%), Gaps = 45/692 (6%)
Query: 6 LSSPIITFIIFFIFYLRHTTSLPSHASLSSCNNTTFNCGHITKLSYPFTGGDRPSYCGPP 65
LSS ++ FI+F +FY LP +S C + F CG+IT S+PF GGDR +CG P
Sbjct: 5 LSSSLMFFILFSLFY-----HLPCDSS--KCESL-FQCGNITA-SFPFWGGDRHKHCGHP 55
Query: 66 QFHLNC-KNNVPELNISSVSYRVLQVNSVTHSLTLARLDLWNETCTHHYVNSTFGGTSFS 124
L C +N L IS + VL V+ ++SLTLAR DL + C+ + N+T F
Sbjct: 56 LLELRCDQNKSTSLFISDQEFYVLHVDQTSYSLTLARPDLLDSFCSLKFTNTTLPPEIFE 115
Query: 125 NGLGNSKITLFYGCQPTSLFTEKPHNLFYCDSNGYKNNSYTLIGPF-----PLDPVLKFV 179
+T FY C P N + +IGP P D F
Sbjct: 116 LSPAYKSVT-FYHCYPV--------------LPDLSNYTCPVIGPISVSGNPEDHETCFA 160
Query: 180 QCDYGVGVPILEEQANRFAGNRSLLREVLMEGFNVNYNNPFENDCLECISSGGQQCGFDS 239
V + ++ N + L VL +GF VN N C EC S + CGFD
Sbjct: 161 NFAANVPKSFVTKEKKL---NITNLESVLEKGFEVNM-NVIMKACQEC-SYSNESCGFDE 215
Query: 240 DENEHICICGNGLCPSGNSSTNGGLIGGVVGGVAALCLLGFVACFVVRRRRKNAKKPISN 299
+ + C P+ + G G V V+ L +L F+ RRR+ +
Sbjct: 216 NFPFEV-KCKPHHSPTDTGLSPGAKAGIAVASVSGLAILILAGLFLYIRRRRKTQDAQYT 274
Query: 300 DLYMPPSSTTSGTNTGTLTSTTNSSQSIPSYPSSKTSTMPKSFYFGVQVFTYEELEEATN 359
+P +S +S + TSTT SS S S S ++ S Y GVQVF+YEELEEAT
Sbjct: 275 SKSLPITSYSSRDTSRNPTSTTISSSSNHSLLPSISNLANGSDYCGVQVFSYEELEEATE 334
Query: 360 NFHTSKELGEGGFGTVYKGDLKDGRVVAVKRHYESNFKRVAQFMNEVEILARLRHKNLVT 419
NF S+ELG+GGFGTVY G LKDGR VAVKR YE + KRV QF NE+EIL L+H NLV
Sbjct: 335 NF--SRELGDGGFGTVYYGVLKDGRAVAVKRLYERSLKRVEQFKNEIEILKSLKHPNLVI 392
Query: 420 LYGCTSKHSRELLLVYEYISNGTVADHLHGDRSSSCLLPWSVRLDIALETAEALAYLHAS 479
LYGCTS+HSRELLLVYEYISNGT+A+HLHG+R+ + L WS RL+IA+ETA AL++LH
Sbjct: 393 LYGCTSRHSRELLLVYEYISNGTLAEHLHGNRAEARPLCWSTRLNIAIETASALSFLHIK 452
Query: 480 DVMHRDVKSNNILLDEKFHVKVADFGLSRLFPNDVTHVSTAPQGTPGYVDPEYYQCYQLT 539
++HRD+K+ NILLD+ + VKVADFGLSRLFP D TH+STAPQGTPGYVDPEYYQCYQL
Sbjct: 453 GIIHRDIKTTNILLDDNYQVKVADFGLSRLFPMDQTHISTAPQGTPGYVDPEYYQCYQLN 512
Query: 540 DKSDVYSFGVVLVELISSLQAVDITRHRNDVNLANMAVNKIQSQELYDLVDPNLGYEKDN 599
+KSDVYSFGVVL ELISS +AVDITRHR+D+NLANMAV+KIQ+ L++LVD +LG++ D
Sbjct: 513 EKSDVYSFGVVLTELISSKEAVDITRHRHDINLANMAVSKIQNNALHELVDSSLGFDNDP 572
Query: 600 SVKRMTTAVAELAFRCLQQQRDLRPSMDEIVEVLRAIKSDEPETQESKVLDVV----VRT 655
V+R AV+ELAFRCLQQ+RD+RP+MDEIVE+LR IK DE + K DVV
Sbjct: 573 EVRRKMMAVSELAFRCLQQERDVRPAMDEIVEILRGIKDDEKKRVLVKSPDVVDIECGGG 632
Query: 656 DELVLLKKG-PYPTSPDSVAEKWVSGSSTSTS 686
D++ LL+ P P SP++ +KW S S T+ S
Sbjct: 633 DDVGLLRNSVPPPISPET--DKWTSSSDTAAS 662
>UniRef100_Q9FF35 Similarity to protein kinase [Arabidopsis thaliana]
Length = 978
Score = 544 bits (1401), Expect = e-153
Identities = 314/668 (47%), Positives = 424/668 (63%), Gaps = 34/668 (5%)
Query: 41 FNCGHITKLSYPFTGGDRPSYCGPPQFHLNCKNNVPELNISSVSYRVLQVNSVTHSLTLA 100
F CG+ ++L YPF +R CG P F +NC + EL +S+V +++L++N + LA
Sbjct: 322 FKCGNQSELYYPFWISERKE-CGHPDFEVNCSGDFAELTVSTVKFQILEMNYEFEIIRLA 380
Query: 101 RLDLWNETCTHHYVNSTFGGTSFSNGLGNSKITLFYGCQPT-----SLFTEKPHNLFYCD 155
R+D N C H N++ T ++ CQ T + + ++ + Y
Sbjct: 381 RMDYRNNLCPQHPANASINHDVLPFSPDTKLSTFYHDCQSTVDVHPNDYIKQLNCEDYIG 440
Query: 156 SNGYKNNSYTLIGPFPLDPVLKFVQCDYGVGVPILEEQANRFAGNRSL--LREVLMEGFN 213
Y +S + G L C+ GV +P+ N+SL +++ + EGF
Sbjct: 441 GKSYFVSSSSHSGDRATLAALS-ASCE-GVDIPVSRSALKTVEKNQSLEAMKKAIDEGFE 498
Query: 214 VNYNNPFENDCLECISSGGQQCGFDSDENEHICICGN----GLCPSGN-SSTNGGL---- 264
+ +NN +C +C SGG CGFD +C C + C S T+ GL
Sbjct: 499 LGFNN----ECSQCAISGGA-CGFDQRLKAFVCYCADEPHESTCYSEIVKKTHAGLSKKG 553
Query: 265 ---IGGVVGGVAALCLLGFVACFVVRRRRKNAKKPISNDLYMPPSSTTSGTNTGTLTSTT 321
IG G + A + G + C +RRR+K A + + L +++ T + T TSTT
Sbjct: 554 KIGIGFASGFLGATLIGGCLLCIFIRRRKKLATQYTNKGLSTTTPYSSNYTMSNTPTSTT 613
Query: 322 NSSQSIPSYPSSKTSTMPKSFYFGVQVFTYEELEEATNNFHTSKELGEGGFGTVYKGDLK 381
S + PS ++ S Y G+QVF+YEELEEAT NF SKELG+GGFGTVY G LK
Sbjct: 614 ISGSNHSLVPSI-SNLGNGSVYSGIQVFSYEELEEATENF--SKELGDGGFGTVYYGTLK 670
Query: 382 DGRVVAVKRHYESNFKRVAQFMNEVEILARLRHKNLVTLYGCTSKHSRELLLVYEYISNG 441
DGR VAVKR +E + KRV QF NE++IL L+H NLV LYGCT++HSRELLLVYEYISNG
Sbjct: 671 DGRAVAVKRLFERSLKRVEQFKNEIDILKSLKHPNLVILYGCTTRHSRELLLVYEYISNG 730
Query: 442 TVADHLHGDRSSSCLLPWSVRLDIALETAEALAYLHASDVMHRDVKSNNILLDEKFHVKV 501
T+A+HLHG+++ S + W RL IA+ETA AL+YLHAS ++HRDVK+ NILLD + VKV
Sbjct: 731 TLAEHLHGNQAQSRPICWPARLQIAIETASALSYLHASGIIHRDVKTTNILLDSNYQVKV 790
Query: 502 ADFGLSRLFPNDVTHVSTAPQGTPGYVDPEYYQCYQLTDKSDVYSFGVVLVELISSLQAV 561
ADFGLSRLFP D TH+STAPQGTPGYVDPEYYQCY+L +KSDVYSFGVVL ELISS +AV
Sbjct: 791 ADFGLSRLFPMDQTHISTAPQGTPGYVDPEYYQCYRLNEKSDVYSFGVVLSELISSKEAV 850
Query: 562 DITRHRNDVNLANMAVNKIQSQELYDLVDPNLGYEKDNSVKRMTTAVAELAFRCLQQQRD 621
DITRHR+D+NLANMA++KIQ+ +++L D +LG+ +D SVK+M ++VAELAFRCLQQ+RD
Sbjct: 851 DITRHRHDINLANMAISKIQNDAVHELADLSLGFARDPSVKKMMSSVAELAFRCLQQERD 910
Query: 622 LRPSMDEIVEVLRAIKSDEPETQESKVLDVVVR-TDELVLLKKG-PYPTSPDSVAEKWVS 679
+RPSMDEIVEVLR I+ D + V+++ V D++ LLK G P P SP++ +K +
Sbjct: 911 VRPSMDEIVEVLRVIQKDGISDSKDVVVEIDVNGGDDVGLLKHGVPPPLSPET--DKTTA 968
Query: 680 GSSTSTSS 687
SS +T+S
Sbjct: 969 SSSNTTAS 976
Score = 77.4 bits (189), Expect = 1e-12
Identities = 67/222 (30%), Positives = 97/222 (43%), Gaps = 20/222 (9%)
Query: 15 IFFIFYLRHTTSLPSHASLSSCNNTTFNCGHITKLSYPFTGGDRPSYCGPPQFHLNCKN- 73
+ +F+L + LP S +T F CG +T +PF G RP CG P L+C+
Sbjct: 16 LVLLFFLSYIHFLPCAQSQREPCDTLFRCGDLTA-GFPFWGVARPQPCGHPSLGLHCQKQ 74
Query: 74 -NVPELNISSVSYRVLQVNSVTHSLTLARLDLWNETCTHHYVNSTFGGTSFSNGLGNSKI 132
N L ISS+ YRVL+VN+ T +L L R D C+ + +T F +
Sbjct: 75 TNSTSLIISSLMYRVLEVNTTTSTLKLVRQDFSGPFCSASFSGATLTPELFELLPDYKTL 134
Query: 133 TLFYGCQPTSLFTEKPHNLFYCDSNGYKNNSYTLIGPFPLDPVLKFVQCD--YGVGVPIL 190
+ +Y C P+ + K F C + G +G D L C + + VPI
Sbjct: 135 SAYYLCNPSLHYPAK----FICPNKG--------VGSIHQDD-LYHNHCGGIFNITVPI- 180
Query: 191 EEQANRFAGNRSLLREVLMEGFNVNYNNPFENDCLECISSGG 232
A N + L VL +GF V + E C EC ++GG
Sbjct: 181 GYAPEEGALNVTNLESVLKKGFEVKLSID-ERPCQECKTNGG 221
>UniRef100_Q9C9P0 Protein kinase, putative [Arabidopsis thaliana]
Length = 1286
Score = 542 bits (1396), Expect = e-152
Identities = 322/669 (48%), Positives = 412/669 (61%), Gaps = 35/669 (5%)
Query: 38 NTTFNCGHITKLSYPFTGGDRPSYCGPPQFHLN-CKNNVPELNISSVSYRVLQVNSVTHS 96
N TF+CG +L YPF R CG P F L+ C EL+ISSV +R+L ++
Sbjct: 632 NGTFSCGDQRELFYPFWTSGRED-CGHPDFKLDDCSGRFAELSISSVKFRILASVYGSNI 690
Query: 97 LTLARLDLWNETCTHHYVNSTFGGTSFSNGLGNSKITLFYGCQPTSLFTEKPHNL--FYC 154
+ L R + + C +N+ F + +T+FY C F ++ N F C
Sbjct: 691 IRLGRSEYIGDLCPQDPINAPFSESVLPFAPNTELLTIFYNCSRD--FPQQVTNFGDFAC 748
Query: 155 DSNGYKNNSYTLIGPFPLDPVLKF--------VQCDYGVGVPILEEQANRFAGNRSL--L 204
+ + SY + P+ + CD V +P N S L
Sbjct: 749 GDDSDDDRSYYVTRNLSFPPLSEINDLLYDFSQSCDRNVSIPASGSTLNILQSTPSNDNL 808
Query: 205 REVLMEGFNVNYNNPFENDCLECISSGGQQCGFDSDENEHICIC-GNGLCPSG-NSSTNG 262
++ L GF + N DC CI S G CG+ + +C P+ N + N
Sbjct: 809 KKALEYGFELELNQ----DCRTCIDSKGA-CGYSQTSSRFVCYSIEEPQTPTPPNPTRNK 863
Query: 263 GLIGGVVGGVAALCLLGFVACFVVRRRRKNAKKPISNDLYMPPSSTTSGTNTGTLTSTTN 322
G+ V G+A L L G C +RRRRK ++ +P +S +S + TSTT
Sbjct: 864 GIAVASVSGLAILLLAGLFLC--IRRRRKTQDAQYTSKS-LPITSYSSRDTSRNPTSTTI 920
Query: 323 SSQSIPSYPSSKTSTMPKSFYFGVQVFTYEELEEATNNFHTSKELGEGGFGTVYKGDLKD 382
SS S S S ++ +S Y GVQVF+YEELEEAT NF S+ELG+GGFGTVY G LKD
Sbjct: 921 SSSSNHSLLPSISNLANRSDYCGVQVFSYEELEEATENF--SRELGDGGFGTVYYGVLKD 978
Query: 383 GRVVAVKRHYESNFKRVAQFMNEVEILARLRHKNLVTLYGCTSKHSRELLLVYEYISNGT 442
GR VAVKR YE + KRV QF NE+EIL L+H NLV LYGCTS+HSRELLLVYEYISNGT
Sbjct: 979 GRAVAVKRLYERSLKRVEQFKNEIEILKSLKHPNLVILYGCTSRHSRELLLVYEYISNGT 1038
Query: 443 VADHLHGDRSSSCLLPWSVRLDIALETAEALAYLHASDVMHRDVKSNNILLDEKFHVKVA 502
+A+HLHG+R+ + L WS RL+IA+ETA AL++LH ++HRD+K+ NILLD+ + VKVA
Sbjct: 1039 LAEHLHGNRAEARPLCWSTRLNIAIETASALSFLHIKGIIHRDIKTTNILLDDNYQVKVA 1098
Query: 503 DFGLSRLFPNDVTHVSTAPQGTPGYVDPEYYQCYQLTDKSDVYSFGVVLVELISSLQAVD 562
DFGLSRLFP D TH+STAPQGTPGYVDPEYYQCYQL +KSDVYSFGVVL ELISS +AVD
Sbjct: 1099 DFGLSRLFPMDQTHISTAPQGTPGYVDPEYYQCYQLNEKSDVYSFGVVLTELISSKEAVD 1158
Query: 563 ITRHRNDVNLANMAVNKIQSQELYDLVDPNLGYEKDNSVKRMTTAVAELAFRCLQQQRDL 622
ITRHR+D+NLANMAV+KIQ+ L++LVD +LGY+ D V+R AVAELAFRCLQQ+RD+
Sbjct: 1159 ITRHRHDINLANMAVSKIQNNALHELVDSSLGYDNDPEVRRKMMAVAELAFRCLQQERDV 1218
Query: 623 RPSMDEIVEVLRAIKSDEPETQESKVLDVV----VRTDELVLLKKG-PYPTSPDSVAEKW 677
RP+MDEIVE+LR IK DE + K DVV D++ LL+ P P SP++ +KW
Sbjct: 1219 RPAMDEIVEILRGIKDDEKKRVLVKSPDVVDIECGGGDDVGLLRNSVPPPISPET--DKW 1276
Query: 678 VSGSSTSTS 686
S S T+ S
Sbjct: 1277 TSSSDTAAS 1285
Score = 85.9 bits (211), Expect = 4e-15
Identities = 64/205 (31%), Positives = 88/205 (42%), Gaps = 27/205 (13%)
Query: 41 FNCGHITKLSYPFTGGDRPSYCGPPQFHLNCKNNVPELNISSVSYRVLQVNSVTHSLTLA 100
F CG+IT +PF+GG+RP CG P L+C NN+ + IS Y VL ++ +++L LA
Sbjct: 416 FQCGNITA-GFPFSGGNRPQICGHPSLELHCYNNMASIIISDHFYNVLHIDQTSNTLRLA 474
Query: 101 RLDLWNETCTHHYVNSTFGGTSFSNGLGNSKITLFYGCQP----TSLFTEKPHNLFYCDS 156
R +L C Y +T F +T+FY C P S +T L
Sbjct: 475 RAELEGSFCNATYTATTLPSKIFEISSTYKSLTVFYLCDPKVSYRSSYTCPGRGLVSVSQ 534
Query: 157 NGYKNNSYTLIGPFPLDPVLKFVQCDYGVGVP--ILEEQANRFAGNRSLLREVLMEGFNV 214
N +NS Q + + VP + E+ N L L EGF V
Sbjct: 535 NSDYHNS---------------CQDSFTINVPKSFVPEEKELDVTN---LESALREGFEV 576
Query: 215 NYNNPFENDCLECISSGGQQCGFDS 239
E C +C SSGG CGF +
Sbjct: 577 KVKVD-EKTCQKCTSSGG-TCGFQN 599
Score = 80.5 bits (197), Expect = 2e-13
Identities = 61/200 (30%), Positives = 87/200 (43%), Gaps = 23/200 (11%)
Query: 43 CGHITKLSYPFTGGDRPSYCGPPQFHLNCKNNVPELNISSVSYRVLQVNSVTHSLTLARL 102
CG+IT +PF GG+R +CG P L+C N+ L ISS + VL +N + +L LAR
Sbjct: 26 CGNITA-GFPFWGGNRLKHCGLPSLELHCSKNITSLFISSQEFYVLHLNQTSKTLKLART 84
Query: 103 DLWNETCTHHYVNSTFGGTSFSNGLGNSKITLFYGCQPTSLFTEKPHNLFYCDSNGY--- 159
DL C + T F ++T+FY C + + + C G+
Sbjct: 85 DLLGSICNSTFTTITLPPNIFELSPTYKRLTVFYYCFLLTHYVSS----YKCPMRGFIFV 140
Query: 160 -KNNSYTLIGPFPLDPVLKFVQCDYGVGVPILEEQANRFAGNRSLLREVLMEGFNVNYNN 218
+N+ Y I D V +G G L N + L VL +GF V
Sbjct: 141 SENHEYHKICG---DTFTVIVPTRFGAGEKEL---------NMTNLESVLSKGFEVKVKI 188
Query: 219 PFENDCLECISSGGQQCGFD 238
+ C EC+SS G CGF+
Sbjct: 189 D-DTSCQECLSSHG-SCGFN 206
Score = 59.7 bits (143), Expect = 3e-07
Identities = 56/191 (29%), Positives = 75/191 (38%), Gaps = 26/191 (13%)
Query: 56 GDRPSYCGPPQFHLNC-KNNVPELNISSVSYRVLQVNSVTHSLTLARLDLWNETCTHHYV 114
GDR +CG P L C +N L IS + VL V+ ++SLTLAR DL + C+ +
Sbjct: 229 GDRHKHCGHPLLELRCDQNKSTSLFISDQEFFVLHVDQTSYSLTLARPDLLHSFCSLTFT 288
Query: 115 NSTFGGTSFSNGLGNSKITLFYGCQPTSLFTEKPHNLFYCDSNGYKNNSYTLIGPF---- 170
N+T F +T FY C P N + +IGP
Sbjct: 289 NTTLPPEIFELSPAYKSVT-FYHCYPV--------------LPDLSNYTCPVIGPISVSG 333
Query: 171 -PLDPVLKFVQCDYGVGVPILEEQANRFAGNRSLLREVLMEGFNVNYNNPFENDCLECIS 229
P D F V + ++ N L VL +GF VN N C C S
Sbjct: 334 NPEDHETCFPNFAANVPTSFVTKEKKLNIAN---LESVLEKGFEVNM-NVIMKACQAC-S 388
Query: 230 SGGQQCGFDSD 240
+ CGFD +
Sbjct: 389 YSNESCGFDEN 399
>UniRef100_Q9C617 Wall-associated kinase, putative [Arabidopsis thaliana]
Length = 907
Score = 542 bits (1396), Expect = e-152
Identities = 322/669 (48%), Positives = 412/669 (61%), Gaps = 35/669 (5%)
Query: 38 NTTFNCGHITKLSYPFTGGDRPSYCGPPQFHLN-CKNNVPELNISSVSYRVLQVNSVTHS 96
N TF+CG +L YPF R CG P F L+ C EL+ISSV +R+L ++
Sbjct: 253 NGTFSCGDQRELFYPFWTSGRED-CGHPDFKLDDCSGRFAELSISSVKFRILASVYGSNI 311
Query: 97 LTLARLDLWNETCTHHYVNSTFGGTSFSNGLGNSKITLFYGCQPTSLFTEKPHNL--FYC 154
+ L R + + C +N+ F + +T+FY C F ++ N F C
Sbjct: 312 IRLGRSEYIGDLCPQDPINAPFSESVLPFAPNTELLTIFYNCSRD--FPQQVTNFGDFAC 369
Query: 155 DSNGYKNNSYTLIGPFPLDPVLKF--------VQCDYGVGVPILEEQANRFAGNRSL--L 204
+ + SY + P+ + CD V +P N S L
Sbjct: 370 GDDSDDDRSYYVTRNLSFPPLSEINDLLYDFSQSCDRNVSIPASGSTLNILQSTPSNDNL 429
Query: 205 REVLMEGFNVNYNNPFENDCLECISSGGQQCGFDSDENEHICIC-GNGLCPSG-NSSTNG 262
++ L GF + N DC CI S G CG+ + +C P+ N + N
Sbjct: 430 KKALEYGFELELNQ----DCRTCIDSKGA-CGYSQTSSRFVCYSIEEPQTPTPPNPTRNK 484
Query: 263 GLIGGVVGGVAALCLLGFVACFVVRRRRKNAKKPISNDLYMPPSSTTSGTNTGTLTSTTN 322
G+ V G+A L L G C +RRRRK ++ +P +S +S + TSTT
Sbjct: 485 GIAVASVSGLAILLLAGLFLC--IRRRRKTQDAQYTSKS-LPITSYSSRDTSRNPTSTTI 541
Query: 323 SSQSIPSYPSSKTSTMPKSFYFGVQVFTYEELEEATNNFHTSKELGEGGFGTVYKGDLKD 382
SS S S S ++ +S Y GVQVF+YEELEEAT NF S+ELG+GGFGTVY G LKD
Sbjct: 542 SSSSNHSLLPSISNLANRSDYCGVQVFSYEELEEATENF--SRELGDGGFGTVYYGVLKD 599
Query: 383 GRVVAVKRHYESNFKRVAQFMNEVEILARLRHKNLVTLYGCTSKHSRELLLVYEYISNGT 442
GR VAVKR YE + KRV QF NE+EIL L+H NLV LYGCTS+HSRELLLVYEYISNGT
Sbjct: 600 GRAVAVKRLYERSLKRVEQFKNEIEILKSLKHPNLVILYGCTSRHSRELLLVYEYISNGT 659
Query: 443 VADHLHGDRSSSCLLPWSVRLDIALETAEALAYLHASDVMHRDVKSNNILLDEKFHVKVA 502
+A+HLHG+R+ + L WS RL+IA+ETA AL++LH ++HRD+K+ NILLD+ + VKVA
Sbjct: 660 LAEHLHGNRAEARPLCWSTRLNIAIETASALSFLHIKGIIHRDIKTTNILLDDNYQVKVA 719
Query: 503 DFGLSRLFPNDVTHVSTAPQGTPGYVDPEYYQCYQLTDKSDVYSFGVVLVELISSLQAVD 562
DFGLSRLFP D TH+STAPQGTPGYVDPEYYQCYQL +KSDVYSFGVVL ELISS +AVD
Sbjct: 720 DFGLSRLFPMDQTHISTAPQGTPGYVDPEYYQCYQLNEKSDVYSFGVVLTELISSKEAVD 779
Query: 563 ITRHRNDVNLANMAVNKIQSQELYDLVDPNLGYEKDNSVKRMTTAVAELAFRCLQQQRDL 622
ITRHR+D+NLANMAV+KIQ+ L++LVD +LGY+ D V+R AVAELAFRCLQQ+RD+
Sbjct: 780 ITRHRHDINLANMAVSKIQNNALHELVDSSLGYDNDPEVRRKMMAVAELAFRCLQQERDV 839
Query: 623 RPSMDEIVEVLRAIKSDEPETQESKVLDVV----VRTDELVLLKKG-PYPTSPDSVAEKW 677
RP+MDEIVE+LR IK DE + K DVV D++ LL+ P P SP++ +KW
Sbjct: 840 RPAMDEIVEILRGIKDDEKKRVLVKSPDVVDIECGGGDDVGLLRNSVPPPISPET--DKW 897
Query: 678 VSGSSTSTS 686
S S T+ S
Sbjct: 898 TSSSDTAAS 906
Score = 90.9 bits (224), Expect = 1e-16
Identities = 73/238 (30%), Positives = 99/238 (40%), Gaps = 29/238 (12%)
Query: 9 PIITFIIFFIFYLRHTT-SLPSHASLSSCNNTTFNCGHITKLSYPFTGGDRPSYCGPPQF 67
P +FF+F H S S L C F CG+IT +PF+GG+RP CG P
Sbjct: 5 PTSCMFLFFLFSSFHPLLSASSKQELGWCE-AKFQCGNITA-GFPFSGGNRPQICGHPSL 62
Query: 68 HLNCKNNVPELNISSVSYRVLQVNSVTHSLTLARLDLWNETCTHHYVNSTFGGTSFSNGL 127
L+C NN+ + IS Y VL ++ +++L LAR +L C Y +T F
Sbjct: 63 ELHCYNNMASIIISDHFYNVLHIDQTSNTLRLARAELEGSFCNATYTATTLPSKIFEISS 122
Query: 128 GNSKITLFYGCQP----TSLFTEKPHNLFYCDSNGYKNNSYTLIGPFPLDPVLKFVQCDY 183
+T+FY C P S +T L N +NS Q +
Sbjct: 123 TYKSLTVFYLCDPKVSYRSSYTCPGRGLVSVSQNSDYHNS---------------CQDSF 167
Query: 184 GVGVP--ILEEQANRFAGNRSLLREVLMEGFNVNYNNPFENDCLECISSGGQQCGFDS 239
+ VP + E+ N L L EGF V E C +C SSGG CGF +
Sbjct: 168 TINVPKSFVPEEKELDVTN---LESALREGFEVKVKVD-EKTCQKCTSSGG-TCGFQN 220
>UniRef100_Q8VYG0 Hypothetical protein At5g38210 [Arabidopsis thaliana]
Length = 686
Score = 533 bits (1374), Expect = e-150
Identities = 326/694 (46%), Positives = 429/694 (60%), Gaps = 46/694 (6%)
Query: 15 IFFIFYLRHTTSLPSHASLSSCNNTTFNCGHITKLSYPFTGGDRPSYCGPPQFHLNCKN- 73
+ +F+L + LP S +T F CG +T +PF G RP CG P L+C+
Sbjct: 16 LVLLFFLSYIHFLPCAQSQREPCDTLFRCGDLTA-GFPFWGVARPQPCGHPSLGLHCQKQ 74
Query: 74 -NVPELNISSVSYRVLQVNSVTHSLTLARLDLWNETCTHHYVNSTFGGTSFSNGLGNSKI 132
N L ISS+ YRVL+VN+ T +L L R D C+ + +T F +
Sbjct: 75 TNSTSLIISSLMYRVLEVNTTTSTLKLVRQDFSGPFCSASFSGATLTPELFELLPDYKTL 134
Query: 133 TLFYGCQPTSLFTEKPHNLFYCDSNGYKNNSYTLIGPFPLDPVLKFVQCD--YGVGVPIL 190
+ +Y C P+ + K F C + G +G D L C + + VPI
Sbjct: 135 SAYYLCNPSLHYPAK----FICPNKG--------VGSIHQDD-LYHNHCGGIFNITVPI- 180
Query: 191 EEQANRFAGNRSLLREVLMEGFNVNYNNPFENDCLECISSGG--------QQCGFDSDEN 242
A N + L VL +GF V + E C EC ++GG C + +
Sbjct: 181 GYAPEEGALNVTNLESVLKKGFEVKLSID-ERPCQECKTNGGICAYHVATPVCCKTNSSS 239
Query: 243 EHICICGNGLCPSGNSSTNGGL-------IGGVVGGVAALCLLGFVACFVVRRRRKNAKK 295
E C + PSG SS + GL IG G + A + G + C +RRR+K A +
Sbjct: 240 EVNC---TPMMPSG-SSAHAGLSKKGKIGIGFASGFLGATLIGGCLLCIFIRRRKKLATQ 295
Query: 296 PISNDLYMPPSSTTSGTNTGTLTSTTNSSQSIPSYPSSKTSTMPKSFYFGVQVFTYEELE 355
+ L +++ T + T TSTT S + PS ++ S Y G+QVF+YEELE
Sbjct: 296 YTNKGLSTTTPYSSNYTMSNTPTSTTISGSNHSLVPSI-SNLGNGSVYSGIQVFSYEELE 354
Query: 356 EATNNFHTSKELGEGGFGTVYKGDLKDGRVVAVKRHYESNFKRVAQFMNEVEILARLRHK 415
EAT NF SKELG+GGFGTVY G LKDGR VAVKR +E + KRV QF NE++IL L+H
Sbjct: 355 EATENF--SKELGDGGFGTVYYGTLKDGRAVAVKRLFERSLKRVEQFKNEIDILKSLKHP 412
Query: 416 NLVTLYGCTSKHSRELLLVYEYISNGTVADHLHGDRSSSCLLPWSVRLDIALETAEALAY 475
NLV LYGCT++HSRELLLVYEYISNGT+A+HLHG+++ S + W RL IA+ETA AL+Y
Sbjct: 413 NLVILYGCTTRHSRELLLVYEYISNGTLAEHLHGNQAQSRPICWPARLQIAIETASALSY 472
Query: 476 LHASDVMHRDVKSNNILLDEKFHVKVADFGLSRLFPNDVTHVSTAPQGTPGYVDPEYYQC 535
LHAS ++HRDVK+ NILLD + VKVADFGLSRLFP D TH+STAPQGTPGYVDPEYYQC
Sbjct: 473 LHASGIIHRDVKTTNILLDSNYQVKVADFGLSRLFPMDQTHISTAPQGTPGYVDPEYYQC 532
Query: 536 YQLTDKSDVYSFGVVLVELISSLQAVDITRHRNDVNLANMAVNKIQSQELYDLVDPNLGY 595
Y+L +KSDVYSFGVVL ELISS +AVDITRHR+D+NLANMA++KIQ+ +++L D +LG+
Sbjct: 533 YRLNEKSDVYSFGVVLSELISSKEAVDITRHRHDINLANMAISKIQNDAVHELADLSLGF 592
Query: 596 EKDNSVKRMTTAVAELAFRCLQQQRDLRPSMDEIVEVLRAIKSDEPETQESKVLDVVVR- 654
+D SVK+M ++VAELAFRCLQQ+RD+RPSMDEIVEVLR I+ D + V+++ V
Sbjct: 593 ARDPSVKKMMSSVAELAFRCLQQERDVRPSMDEIVEVLRVIQKDGISDSKDVVVEIDVNG 652
Query: 655 TDELVLLKKG-PYPTSPDSVAEKWVSGSSTSTSS 687
D++ LLK G P P SP++ +K + SS +T+S
Sbjct: 653 GDDVGLLKHGVPPPLSPET--DKTTASSSNTTAS 684
>UniRef100_Q9LPQ6 F15H18.11 [Arabidopsis thaliana]
Length = 1154
Score = 497 bits (1279), Expect = e-139
Identities = 298/649 (45%), Positives = 391/649 (59%), Gaps = 58/649 (8%)
Query: 42 NCGHITKLSYPF-TGGDRPSYCGPPQFHLNC--KNNVPELNISSVSYRVLQVNSVTHSLT 98
+CG ++SYPF G + S+CG P F L C + +P L IS Y + ++ +T S
Sbjct: 267 SCGKGPQISYPFYLSGKQESFCGYPSFELTCDDEEKLPVLGISGEEYVIKNISYLTQSFQ 326
Query: 99 LARLDLWNETCTHHYVNSTFGGTSFSNGLGNSKITLFYGCQPTSLFTEKPHNLFYCDSNG 158
+ ++ C N T T F + T+ Y C L + + L C N
Sbjct: 327 VVNSKASHDPCPRPLNNLTLHRTPFFVNPSHINFTILYNCSDHLLEDFRTYPLT-CARNT 385
Query: 159 YKNNSYTLIGPFPL--DPVLKFVQCDYGVGVPILEEQANRFAGNRSLLREVLMEGFNVNY 216
S+ + L + + + C V VP+L + G + E+L GF +N+
Sbjct: 386 SLLRSFGVFDRKKLGKEKQIASMSCQKLVDVPVLASNESDVMGMTYV--EILKRGFVLNW 443
Query: 217 NNPFENDCLECISSGGQQCGFDSDENEHICICGNG-----LCPSGNSSTNGGLIGGVVGG 271
N C CI+SGG+ CG +D+ E +C+C +G C +G + +I V+ G
Sbjct: 444 T---ANSCFRCITSGGR-CG--TDQQEFVCLCPDGPKLHDTCTNGKNDKRRRVIVKVLIG 497
Query: 272 VAALCLLGFVACFVVRRRRKNAKKPISNDLYMPPSSTTSGTNTGTLTSTTNSSQSIPSYP 331
+ A + +V RRK T S N+ L ++I S P
Sbjct: 498 LIAASIFWYVY-----HRRK----------------TKSYRNSSALLP-----RNISSDP 531
Query: 332 SSKTSTMPKS--FYFGVQVFTYEELEEATNNFHTSKELGEGGFGTVYKGDLKDGRVVAVK 389
S+K+ + K+ GV +F+YEELEEATNNF SKELG+GGFGTVY G LKDGR VAVK
Sbjct: 532 SAKSFDIEKAEELLVGVHIFSYEELEEATNNFDPSKELGDGGFGTVYYGKLKDGRSVAVK 591
Query: 390 RHYESNFKRVAQFMNEVEILARLRHKNLVTLYGCTSKHSRELLLVYEYISNGTVADHLHG 449
R Y++NFKR QF NEVEIL LRH NLV L+GC+SK SR+LLLVYEY++NGT+ADHLHG
Sbjct: 592 RLYDNNFKRAEQFRNEVEILTGLRHPNLVALFGCSSKQSRDLLLVYEYVANGTLADHLHG 651
Query: 450 DRSSSCLLPWSVRLDIALETAEALAYLHASDVMHRDVKSNNILLDEKFHVKVADFGLSRL 509
+++ LPWS+RL IA+ETA AL YLHAS ++HRDVKSNNILLD+ F+VKVADFGLSRL
Sbjct: 652 PQANPSSLPWSIRLKIAVETASALKYLHASKIIHRDVKSNNILLDQNFNVKVADFGLSRL 711
Query: 510 FPNDVTHVSTAPQGTPGYVDPEYYQCYQLTDKSDVYSFGVVLVELISSLQAVDITRHRND 569
FP D THVSTAPQGTPGYVDP+Y+ CYQL++KSDVYSF VVL+ELISSL AVDITR R +
Sbjct: 712 FPMDKTHVSTAPQGTPGYVDPDYHLCYQLSNKSDVYSFAVVLMELISSLPAVDITRPRQE 771
Query: 570 VNLANMAVNKIQSQELYDLVDPNLGYEKDNSVKRMTTAVAELAFRCLQQQRDLRPSMDEI 629
+NL+NMAV KIQ+ EL D+VDP+LG++ D V++ AVAELAF+CLQ +DLRP M +
Sbjct: 772 INLSNMAVVKIQNHELRDMVDPSLGFDTDTRVRQTVIAVAELAFQCLQSDKDLRPCMSHV 831
Query: 630 VEVLRAIKSDEPETQESKVLDVVVRTDELVLLKKGP-YPTSPDSVAEKW 677
+ L I+++ E V+DV K GP SPDSV KW
Sbjct: 832 QDTLTRIQNN-GFGSEMDVVDV---------NKSGPLVAQSPDSVIVKW 870
Score = 60.1 bits (144), Expect = 2e-07
Identities = 56/245 (22%), Positives = 99/245 (39%), Gaps = 21/245 (8%)
Query: 11 ITFIIFFIFYLRHTTSLPSHASLSSCNNTTFNCGH--ITKLSYPFTGG-DRPSYCGPPQF 67
I+F++F + L S N T GH +YPF GG ++P +CG F
Sbjct: 8 ISFVVFSVADLPSCFSADQQYEECRSRNLTCGSGHRVFESTTYPFWGGFNKPKFCGHSSF 67
Query: 68 HLNCKNNVP-ELNISSVSYRVLQVNSVTHSLTLARLDLWNETCTHHYVNSTFGGTSFSNG 126
L+C+ + L I +++ RV+ N H +++A L + C + + + G F+
Sbjct: 68 KLSCEGDQNLTLAIGNITLRVVSANLEDHKISVADDSLLDGGCLN--IWNFNGKNQFTLD 125
Query: 127 LGNSKITLFYGCQPTSLFTEKPHNLFYCDSNGYKNNSYTLIGPFPLDP-VLKFVQCDYGV 185
I +F C + C+ + +Y ++ D +K+ +
Sbjct: 126 SNTETIDVFVNCSGVAPLQ------ISCEESYEDPVTYHVLRSSDSDEGCMKYAE----- 174
Query: 186 GVPILEEQANRFAGNRSLLREVLMEGFNVNYNNPFENDCLECISSGGQQCGFDSDENEHI 245
+P+L + + E L +GF++ Y + C CI SGG CG D
Sbjct: 175 -IPMLRSAKDELQRSELTFVEALRKGFDLRYIME-DKACRRCIDSGG-ICGSALDSESFR 231
Query: 246 CICGN 250
C+C +
Sbjct: 232 CLCAD 236
>UniRef100_Q5ZC67 Serine/threonine-specific protein kinase-like [Oryza sativa]
Length = 668
Score = 466 bits (1200), Expect = e-130
Identities = 295/681 (43%), Positives = 385/681 (56%), Gaps = 69/681 (10%)
Query: 30 HASLSSCNNTTFNCGHITKLSYPFTGGDRPSY--------------CGPPQFHLNCKNNV 75
H + + C T CG++T + YPF G P++ CG P F + C V
Sbjct: 19 HRARAECEPAT--CGNLT-VRYPFWLGG-PNFNQSNQSSPSSALASCGHPAFEVWCNGGV 74
Query: 76 PELNISSVSYRVLQVNSVTHSLTLARL-DLWNETC-THHYVNSTFGGTSFSNGLGNSKIT 133
L S + + NS + R+ D + C T ++S+ + F+ N I
Sbjct: 75 ASLRGSQILVLSIDYNSSSFVAAHKRVADGGDGVCRTDFNISSSLALSPFTISSSNRAIC 134
Query: 134 LFYGCQPTSLFTEKPH--NLFYCDSNGYKNNSYTLIGPF---PLDPVLKFVQCDYGVGVP 188
Y C T E P L + Y +G P +K C Y +P
Sbjct: 135 FLYSCNGT----EPPEIDGLVNATISSCSKPIYAYLGGIYDRDNPPAIKAGNCTYSY-LP 189
Query: 189 IL--EEQANRFAGNRSLLREVLMEGFNVNYNNPFENDCLECISSGGQQCGFDSDENEHIC 246
+L + AN AG +GF + + DC C SGGQ + C
Sbjct: 190 VLWPDSPANLTAGTN--YSPQFKKGFVLEWQKNGFGDCDACNGSGGQCRYINDSAAAFAC 247
Query: 247 ICGNG-----LCPSGNSSTNGGLIGGVVGGVAALCLLGFVACFVVRRRRKNAKKPISNDL 301
+C +G CP S ++ +IG G + L+ + F +R+K +
Sbjct: 248 LCSDGKLRRSTCPGSRSKSH--IIGIACGSSGGILLIVSIFIFAWHKRKKRKQ------- 298
Query: 302 YMPPSSTTSGTNTGTLTSTTNSSQSIPSYPSSKTSTMPKSFYFGVQVFTYEELEEATNNF 361
T L +SS S+ SY SK + S + +FTYEELEEAT F
Sbjct: 299 ------------TRDLKDLMHSSSSMQSY--SKDLELGGSPH----IFTYEELEEATAGF 340
Query: 362 HTSKELGEGGFGTVYKGDLKDGRVVAVKRHYESNFKRVAQFMNEVEILARLRHKNLVTLY 421
S+ELG+GGFGTVYKG L+DGRVVAVKR Y++N++RV QF+NEV+IL+RL H+NLV LY
Sbjct: 341 SASRELGDGGFGTVYKGKLRDGRVVAVKRLYKNNYRRVEQFLNEVDILSRLLHQNLVILY 400
Query: 422 GCTSKHSRELLLVYEYISNGTVADHLHGDRSSSCLLPWSVRLDIALETAEALAYLHASDV 481
GCTS+ SR+LLLVYEYI NGTVADHLHG R+ L W VR+ IA+ETAEALAYLHA ++
Sbjct: 401 GCTSRSSRDLLLVYEYIPNGTVADHLHGPRAGERGLTWPVRMTIAIETAEALAYLHAVEI 460
Query: 482 MHRDVKSNNILLDEKFHVKVADFGLSRLFPNDVTHVSTAPQGTPGYVDPEYYQCYQLTDK 541
+HRDVK+NNILLD FHVKVADFGLSRLFP +VTHVST PQGTPGYVDP Y+QCY+LTDK
Sbjct: 461 IHRDVKTNNILLDNNFHVKVADFGLSRLFPLEVTHVSTVPQGTPGYVDPVYHQCYKLTDK 520
Query: 542 SDVYSFGVVLVELISSLQAVDITRHRNDVNLANMAVNKIQSQELYDLVDPNLGYEKDNSV 601
SDVYSFGVVL+ELISS AVD++R +D+NLANMA+N+IQ+ E+ LVDP +GYE D+
Sbjct: 521 SDVYSFGVVLIELISSKPAVDMSRSHSDINLANMALNRIQNHEVDQLVDPEIGYETDSET 580
Query: 602 KRMTTAVAELAFRCLQQQRDLRPSMDEIVEVLRAIKSDEPETQESKVLDVVVRTDELVLL 661
KRM VAELAF+CLQ R+ RP + E+VEVL IK+ E ++ ++ E L
Sbjct: 581 KRMVDLVAELAFQCLQMDRESRPPIKEVVEVLNCIKNGECPAEK---MNKNASPKEDSHL 637
Query: 662 KKGPYPTSPDSVAEKWVSGSS 682
K SPDSV ++ S S+
Sbjct: 638 LKDSLQYSPDSVIHRFHSQST 658
>UniRef100_Q8S011 P0034C09.1 protein [Oryza sativa]
Length = 687
Score = 459 bits (1180), Expect = e-127
Identities = 289/652 (44%), Positives = 387/652 (59%), Gaps = 59/652 (9%)
Query: 60 SYCGPPQFHLNCKNN---VPELNISSVSYRVLQVNSVTHSLTLARLDLWNETCTHHYVNS 116
++CG P + C + + + SY V +++ + +++LA D+ N TC N
Sbjct: 70 TFCGYPGLEIICDGGGGGKAVMMLGNDSYTVSRIDYASLTVSLADADVANGTCPVVSHNV 129
Query: 117 TFGGTSFSNGLGNS--KITLFYGCQ--PTSLFTEKPHNL--FYCDSNGYKNNSYTLIGPF 170
T S L ++ + F+ C P + KP ++ C N + + + P
Sbjct: 130 TIPPAPSSLHLADTVGMLIFFFRCAFGPAANAPPKPPSIHPLTCGENSEDAPTQSFLLPA 189
Query: 171 -PLDPV-LKFVQCDYGVGVPIL----EEQANRFAGNRSLLREVLMEGFNVNYNNPFENDC 224
PL P L C GVP+L AN A + L +GF ++++ + C
Sbjct: 190 SPLPPGDLWHRGCSAVYGVPVLGGSLPSDANDPAWRKDGYIASLRKGFQMSWDR--SDRC 247
Query: 225 LECISSGGQQCGFDSDENEHICICGNGLCPSGN----SSTNGGLIGGVVGGVAALCLLGF 280
C + G+ CG++ + C+C NGL S S + L G V GG++A+ LG
Sbjct: 248 SRCELTSGK-CGYNQNGKFLGCLCANGLVDSDACSKISDSTLRLAGVVGGGLSAVFALGL 306
Query: 281 VACFVVRRRRKNAKKPISNDLYMPPSSTTSGTNTGTLTSTTNSSQSIPSYPSSKTSTMPK 340
+A R+RK+ K S+ L S + GT P S M
Sbjct: 307 IATVFFVRKRKHKKVNSSSKLLK--YSGSGGT------------------PRSMGGDMES 346
Query: 341 SFYFGVQ--VFTYEELEEATNNFHTSKELGEGGFGTVYKGDLKDGRVVAVKRHYESNFKR 398
+Q +F+YEELEEAT++F+ ++ELG+GGFGTVYKG L+DGRVVAVKR Y ++++R
Sbjct: 347 GSVKDLQTHLFSYEELEEATDSFNENRELGDGGFGTVYKGILRDGRVVAVKRLYNNSYRR 406
Query: 399 VAQFMNEVEILARLRHKNLVTLYGCTSKHSRELLLVYEYISNGTVADHLHGDRSSSCLLP 458
V QF+NE IL+RLRH NLV YGCTS SRELLLVYE+++NGTVADHLHG R+ L
Sbjct: 407 VEQFVNEAAILSRLRHPNLVMFYGCTSSQSRELLLVYEFVANGTVADHLHGHRAQERALS 466
Query: 459 WSVRLDIALETAEALAYLHASD--VMHRDVKSNNILLDEKFHVKVADFGLSRLFPNDVTH 516
W +RL+IA+E+A AL YLHA + ++HRDVK+ NILLD FHVKVADFGLSRLFP DVTH
Sbjct: 467 WPLRLNIAVESAAALTYLHAIEPPIVHRDVKTTNILLDADFHVKVADFGLSRLFPLDVTH 526
Query: 517 VSTAPQGTPGYVDPEYYQCYQLTDKSDVYSFGVVLVELISSLQAVDITRHRNDVNLANMA 576
VSTAPQGTPGYVDPEY+QCYQLTDKSDVYSFGVVLVELISS AVDITR RN++NLA MA
Sbjct: 527 VSTAPQGTPGYVDPEYHQCYQLTDKSDVYSFGVVLVELISSKPAVDITRQRNEINLAGMA 586
Query: 577 VNKIQSQELYDLVDPNLGYEKDNSVKRMTTAVAELAFRCLQQQRDLRPSMDEIVEVLRAI 636
+N+IQ +L +LVD LGYE D + K+M T VAELAFRCLQQ ++RP + E++E L+ +
Sbjct: 587 INRIQKSQLEELVDLELGYESDPATKKMMTMVAELAFRCLQQNGEMRPPIKEVLEGLKGV 646
Query: 637 KSDEPETQESKVLDVVVRTDELVLLKKGP-YPTSPDSVAEKWVSGSSTSTSS 687
+ D+ V + KKGP P SPD+V +W S +T +S
Sbjct: 647 Q------------DLCVMEKDGGKDKKGPDPPLSPDTVHAQWDSRQTTPNTS 686
>UniRef100_Q9C6K9 Wall-associated kinase, putative [Arabidopsis thaliana]
Length = 629
Score = 443 bits (1140), Expect = e-123
Identities = 278/694 (40%), Positives = 398/694 (57%), Gaps = 89/694 (12%)
Query: 11 ITFIIFFIFYLRHTTSLPSHASLSSCNNTTFNCGHITKLSYPFTGGDRPSYCGPPQFHLN 70
++ ++FF +L + + + C + F CG +PF D PS CG F LN
Sbjct: 4 VSVLLFFFLFLLAAEARSTKRT--GCKD--FTCGE-HDFKFPFFRTDMPSRCG--LFKLN 56
Query: 71 CKNNVPELNISSVSYRVLQVNSVTHSLTLARLD-LWNETCTHHYVNSTFGGTSFSN-GLG 128
C N+PE+ + + V SV+ + T+ +D N++ T T G + S+ L
Sbjct: 57 CSANIPEIQLEKDG-KWYTVKSVSQANTITIIDPRLNQSLT------TGGCSDLSSFSLP 109
Query: 129 NS---KITLFYGCQPTSLFTEKPHNLFYCDSNGYKNNSYTLIGPFPLDPVLKFVQCDYGV 185
+S K+ Y C +S + + Y + G ++ Y +G D+ V
Sbjct: 110 DSPWLKLNTLYKCNNSS----RKNGFSYANCRGEGSSLYYNLGD------------DHDV 153
Query: 186 G----VPILEEQANRFAGNRSLLREVLMEGFNVNYNNPFENDCLECISSGGQQCGFDSDE 241
+ E GN S + F+++ P +C C ++GG+ +
Sbjct: 154 SGCSPIKTPESWVTPKNGNLSDVNAT----FSLHIELP--GNCFRCHNNGGE---CTKVK 204
Query: 242 NEHICICGNGLCPSGNSSTNGGLIGGVVGGVAALCLLGFVACFVVRRRRKNAKKPISNDL 301
N + C+ N + ++ GL G+ G V + +L + + R R+ +S D
Sbjct: 205 NNYRCVGANTEPNNYHAEMRLGL--GIGGSVILIIILVALFAVIHRNYRRKDGSELSRD- 261
Query: 302 YMPPSSTTSGTNTGTLTSTTNSSQSIPSYPSSKTSTMPKSFYFGVQVFTYEELEEATNNF 361
+SK+ +F + +F+Y+EL+ AT+NF
Sbjct: 262 ------------------------------NSKSDVEFSQVFFKIPIFSYKELQAATDNF 291
Query: 362 HTSKELGEGGFGTVYKGDLKDGRVVAVKRHYESNFKRVAQFMNEVEILARLRHKNLVTLY 421
+ LG+GGFGTVY G ++DGR VAVKR YE N++R+ QFMNE+EIL RL HKNLV+LY
Sbjct: 292 SKDRLLGDGGFGTVYYGKVRDGREVAVKRLYEHNYRRLEQFMNEIEILTRLHHKNLVSLY 351
Query: 422 GCTSKHSRELLLVYEYISNGTVADHLHGDRSS-SCLLPWSVRLDIALETAEALAYLHASD 480
GCTS+ SRELLLVYE+I NGTVADHL+G+ + L WS+RL IA+ETA ALAYLHASD
Sbjct: 352 GCTSRRSRELLLVYEFIPNGTVADHLYGENTPHQGFLTWSMRLSIAIETASALAYLHASD 411
Query: 481 VMHRDVKSNNILLDEKFHVKVADFGLSRLFPNDVTHVSTAPQGTPGYVDPEYYQCYQLTD 540
++HRDVK+ NILLD F VKVADFGLSRL P+DVTHVSTAPQGTPGYVDPEY++CY LTD
Sbjct: 412 IIHRDVKTTNILLDRNFGVKVADFGLSRLLPSDVTHVSTAPQGTPGYVDPEYHRCYHLTD 471
Query: 541 KSDVYSFGVVLVELISSLQAVDITRHRNDVNLANMAVNKIQSQELYDLVDPNLGYEKDNS 600
KSDVYSFGVVLVELISS AVDI+R ++++NL+++A+NKIQ+ ++L+D NLGY +
Sbjct: 472 KSDVYSFGVVLVELISSKPAVDISRCKSEINLSSLAINKIQNHATHELIDQNLGYATNEG 531
Query: 601 VKRMTTAVAELAFRCLQQQRDLRPSMDEIVEVLRAIKSDEP-----ETQESKVLDVVVRT 655
V++MTT VAELAF+CLQQ +RP+M+++V L+ I+++E + +E ++
Sbjct: 532 VRKMTTMVAELAFQCLQQDNTMRPTMEQVVHELKGIQNEEQKCPTYDYREETIIPHPSPP 591
Query: 656 D--ELVLLKKGPYPTSPDSVAEKWVSGSSTSTSS 687
D E LLK +P SP SV ++W S S+T +S
Sbjct: 592 DWGEAALLKNMKFPRSPVSVTDQWTSKSTTPNTS 625
>UniRef100_Q6L4G6 Putative receptor-like protein kinase [Oryza sativa]
Length = 912
Score = 442 bits (1138), Expect = e-122
Identities = 274/664 (41%), Positives = 368/664 (55%), Gaps = 78/664 (11%)
Query: 34 SSCNNTTFNCGHIT-KLSYPFT--GGDRPSYCGPPQFHLNCK-NNVPELNISSVSYRVLQ 89
++C ++ CG + + YPF+ G RP YC P + L C +N ++++S +++V
Sbjct: 316 TNCTPASYQCGSLKFDVDYPFSANGVHRPDYCSYPGYRLICSPDNKLMIHMNSTAFQVTD 375
Query: 90 VNSVTHSLTLARLDLWNETCTHHYVNSTFGGTSFSNGLGNSKITLFYGCQPTSLFTEKPH 149
++ L + E C Y N+T + F + +T++ C +
Sbjct: 376 IDYGNKFLAVIDQTQPQEACLDRYHNTTIDESKFMYTDRDQFLTVYVNCSANFSSLPLIY 435
Query: 150 NLFYCDSNGYKNNSYTLIGPFPLDPVLKFV--QCDYGVGVPILEEQANRFAGNRSLLREV 207
+L C S G +SY + D + + C + VP A A S L +V
Sbjct: 436 DLVSCVSGG---SSYYRLHKNKDDSLESDILGSCSSTIVVPCNSTMAGSLAAGNSSLADV 492
Query: 208 LMEGFNVNYNNPFENDCLECISSGGQQCGFDSDENEHICICGNGLCPSGNSSTNGGLIGG 267
+ GF + + G S +++ I +
Sbjct: 493 IRGGFTARW-----------------KVGLGSKKSKKKAIA---------------IATS 520
Query: 268 VVGGVAALCLLGFVACFVVRRRRKNAKKPISNDLYMPPSSTTSGTNTGTLTSTTNSSQSI 327
+ GV L LL V F+ R+R+ K T++ L T S ++
Sbjct: 521 IASGVLFLLLL--VVSFLYIRKRRQYKM----------------TSSSRLLKYTTSGRTP 562
Query: 328 PSYPSSKTSTMPKSF-YFGVQVFTYEELEEATNNFHTSKELGEGGFGTVYKGDLKDGRVV 386
S SS SF Y F YEELEEAT+ F ++ELG+GGFGTVYKG+L+DGRVV
Sbjct: 563 RSKGSSDKFVESGSFHYLQTHHFAYEELEEATDGFSDARELGDGGFGTVYKGELRDGRVV 622
Query: 387 AVKRHYESNFKRVAQFMNEVEILARLRHKNLVTLYGCTSKHSRELLLVYEYISNGTVADH 446
AVKR Y ++ +RV QF+NE IL+RLRH NLV YGCTS SRELLLVYE++ NGTVADH
Sbjct: 623 AVKRLYNNSCRRVEQFVNEAAILSRLRHPNLVLFYGCTSSRSRELLLVYEFVPNGTVADH 682
Query: 447 LHGDRSSSCLLPWSVRLDIALETAEALAYLHASD---VMHRDVKSNNILLDEKFHVKVAD 503
LHG R+ L W +RL++A+E A ALAYLHA + ++HRDVK+NNILLD FHVKVAD
Sbjct: 683 LHGHRAPERALTWPLRLNVAVEAAAALAYLHAVEPAPIVHRDVKTNNILLDANFHVKVAD 742
Query: 504 FGLSRLFPNDVTHVSTAPQGTPGYVDPEYYQCYQLTDKSDVYSFGVVLVELISSLQAVDI 563
FGLSRLFP D THVSTAPQGTPGYVDPEY+QCYQLTDKSDVYSFGVVLVELISS AVD+
Sbjct: 743 FGLSRLFPRDATHVSTAPQGTPGYVDPEYHQCYQLTDKSDVYSFGVVLVELISSKPAVDV 802
Query: 564 TRHRNDVNLANMAVNKIQSQELYDLVDPNLGYEKDNSVKRMTTAVAELAFRCLQQQRDLR 623
TR R+++NLA MAVNKIQ ++ LVD LGY D + ++ T VAELAFRCLQ ++R
Sbjct: 803 TRDRDEINLAGMAVNKIQRCQVDQLVDDELGYSSDEATRKTMTMVAELAFRCLQHNGEMR 862
Query: 624 PSMDEIVEVLRAIKSDEPETQESKVLDVVVRTDELVLLKKGPYPTSPDSVAEKWVSGSST 683
P + E+ +VLR I+ + ++ K+G P SP++V W S S+T
Sbjct: 863 PPIKEVADVLRGIQDECRAAEKGG--------------KRGS-PCSPNTVHAPWDSMSTT 907
Query: 684 STSS 687
+S
Sbjct: 908 PNTS 911
Score = 54.7 bits (130), Expect = 9e-06
Identities = 59/234 (25%), Positives = 92/234 (39%), Gaps = 29/234 (12%)
Query: 43 CGHITKLSYPF------------TGGDRPSYCGPPQFHLNCKNNVPELNISSVSYRVLQV 90
CG ++ YPF G R YCG P + C ++ L + + +Y VL++
Sbjct: 41 CGEQVEIKYPFYLSNTTDQVVVVDGNTR--YCGYPWLGIICDHDRAILRLGNYNYTVLEI 98
Query: 91 NSVTHSLTLARLD-LWNETCTHHYVNSTFGGTSFSNGLGNSKITLFYGCQPTSLFTEKPH 149
N H++T+A D L C N T GN IT F+ C T+ +P
Sbjct: 99 NHGNHTVTVADSDALDGGDCPRVKHNVTLPEVLTFPSPGNDSITFFFDCNSTANVVLRPP 158
Query: 150 ------NLFYCDSNGYKNN--SYTLIGPFPLDPVLKFVQCDYGVGVPILEE--QANRFAG 199
N D G ++ S+ P C V VP+L++ ++ G
Sbjct: 159 PYIRPINCSTFDFPGRRDTAPSFVATQPDVAGETEWLGLCKEVVMVPVLKDWLMNEKYYG 218
Query: 200 --NRSLLREVLMEGFNVNYNNPFENDCLECISSGGQQCGFDSDENEHICICGNG 251
VL GF +++ +P C EC SGG +C + + C+C +G
Sbjct: 219 KLGDDGYGAVLKRGFQLSW-DPTAGMCHECEVSGG-RCSYGTKNEFLGCLCSDG 270
>UniRef100_Q941D0 At1g25390/F2J7_14 [Arabidopsis thaliana]
Length = 443
Score = 412 bits (1060), Expect = e-113
Identities = 225/469 (47%), Positives = 305/469 (64%), Gaps = 44/469 (9%)
Query: 208 LMEGFNVNYNNPFENDCLECISSGGQQCGFDSDENEHICICGNGLCPSGNSSTNGGLIGG 267
LME + + +C C ++GG+ +N + C+ N + ++ GL G
Sbjct: 5 LMEMLPSSLHIELPGNCFRCHNNGGE---CTKVKNNYRCVGANTEPNNYHAEMRLGL--G 59
Query: 268 VVGGVAALCLLGFVACFVVRRRRKNAKKPISNDLYMPPSSTTSGTNTGTLTSTTNSSQSI 327
+ G V + +L + + R R+ +S D
Sbjct: 60 IGGSVILIIILVALFAVIHRNYRRKDGSELSRD--------------------------- 92
Query: 328 PSYPSSKTSTMPKSFYFGVQVFTYEELEEATNNFHTSKELGEGGFGTVYKGDLKDGRVVA 387
+SK+ +F + +F+Y+EL+ AT+NF + LG+GGFGTVY G ++DGR VA
Sbjct: 93 ----NSKSDVEFSQVFFKIPIFSYKELQAATDNFSKDRLLGDGGFGTVYYGKVRDGREVA 148
Query: 388 VKRHYESNFKRVAQFMNEVEILARLRHKNLVTLYGCTSKHSRELLLVYEYISNGTVADHL 447
VKR YE N++R+ QFMNE+EIL RL HKNLV+LYGCTS+ SRELLLVYE+I NGTVADHL
Sbjct: 149 VKRLYEHNYRRLEQFMNEIEILTRLHHKNLVSLYGCTSRRSRELLLVYEFIPNGTVADHL 208
Query: 448 HGDRSS-SCLLPWSVRLDIALETAEALAYLHASDVMHRDVKSNNILLDEKFHVKVADFGL 506
+G+ + L WS+RL IA+ETA ALAYLHASD++HRDVK+ NILLD F VKVADFGL
Sbjct: 209 YGENTPHQGFLTWSMRLSIAIETASALAYLHASDIIHRDVKTTNILLDRNFGVKVADFGL 268
Query: 507 SRLFPNDVTHVSTAPQGTPGYVDPEYYQCYQLTDKSDVYSFGVVLVELISSLQAVDITRH 566
SRL P+DVTHVSTAPQGTPGYVDPEY++CY LTDKSDVYSFGVVLVELISS AVDI+R
Sbjct: 269 SRLLPSDVTHVSTAPQGTPGYVDPEYHRCYHLTDKSDVYSFGVVLVELISSKPAVDISRC 328
Query: 567 RNDVNLANMAVNKIQSQELYDLVDPNLGYEKDNSVKRMTTAVAELAFRCLQQQRDLRPSM 626
++++NL+++A+NKIQ+ ++L+D NLGY + V++MTT VAELAF+CLQQ +RP+M
Sbjct: 329 KSEINLSSLAINKIQNHATHELIDQNLGYATNEGVRKMTTMVAELAFQCLQQDNTMRPTM 388
Query: 627 DEIVEVLRAIKSDEP-----ETQESKVLDVVVRTD--ELVLLKKGPYPT 668
+++V L+ I+++E + +E ++ D E LLKK PT
Sbjct: 389 EQVVHELKGIQNEEQKCPTYDYREETIIPHPSPPDWGEAALLKKYEIPT 437
>UniRef100_Q5ZC64 Putative receptor protein kinase CRINKLY4 [Oryza sativa]
Length = 666
Score = 410 bits (1054), Expect = e-113
Identities = 279/701 (39%), Positives = 376/701 (52%), Gaps = 82/701 (11%)
Query: 21 LRHTTSLPSHASLSSCNNTTFN---CGHITKLSYPFT-GGDRPSYCGPPQFHLNCKNNVP 76
L H + H S+ NT+ CG++T +SYPF+ G +P CG P L C N
Sbjct: 13 LLHAAAAIGHNETSTSGNTSCTPARCGNLT-ISYPFSLSGVQPVSCGYPVLDLTCDNRTG 71
Query: 77 ELNISSVS----YRVLQVNSVTHSLTLARLDLW--NETCT--HHYVNSTFGGTSFSNGLG 128
+S +RV + +SL A + + C V S+ F
Sbjct: 72 RAFLSRTFRDHLFRVDSIFYENNSLVAAVETTFAGDADCPVPDFNVTSSLSPYPFIISNT 131
Query: 129 NSKITLFYGCQPTSLFTEKPHNLFYCDSNGYKNNSYTLIGPFPLDPVLKFVQCDYGVGVP 188
N + Y C + Y ++ + P + C+ V VP
Sbjct: 132 NKYLAFIYDCSIPEHVGQLQRPCGNRTMGAYISDKWNSTPPSGVRG-----NCN-SVSVP 185
Query: 189 ILEEQANRFAGNRSL---LREVLMEGFNVNYNNPF--ENDCLECISSGGQQCGFDSDENE 243
+ + G + + +++ +GF + + + DC C GG+ C F+ +
Sbjct: 186 V----RGYYDGMKPVSGHYEQLIKDGFVLEWMRSVMGDQDCDGCRRRGGE-CRFEQLSFQ 240
Query: 244 HICICGNGLCPSGNSSTNGG-------------LIGGVVGGVAALCLLGFVACFVVRRRR 290
C C +GL S ++ TN + G V L +LG + + RR+
Sbjct: 241 --CFCPDGLLCSNSTRTNTTSSHPSGKVNRGIKIAAGTAAAVVCLGILGVGSTVLYTRRK 298
Query: 291 KNAKKPISNDLYMPPSSTTSGTNTGTLTSTTNSSQSIPSYPSSKTSTMPKSFYFGVQVFT 350
+ ++ GT +LT K ++ Y +FT
Sbjct: 299 RKRSASFEGLIH-------GGTPLPSLT---------------KEFSLAGLAY--THIFT 334
Query: 351 YEELEEATNNFHTSKELGEGGFGTVYKGDLKDGRVVAVKRHYESNFKRVAQFMNEVEILA 410
YEEL+EAT+ F ++ELG GGFGTVYKG L++G VAVKR Y++++K V QF NEV IL+
Sbjct: 335 YEELDEATDGFSDARELGVGGFGTVYKGILRNGDTVAVKRLYKNSYKSVEQFQNEVGILS 394
Query: 411 RLRHKNLVTLYGCTSK-HSRELLLVYEYISNGTVADHLHGDRSS-SCLLPWSVRLDIALE 468
RLRH NLVTL+GCTS+ +SR+LLLVYE++ NGT+ADHLHG ++ S L W RL IA+E
Sbjct: 395 RLRHPNLVTLFGCTSQTNSRDLLLVYEFVPNGTLADHLHGGAAARSSSLDWPTRLGIAVE 454
Query: 469 TAEALAYLHASD--VMHRDVKSNNILLDEKFHVKVADFGLSRLFPNDVTHVSTAPQGTPG 526
TA AL YLH + V+HRDVK+NNILLDE FHVKVADFGLSRLFP D THVSTAPQGTPG
Sbjct: 455 TASALEYLHTVEPQVVHRDVKTNNILLDEGFHVKVADFGLSRLFPADATHVSTAPQGTPG 514
Query: 527 YVDPEYYQCYQLTDKSDVYSFGVVLVELISSLQAVDITRHRNDVNLANMAVNKIQSQELY 586
Y+DP Y+QCYQLTDKSDVYSFGVVLVELISS AVD+ R DVNLANMAV+ IQS E+
Sbjct: 515 YLDPMYHQCYQLTDKSDVYSFGVVLVELISSKPAVDMNRRGGDVNLANMAVHMIQSYEME 574
Query: 587 DLVDPNLGYEKDNSVKRMTTAVAELAFRCLQQQRDLRPSMDEIVEVLRAIKSDEPETQES 646
LVDP LGY D +R VAE+AFRCLQ ++D+RP + E+++ LR E+
Sbjct: 575 QLVDPQLGYASDGETRRTVDLVAEVAFRCLQPEQDVRPPIGEVLDALR----------EA 624
Query: 647 KVLDVVVRTDELVLLKKGPYPTSPDSVAEKWVSGSSTSTSS 687
+ +D V + L K SPD V +W+S S+TS +S
Sbjct: 625 QRMDKVGYVKDDAGLVKKSRDGSPDCVMYQWISPSTTSNNS 665
>UniRef100_Q6QLL5 WAK-like kinase [Lycopersicon esculentum]
Length = 703
Score = 297 bits (760), Expect = 8e-79
Identities = 223/647 (34%), Positives = 332/647 (50%), Gaps = 97/647 (14%)
Query: 31 ASLSSCNNTTFN--CG----HITKLSYPFTGGDRPSYCGPPQFHLNCKNNVPELNIS--- 81
+S+ + N+T N CG + ++SYPF + CG L+C + E+ I
Sbjct: 22 SSVEATNSTKCNQYCGAAGSYSPRVSYPFGFSEG---CG---IRLDCTESTGEIRIGEYI 75
Query: 82 --SVSYRVLQVN-SVTHSLTLARLDLWNETCTHHYVNSTFGGTSFSNGL--GNSKITLFY 136
+V+ L VN S+ S + L ++ T FG T + NGL N K+
Sbjct: 76 IQNVTSETLMVNFSMNCSRPIEDLQQFDRT--------NFGMT-WRNGLLLHNCKVPKSE 126
Query: 137 GCQPTSLFTEKPHNLFYCDSNGYKNNSYTLIGPFPLD-PVLKFVQCDYGVGVPILEEQAN 195
P+ + + + N+ CDS + Y+ LD LK C + ++ +
Sbjct: 127 CTIPSEILSTRL-NIQSCDSKKENVSCYSEARADYLDYQKLKNTGCGTVISSILIGMDND 185
Query: 196 RFAGNRSLLREVLME-GFNVNYNNPFENDCLEC--ISSGGQQCGF--------------D 238
+ + ME + + + ND C +S G + GF D
Sbjct: 186 TMKSSAMFIEFQTMELAWGLEGDCACHNDA-NCTNVSLPGNRKGFRCRCKDGFVGDGFSD 244
Query: 239 SDENEHICICG-----NGLCPSGNSSTNGGLIGGVVGGVAALCLLGFVACFVVRRRRKNA 293
D + C +G C G ++ G L+GG++ G + L V C+ +RRR +
Sbjct: 245 GDGCRKVSRCNPSRYLSGRC--GGTTRIGVLVGGIIAGAGLMAALA-VLCYCIRRRSASL 301
Query: 294 KKPISNDLYMPPSSTTSGTNTGTLTSTTNSSQSIPSYPSSKTSTMPKSFYFGVQVFTYEE 353
KK +S + S +G+N+ V VF Y+E
Sbjct: 302 KKRMSARRLL---SEAAGSNS-------------------------------VHVFQYKE 327
Query: 354 LEEATNNFHTSKELGEGGFGTVYKGDLKDGRVVAVKRHYESNFKRVAQFMNEVEILARLR 413
+E ATN+F + LG G +GTVY G L VA+K+ + V Q MNEV++L+ +
Sbjct: 328 IERATNSFSEKQRLGIGAYGTVYAGKLHSDEWVAIKKLRHRDPDGVEQVMNEVKLLSSVS 387
Query: 414 HKNLVTLYGCTSKHSRELLLVYEYISNGTVADHLHGDRSSSCLLPWSVRLDIALETAEAL 473
H NLV L GC ++ E +LVYE++ NGT+A HL +RSS LPW++RL IA ETA A+
Sbjct: 388 HPNLVRLLGCCIENG-EQILVYEFMPNGTLAQHLQRERSSG--LPWTIRLTIATETAHAI 444
Query: 474 AYLHAS---DVMHRDVKSNNILLDEKFHVKVADFGLSRLFPNDVTHVSTAPQGTPGYVDP 530
A+LH++ + HRD+KS+NILLD F+ KVADFGLSR D +H+STAPQGTPGYVDP
Sbjct: 445 AHLHSAMNPPIYHRDIKSSNILLDYNFNSKVADFGLSRFGMTDDSHISTAPQGTPGYVDP 504
Query: 531 EYYQCYQLTDKSDVYSFGVVLVELISSLQAVDITRHRNDVNLANMAVNKIQSQELYDLVD 590
+Y+Q Y L+DKSDVYSFGVVLVE+I++++ VD +R +++NLA +A+++I + +++D
Sbjct: 505 QYHQNYHLSDKSDVYSFGVVLVEIITAMKVVDFSRSHSEINLAALAIDRIGKGRVDEIID 564
Query: 591 PNLGYEKDNSVKRMTTAVAELAFRCLQQQRDLRPSMDEIVEVLRAIK 637
P L +D VAELAFRCL RD+RPSM E+ + L I+
Sbjct: 565 PFLEPHRDAWTLSSVHRVAELAFRCLAFHRDMRPSMTEVADELEQIR 611
>UniRef100_Q8RY67 At2g23450/F26B6.10 [Arabidopsis thaliana]
Length = 708
Score = 288 bits (738), Expect = 3e-76
Identities = 175/453 (38%), Positives = 255/453 (55%), Gaps = 67/453 (14%)
Query: 264 LIGGVVGGVAALCLLGFVACFVVRRRRKNAKKPISNDLYMPPSSTTSGTNTGTLTSTTNS 323
++GG VGG L L F F +RRR
Sbjct: 286 IVGGTVGGAFLLAALAFF--FFCKRRR--------------------------------- 310
Query: 324 SQSIPSYPSSKTSTMPKSFYFGVQVFTYEELEEATNNFHTSKELGEGGFGTVYKGDLKDG 383
S + S+ S+K + V F Y+E+E+AT+ F ++LG G +GTVY+G L++
Sbjct: 311 STPLRSHLSAKRLLSEAAGNSSVAFFPYKEIEKATDGFSEKQKLGIGAYGTVYRGKLQND 370
Query: 384 RVVAVKRHYESNFKRVAQFMNEVEILARLRHKNLVTLYGCTSKHSRELLLVYEYISNGTV 443
VA+KR + + + Q MNE+++L+ + H NLV L GC + + +LVYEY+ NGT+
Sbjct: 371 EWVAIKRLRHRDSESLDQVMNEIKLLSSVSHPNLVRLLGCCIEQG-DPVLVYEYMPNGTL 429
Query: 444 ADHLHGDRSSSCLLPWSVRLDIALETAEALAYLHAS---DVMHRDVKSNNILLDEKFHVK 500
++HL DR S LPW++RL +A +TA+A+AYLH+S + HRD+KS NILLD F+ K
Sbjct: 430 SEHLQRDRGSG--LPWTLRLTVATQTAKAIAYLHSSMNPPIYHRDIKSTNILLDYDFNSK 487
Query: 501 VADFGLSRLFPNDVTHVSTAPQGTPGYVDPEYYQCYQLTDKSDVYSFGVVLVELISSLQA 560
VADFGLSRL + +H+STAPQGTPGY+DP+Y+QC+ L+DKSDVYSFGVVL E+I+ L+
Sbjct: 488 VADFGLSRLGMTESSHISTAPQGTPGYLDPQYHQCFHLSDKSDVYSFGVVLAEIITGLKV 547
Query: 561 VDITRHRNDVNLANMAVNKIQSQELYDLVDPNLGYEKDNSVKRMTTAVAELAFRCLQQQR 620
VD TR ++NLA +AV+KI S + +++DP L + D VAELAFRCL
Sbjct: 548 VDFTRPHTEINLAALAVDKIGSGCIDEIIDPILDLDLDAWTLSSIHTVAELAFRCLAFHS 607
Query: 621 DLRPSMDEIVEVLRAIK---------SDEP--------ETQESKVLDVVVRTDELVLLKK 663
D+RP+M E+ + L I+ D P E V + + +V+ +K
Sbjct: 608 DMRPTMTEVADELEQIRLSGWIPSMSLDSPAGSLRSSDRGSERSVKQSSIGSRRVVIPQK 667
Query: 664 GP---------YPTSPDSVAEKWVSGSSTSTSS 687
P +SP SV + W+S S+ +++
Sbjct: 668 QPDCLASVEEISDSSPISVQDPWLSAQSSPSTN 700
>UniRef100_O80461 Hypothetical protein At2g23450 [Arabidopsis thaliana]
Length = 708
Score = 288 bits (738), Expect = 3e-76
Identities = 175/453 (38%), Positives = 255/453 (55%), Gaps = 67/453 (14%)
Query: 264 LIGGVVGGVAALCLLGFVACFVVRRRRKNAKKPISNDLYMPPSSTTSGTNTGTLTSTTNS 323
++GG VGG L L F F +RRR
Sbjct: 286 IVGGTVGGAFLLAALAFF--FFCKRRR--------------------------------- 310
Query: 324 SQSIPSYPSSKTSTMPKSFYFGVQVFTYEELEEATNNFHTSKELGEGGFGTVYKGDLKDG 383
S + S+ S+K + V F Y+E+E+AT+ F ++LG G +GTVY+G L++
Sbjct: 311 STPLRSHLSAKRLLSEAAGNSSVAFFPYKEIEKATDGFSEKQKLGIGAYGTVYRGKLQND 370
Query: 384 RVVAVKRHYESNFKRVAQFMNEVEILARLRHKNLVTLYGCTSKHSRELLLVYEYISNGTV 443
VA+KR + + + Q MNE+++L+ + H NLV L GC + + +LVYEY+ NGT+
Sbjct: 371 EWVAIKRLRHRDSESLDQVMNEIKLLSSVSHPNLVRLLGCCIEQG-DPVLVYEYMPNGTL 429
Query: 444 ADHLHGDRSSSCLLPWSVRLDIALETAEALAYLHAS---DVMHRDVKSNNILLDEKFHVK 500
++HL DR S LPW++RL +A +TA+A+AYLH+S + HRD+KS NILLD F+ K
Sbjct: 430 SEHLQRDRGSG--LPWTLRLTVATQTAKAIAYLHSSMNPPIYHRDIKSTNILLDYDFNSK 487
Query: 501 VADFGLSRLFPNDVTHVSTAPQGTPGYVDPEYYQCYQLTDKSDVYSFGVVLVELISSLQA 560
VADFGLSRL + +H+STAPQGTPGY+DP+Y+QC+ L+DKSDVYSFGVVL E+I+ L+
Sbjct: 488 VADFGLSRLGMTESSHISTAPQGTPGYLDPQYHQCFHLSDKSDVYSFGVVLAEIITGLKV 547
Query: 561 VDITRHRNDVNLANMAVNKIQSQELYDLVDPNLGYEKDNSVKRMTTAVAELAFRCLQQQR 620
VD TR ++NLA +AV+KI S + +++DP L + D VAELAFRCL
Sbjct: 548 VDFTRPHTEINLAALAVDKIGSGCIDEIIDPILDLDLDAWTLSSIHTVAELAFRCLAFHS 607
Query: 621 DLRPSMDEIVEVLRAIK---------SDEP--------ETQESKVLDVVVRTDELVLLKK 663
D+RP+M E+ + L I+ D P E V + + +V+ +K
Sbjct: 608 DMRPTMTEVADELEQIRLSGWIPSMSLDSPAGSLRSSDRGSERSVKQSSIGSRRVVIPQK 667
Query: 664 GP---------YPTSPDSVAEKWVSGSSTSTSS 687
P +SP SV + W+S S+ +++
Sbjct: 668 QPDCLASVEEISDSSPISVQDPWLSAQSSPSTN 700
>UniRef100_Q9CAS4 Hypothetical protein T17F3.6 [Arabidopsis thaliana]
Length = 625
Score = 284 bits (727), Expect = 5e-75
Identities = 222/671 (33%), Positives = 333/671 (49%), Gaps = 96/671 (14%)
Query: 6 LSSPIITFIIFFIFYLRHTTSLPSHASLSSCNNTTFNCGHITKLS-YPFTGGDRPSYCGP 64
+S P IF L ++ PS AS +SC+++ F+C YPF+ CG
Sbjct: 1 MSQPPWRCFSLLIFVLTIFSTKPSSAS-TSCSSS-FHCPPFNSSPPYPFSASPG---CGH 55
Query: 65 PQFHLNCKNNVPELNISSVSYRVLQVNSVTHSLTLARLDLWNET---CTHHYVNST---- 117
P F + C ++ + I ++++ +L +S++ SLTL+ + N C+ +S+
Sbjct: 56 PNFQIQCSSSRATITIKNLTFSILHYSSISSSLTLSPITNTNRNTNNCSSLRFSSSPNRF 115
Query: 118 --FGGTSFS-NGLGNSKITLFYGCQPTSLFTEKPHNLFYC--DSNGYKNNSYTLIGPFPL 172
G+ F + S+++L C P +L N C D KN L G
Sbjct: 116 IDLTGSPFRVSDSSCSRLSLLRPCSPFTL-----PNCSRCPWDCKLLKNPGRILHGCEST 170
Query: 173 DPVLKFVQCDYGVGVPILEEQANRFAGNRSLLREVLMEGFNVNYN---NPFENDCLECIS 229
L C G + L++ RF GF V ++ +P+ C +C
Sbjct: 171 HGSLSEQGCQ-GDLLGFLQDFFTRF-------------GFEVEWDESQDPYFIKCRDCQI 216
Query: 230 SGGQQCGFDSDENEHICICGNGLCPS------GNSSTNGGLIGGVVGGVAALCLLGFVAC 283
G CGF+S IC + N N + ++ + L L+ VA
Sbjct: 217 KNGV-CGFNSTHPNQDFICFHKSRSELVTQRDNNKRVNHIAVLSLIFALTCLLLVFSVAV 275
Query: 284 FVVRRRRKNAKKPISNDLYMPPSSTTSGTNTGTLTSTTNSSQSIPSYPSSKTSTMPKSFY 343
+ R RR + I+ + P++ +S
Sbjct: 276 AIFRSRRASFLSSINEE-----------------------------DPAALFLRHHRSAA 306
Query: 344 FGVQVFTYEELEEATNNFHTSKELGEGGFGTVYKGDLKDGRVVAVKRHYESNFKRVAQFM 403
VFT+EELE ATN F +++G+GGFG+VY G L DG+++AVK + + + F
Sbjct: 307 LLPPVFTFEELESATNKFDPKRKIGDGGFGSVYLGQLSDGQLLAVKFLHHHHAFSMKSFC 366
Query: 404 NEVEILARLRHKNLVTLYGCTSKHSRELLLVYEYISNGTVADHLHGDRSSSCLLPWSVRL 463
NE+ IL+ + H NLV L+G S R LLLV++Y++NGT+ADHLHG + W VRL
Sbjct: 367 NEILILSSINHPNLVKLHGYCSD-PRGLLLVHDYVTNGTLADHLHGRGPK---MTWRVRL 422
Query: 464 DIALETAEALAYLH---ASDVMHRDVKSNNILLDEKFHVKVADFGLSRLFPNDVT----- 515
DIAL+TA A+ YLH V+HRD+ S+NI +++ +KV DFGLSRL T
Sbjct: 423 DIALQTALAMEYLHFDIVPPVVHRDITSSNIFVEKDMKIKVGDFGLSRLLVFSETTVNSA 482
Query: 516 ----HVSTAPQGTPGYVDPEYYQCYQLTDKSDVYSFGVVLVELISSLQAVDITRHRNDVN 571
+V T PQGTPGY+DP+Y++ ++LT+KSDVYS+GVVL+ELI+ ++AVD R + D+
Sbjct: 483 TSSDYVCTGPQGTPGYLDPDYHRSFRLTEKSDVYSYGVVLMELITGMKAVDQRREKRDMA 542
Query: 572 LANMAVNKIQSQELYDLVDPNLGYEKDN----SVKRMTTAVAELAFRCLQQQRDLRPSMD 627
LA++ V+KIQ L ++DP L + D+ S AVAELAFRC+ +D RP
Sbjct: 543 LADLVVSKIQMGLLDQVIDPLLALDGDDVAAVSDGFGVAAVAELAFRCVATDKDDRPDAK 602
Query: 628 EIVEVLRAIKS 638
EIV+ LR I+S
Sbjct: 603 EIVQELRRIRS 613
>UniRef100_Q8H7R6 Hypothetical protein OSJNBa0081P02.15 [Oryza sativa]
Length = 704
Score = 281 bits (720), Expect = 3e-74
Identities = 147/297 (49%), Positives = 210/297 (70%), Gaps = 8/297 (2%)
Query: 346 VQVFTYEELEEATNNFHTSKELGEGGFGTVYKGDLKDGRVVAVKRHYESNFKRVAQFMNE 405
V +TY E++ ATN F + LG G +GTVY G L + R+VAVKR + + + + MNE
Sbjct: 324 VPFYTYREIDRATNGFAEDQRLGTGAYGTVYAGRLSNNRLVAVKRIKQRDNAGLDRVMNE 383
Query: 406 VEILARLRHKNLVTLYGCTSKHSRELLLVYEYISNGTVADHLHGDRSSSCLLPWSVRLDI 465
V++++ + H+NLV L GC +H +++L VYE++ NGT+A HL +R + +PW+VRL I
Sbjct: 384 VKLVSSVSHRNLVRLLGCCIEHGQQIL-VYEFMPNGTLAQHLQRERGPA--VPWTVRLRI 440
Query: 466 ALETAEALAYLHAS---DVMHRDVKSNNILLDEKFHVKVADFGLSRLFPNDV--THVSTA 520
A+ETA+A+AYLH+ + HRD+KS+NILLD +++ KVADFGLSR+ V +H+STA
Sbjct: 441 AVETAKAIAYLHSEVHPPIYHRDIKSSNILLDHEYNSKVADFGLSRMGMTSVDSSHISTA 500
Query: 521 PQGTPGYVDPEYYQCYQLTDKSDVYSFGVVLVELISSLQAVDITRHRNDVNLANMAVNKI 580
PQGTPGYVDP+Y+Q + L+DKSDVYSFGVVLVE+I++++AVD +R ++VNLA +AV++I
Sbjct: 501 PQGTPGYVDPQYHQNFHLSDKSDVYSFGVVLVEIITAMKAVDFSRVGSEVNLAQLAVDRI 560
Query: 581 QSQELYDLVDPNLGYEKDNSVKRMTTAVAELAFRCLQQQRDLRPSMDEIVEVLRAIK 637
L D+VDP L +D VAELAFRCL ++RPSM E+ + L I+
Sbjct: 561 GKGSLDDIVDPYLDPHRDAWTLTSIHKVAELAFRCLAFHSEMRPSMAEVADELEQIQ 617
>UniRef100_Q9FL01 Similarity to protein kinase [Arabidopsis thaliana]
Length = 622
Score = 265 bits (677), Expect = 3e-69
Identities = 142/308 (46%), Positives = 204/308 (66%), Gaps = 8/308 (2%)
Query: 346 VQVFTYEELEEATNNFHTSKELGEGGFGTVYKGDLKDGRVVAVKRHYESNFKRVAQFMNE 405
V +TY+E+E+AT++F LG G +GTVY G+ + VA+KR + + Q +NE
Sbjct: 299 VPFYTYKEIEKATDSFSDKNMLGTGAYGTVYAGEFPNSSCVAIKRLKHKDTTSIDQVVNE 358
Query: 406 VEILARLRHKNLVTLYGCTSKHSRELLLVYEYISNGTVADHLHGDRSSSCLLPWSVRLDI 465
+++L+ + H NLV L GC E LVYE++ NGT+ HL +R L W +RL I
Sbjct: 359 IKLLSSVSHPNLVRLLGCCFADG-EPFLVYEFMPNGTLYQHLQHERGQPPL-SWQLRLAI 416
Query: 466 ALETAEALAYLHAS---DVMHRDVKSNNILLDEKFHVKVADFGLSRLFPN---DVTHVST 519
A +TA A+A+LH+S + HRD+KS+NILLD +F+ K++DFGLSRL + + +H+ST
Sbjct: 417 ACQTANAIAHLHSSVNPPIYHRDIKSSNILLDHEFNSKISDFGLSRLGMSTDFEASHIST 476
Query: 520 APQGTPGYVDPEYYQCYQLTDKSDVYSFGVVLVELISSLQAVDITRHRNDVNLANMAVNK 579
APQGTPGY+DP+Y+Q +QL+DKSDVYSFGVVLVE+IS + +D TR ++VNLA++AV++
Sbjct: 477 APQGTPGYLDPQYHQDFQLSDKSDVYSFGVVLVEIISGFKVIDFTRPYSEVNLASLAVDR 536
Query: 580 IQSQELYDLVDPNLGYEKDNSVKRMTTAVAELAFRCLQQQRDLRPSMDEIVEVLRAIKSD 639
I + D++DP L E + + +AELAFRCL R++RP+M EI E L IK
Sbjct: 537 IGRGRVVDIIDPCLNKEINPKMFASIHNLAELAFRCLSFHRNMRPTMVEITEDLHRIKLM 596
Query: 640 EPETQESK 647
T+ K
Sbjct: 597 HYGTESGK 604
>UniRef100_Q8GYF5 Hypothetical protein At5g66790/MUD21_3 [Arabidopsis thaliana]
Length = 620
Score = 265 bits (677), Expect = 3e-69
Identities = 142/308 (46%), Positives = 204/308 (66%), Gaps = 8/308 (2%)
Query: 346 VQVFTYEELEEATNNFHTSKELGEGGFGTVYKGDLKDGRVVAVKRHYESNFKRVAQFMNE 405
V +TY+E+E+AT++F LG G +GTVY G+ + VA+KR + + Q +NE
Sbjct: 297 VPFYTYKEIEKATDSFSDKNMLGTGAYGTVYAGEFPNSSCVAIKRLKHKDTTSIDQVVNE 356
Query: 406 VEILARLRHKNLVTLYGCTSKHSRELLLVYEYISNGTVADHLHGDRSSSCLLPWSVRLDI 465
+++L+ + H NLV L GC E LVYE++ NGT+ HL +R L W +RL I
Sbjct: 357 IKLLSSVSHPNLVRLLGCCFADG-EPFLVYEFMPNGTLYQHLQHERGQPPL-SWQLRLAI 414
Query: 466 ALETAEALAYLHAS---DVMHRDVKSNNILLDEKFHVKVADFGLSRLFPN---DVTHVST 519
A +TA A+A+LH+S + HRD+KS+NILLD +F+ K++DFGLSRL + + +H+ST
Sbjct: 415 ACQTANAIAHLHSSVNPPIYHRDIKSSNILLDHEFNSKISDFGLSRLGMSTDFEASHIST 474
Query: 520 APQGTPGYVDPEYYQCYQLTDKSDVYSFGVVLVELISSLQAVDITRHRNDVNLANMAVNK 579
APQGTPGY+DP+Y+Q +QL+DKSDVYSFGVVLVE+IS + +D TR ++VNLA++AV++
Sbjct: 475 APQGTPGYLDPQYHQDFQLSDKSDVYSFGVVLVEIISGFKVIDFTRPYSEVNLASLAVDR 534
Query: 580 IQSQELYDLVDPNLGYEKDNSVKRMTTAVAELAFRCLQQQRDLRPSMDEIVEVLRAIKSD 639
I + D++DP L E + + +AELAFRCL R++RP+M EI E L IK
Sbjct: 535 IGRGRVVDIIDPCLNKEINPKMFASIHNLAELAFRCLSFHRNMRPTMVEITEDLHRIKLM 594
Query: 640 EPETQESK 647
T+ K
Sbjct: 595 HYGTESGK 602
>UniRef100_Q5W6V9 Hypothetical protein OSJNBb0059K16.4 [Oryza sativa]
Length = 640
Score = 256 bits (653), Expect = 2e-66
Identities = 149/307 (48%), Positives = 192/307 (62%), Gaps = 11/307 (3%)
Query: 349 FTYEELEEATNNFHTSKELGEGGFGTVYKGDLKDGRVVAVKRHYESNFKRVAQFMNEVEI 408
F+ EL AT NF LG GG+G VY+G L DG VVAVK N K Q +NEV +
Sbjct: 336 FSGRELRRATANFSRDNLLGAGGYGEVYRGVLADGTVVAVKCAKLGNTKSTEQVLNEVRV 395
Query: 409 LARLRHKNLVTLYGCTSKHSRELLLVYEYISNGTVADHLHGDRSSSCLLPWSVRLDIALE 468
L+++ H++LV L GC + L+ VYE+I NGT+ADHL+G S L PW RL IA
Sbjct: 396 LSQVNHRSLVRLLGCCVDLEQPLM-VYEFIPNGTLADHLYGPLSHPPL-PWRRRLAIAHH 453
Query: 469 TAEALAYLHASDV---MHRDVKSNNILLDEKFHVKVADFGLSRLFPNDVTHVSTAPQGTP 525
TA+ +AYLH S V HRD+KS+NILLDE+ KV+DFGLSRL ++HVST QGT
Sbjct: 454 TAQGIAYLHFSAVPPIYHRDIKSSNILLDERMDGKVSDFGLSRLAEQGLSHVSTCAQGTL 513
Query: 526 GYVDPEYYQCYQLTDKSDVYSFGVVLVELISSLQAVDITRHRNDVNLANMAVNKIQSQEL 585
GY+DPEYY+ YQLTDKSDVYSFGVVL+EL++ +A+D R +DVNLA + + L
Sbjct: 514 GYLDPEYYRNYQLTDKSDVYSFGVVLLELLTCKRAIDFGRGADDVNLAVHVQRAAEEERL 573
Query: 586 YDLVDPNLGYEKDNSVKRMTTAVAELAF---RCLQQQRDLRPSMDEIVEVLRAIKSDEPE 642
D+VDP L KDN+ + + L F CL+++R RPSM E+ E + I + E
Sbjct: 574 MDVVDPVL---KDNATQLQCDTIKALGFLALGCLEERRQNRPSMKEVAEEIEYIMNIEAG 630
Query: 643 TQESKVL 649
K L
Sbjct: 631 NAHLKEL 637
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.317 0.134 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,208,129,161
Number of Sequences: 2790947
Number of extensions: 54415641
Number of successful extensions: 231493
Number of sequences better than 10.0: 18912
Number of HSP's better than 10.0 without gapping: 12586
Number of HSP's successfully gapped in prelim test: 6327
Number of HSP's that attempted gapping in prelim test: 184537
Number of HSP's gapped (non-prelim): 23706
length of query: 687
length of database: 848,049,833
effective HSP length: 134
effective length of query: 553
effective length of database: 474,062,935
effective search space: 262156803055
effective search space used: 262156803055
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 78 (34.7 bits)
Medicago: description of AC147875.4


