

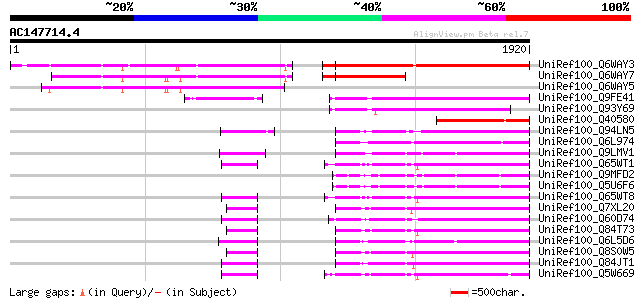
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147714.4 - phase: 0 /pseudo
(1920 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q6WAY3 Gag/pol polyprotein [Pisum sativum] 1070 0.0
UniRef100_Q6WAY7 Gag/pol polyprotein [Pisum sativum] 541 e-151
UniRef100_Q6WAY5 Gag/pol polyprotein [Pisum sativum] 535 e-150
UniRef100_Q9FE41 Oryza sativa (japonica cultivar-group) genomic ... 510 e-142
UniRef100_Q93Y69 Putative gag-pol [Oryza sativa] 446 e-123
UniRef100_Q40580 Integrase [Nicotiana tabacum] 414 e-113
UniRef100_Q94LN5 Putative retroelement pol polyprotein [Oryza sa... 410 e-112
UniRef100_Q6L974 GAG-POL [Vitis vinifera] 410 e-112
UniRef100_Q9LMV1 F5M15.26 [Arabidopsis thaliana] 382 e-104
UniRef100_Q65WT1 Putative polyprotein [Oryza sativa] 381 e-103
UniRef100_Q9MFD2 Orf764 protein [Beta vulgaris subsp. vulgaris] 381 e-103
UniRef100_Q5U6F6 Orf764 protein [Beta vulgaris subsp. vulgaris] 381 e-103
UniRef100_Q65WT8 Putative polyprotein [Oryza sativa] 380 e-103
UniRef100_Q7XL20 OSJNBa0079M09.11 protein [Oryza sativa] 380 e-103
UniRef100_Q60D74 Putative polyprotein [Oryza sativa] 379 e-103
UniRef100_Q84T73 Putative polyprotein [Oryza sativa] 379 e-103
UniRef100_Q6L5D6 Putative polyprotein [Oryza sativa] 378 e-102
UniRef100_Q8S0W5 Putative polyprotein [Oryza sativa] 377 e-102
UniRef100_Q84JT1 Putative polyprotein [Oryza sativa] 372 e-101
UniRef100_Q5W669 Putatve polyprotein [Oryza sativa] 372 e-101
>UniRef100_Q6WAY3 Gag/pol polyprotein [Pisum sativum]
Length = 2262
Score = 1070 bits (2767), Expect = 0.0
Identities = 491/718 (68%), Positives = 611/718 (84%), Gaps = 8/718 (1%)
Query: 1204 AIYYLSKKLTDCETRYTMLEKTCCALAWAAKRLRHYLVNHTTWLISRMDPIKYIFEKAVV 1263
AIYYLSKK TDCETRY++LEKTCCALAWAA+RLR Y++NHTT LIS+MDP+KYIFEK +
Sbjct: 1551 AIYYLSKKFTDCETRYSLLEKTCCALAWAARRLRQYMLNHTTLLISKMDPVKYIFEKPAL 1610
Query: 1264 TGKIARWQVLLSEYDIVFKTQKAIKGSILADHPAYQPLDDYQPIESDFPNEEIVYLKSKD 1323
TG++ARWQ++L+EYDI + +QKAIKGSIL+D+ A QP++DYQP+ +FP+E+I+YLK KD
Sbjct: 1611 TGRVARWQMILTEYDIQYTSQKAIKGSILSDYLAEQPIEDYQPMMFEFPDEDIMYLKMKD 1670
Query: 1324 CEEPLIGEGPDPNSKWGLVFDGAANAYGKGIGAVIVSPQGHHIPFTAQILFECTNNMVEY 1383
C+EPL+ EGPDP+ KW L+FDGA N G G+GAV+++P+G H+PF+A++ F+ TNN EY
Sbjct: 1671 CKEPLVEEGPDPDDKWTLMFDGAVNMNGNGVGAVLINPKGAHMPFSARLTFDVTNNEAEY 1730
Query: 1384 EACIFGIEEAIGMRIKHLDIYGDSALIINQIKGEWETHHAKLIPYRDYARRLLTYFTKVE 1443
EACI GIEEAI +RIK LDI+GDSAL++NQ+ G+W T+ LIPYRDY RR+LT+F KV+
Sbjct: 1731 EACIMGIEEAIDLRIKTLDIFGDSALVVNQVNGDWNTNQPHLIPYRDYTRRILTFFKKVK 1790
Query: 1444 LHHIPRDENQMADALATLSSMFRVNHWNDVPIIKVQRLERPSHVFTIGDVIDQAGENMVD 1503
L+H+PRDENQMADALATLSSM +VN WN VP + V RLERP++VF V+ +D
Sbjct: 1791 LYHVPRDENQMADALATLSSMIKVNWWNHVPHVAVNRLERPAYVFAAESVV-------ID 1843
Query: 1504 NKPWYYDIKQCLLSREYPPGASNKDKKTLRRLASRFLLD-GDILYKRNYDMVLLRCVDEH 1562
KPWYYDIK L ++EYP GAS DKKTLRRLA F L+ D+LYKRN+DMVLLRC+D
Sbjct: 1844 EKPWYYDIKNFLKTQEYPEGASKNDKKTLRRLAGSFYLNQDDVLYKRNFDMVLLRCMDRP 1903
Query: 1563 EAEQLMHDVHDGTFGTHATGHTMSRKLLRAGYYWLTMEHDCYQHARKCHKCQIYADKIHV 1622
EA+ LM +VH+G+FGTHA GH M++KLLRAGYYW+TME DC+++ARKCHKCQIYAD++HV
Sbjct: 1904 EADMLMQEVHEGSFGTHAGGHAMAKKLLRAGYYWMTMESDCFKYARKCHKCQIYADRVHV 1963
Query: 1623 PPHALNVISSPWPFSMVGIDMIGRIEPKASNGHRFILVAIDYFTKWVEAASYTNVTKQVV 1682
PP LNV++SPWPF+M GIDMIG+IEP ASNGHRFILVAIDYFTKWVEAASY N+TKQVV
Sbjct: 1964 PPSPLNVMNSPWPFAMWGIDMIGKIEPTASNGHRFILVAIDYFTKWVEAASYANITKQVV 2023
Query: 1683 AKFIKNNIICRYGVPSKIITDNGTNLNNNVVQALCEEFKIEHHNSSPYRPQINGVVEAAN 1742
+FIK IICRYGVP +IITDNG+NLNN +++ LC++FKIEHHNSSPYRP++NG VEAAN
Sbjct: 2024 TRFIKKEIICRYGVPERIITDNGSNLNNKMMKELCKDFKIEHHNSSPYRPKMNGAVEAAN 2083
Query: 1743 KNIKRIVQKMVTTYRDWHEMLPYALHGYRTTVRSSTGATPFSLVYGMEAVLPLEVEIPSL 1802
KNIK+IV+KMV TY+DWHEMLP+ALHGYRT+VR+STGATP+SLVYGMEAVLP+EVEIPSL
Sbjct: 2084 KNIKKIVRKMVVTYKDWHEMLPFALHGYRTSVRTSTGATPYSLVYGMEAVLPVEVEIPSL 2143
Query: 1803 RVIMEAKLSEAEWCQSRYDQLNLVEEKRMDAMARGQSYQARMKTAFDKKVHPREFKEGEL 1862
RV+++ KL EAEW ++R+++L+L+EE+R+ + GQ YQ RMK AFD+KV PR ++ G+L
Sbjct: 2144 RVLLDVKLDEAEWIRTRFNELSLIEERRLAVVCHGQLYQRRMKRAFDQKVRPRSYQIGDL 2203
Query: 1863 VLKRRISQQPDPRGKWTPNYEGSYVVKKAFSGGALILTHMDGVELPNPVNADLVKKYF 1920
VLKR + D RGKWTPNYEG YVVKK FSGGAL+LT MDG + P+PVN+D+VKKYF
Sbjct: 2204 VLKRILPPGTDNRGKWTPNYEGPYVVKKVFSGGALMLTTMDGEDFPSPVNSDVVKKYF 2261
Score = 544 bits (1401), Expect = e-152
Identities = 387/1135 (34%), Positives = 569/1135 (50%), Gaps = 124/1135 (10%)
Query: 3 QLRTELATLREELAKANDVMTALLAAQEQSATAIPIATAIPVTTSMLPTASADARFAMPA 62
Q++TELA +R +A+ +M + QE+ + A+ +
Sbjct: 3 QVQTELAEMRANMAQFMHMMQGVAQGQEELRALVQRQEAVTPPPNQ-------------- 48
Query: 63 GFPYGLPPFFTPSTAAGTSGTANNVLIPATNAV-------SINATLPQTTAAVTEPLVHA 115
P G P TP+ A + A + + NA + A P+V
Sbjct: 49 ALPEGNPVHDTPAAAIPVNNYAVGEELMGIRVDGQPIAPDAANARVIHAPARNRIPIVDR 108
Query: 116 IPQGVNINTQHGSIPVTK-TMEEMMEELAKELRHEIKANRGNADSFKTQDLCLVSKVDVP 174
+ ++ IP + ++ LA+++R N+ F ++ LV + +P
Sbjct: 109 QEDLFTMFSEDEDIPGRNDARDRKVDALAEKIR---AMECQNSLGFDVTNMGLVEGLRIP 165
Query: 175 KKFKIPDFDRYNGLTCPQNHIIKYVRKMGNYKDNDSLMIHCFQDSLMEDAAEWYTSLSKN 234
KFK P FD+YNG +CP+ H+ Y RK+ Y D++ + ++ FQDSL + +WY L +
Sbjct: 166 YKFKAPSFDKYNGTSCPRTHVQAYYRKISAYTDDEKMWMYFFQDSLSGASLDWYMELKSD 225
Query: 235 DIHTFDELAAAFKSHYGFNTRLKPNREFLRSLSQKKEESFREYAQRWRGAAARITPALDE 294
I + +L AF Y N + P+R L+SL QK ESF+EYAQRWR AAR+ P + E
Sbjct: 226 SIRCWRDLGEAFLRQYKHNMDMAPSRTQLQSLCQKSNESFKEYAQRWRELAARVQPPMLE 285
Query: 295 EEMTQTFLKTLKKDYVERMIIAAPNNFSEMVTMGTRLEEAVREGIIVFEKAESSVNASKR 354
E+T F+ TL+ +++RM +FS++V G R E ++ G I + ++SK+
Sbjct: 286 RELTDMFIGTLQGVFMDRMGSCPFGSFSDVVICGERTESLIKTGKI----QDVGSSSSKK 341
Query: 355 YGNGHHKKKETEVGMVSAGAGQSMATVAPINATQMPPSYPYQKQQLPVQQQQQNQQARP- 413
G +++E E V Q+ A +P P Q+QQ VQQ QQ QQ RP
Sbjct: 342 PFAGAPRRREGETNAVQHRRDQNRIEYRQAAAVTIPAPQPRQQQQQRVQQPQQQQQQRPY 401
Query: 414 ---------TFPPIPMLYAELLPTLLHRGH---CTTRQGKPPPDPLPPRFRSDLKCDFHQ 461
F +PM YAELLP LL G CT PP LPP + ++++CDFH
Sbjct: 402 QPRQRMPDRRFDSLPMSYAELLPELLRLGLVELCTMA----PPTVLPPGYDANVRCDFHS 457
Query: 462 GALGHDVEGCYALKYIVKKLIDQGKLTFKNNVPHVLDNPLPNHAA--VNMIEVCEEAPRL 519
GA GH E C AL++ V+ LID + F VP+V++NP+P H VN IE E +
Sbjct: 458 GAPGHHTEKCRALQHKVQDLIDANAINFAP-VPNVVNNPMPQHGGHRVNNIEGKEAEDLV 516
Query: 520 D-VRNVATPLVPLHIKLCKASLFSHDHAKCLGCLRNPLGCYTVQDDIQSLMNDNFLTVS- 577
D V +V T L+ + +L ++S CLGC + GC ++ IQ +M++ L S
Sbjct: 517 DNVDDVQTSLLVVKSRLLNEGVYSGCDEDCLGCAESENGCDQLRAGIQGMMDEGCLQFSR 576
Query: 578 ------DVCVIVPVF-----HDPPVKSMPLKENAEPLVIRLPGPV--PYT---------- 614
V I F H V S P N P+ I +P + P T
Sbjct: 577 AVKDRGTVSTITIYFKPSEGHGQRVVSAPAT-NGTPVTIPVPVTISAPTTIVASGRRAVE 635
Query: 615 SDRAIPYKY-----------------NATIIENGV--EVPLVSLAVVNNTAEGTSAALRS 655
+ RA+P+KY N T + G+ VP V+N G RS
Sbjct: 636 NSRAVPWKYDNAYRSNRRVESQTKPVNQTPVTIGLANRVPATVGPAVDNVG-GPGGFTRS 694
Query: 656 GRV-RPPLFQKKAATPTVPPIDKPTPTDVSPVNRDASQPGRSIEDSNLDEILRLIKRSDY 714
GR+ P + A K + PV ++A P S E +++E +++IK+SDY
Sbjct: 695 GRLFAPQPLRDNNAEALAKAKGKQAVVEEEPVQKEA--PEGSFE-KDVEEFMKMIKKSDY 751
Query: 715 KIVDQLLQTPSKISILSLLLSSAAHRDTLLKVLEQAYVDHEVTLDHFGSIVGNITACSNL 774
KIVDQL QTPSKISILSLLL S AHR+ LLK+L AYV E++++ ++ N++ +
Sbjct: 752 KIVDQLNQTPSKISILSLLLCSEAHRNALLKMLNLAYVPQEISVNQLEGVMANVSTRHGV 811
Query: 775 WFSENELPEAGKHHNLALHISVNCTPDIISNVLVDTGSSLNVMPKSTLDQLSYRGTPLRR 834
F+ +LP G++HN ALHI++ C ++S+VLVDTGSSLNV+PK L ++ G L
Sbjct: 812 GFTNLDLPPEGRNHNKALHITMECKGAVLSHVLVDTGSSLNVLPKQILKKIDVEGFVLTP 871
Query: 835 STFLVKAFDGTRKSVLGEIDLPITIGPETFLITFQVMDINASYNCLLGRP*IHDAGAVTS 894
S +V+AFD +++SV GE+ LP+ IGPE F I F VMDI +Y+CLLGRP IH AGAV+S
Sbjct: 872 SDLIVRAFDRSKRSVCGEVTLPVKIGPEVFDIIFYVMDIQPAYSCLLGRPWIHAAGAVSS 931
Query: 895 TLHQKLKFAKSGKLVTIHGEEAYLVSQLSSFSCIEA-GSAEGT---AFQGLTIEG---TE 947
TLHQKLK+ +G++VT+ GEE LVS LSSF +E G T F+ + +E E
Sbjct: 932 TLHQKLKYVWNGQIVTVCGEEEILVSHLSSFKYVEVDGEIHETLCQVFETVALEKVAYAE 991
Query: 948 PKRDGTAMASLKDAQRAVQEGQAAG*GRLIQLRENKHKEGLGFSPTSGVSTG-----AFY 1002
++ G ++ S K A+ V G+A G G+++ L + K G+G+ P G F
Sbjct: 992 QRKPGVSITSYKQAKEVVDSGKAEGWGKMVDLPVKEDKFGVGYEPLQAEQNGQAGPSTFT 1051
Query: 1003 SAGFVNAITEEATGFGP-----------RPVFVIPGGIARDWDAIDIPSIMHVSE 1046
SAG +N ATG RP PGG +W A ++ + ++E
Sbjct: 1052 SAGLMNHGDVSATGSEDCDSDCDLDNWVRP--CAPGGSINNWTAEEVVQVTLLTE 1104
Score = 60.1 bits (144), Expect = 7e-07
Identities = 27/51 (52%), Positives = 39/51 (75%), Gaps = 2/51 (3%)
Query: 1156 NFEFPVYEAEDEEGDD--IPYEITRLLEQEGRAIQPHQEEIEIINLGIPTA 1204
+F+ P+Y+AE+E +D +P E+ RLL+QE R IQPHQEE+E++NLG A
Sbjct: 1121 DFDNPIYQAEEESEEDCELPAELVRLLKQEERVIQPHQEELEVVNLGTEDA 1171
>UniRef100_Q6WAY7 Gag/pol polyprotein [Pisum sativum]
Length = 1814
Score = 541 bits (1393), Expect = e-151
Identities = 357/972 (36%), Positives = 521/972 (52%), Gaps = 94/972 (9%)
Query: 156 NADSFKTQDLCLVSKVDVPKKFKIPDFDRYNGLTCPQNHIIKYVRKMGNYKDNDSLMIHC 215
N+ F ++ LV + +P KFK P FD+YNG +CP+ H+ Y RK+ Y D++ + ++
Sbjct: 148 NSLGFDVTNMGLVEGLRIPYKFKAPSFDKYNGTSCPRTHVQAYYRKISAYTDDEKMWMYF 207
Query: 216 FQDSLMEDAAEWYTSLSKNDIHTFDELAAAFKSHYGFNTRLKPNREFLRSLSQKKEESFR 275
FQDSL + +WY L ++ I + +L AF Y N + P+R L+SL QK ESF+
Sbjct: 208 FQDSLSGASLDWYMELKRDSIRCWKDLGEAFLRQYKHNMDMAPSRTQLQSLCQKSGESFK 267
Query: 276 EYAQRWRGAAARITPALDEEEMTQTFLKTLKKDYVERMIIAAPNNFSEMVTMGTRLEEAV 335
EYAQRWR AAR+ P + E E+T F+ TL+ +++RM +FS++V G R E +
Sbjct: 268 EYAQRWRELAARVQPPMLERELTDMFIGTLQGVFMDRMGSCPFVSFSDVVICGERTESLI 327
Query: 336 REGIIVFEKAESSVNASKRYGNGHHKKKETEVGMVSAGAGQSMATVAPINATQMPPSYPY 395
+ G I ++ ++SK+ G +++E E V Q+ + + A +P P
Sbjct: 328 KTGKI----QDAGSSSSKKPFAGAPRRREGETNAVQYRRDQNRSQRCQVAAVTIPAPQPR 383
Query: 396 QKQQLPVQQ-QQQNQQARP----------TFPPIPMLYAELLPTLLHRGHCTTRQGKPPP 444
Q+QQ VQQ QQQ QQ RP F +PM YAELLP LL G R PP
Sbjct: 384 QQQQQRVQQPQQQQQQQRPYQPRQRMPDRRFDSLPMSYAELLPELLRLGMVELRT-MAPP 442
Query: 445 DPLPPRFRSDLKCDFHQGALGHDVEGCYALKYIVKKLIDQGKLTFKNNVPHVLDNPLPNH 504
LPP + ++++CDFH GA GH E C AL++ V+ LID + F VP+V++NP+P H
Sbjct: 443 TVLPPGYDANVRCDFHSGAPGHHTEKCRALQHKVQDLIDAKAINFAP-VPNVVNNPMPQH 501
Query: 505 AA--VNMIEVCE-EAPRLDVRNVATPLVPLHIKLCKASLFSHDHAKCLGCLRNPLGCYTV 561
VN IE E E ++V +V T L+ + +L ++S CLGC + GC +
Sbjct: 502 GGHRVNNIEGKEAEDLVVNVDDVQTSLLVVKGRLLNGGVYSGCDEDCLGCAESENGCDQL 561
Query: 562 QDDIQSLMNDNFL----------TVSDVCVI-------------VPVFHDPPVK-SMPLK 597
+ IQ +M++ L TVS + + P + PV S+P+
Sbjct: 562 RTGIQGMMDEGCLQFSRAVKDRGTVSTITIYFKPSEGRGQRVVSAPATNGTPVTISVPVT 621
Query: 598 ENAEPLVIRLPGPVPYTSDRAIPYKYNATIIEN-----------------GV--EVPLVS 638
+A P I G + RA+P+KY+ N G+ VP
Sbjct: 622 ISA-PTTIAASGRRAVENSRAVPWKYDNAYRSNRRAESQTRPVNQAPVTIGLPNRVPATV 680
Query: 639 LAVVNNTAEGTSAALRSGRV-RPPLFQKKAATPTVPPIDKPTPTDVSPVNRDASQPGRSI 697
V+N G RSGR+ P + A K + PV ++A P S
Sbjct: 681 GPAVDNVG-GPGGFTRSGRLFAPQPLRDNNAEALAKAKGKQAVVEEEPVQKEA--PEGSF 737
Query: 698 EDSNLDEILRLIKRSDYKIVDQLLQTPSKISILSLLLSSAAHRDTLLKVLEQAYVDHEVT 757
E +++E +++IK+SDYKIVDQL QTPSKISILSLLL S AHR+ LLK+L AYV E++
Sbjct: 738 E-KDVEEFMKIIKKSDYKIVDQLNQTPSKISILSLLLCSEAHRNALLKMLNLAYVPQEIS 796
Query: 758 LDHFGSIVGNITACSNLWFSENELPEAGKHHNLALHISVNCTPDIISNVLVDTGSSLNVM 817
++ ++ N++ + F+ +L G++HN ALHI++ C ++S+VLVDTGSSLNV+
Sbjct: 797 VNQLEGVMANVSTRHGVGFTNLDLTPEGRNHNKALHITMECKGAVLSHVLVDTGSSLNVL 856
Query: 818 PKSTLDQLSYRGTPLRRSTFLVKAFDGTRKSVLGEIDLPITIGPETFLITFQVMDINASY 877
PK L ++ G L S +V+AFDG+++SV GE+ LP+ IGPE F I F VMDI +Y
Sbjct: 857 PKQILKKIDVEGFVLTPSDLIVRAFDGSKRSVCGEVTLPVKIGPEVFDIIFYVMDIQPAY 916
Query: 878 NCLLGRP*IHDAGAVTSTLHQKLKFAKSGKLVTIHGEEAYLVSQLSSFSCIEA-GSAEGT 936
+CLLGRP IH AGAV+STLHQKLK+ +G++VT+ GEE LVS LSSF +E G T
Sbjct: 917 SCLLGRPWIHAAGAVSSTLHQKLKYVWNGQIVTVCGEEEILVSHLSSFKYVEVDGEIHET 976
Query: 937 ---AFQGLTIEG---TEPKRDGTAMASLKDAQRAVQEGQAAG*GRLIQLRENKHKEGLGF 990
AF+ + +E E ++ G ++ S K A+ V G+A G G+++ L + K G+G+
Sbjct: 977 LCQAFETVALEKVAYAEQRKPGASITSYKQAKEVVDSGKAEGWGKMVYLPVKEDKFGVGY 1036
Query: 991 SPTSGVSTG-----AFYSAGFVNAITEEATGFGP-----------RPVFVIPGGIARDWD 1034
P G F SAG +N ATG RP PGG +W
Sbjct: 1037 EPLQAEQNGQAGPSTFTSAGLMNHGDVSATGSEDCDSDCDLDNWVRP--CAPGGSINNWT 1094
Query: 1035 AIDIPSIMHVSE 1046
A ++ + ++E
Sbjct: 1095 AEEVVQVTLLTE 1106
Score = 400 bits (1029), Expect = e-109
Identities = 180/261 (68%), Positives = 226/261 (85%)
Query: 1204 AIYYLSKKLTDCETRYTMLEKTCCALAWAAKRLRHYLVNHTTWLISRMDPIKYIFEKAVV 1263
AIYYLSKK TDCETRY++LEKTCCALAWAA+RLR Y++NHTT LIS+MDP+KYIFEK +
Sbjct: 1553 AIYYLSKKFTDCETRYSLLEKTCCALAWAARRLRQYMLNHTTLLISKMDPVKYIFEKPAL 1612
Query: 1264 TGKIARWQVLLSEYDIVFKTQKAIKGSILADHPAYQPLDDYQPIESDFPNEEIVYLKSKD 1323
TG++ARWQ++L+EYDI + +QKAIKGSIL+D+ A QP++DYQP+ +FP+E+I+YLK KD
Sbjct: 1613 TGRVARWQMILTEYDIQYTSQKAIKGSILSDYLAEQPIEDYQPMMFEFPDEDIMYLKVKD 1672
Query: 1324 CEEPLIGEGPDPNSKWGLVFDGAANAYGKGIGAVIVSPQGHHIPFTAQILFECTNNMVEY 1383
CEEPL+ EGPDP+ KW L+FDGA N G G+GAV+++P+G HIPF+A++ F+ TNN EY
Sbjct: 1673 CEEPLVEEGPDPDDKWTLMFDGAVNMNGNGVGAVLINPKGAHIPFSARLTFDVTNNEAEY 1732
Query: 1384 EACIFGIEEAIGMRIKHLDIYGDSALIINQIKGEWETHHAKLIPYRDYARRLLTYFTKVE 1443
EACI GIEEAI +RIK LDIYGDSAL++NQ+ G+W T+ LIPYRDY RR+LT+F KV
Sbjct: 1733 EACIMGIEEAIDLRIKTLDIYGDSALVVNQVNGDWNTNQPHLIPYRDYTRRILTFFKKVR 1792
Query: 1444 LHHIPRDENQMADALATLSSM 1464
L+H+PRDENQMADALATLSSM
Sbjct: 1793 LYHVPRDENQMADALATLSSM 1813
Score = 58.2 bits (139), Expect = 3e-06
Identities = 27/51 (52%), Positives = 37/51 (71%), Gaps = 2/51 (3%)
Query: 1156 NFEFPVYEAEDEEGDD--IPYEITRLLEQEGRAIQPHQEEIEIINLGIPTA 1204
+F P+Y+AE+E +D +P E+ RLL QE R IQPHQEE+E++NLG A
Sbjct: 1123 DFGNPIYQAEEESEEDCELPAELVRLLRQEERVIQPHQEELEVVNLGTEDA 1173
>UniRef100_Q6WAY5 Gag/pol polyprotein [Pisum sativum]
Length = 1105
Score = 535 bits (1379), Expect = e-150
Identities = 355/992 (35%), Positives = 523/992 (51%), Gaps = 102/992 (10%)
Query: 116 IPQGVNINT----QHGSIPVTKTMEEMMEELAKE----------------LRHEIKANR- 154
+P N T +P+ E+M L+++ L +I+A
Sbjct: 87 VPDAANARTIRAPARNPVPIVDRQEDMFTLLSEDEDVLGRVDERDRKVDALAEKIRAMEC 146
Query: 155 GNADSFKTQDLCLVSKVDVPKKFKIPDFDRYNGLTCPQNHIIKYVRKMGNYKDNDSLMIH 214
N+ SF ++ LV + +P KFK P FD+YN +CP+ H+ Y RK+ Y D++ + ++
Sbjct: 147 QNSLSFDVTNMGLVEGLRIPYKFKAPSFDKYNDTSCPRTHVQAYYRKISAYTDDEKMWMY 206
Query: 215 CFQDSLMEDAAEWYTSLSKNDIHTFDELAAAFKSHYGFNTRLKPNREFLRSLSQKKEESF 274
FQDSL + +WY L ++ I + +L AF Y N + P+R L+SL QK ESF
Sbjct: 207 FFQDSLSGASLDWYMELKRDSIRCWRDLGEAFLRQYKHNMDMAPSRTQLQSLCQKSGESF 266
Query: 275 REYAQRWRGAAARITPALDEEEMTQTFLKTLKKDYVERMIIAAPNNFSEMVTMGTRLEEA 334
+EYAQRWR AAR+ P + E E+T F+ TL+ +++RM +FS++V G R E
Sbjct: 267 KEYAQRWRELAARVQPPMLERELTDMFIGTLQGVFMDRMGSCPFGSFSDVVICGERTESL 326
Query: 335 VREGIIVFEKAESSVNASKRYGNGHHKKKETEVGMVSAGAGQSMATVAPINATQMPPSYP 394
++ G I + ++SK+ G +++E E +V Q+ A +P P
Sbjct: 327 IKTGKI----QDVGSSSSKKPFAGAPRRREGETNVVQHRRDQNRIEYRQAAAVTIPAPQP 382
Query: 395 YQKQQLPVQQ-QQQNQQARP----------TFPPIPMLYAELLPTLLHRGHCTTRQGKPP 443
Q+QQ VQQ QQQ QQ RP F +PM YAELLP LL G R P
Sbjct: 383 RQQQQQRVQQPQQQQQQQRPYQPRQRMPDRRFDSLPMSYAELLPELLRLGMVELRT-MAP 441
Query: 444 PDPLPPRFRSDLKCDFHQGALGHDVEGCYALKYIVKKLIDQGKLTFKNNVPHVLDNPLPN 503
P LPP + ++++CDFH GA GH E C AL++ V+ LID + F VP+V++NP+P
Sbjct: 442 PTVLPPGYDANVRCDFHSGAPGHHTEKCRALQHKVQDLIDAKAINFAP-VPNVVNNPMPQ 500
Query: 504 HAA--VNMIEVCE-EAPRLDVRNVATPLVPLHIKLCKASLFSHDHAKCLGCLRNPLGCYT 560
H VN IE E E ++V +V T L+ + +L ++S CLGC + GC
Sbjct: 501 HGGHRVNNIEGKEAEDLVVNVDDVQTSLLVVKGRLLNGGVYSGCDEDCLGCAESENGCDQ 560
Query: 561 VQDDIQSLMNDNFL----------TVSDVCVI-------------VPVFHDPPVK-SMPL 596
++ IQ +M++ L TVS + + P +D PV S+P+
Sbjct: 561 LRAGIQGMMDEGCLQFSRAVKDRGTVSTITIYFKPTEGRGQRVVSAPATNDTPVTISVPV 620
Query: 597 KENAEPLVIRLPGPVPYTSDRAIPYKYNATIIEN-----------------GV--EVPLV 637
+A P I G + R +P+KY+ N G+ VP
Sbjct: 621 TISA-PTTIVASGRRAVENSRVVPWKYDNAYRSNRRAESQTRPVNQAPVTIGLPNRVPAT 679
Query: 638 SLAVVNNTAEGTSAALRSGRV-RPPLFQKKAATPTVPPIDKPTPTDVSPVNRDASQPGRS 696
V+N G RSGR+ P + A K + PV ++A +
Sbjct: 680 VGPAVDNVG-GPGGFTRSGRLFAPQPLRDNNAEALAKAKGKQAVVEEEPVQKEAPE---G 735
Query: 697 IEDSNLDEILRLIKRSDYKIVDQLLQTPSKISILSLLLSSAAHRDTLLKVLEQAYVDHEV 756
+ +++E +++IK+SDYKIVDQL QTPSKISILSLL+ S AHR+ LLK+L AYV E+
Sbjct: 736 TFEKDVEEFMKIIKKSDYKIVDQLNQTPSKISILSLLMCSEAHRNALLKMLNLAYVPQEI 795
Query: 757 TLDHFGSIVGNITACSNLWFSENELPEAGKHHNLALHISVNCTPDIISNVLVDTGSSLNV 816
+++ ++ N++ + F+ +LP G++HN ALHI++ C ++S+VLVDTGSSLNV
Sbjct: 796 SVNQLEGVMANVSTRHGVGFTNLDLPPEGRNHNKALHITMECKGAVLSHVLVDTGSSLNV 855
Query: 817 MPKSTLDQLSYRGTPLRRSTFLVKAFDGTRKSVLGEIDLPITIGPETFLITFQVMDINAS 876
+P L ++ G L S +V+AFDG+++SV GE+ LP+ IGPE F I F VMDI +
Sbjct: 856 LPNQILKKIDVEGFVLTPSDLIVRAFDGSKRSVCGEVTLPVKIGPEVFDIIFYVMDIQPA 915
Query: 877 YNCLLGRP*IHDAGAVTSTLHQKLKFAKSGKLVTIHGEEAYLVSQLSSFSCIEA-GSAEG 935
Y+CLLGRP IH AGAV+STLHQKLK+ +G++VT+ GEE LVS LSSF +E G
Sbjct: 916 YSCLLGRPWIHAAGAVSSTLHQKLKYVWNGQIVTVCGEEEILVSHLSSFKYVEVDGEIHE 975
Query: 936 T---AFQGLTIEG---TEPKRDGTAMASLKDAQRAVQEGQAAG*GRLIQLRENKHKEGLG 989
T AF+ + +E E ++ G ++ S K A+ V G+A G G+++ L + K G+G
Sbjct: 976 TLCQAFETVALEKVAYAEQRKPGASITSYKQAKEVVDSGKAEGWGKMVDLPVKEDKFGVG 1035
Query: 990 FSPTSGVSTG-----AFYSAGFVNAITEEATG 1016
+ P G F SAG +N ATG
Sbjct: 1036 YEPLRAEQNGQAGPSTFTSAGLMNHGDVSATG 1067
>UniRef100_Q9FE41 Oryza sativa (japonica cultivar-group) genomic DNA, chromosome 1, PAC
clone:P0433F09 [Oryza sativa]
Length = 2876
Score = 510 bits (1314), Expect = e-142
Identities = 290/751 (38%), Positives = 424/751 (55%), Gaps = 36/751 (4%)
Query: 1184 GRAIQPHQEEIEIINLGIPTAIYYLSKKLTDCETRYTMLEKTCCALAWAAKRLRHYLVNH 1243
G + H +E G A YYLS+ + E Y+ +EK C AL +A K+LRHY++ H
Sbjct: 2147 GALLAQHNDE------GKEVACYYLSRTMVGAEQNYSPIEKLCLALIFALKKLRHYMLAH 2200
Query: 1244 TTWLISRMDPIKYIFEKAVVTGKIARWQVLLSEYDIVFKTQKAIKGSILADHPAYQPLDD 1303
LI+R DPI+Y+ + V+TG++ +W +L+ EYDI F QKAIKG LA+ A P+ D
Sbjct: 2201 QIQLIARADPIRYVLSQPVLTGRLGKWALLMMEYDITFVPQKAIKGQALAEFLATHPMPD 2260
Query: 1304 YQPIESDFPNEEIVYLKSKDCEEPLIGEGPDPNSKWGLVFDGAA----NAYGK-----GI 1354
P+ ++ P+EEI + ++ +W L FDGA+ N G G
Sbjct: 2261 DSPLIANLPDEEIFTAELQE--------------QWELYFDGASRKDINPDGTPRRRAGA 2306
Query: 1355 GAVIVSPQGHHIPFTAQILFE-CTNNMVEYEACIFGIEEAIGMRIKHLDIYGDSALIINQ 1413
G V +PQG I + +L E C+NN EYEA IFG+ A+ M ++ L +GDS LII Q
Sbjct: 2307 GLVFKTPQGGVIYHSFSLLKEECSNNEAEYEALIFGLLLALSMEVRSLRAHGDSRLIIRQ 2366
Query: 1414 IKGEWETHHAKLIPYRDYARRLLTYFTKVELHHIPRDENQMADALATLSSMFRVNHWNDV 1473
I +E +L+PY ARRL+ F +E+ H+PR +N ADALA L++ N
Sbjct: 2367 INNIYEVRKPELVPYYTVARRLMDKFEHIEVIHVPRSKNAPADALAKLAAALVFQGDNPA 2426
Query: 1474 PIIKVQRLERPSHVFTIGDVIDQAGENMVDNKPWYYDIKQCLLSREYPPGASNKDKKTLR 1533
I+ +R P+ + I + ++ N + + W P +++ L+
Sbjct: 2427 QIVVEERWLLPAVLELIPEEVNIIITNSAEEEDWRQPFLDYFKHGSLPEDPV--ERRQLQ 2484
Query: 1534 RLASRFLLDGDILYKRNYDM-VLLRCVDEHEAEQLMHDVHDGTFGTHATGHTMSRKLLRA 1592
R ++ +LYKR+Y VLLRCVD EA +++ +VH G G H +G M +
Sbjct: 2485 RRLPSYIYKAGVLYKRSYGQEVLLRCVDRSEANRVLQEVHHGVCGGHQSGPKMYHSIRLV 2544
Query: 1593 GYYWLTMEHDCYQHARKCHKCQIYADKIHVPPHALNVISSPWPFSMVGIDMIGRIEPKAS 1652
GYYW + DC + A+ CH CQI+ + H PP L+ WPF GID+IG I P +S
Sbjct: 2545 GYYWPGIMADCLKTAKTCHGCQIHDNFKHQPPAPLHPTVPSWPFDAWGIDVIGLINPPSS 2604
Query: 1653 NGHRFILVAIDYFTKWVEAASYTNVTKQVVAKFIKNNIICRYGVPSKIITDNGTNLNNNV 1712
GHRFIL A DYF+KW EA V V F++ +II R+GVP +I +DN +
Sbjct: 2605 RGHRFILTATDYFSKWAEAVPLREVKSSDVINFLERHIIYRFGVPHRITSDNAKAFKSQK 2664
Query: 1713 VQALCEEFKIEHHNSSPYRPQINGVVEAANKNIKRIVQKMVTTY-RDWHEMLPYALHGYR 1771
+ E++KI+ + S+ Y PQ NG+ EA NK + +I++K V + RDWH+ L AL YR
Sbjct: 2665 IYRFMEKYKIKWNYSTGYYPQANGMAEAFNKTLGKILKKTVDKHRRDWHDRLYEALWAYR 2724
Query: 1772 TTVRSSTGATPFSLVYGMEAVLPLEVEIPSLRVIMEAKLSEAEWCQSRYDQLNLVEEKRM 1831
TVR+ T ATP+SLVYG EAVLPLE+++PSLRV + +L++ E + R+ +L+ VEE+R+
Sbjct: 2725 VTVRTPTQATPYSLVYGNEAVLPLEIQLPSLRVAIHDELTKDEQIRLRFQELDAVEEERL 2784
Query: 1832 DAMARGQSYQARMKTAFDKKVHPREFKEGE--LVLKRRISQQPDPRGKWTPNYEGSYVVK 1889
A+ + Y+ M A+DK V R F++GE LVL+R I +GK+ P +EG YV++
Sbjct: 2785 GALQNLELYRQNMVRAYDKLVKQRVFRKGELVLVLRRPIVVTHKMKGKFEPKWEGPYVIE 2844
Query: 1890 KAFSGGALILTHMDGVELPNPVNADLVKKYF 1920
+A+ GGA L G + P+N +KKYF
Sbjct: 2845 QAYDGGAYQLIDHQGSQPMPPINGRFLKKYF 2875
Score = 104 bits (260), Expect = 2e-20
Identities = 79/288 (27%), Positives = 130/288 (44%), Gaps = 21/288 (7%)
Query: 647 EGTSAALRSGRVRPPLFQKKAATPTVPPIDKPTPTDVSPVNRDASQPGRSIEDSNLDEIL 706
E + +LR G+ P + K VP +DKP + AS PG + E
Sbjct: 1252 EEVNISLRGGKTLPDPHKSK-----VPNVDKPA--------KKASPPGEAPEAPETKTGS 1298
Query: 707 RLIKRSDYKIVDQLLQTPSKISILSLLLSSAAHRDTLLKVLEQAYVDHEVTLDHFGSIVG 766
+ DYK++ L + P+ +S+ L+ R+ L+K L+ V H +
Sbjct: 1299 KEKPAVDYKVLAHLKRIPALLSVYDALMMVPDLREALIKALQAPEVYEVDMAKH--RLYD 1356
Query: 767 NITACSNLWFSENELPEAGKHHNLALHISVNCTPDIISNVLVDTGSSLNVMPKSTLDQLS 826
N + + F++ + G HN L+I N + +L+D GS++N++P +L +
Sbjct: 1357 NPLFVNEITFADEDNIIKGGDHNRPLYIEGNIGSAHLRRILIDLGSAVNILPVRSLTRAG 1416
Query: 827 YRGTPLRRSTFLVKAFDGTRKSVLGEIDLPITIGPETFLITFQVMDINASYNCLLGRP*I 886
+ L ++ FD K LG I + I + +F + F V++ N SY+ LLGRP I
Sbjct: 1417 FTTKDLEPIDVVICGFDNQGKPTLGAITIKIQMSTFSFKVRFFVIEANTSYSALLGRPWI 1476
Query: 887 HDAGAVTSTLHQKLKFAKSGKLVTIHGEEAYLVSQLSSFSCIEAGSAE 934
H V STLHQ LKF +G + + S S ++ E+ A+
Sbjct: 1477 HKYRVVPSTLHQCLKFLDG------NGVQQRITSNFSPYTIQESYHAD 1518
>UniRef100_Q93Y69 Putative gag-pol [Oryza sativa]
Length = 1262
Score = 446 bits (1147), Expect = e-123
Identities = 248/683 (36%), Positives = 381/683 (55%), Gaps = 36/683 (5%)
Query: 1184 GRAIQPHQEEIEIINLGIPTAIYYLSKKLTDCETRYTMLEKTCCALAWAAKRLRHYLVNH 1243
G + H +E G A YYLS+ + E Y+ +EK C AL +A K+LRHY++ H
Sbjct: 601 GALLAQHNDE------GKEVACYYLSRTMVGAERNYSPIEKLCLALIFALKKLRHYMLTH 654
Query: 1244 TTWLISRMDPIKYIFEKAVVTGKIARWQVLLSEYDIVFKTQKAIKGSILADHPAYQPLDD 1303
LI+ +DPI+Y+ + ++ G++ +W +L+ E+DI + QKA+KG LA+ A P+ D
Sbjct: 655 QIQLIATVDPIRYVLSQPLLAGRLGKWALLMMEFDITYVPQKAVKGQALAEFLAAHPVPD 714
Query: 1304 YQPIESDFPNEEIVYLKSKDCEEPLIGEGPDPNSKWGLVFDGAANAYGK---------GI 1354
P+ ++ P+E++ +++ EP W L FDGA+ G
Sbjct: 715 DSPLITELPDEDVFTIET----EP----------SWELCFDGASRTENDRDGTPRKRAGA 760
Query: 1355 GAVIVSPQGHHIPFTAQILFE-CTNNMVEYEACIFGIEEAIGMRIKHLDIYGDSALIINQ 1413
G V +PQG I + +L E C+NN EYEA IFG+ A+ M ++ + +YGDS LI+ Q
Sbjct: 761 GLVFKTPQGGVIYHSFSLLKEECSNNEAEYEALIFGLLLALSMEVRSIRVYGDSQLIVQQ 820
Query: 1414 IKGEWETHHAKLIPYRDYARRLLTYFTKVELHHIPRDENQMADALATLSSMFRVNHWNDV 1473
I +E +L+PY ARRL+ F +E+ H+PR N ADALA L++ +
Sbjct: 821 INDIYEVLKQELVPYYSAARRLMEMFGHIEVMHVPRSRNAPADALAKLAAALVLPQGGPT 880
Query: 1474 PIIKVQRLERPSHVFTIGDV--IDQAGENMVDNKPWYYDIKQCLLSREYPPGASNKDKKT 1531
+ +R P+ + + + +D W P ++ +++
Sbjct: 881 QVNVEERWLLPAVLELLPNEYEVDTVMAAAAKEDDWRVPFLNYFRHGSLPD--NSVERRQ 938
Query: 1532 LRRLASRFLLDGDILYKRNYDM-VLLRCVDEHEAEQLMHDVHDGTFGTHATGHTMSRKLL 1590
L+R ++ + Y+R+Y VLLRCVD EA++ + +VH G G H +G M +
Sbjct: 939 LQRRLPSYVYKSGVSYRRSYGQEVLLRCVDRLEADKALQEVHHGVCGGHQSGPKMYHSIR 998
Query: 1591 RAGYYWLTMEHDCYQHARKCHKCQIYADKIHVPPHALNVISSPWPFSMVGIDMIGRIEPK 1650
AGYYW + DC + A+ CH CQI+ D H+PP L+ WPF GID+IG I+P
Sbjct: 999 LAGYYWPEIMADCLKVAKSCHGCQIHGDFKHLPPVPLHPTVPAWPFEAWGIDVIGPIDPP 1058
Query: 1651 ASNGHRFILVAIDYFTKWVEAASYTNVTKQVVAKFIKNNIICRYGVPSKIITDNGTNLNN 1710
+S GHRFI DYF+KW EA S V V F++ +II R+GVP +I +DNG +
Sbjct: 1059 SSRGHRFIFAITDYFSKWAEAVSLREVKTDNVISFLERHIIYRFGVPHRISSDNGKAFKS 1118
Query: 1711 NVVQALCEEFKIEHHNSSPYRPQINGVVEAANKNIKRIVQKMVTTY-RDWHEMLPYALHG 1769
+ +Q ++KI + S+ Y PQ NG++EA NK + +I++K+V + RDWH+ L AL
Sbjct: 1119 HKMQRFIAKYKIRWNYSTGYYPQANGMIEAFNKTLGKILKKVVNRHRRDWHDHLFEALWA 1178
Query: 1770 YRTTVRSSTGATPFSLVYGMEAVLPLEVEIPSLRVIMEAKLSEAEWCQSRYDQLNLVEEK 1829
YR TVR+ T TP+SLVYG EAVLPLEVE+PSLRV + ++++ E + R+ +L+ +EE
Sbjct: 1179 YRVTVRTPTQCTPYSLVYGSEAVLPLEVEVPSLRVAIHEEITQDEQVRLRFQELDTLEEG 1238
Query: 1830 RMDAMARGQSYQARMKTAFDKKV 1852
R+ A+ + Y+ M A++K V
Sbjct: 1239 RLQAVQNLELYRQNMVRAYNKLV 1261
>UniRef100_Q40580 Integrase [Nicotiana tabacum]
Length = 382
Score = 414 bits (1065), Expect = e-113
Identities = 192/342 (56%), Positives = 263/342 (76%), Gaps = 2/342 (0%)
Query: 1579 HATGHTMSRKLLRAGYYWLTMEHDCYQHARKCHKCQIYADKIHVPPHALNVISSPWPFSM 1638
H G +S+K+LRAGY+W+TME DC ++ +KCH+CQIYAD + VPP+ LN S+PWPF+
Sbjct: 26 HMNGFVLSKKILRAGYFWMTMETDCIRYVQKCHQCQIYADMMRVPPNKLNATSAPWPFAA 85
Query: 1639 VGIDMIGRIEPKASNGHRFILVAIDYFTKWVEAASYTNVTKQVVAKFIKNNIICRYGVPS 1698
G+D+IG IEP ASN H+FILVAIDYFTKWVEAASY VTK+VV F+++ I+CR+ VP
Sbjct: 86 WGMDIIGPIEPTASNWHKFILVAIDYFTKWVEAASYKVVTKKVVTDFVRDCIVCRFEVPE 145
Query: 1699 KIITDNGTNLNNNVVQALCEEFKIEHHNSSPYRPQINGVVEAANKNIKRIVQKMVTTYRD 1758
IITDN NL++ +++A+C+ FKI+H NS+ Y PQ+NGVVEAANKNIK+I++KMV +++
Sbjct: 146 SIITDNAANLHSALMKAMCDTFKIKHKNSTAYMPQMNGVVEAANKNIKKILRKMVDNHKE 205
Query: 1759 WHEMLPYALHGYRTTVRSSTGATPFSLVYGMEAVLPLEVEIPSLRVIMEAKLSEAEWCQS 1818
WHE LP+AL GY T V +S GATP+ LVY EAV+ +EVEI SLR+I E LS+AEW +S
Sbjct: 206 WHEKLPFALLGYCTIVCTSIGATPYLLVYSTEAVIHVEVEITSLRIIQEDVLSDAEWIRS 265
Query: 1819 RYDQLNLVEEKRMDAMARGQSYQARMKTAFDKKVHPREFKEGELVLKRRISQQPDPRGKW 1878
RY+QL+L++ KR+ + GQ Y RM AF+K+V PR+F G+LVLKR Q + +GK+
Sbjct: 266 RYEQLSLIDGKRV--VCHGQLYHNRMSRAFNKRVKPRQFASGQLVLKRIFPHQDEAKGKF 323
Query: 1879 TPNYEGSYVVKKAFSGGALILTHMDGVELPNPVNADLVKKYF 1920
+PN++G Y+V + +GG LIL +DG P P+N+D+VK+Y+
Sbjct: 324 SPNWQGPYMVHRVLTGGTLILAEIDGEVWPKPINSDVVKRYY 365
>UniRef100_Q94LN5 Putative retroelement pol polyprotein [Oryza sativa]
Length = 1580
Score = 410 bits (1054), Expect = e-112
Identities = 243/719 (33%), Positives = 378/719 (51%), Gaps = 59/719 (8%)
Query: 1205 IYYLSKKLTDCETRYTMLEKTCCALAWAAKRLRHYLVNHTTWLISRMDPIKYIFEKAVVT 1264
++YLS++L + ETRY+ +EK C L ++ RLRHYL+++ +I + D +KY+ ++
Sbjct: 911 VFYLSRRLLNAETRYSPVEKLCLCLYFSCTRLRHYLLSNECTVICKADVVKYMLSAPILK 970
Query: 1265 GKIARWQVLLSEYDIVFKTQKAIKGSILADHPAYQPLDDYQPIESDFPNEEIVYLKSKDC 1324
G+I +W L+E+D+ +++ KAIKG +A+ + DY+ + + E+V
Sbjct: 971 GRIGKWIFSLTEFDLRYESPKAIKGQAIANF-----IVDYR--DDSIGSVEVV------- 1016
Query: 1325 EEPLIGEGPDPNSKWGLVFDGAANAYGKGIGAVIVSPQGHHIPFTAQILFECTNNMVEYE 1384
W L FDG+ +G GIG VI+SP+G F I TNN EYE
Sbjct: 1017 -------------PWTLFFDGSVCTHGCGIGLVIISPRGACFEFAYTIKPYATNNQAEYE 1063
Query: 1385 ACIFGIEEAIGMRIKHLDIYGDSALIINQIKGEWETHHAKLIPYRDYARRLLTYFTKVEL 1444
A + G++ + ++I GDS L+I+Q+ GE+E + LI Y + + L+ F V L
Sbjct: 1064 AVLKGLQLLKEVEADTIEIMGDSLLVISQLAGEYECKNDTLIVYNEKCQELMREFRLVTL 1123
Query: 1445 HHIPRDENQMADALATLSSMFRVNHWNDVPIIKVQRLERPSHVFTIGDVIDQAGENMVDN 1504
H+ R++N A+ LA +S ++ + ++ ++ V T D
Sbjct: 1124 KHVSREQNIEANDLAQGASGYK---------LMIKHVQIEVAVITADD------------ 1162
Query: 1505 KPWYYDIKQCLLSREYPPGASNKDKKTLRRLASRFLLDGDILYKRNYDMVLLRCVDEHEA 1564
W Y + Q Y S + LR A +++L D LY R D VLL+C+ +A
Sbjct: 1163 --WRYYVYQ------YLQDPSQSASRKLRYKALKYILLDDELYYRTIDGVLLKCLSTDQA 1214
Query: 1565 EQLMHDVHDGTFGTHATGHTMSRKLLRAGYYWLTMEHDCYQHARKCHKCQIYADKIHVPP 1624
+ + +VH+G GTH + H M L AGY+W TM DC+++ + C CQ + P
Sbjct: 1215 KVAIGEVHEGICGTHQSAHKMKWLLRCAGYFWPTMLEDCFRYYKGCQDCQKFGAIQRAPA 1274
Query: 1625 HALNVISSPWPFSMVGIDMIGRIEPKASNGHRFILVAIDYFTKWVEAASYTNVTKQVVAK 1684
A+N I PWPF GIDMIG I P +S GH+FILVA DYFTKWVEA V +
Sbjct: 1275 SAMNPIIKPWPFRGWGIDMIGMINPPSSKGHKFILVATDYFTKWVEAIPLKKVDSGDAIQ 1334
Query: 1685 FIKNNIICRYGVPSKIITDNGTNLNNNVVQALCEEFKIEHHNSSPYRPQINGVVEAANKN 1744
F++ +II R G+P I TD G+ ++ + I+ NSSPY Q NG EA+NK+
Sbjct: 1335 FVQEHIIYRVGIPQTITTDQGSIFVSDEFIQFADSMGIKLLNSSPYYAQANGQAEASNKS 1394
Query: 1745 IKRIVQKMVTTY-RDWHEMLPYALHGYRTTVRSSTGATPFSLVYGMEAVLPLEVEIPSLR 1803
+ +++++ ++ Y R WH L AL YR S P+ LVY +A+LP EV I S R
Sbjct: 1395 LIKLIKRKISDYPRQWHTRLAEALWSYRMACHGSIQVPPYKLVYRHKAILPWEVRIGSRR 1454
Query: 1804 VIMEAKLSEAEWCQSRYDQLNLVEEKRMDAMARGQSYQARMKTAFDKKVHPREFKEGELV 1863
++ L+ E+ D+ + + R+ A+A+ + R+ ++KKV P++F EGEL+
Sbjct: 1455 TELQNDLTADEYSNLMADEREDLVQSRLRALAKVIKDKERVTRHYNKKVVPKDFSEGELI 1514
Query: 1864 LK--RRISQQPDPRGKWTPNYEGSYVVKKAFSGGALILTHMDGVELPNPVNADLVKKYF 1920
K I + GKW+PN+EG + + K S GA +L +DG +N +KKY+
Sbjct: 1515 WKLILPIGTRDSKFGKWSPNWEGPFQIHKVVSKGAYMLQGLDGEVYDRALNGKYLKKYY 1573
Score = 72.8 bits (177), Expect = 1e-10
Identities = 58/203 (28%), Positives = 101/203 (49%), Gaps = 7/203 (3%)
Query: 780 ELPEAGKHHNLA-LHISVNCTPDIISNVLVDTGSSLNVMPKSTLDQLSYRGTPLRRSTFL 838
E PE ++ +L L+I+ +S ++VD G+++N+MP +T +L L ++ +
Sbjct: 351 EKPEGTENRHLKPLYINGYVNGKPMSKMMVDGGAAVNLMPYATFRKLGRNAEDLIKTNMV 410
Query: 839 VKAFDGTRKSVLGEIDLPITIGPETFLITFQVMDINASYNCLLGRP*IHDAGAVTSTLHQ 898
+K F G +G +++ +T+G +T TF V+D SY+ LLGR IH + ST+HQ
Sbjct: 411 LKDFGGNPSETMGVLNVELTVGSKTIPTTFFVIDGKGSYSLLLGRDWIHANCCIPSTIHQ 470
Query: 899 KLKFAKSGKLVTIHGEEAYLVSQLSSFSCIEAGSAEGTAFQGLTIEGTEPKRDGTAMASL 958
L G + I ++ L + S+ C E + T++ + K+ G + S
Sbjct: 471 CL-IQWQGDKIEIVPADSQLKMENPSY-CFEGVMEGSNVYTKDTVDDLDDKQ-GQGLMSA 527
Query: 959 KDAQRAVQEGQAAG*GR--LIQL 979
D + + A G GR LI+L
Sbjct: 528 DDLEE-IDISPAIGQGRHLLIEL 549
>UniRef100_Q6L974 GAG-POL [Vitis vinifera]
Length = 1027
Score = 410 bits (1053), Expect = e-112
Identities = 243/718 (33%), Positives = 377/718 (51%), Gaps = 38/718 (5%)
Query: 1205 IYYLSKKLTDCETRYTMLEKTCCALAWAAKRLRHYLVNHTTWLISRMDPIKYIFEKAVVT 1264
IYY+S+ L D ETRY+ +E AL AA++LR Y H +++ P++ I K +T
Sbjct: 345 IYYVSRALADVETRYSKMELISLALRSAAQKLRPYFQAHPVIVLTDQ-PLRNILHKPDLT 403
Query: 1265 GKIARWQVLLSEYDIVFKTQKAIKGSILADHPAYQPLDDYQPIESDFPNEEIVYLKSKDC 1324
G++ +W + LSE+ I F+ + ++KG ++AD +Y
Sbjct: 404 GRMLQWAIELSEFGIEFQPRLSMKGQVMADFVL-----EYS------------------- 439
Query: 1325 EEPLIGEGPDPNSKWGLVFDGAANAYGKGIGAVIVSPQGHHIPFTAQILFECTNNMVEYE 1384
+P EG W L DGA+ + G G+G ++ SP G H+ ++ F +NN EYE
Sbjct: 440 RKPGQHEGSRKKEWWTLRVDGASRSSGSGVGLLLQSPTGEHLEQAIRLGFSASNNEAEYE 499
Query: 1385 ACIFGIEEAIGMRIKHLDIYGDSALIINQIKGEWETHHAKLIPYRDYARRLLTYFTKVEL 1444
A + G++ A+ + + L I+ DS L++ ++ E+E A++ Y R L FT+ +
Sbjct: 500 AILSGLDLALALSVSKLRIFSDSQLVVKHVQEEYEAKDARMARYLAKVRNTLQQFTEWTI 559
Query: 1445 HHIPRDENQMADALATLSSMFRVNHWNDVPIIKVQRLERPSHVFTIGDVIDQAGENMVDN 1504
I R +N+ ADALA +++ + +PI VQ S + + D+
Sbjct: 560 EKIKRADNRRADALAGIAASLSIKEAILLPI-HVQTNPSVSEI----SICSTTEAPQADD 614
Query: 1505 KPWYYDIKQCLLSREYPPGASNKDKKTLRRLASRFLLDGDILYKRNYDMVLLRCVDEHEA 1564
+ W DI + + + P K +R A+RF L G LYKR++ LRC+ EA
Sbjct: 615 QEWMNDITEYIRTGTLP--GDPKQAHKVRVQAARFTLIGGHLYKRSFTGPYLRCLGHSEA 672
Query: 1565 EQLMHDVHDGTFGTHATGHTMSRKLLRAGYYWLTMEHDCYQHARKCHKCQIYADKIHVPP 1624
+ ++ ++H+G +G H+ G +++ + GYYW TM+ + + ++C KCQ YA H+P
Sbjct: 673 QYVLAELHEGIYGNHSGGRSLAHRAHSQGYYWPTMKKEAAAYVKRCDKCQRYAPIPHMPS 732
Query: 1625 HALNVISSPWPFSMVGIDMIGRIEPKASNGHRFILVAIDYFTKWVEAASYTNVTKQVVAK 1684
L IS PWPF+ G+D++ R P A +F+LVA DYF+KWVEA +Y + + V K
Sbjct: 733 TTLKSISGPWPFAQWGMDIV-RPLPTAPAQKKFLLVATDYFSKWVEAEAYASTKDKDVTK 791
Query: 1685 FIKNNIICRYGVPSKIITDNGTNLNNNVVQALCEEFKIEHHNSSPYRPQINGVVEAANKN 1744
F+ NIICR+G+P II DNG ++ + C E I + S+P PQ NG EA NK
Sbjct: 792 FVWKNIICRFGIPQTIIADNGPQFDSIAFRNFCSELNIRNSYSTPRYPQSNGQAEATNKT 851
Query: 1745 IKRIVQKMVTTYR-DWHEMLPYALHGYRTTVRSSTGATPFSLVYGMEAVLPLEVEIPSLR 1803
+ ++K + + W E LP L YRTT TG TPF+L YGM+AV+P+E+ +P++
Sbjct: 852 LITALKKRLEQAKGKWVEELPGVLWAYRTTPGRPTGNTPFALAYGMDAVIPIEIGLPTIW 911
Query: 1804 VIMEAKLSEAEWCQSRYDQLNLVEEKRMDAMARGQSYQARMKTAFDKKVHPREFKEGELV 1863
AK S+A R L+ +E R A R YQ R +++KV PR K G LV
Sbjct: 912 T-NAAKQSDANMQLGR--NLDWTDEVRESASIRMADYQQRASAHYNRKVRPRSLKNGTLV 968
Query: 1864 LKRRISQQPD-PRGKWTPNYEGSYVVKKAFSGGALILTHMDGVELPNPVNADLVKKYF 1920
L++ + GK+ N+EG Y+V KA GA L +DG L P N +K+Y+
Sbjct: 969 LRKFFENTTEVGAGKFQANWEGPYIVSKASDNGAYHLQKLDGTPLLRPWNVSNLKQYY 1026
>UniRef100_Q9LMV1 F5M15.26 [Arabidopsis thaliana]
Length = 1838
Score = 382 bits (980), Expect = e-104
Identities = 250/727 (34%), Positives = 369/727 (50%), Gaps = 47/727 (6%)
Query: 1205 IYYLSKKLTDCETRYTMLEKTCCALAWAAKRLRHYLVNHTTWLISRMDPIKYIFEKAVVT 1264
I+Y SK L + ETRY ++EK A+ +A++LR Y +HT +++ P++ +
Sbjct: 1147 IFYTSKSLVEAETRYPVIEKAALAVVTSARKLRPYFQSHTIAVLTDQ-PLRVALHSPSQS 1205
Query: 1265 GKIARWQVLLSEYDIVFKTQKAIKGSILADHPAYQPLDDYQPIESDFPNEEIVYLKSKDC 1324
G++ +W V LSEYDI F+ + A+K +LAD PL + S EE
Sbjct: 1206 GRMTKWAVELSEYDIDFRPRPAMKSQVLADFLIELPLQSAERAVSGNRGEE--------- 1256
Query: 1325 EEPLIGEGPDPNSKWGLVFDGAANAYGKGIGAVIVSPQGHHIPFTAQILFECTNNMVEYE 1384
W L DG+++A G GIG +VSP + + ++ F TNN+ EYE
Sbjct: 1257 --------------WSLYVDGSSSARGSGIGIRLVSPTAEVLEQSFRLRFVATNNVAEYE 1302
Query: 1385 ACIFGIEEAIGMRIKHLDIYGDSALIINQIKGEWETHHAKLIPYRDYARRLLTYFTKVEL 1444
I G+ A GM+I + + DS LI Q+ GE+E + K+ Y + + F +L
Sbjct: 1303 VLIAGLRLAAGMQITTIHAFTDSQLIAGQLSGEYEAKNEKMDAYLKIVQLMTKDFENFKL 1362
Query: 1445 HHIPRDENQMADALATLSSMFRVNHWNDVP-IIKVQRLERPS-----HVFTIGDVIDQAG 1498
IPR +N ADALA L+ + +D+ II V+ +++PS V + +
Sbjct: 1363 SKIPRGDNAPADALAALA----LTSDSDLRRIIPVESIDKPSIDSTDAVEIVNTIRSSNA 1418
Query: 1499 ENMVDNKPWYYDIKQCLLSREYPPGASNKDKKTLRRL---ASRFLLDGDILYKRNYDMVL 1555
+ D W +I+ L P DK T RRL A+++ L + L K + +
Sbjct: 1419 PDPADPTDWRVEIRDYLSDGTLP-----SDKWTARRLRIKAAKYTLMKEHLLKVSAFGAM 1473
Query: 1556 LRCVDEHEAEQLMHDVHDGTFGTHATGHTMSRKLLRAGYYWLTMEHDCYQHARKCHKCQI 1615
L C+ E ++M + H+G G H+ G ++ KL + G+YW TM DC KC +CQ
Sbjct: 1474 LNCLHGTEINEIMKETHEGAAGNHSGGRALALKLKKLGFYWPTMISDCKTFTAKCEQCQR 1533
Query: 1616 YADKIHVPPHALNVISSPWPFSMVGIDMIGRIEPKASNGHRFILVAIDYFTKWVEAASYT 1675
+A IH P L +P+PF +D++G + AS RFILV DYFTKWVEA SY
Sbjct: 1534 HAPTIHQPTELLRAGVAPYPFMRWAMDIVGPMP--ASRQKRFILVMTDYFTKWVEAESYA 1591
Query: 1676 NVTKQVVAKFIKNNIICRYGVPSKIITDNGTNLNNNVVQALCEEFKIEHHNSSPYRPQIN 1735
+ V F+ IICR+G+P +IITDNG+ + + C +KI + S+P PQ N
Sbjct: 1592 TIRANDVQNFVWKFIICRHGLPYEIITDNGSQFISLSFENFCASWKIRLNKSTPRYPQGN 1651
Query: 1736 GVVEAANKNIKRIVQKMVTTYRD-WHEMLPYALHGYRTTVRSSTGATPFSLVYGMEAVLP 1794
G EA NK I ++K + + W + L L YRTT RS+T TPF+ YGMEA+ P
Sbjct: 1652 GQAEATNKTILSGLKKRLDEKKGAWADELDGVLWSYRTTPRSATDQTPFAHAYGMEAMAP 1711
Query: 1795 LEVEIPSLRVIMEAKLSEAEWCQSRYDQLNLVEEKRMDAMARGQSYQARMKTAFDKKVHP 1854
EV SLR M K E + D+L+ +EE R A+ R Q+YQ +++KVH
Sbjct: 1712 AEVGYSSLRRSMMVKNPELN-DRMMLDRLDDLEEIRNAALCRIQNYQLAAAKHYNQKVHN 1770
Query: 1855 REFKEGELVLKRRISQQPD-PRGKWTPNYEGSYVVKKAFSGGALILTHMDGVELPNPVNA 1913
R F G+LVL++ + GK N+EGSY V K G L M G +P N+
Sbjct: 1771 RHFDVGDLVLRKVFENTAEINAGKLGANWEGSYQVSKIVRPGDYELLTMSGTAVPRTWNS 1830
Query: 1914 DLVKKYF 1920
+K+Y+
Sbjct: 1831 MHLKRYY 1837
Score = 68.6 bits (166), Expect = 2e-09
Identities = 47/176 (26%), Positives = 81/176 (45%), Gaps = 7/176 (3%)
Query: 776 FSENELPEAGKHHNLALHISVNCTPDIISNVLVDTGSSLNVMPKSTLDQLSYRGTPLRRS 835
F++ +L HN L + + + ++ VL+DTGSS++++ K L ++ ++
Sbjct: 602 FTDVDLEGLDTPHNDPLVVELIISDSRVTRVLIDTGSSVDLIFKDVLTAMNITDRQIKPV 661
Query: 836 TFLVKAFDGTRKSVLGEIDLPITIGPETFLITFQVMDINASYNCLLGRP*IHDAGAVTST 895
+ + FDG +G I LPI +G + F V+ A YN +LG P IH A+ ST
Sbjct: 662 SKPLAGFDGDFVMTIGTIKLPIFVGGLIAWVKFVVIGKPAVYNVILGTPWIHQMQAIPST 721
Query: 896 LHQKLKFAKSGKLVTIHG-------EEAYLVSQLSSFSCIEAGSAEGTAFQGLTIE 944
HQ +KF + T+ +Y S+L + ++ T G+ E
Sbjct: 722 YHQCVKFPTHNGIFTLRAPKEAKTPSRSYEESELCRTEMVNIDESDPTRCVGVGAE 777
Score = 48.5 bits (114), Expect = 0.002
Identities = 45/220 (20%), Positives = 92/220 (41%), Gaps = 5/220 (2%)
Query: 133 KTMEEMMEELAKELRHEIKANRGNADSFKTQDLCLVSKVDVPKKFKIPDFDRYNGLTCPQ 192
+T+ EM ++ + + +R ++ +T ++ V + KI + YNG P+
Sbjct: 173 RTISEMSSKIHRATSTAPELDRVLEETQQTPFTRRITNVSIRGAQKIK-LESYNGRNDPK 231
Query: 193 NHIIKY---VRKMGNYKDN-DSLMIHCFQDSLMEDAAEWYTSLSKNDIHTFDELAAAFKS 248
+ + + + DN D+ F + L A W++ L N I +F +L ++F
Sbjct: 232 EFLTSFNVAINRAELTIDNFDAGRCQIFIEHLTGPAHNWFSRLKPNSIDSFHQLTSSFLK 291
Query: 249 HYGFNTRLKPNREFLRSLSQKKEESFREYAQRWRGAAARITPALDEEEMTQTFLKTLKKD 308
HY + + L S+SQ +ES R + R++ IT + +
Sbjct: 292 HYAPLIENQTSNADLWSISQGAKESLRSFVDRFKLVVTNITVPDEAAIVALRNAVWYDSR 351
Query: 309 YVERMIIAAPNNFSEMVTMGTRLEEAVREGIIVFEKAESS 348
+ + + + AP+ + + +R E E +I+ K S+
Sbjct: 352 FRDDITLHAPSTLEDALHRASRFIELEEEKLILARKHNST 391
>UniRef100_Q65WT1 Putative polyprotein [Oryza sativa]
Length = 1992
Score = 381 bits (979), Expect = e-103
Identities = 250/784 (31%), Positives = 386/784 (48%), Gaps = 81/784 (10%)
Query: 1165 EDEEGDDIPYEITRLLEQEGRAIQPHQEEIEIINLGIPTAIYYLSKKLTDCETRYTMLEK 1224
E+ GD P E R PH++ + +Y++S+ L D +TRY +K
Sbjct: 1261 EERPGDTAPSEEDR----------PHRK--------VQRPVYFVSEALRDAKTRYPQAQK 1302
Query: 1225 TCCALAWAAKRLRHYLVNHTTWLISRMDPIKYIFEKAVVTGKIARWQVLLSEYDIVFKTQ 1284
A+ A+++LRHY H +++ P+ I TG++ +W + LSE+D+ F+ +
Sbjct: 1303 MLYAVLMASRKLRHYFQAHRVTVVTSY-PLGQILHNREGTGRVVKWAIELSEFDLHFEPR 1361
Query: 1285 KAIKGSILADHPAYQPLDDYQPIESDFPNEEIVYLKSKDCEEPLIGEGPDPNSKWGLVFD 1344
AIK LAD A ++ P E+ S + P G W + FD
Sbjct: 1362 HAIKSQALADFVA-----EWTPAPEPVSIPEV---NSDPSQPPHTGH-------WVMQFD 1406
Query: 1345 GAANAYGKGIGAVIVSPQGHHIPFTAQILFECTNNMVEYEACIFGIEEAIGMRIKHLDIY 1404
G+ + G G G + SP G + + ++ F TNNM EYE + G+ A G+ I+ L +
Sbjct: 1407 GSLSLQGAGAGVTLTSPTGDVLRYLDRLDFRATNNMAEYEGLLAGLRVAAGLGIRRLLVL 1466
Query: 1405 GDSALIINQIKGEWETHHAKLIPYRDYARRLLTYFTKVELHHIPRDENQMADALATLSSM 1464
GDS L++NQ+ E+ ++ Y RR+ +F +EL H+PR +N +AD L+ L+S
Sbjct: 1467 GDSQLVVNQVCKEYRCSDPQMDAYVRQVRRMEHHFDGIELRHVPRRDNMIADELSRLAS- 1525
Query: 1465 FRVNHWNDVPIIKVQRLERPSHVFTIGDVIDQAGENMVDNKP------------------ 1506
+ P +RL +PS D GE V ++P
Sbjct: 1526 ---SRAQTPPGAFEERLTQPSAR------PDPLGETGVPDRPPRPVGVQASGPEGSAPSS 1576
Query: 1507 -----WYYDIKQCLLSREYPPGASNKDKKTLRRLASRFLLDGDILYKRNYDMVLLRCVDE 1561
W +I+ L + P ++ ++R++ R++L LY+R + +LL+C+ +
Sbjct: 1577 LRLIAWISEIQAYLTDKTLPEDREGSER--VQRISKRYVLVEGTLYRRAANGILLKCIPQ 1634
Query: 1562 HEAEQLMHDVHDGTFGTHATGHTMSRKLLRAGYYWLTMEHDCYQHARKCHKCQIYADKIH 1621
+ +L+ D+H+G G H+ T+ K R G+YW T +D R+C CQ +A +IH
Sbjct: 1635 EQGVELLADIHEGECGAHSASRTLVGKAFRQGFYWPTALNDAVDLVRRCRACQFHAKQIH 1694
Query: 1622 VPPHALNVISSPWPFSMVGIDMIGRIEPKASNGHRFILVAIDYFTKWVEAASYTNVTKQV 1681
P AL +I WPF++ G+D++G +A G ++ VAID FTKW EA + K
Sbjct: 1695 QPAQALQIIPLSWPFAVWGLDILGPFR-RAPGGFEYLYVAIDKFTKWPEAYPVIKIDKHS 1753
Query: 1682 VAKFIKNNIICRYGVPSKIITDNGTNLNNNVVQALCEEFKIEHHNSSPYRPQINGVVEAA 1741
KFIK I R+GVP++IITDNGT + + CE+ I+ +SP P+ NG VE A
Sbjct: 1754 ALKFIK-GITARFGVPNRIITDNGTQFTSELFGDYCEDMGIKLCFASPAHPRSNGQVERA 1812
Query: 1742 N----KNIKRIVQKMVTTYRD-WHEMLPYALHGYRTTVRSSTGATPFSLVYGMEAVLPLE 1796
N K +K ++ + D W E LP L RTT +TG TPF LVYG EAVLP E
Sbjct: 1813 NAEILKGLKTKTFNILKKHGDSWIEELPAVLWANRTTPSRATGETPFFLVYGAEAVLPSE 1872
Query: 1797 VEIPSLRVIMEAKLSEAEWCQSRYDQLNLVEEKRMDAMARGQSYQARMKTAFDKKVHPRE 1856
+ + S R M EA+ Q R D L+ +EE+R A R YQ ++ + V R
Sbjct: 1873 LTLRSPRATM---YCEADQDQLRRDDLDYLEERRRRAALRAARYQQSLRRYHQRHVRARS 1929
Query: 1857 FKEGELVLKRRISQQPDPRGKWTPNYEGSYVVKKAFSGGALILTHMDGVELPNPVNADLV 1916
+LVL+R Q K +P +EG Y V G++ L DG ELPNP N + +
Sbjct: 1930 LCVDDLVLRR--VQTRAGLSKLSPMWEGPYRVVGVPRPGSIRLATGDGTELPNPWNIEHL 1987
Query: 1917 KKYF 1920
++++
Sbjct: 1988 RRFY 1991
Score = 56.2 bits (134), Expect = 1e-05
Identities = 38/137 (27%), Positives = 65/137 (46%), Gaps = 5/137 (3%)
Query: 782 PEAGKHHNLALHISVNCTPDIISNVLVDTGSSLNVMPKSTLDQLSYRGTPLRRSTFLVKA 841
P + +A+ +S + VL+D G++LN++ + D + G LR S ++
Sbjct: 568 PCVARGGQIAMVVSPTICNVKLGRVLIDGGAALNILSPAAFDAIKAPGMVLRPSQPIIGV 627
Query: 842 FDGTRKSVLGEIDLPITIG-PETFL---ITFQVMDINASYNCLLGRP*IHDAGAVTSTLH 897
G LG IDLP+T G P F + F V D++ YN +LGRP + A +
Sbjct: 628 TPG-HTWPLGHIDLPVTFGGPANFRTERVNFDVADLSLPYNAVLGRPALVKFMAAVHYAY 686
Query: 898 QKLKFAKSGKLVTIHGE 914
++K +++HG+
Sbjct: 687 LQMKMPGPDGPISVHGD 703
>UniRef100_Q9MFD2 Orf764 protein [Beta vulgaris subsp. vulgaris]
Length = 764
Score = 381 bits (978), Expect = e-103
Identities = 240/730 (32%), Positives = 387/730 (52%), Gaps = 52/730 (7%)
Query: 1194 IEIINLGIPTAIYYLSKKLTDCETRYTMLEKTCCALAWAAKRLRHYLVNHTTWLISRMDP 1253
+E I +P IY++S L + E RY +LEK A+ AA++LR Y HT ++S P
Sbjct: 83 VERIKTQLP--IYFVSHVLNEAEQRYPLLEKMSLAVMIAARKLRPYFDAHTIQVLSN-HP 139
Query: 1254 IKYIFEKAVVTGKIARWQVLLSEYDIVFKTQKAIKGSILADHPAYQPLDDYQPIESDFPN 1313
++ +K +G++ RW V LSE+++ +K + AIK LAD +E+ +
Sbjct: 140 LEKALQKLDTSGRLLRWAVELSEFEVEYKPRTAIKAQALADFI----------VEASYEE 189
Query: 1314 EEIVYLKSKDCEEPLIGEGPDPNSKWGLVFDGAANAYGKGIGAVIVSPQGHHIPFTAQIL 1373
EE +P W L DG+A G G G ++ SP+G+ F I
Sbjct: 190 EE------------------EPVGVWKLAVDGSAAQTGSGAGIIMTSPEGN--VFEYAIK 229
Query: 1374 FECTNNMVEYEACIFGIEEAIGMRIKHLDIYGDSALIINQIKGEWETHHAKLIPYRDYAR 1433
F+ +NN EYEA I GI+ + K + + DS L+ +QI+GE+E + Y +
Sbjct: 230 FKASNNEAEYEAAIAGIKMCMAADAKKIRLQTDSQLVASQIRGEYEAREPAMQKYLQMIK 289
Query: 1434 RLLTYFTKVELHHIPRDENQMADALATLSSMFRVNHWNDVPIIKVQRLERPSHVFTIGDV 1493
L +E+ +PR +N ADAL+ L+S + N +++V I V
Sbjct: 290 DLAAQLISLEVVLVPRADNSQADALSKLASS-TLQDLNRTVMVEVMEQRSIDKAERINCV 348
Query: 1494 IDQAGENMVDNKPWYYDIKQCLLSREYPPGASNKDKKTLRRLASRFLLDGDILYKRNYDM 1553
Q K WY DI++ L+ + P + K ++R + +++ LYKR+
Sbjct: 349 TAQ--------KEWYDDIQEYKLTGKLPDDQTIA--KAVKRDSHWYIIYQGQLYKRSDSY 398
Query: 1554 VLLRCVDEHEAEQLMHDVHDGTFGTHATGHTMSRKLLRAGYYWLTMEHDCYQHARKCHKC 1613
LLRC E E M + +G G H G +S +++R G YW TM DC ++ +KC KC
Sbjct: 399 PLLRCTSESEKLPAMEEAPEGICGNHIGGKALSHEVMRRGNYWPTMVKDCLRYVKKCDKC 458
Query: 1614 QIYADKIHVPPHALNVISSPWPFSMVGIDMIGRIEPKASNGHRFILVAIDYFTKWVEAAS 1673
Q +A ++ P + L I +P PF+ G+D++G AS G ++++VA+DYFTKW+EA
Sbjct: 459 QKFAPVMNQPANDLCPILNPIPFAQWGMDILGPFTT-ASGGRKYLIVAVDYFTKWIEAEP 517
Query: 1674 YTNVTKQVVAKFIKNNIICRYGVPSKIITDNGTNLNNNVVQALCEEFKIEHHNSSPYRPQ 1733
+ + V FI NII R+G+P I+ D+GT + + ++ C EFKI+ +++ PQ
Sbjct: 518 TKFIKARQVRSFIWKNIITRFGIPQCIVFDHGTQFDCDTIKDYCAEFKIKFASAAVCHPQ 577
Query: 1734 INGVVEAANKNIKRIVQKMVTTYRD-WHEMLPYALHGYRTTVRSSTGATPFSLVYGMEAV 1792
NG EAANK I +QK + ++ W +++P L RTT + +TG +PFSL +G EAV
Sbjct: 578 SNGQAEAANKQILLALQKKLEEHKGLWADLVPEVLWANRTTEKGATGKSPFSLAFGAEAV 637
Query: 1793 LPLEVEIPSLRV-IMEAKLSEAEWCQSRYDQLNLVEEKRMDAMARGQSYQARMKTAFDKK 1851
+P+EV +PS RV + +++E Q + L+ + E R+ A + + + RM ++++
Sbjct: 638 VPIEVGLPSFRVQHYDPEVNE----QLMREALDQLPEMRLQAALKLAAQKNRMSRIYNRR 693
Query: 1852 VHPREFKEGELVLKRRIS-QQPDPRGKWTPNYEGSYVVKKAFSGGALILTHMDGVELPNP 1910
V R EG+LVL+R + + + GK+T N+EG Y + + + G+ L ++G L N
Sbjct: 694 VRHRPLTEGDLVLRRTAAVGKGNVHGKFTANWEGPYQICEQVAPGSYKLMTVEGEPLKNS 753
Query: 1911 VNADLVKKYF 1920
NAD++KKYF
Sbjct: 754 FNADVLKKYF 763
>UniRef100_Q5U6F6 Orf764 protein [Beta vulgaris subsp. vulgaris]
Length = 764
Score = 381 bits (978), Expect = e-103
Identities = 240/730 (32%), Positives = 387/730 (52%), Gaps = 52/730 (7%)
Query: 1194 IEIINLGIPTAIYYLSKKLTDCETRYTMLEKTCCALAWAAKRLRHYLVNHTTWLISRMDP 1253
+E I +P IY++S L + E RY +LEK A+ AA++LR Y HT ++S P
Sbjct: 83 VERIKTQLP--IYFVSHVLNEAEQRYPLLEKMSLAVMIAARKLRPYFDAHTIQVLSN-HP 139
Query: 1254 IKYIFEKAVVTGKIARWQVLLSEYDIVFKTQKAIKGSILADHPAYQPLDDYQPIESDFPN 1313
++ +K +G++ RW V LSE+++ +K + AIK LAD +E+ +
Sbjct: 140 LEKALQKLDTSGRLLRWAVELSEFEVEYKPRTAIKAQALADFI----------VEASYEE 189
Query: 1314 EEIVYLKSKDCEEPLIGEGPDPNSKWGLVFDGAANAYGKGIGAVIVSPQGHHIPFTAQIL 1373
EE +P W L DG+A G G G ++ SP+G+ F I
Sbjct: 190 EE------------------EPVGVWKLAVDGSAAQTGSGAGIIMTSPEGN--VFEYAIK 229
Query: 1374 FECTNNMVEYEACIFGIEEAIGMRIKHLDIYGDSALIINQIKGEWETHHAKLIPYRDYAR 1433
F+ +NN EYEA I GI+ + K + + DS L+ +QI+GE+E + Y +
Sbjct: 230 FKASNNEAEYEAAIAGIKMCMAADAKKIRLQTDSQLVASQIRGEYEAREPAMQKYLQMIK 289
Query: 1434 RLLTYFTKVELHHIPRDENQMADALATLSSMFRVNHWNDVPIIKVQRLERPSHVFTIGDV 1493
L +E+ +PR +N ADAL+ L+S + N +++V I V
Sbjct: 290 DLAAQLISLEVVLVPRADNSQADALSKLASS-TLQDLNRTVMVEVMEQRSIDKAERINCV 348
Query: 1494 IDQAGENMVDNKPWYYDIKQCLLSREYPPGASNKDKKTLRRLASRFLLDGDILYKRNYDM 1553
Q K WY DI++ L+ + P + K ++R + +++ LYKR+
Sbjct: 349 TAQ--------KEWYDDIQEYKLTGKLPDDQTIA--KAVKRDSHWYIIYQGQLYKRSDSY 398
Query: 1554 VLLRCVDEHEAEQLMHDVHDGTFGTHATGHTMSRKLLRAGYYWLTMEHDCYQHARKCHKC 1613
LLRC E E M + +G G H G +S +++R G YW TM DC ++ +KC KC
Sbjct: 399 PLLRCTSESEKLPAMEEAPEGICGNHIGGKALSHEVMRRGNYWPTMVKDCLRYVKKCDKC 458
Query: 1614 QIYADKIHVPPHALNVISSPWPFSMVGIDMIGRIEPKASNGHRFILVAIDYFTKWVEAAS 1673
Q +A ++ P + L I +P PF+ G+D++G AS G ++++VA+DYFTKW+EA
Sbjct: 459 QKFAPVMNQPANDLCPILNPIPFAQWGMDILGPFTT-ASGGRKYLIVAVDYFTKWIEAEP 517
Query: 1674 YTNVTKQVVAKFIKNNIICRYGVPSKIITDNGTNLNNNVVQALCEEFKIEHHNSSPYRPQ 1733
+ + V FI NII R+G+P I+ D+GT + + ++ C EFKI+ +++ PQ
Sbjct: 518 TKFIKARQVRSFIWKNIITRFGIPQCIVFDHGTQFDCDTIKDYCAEFKIKFASAAVCHPQ 577
Query: 1734 INGVVEAANKNIKRIVQKMVTTYRD-WHEMLPYALHGYRTTVRSSTGATPFSLVYGMEAV 1792
NG EAANK I +QK + ++ W +++P L RTT + +TG +PFSL +G EAV
Sbjct: 578 SNGQAEAANKQILLALQKKLEEHKGLWADLVPEVLWANRTTEKGATGKSPFSLAFGAEAV 637
Query: 1793 LPLEVEIPSLRV-IMEAKLSEAEWCQSRYDQLNLVEEKRMDAMARGQSYQARMKTAFDKK 1851
+P+EV +PS RV + +++E Q + L+ + E R+ A + + + RM ++++
Sbjct: 638 VPIEVGLPSFRVQHYDPEVNE----QLMREALDQLPEMRLQAALKLAAQKNRMSRIYNRR 693
Query: 1852 VHPREFKEGELVLKRRIS-QQPDPRGKWTPNYEGSYVVKKAFSGGALILTHMDGVELPNP 1910
V R EG+LVL+R + + + GK+T N+EG Y + + + G+ L ++G L N
Sbjct: 694 VRHRPLTEGDLVLRRTAAVGKGNVHGKFTANWEGPYQICEQVAPGSYKLMTVEGEPLKNS 753
Query: 1911 VNADLVKKYF 1920
NAD++KKYF
Sbjct: 754 FNADVLKKYF 763
>UniRef100_Q65WT8 Putative polyprotein [Oryza sativa]
Length = 2027
Score = 380 bits (976), Expect = e-103
Identities = 249/784 (31%), Positives = 385/784 (48%), Gaps = 81/784 (10%)
Query: 1165 EDEEGDDIPYEITRLLEQEGRAIQPHQEEIEIINLGIPTAIYYLSKKLTDCETRYTMLEK 1224
E+ GD P E R PH++ + +Y++S+ L D +TRY +K
Sbjct: 1296 EERPGDTAPSEEDR----------PHRK--------VQRPVYFVSEALRDAKTRYPQAQK 1337
Query: 1225 TCCALAWAAKRLRHYLVNHTTWLISRMDPIKYIFEKAVVTGKIARWQVLLSEYDIVFKTQ 1284
A+ A+++LRHY H +++ P+ I TG++ +W + LSE+D+ F+ +
Sbjct: 1338 MLYAVLMASRKLRHYFQAHRVTVVTSY-PLGQILHNREGTGRVVKWTIELSEFDLHFEPR 1396
Query: 1285 KAIKGSILADHPAYQPLDDYQPIESDFPNEEIVYLKSKDCEEPLIGEGPDPNSKWGLVFD 1344
AIK LAD A ++ P E+ S + P G W + FD
Sbjct: 1397 HAIKSQALADFVA-----EWTPAPEPVSIPEV---NSDPSQPPYTGH-------WVMQFD 1441
Query: 1345 GAANAYGKGIGAVIVSPQGHHIPFTAQILFECTNNMVEYEACIFGIEEAIGMRIKHLDIY 1404
G+ + G G G + SP G + + ++ F TNNM EYE + G+ A G+ I+ L +
Sbjct: 1442 GSLSLQGAGAGVTLTSPTGDVLRYLVRLDFRATNNMAEYEGLLAGLRVAAGLGIRRLLVL 1501
Query: 1405 GDSALIINQIKGEWETHHAKLIPYRDYARRLLTYFTKVELHHIPRDENQMADALATLSSM 1464
GDS L++NQ+ E+ ++ Y RR+ +F +EL H+PR +N +AD L+ L+S
Sbjct: 1502 GDSQLVVNQVCKEYRCSDPQMDAYVRQVRRMERHFDGIELRHVPRRDNMIADELSRLAS- 1560
Query: 1465 FRVNHWNDVPIIKVQRLERPSHVFTIGDVIDQAGENMVDNKP------------------ 1506
+ P +RL +PS D GE V ++P
Sbjct: 1561 ---SRAQTPPGAFEERLTQPSAR------PDPLGETGVPDRPPRPVGVQASGPEGSAPSS 1611
Query: 1507 -----WYYDIKQCLLSREYPPGASNKDKKTLRRLASRFLLDGDILYKRNYDMVLLRCVDE 1561
W +I+ L + P ++ ++R++ R++L LY+R + +LL+C+ +
Sbjct: 1612 LRLIAWISEIQAYLTDKTLPEDREGSER--VQRISKRYVLVEGTLYRRAANGILLKCIPQ 1669
Query: 1562 HEAEQLMHDVHDGTFGTHATGHTMSRKLLRAGYYWLTMEHDCYQHARKCHKCQIYADKIH 1621
+ +L+ D+H+G G H+ T+ K R G+YW T +D R+C CQ +A +IH
Sbjct: 1670 EQGVELLADIHEGECGAHSASRTLVGKAFRQGFYWPTALNDAVDLVRRCRACQFHAKQIH 1729
Query: 1622 VPPHALNVISSPWPFSMVGIDMIGRIEPKASNGHRFILVAIDYFTKWVEAASYTNVTKQV 1681
P AL +I WPF++ G+D++G +A G ++ VAID FTKW EA + K
Sbjct: 1730 QPAQALQIIPLSWPFAVWGLDILGPFR-RAPGGFEYLYVAIDKFTKWPEAYPVIKIDKHS 1788
Query: 1682 VAKFIKNNIICRYGVPSKIITDNGTNLNNNVVQALCEEFKIEHHNSSPYRPQINGVVEAA 1741
KFIK I R+GVP++IITDNGT + + CE+ I+ +SP P+ NG VE A
Sbjct: 1789 ALKFIK-GITARFGVPNRIITDNGTQFTSELFGDYCEDMGIKLCFASPAHPRSNGQVERA 1847
Query: 1742 N----KNIKRIVQKMVTTYRD-WHEMLPYALHGYRTTVRSSTGATPFSLVYGMEAVLPLE 1796
N K +K ++ + D W E LP L RTT +T TPF LVYG EAVLP E
Sbjct: 1848 NAEILKGLKTKTFNILKKHGDSWIEELPVVLWANRTTPSRATRETPFFLVYGAEAVLPSE 1907
Query: 1797 VEIPSLRVIMEAKLSEAEWCQSRYDQLNLVEEKRMDAMARGQSYQARMKTAFDKKVHPRE 1856
+ + S R M EA+ Q R D L+ +EE+R A R YQ ++ + V R
Sbjct: 1908 LTLRSPRATM---YCEADQDQLRRDDLDYLEERRRRAALRAARYQQSLRRYHQRHVRARS 1964
Query: 1857 FKEGELVLKRRISQQPDPRGKWTPNYEGSYVVKKAFSGGALILTHMDGVELPNPVNADLV 1916
+LVL+R Q K +P +EG Y V G++ L DG ELPNP N + +
Sbjct: 1965 LCVDDLVLRR--VQTRAGLSKLSPMWEGPYRVIGVPRPGSIRLATGDGTELPNPWNIEHL 2022
Query: 1917 KKYF 1920
++++
Sbjct: 2023 RRFY 2026
Score = 57.0 bits (136), Expect = 6e-06
Identities = 39/137 (28%), Positives = 65/137 (46%), Gaps = 5/137 (3%)
Query: 782 PEAGKHHNLALHISVNCTPDIISNVLVDTGSSLNVMPKSTLDQLSYRGTPLRRSTFLVKA 841
P + +A+ +S + VL+D G++LNV+ + D + G LR S ++
Sbjct: 603 PCVARGGQIAMVVSPTVCNVKLGRVLIDGGAALNVLSPAAFDAIKAPGMMLRPSQPIIGV 662
Query: 842 FDGTRKSVLGEIDLPITIG-PETFL---ITFQVMDINASYNCLLGRP*IHDAGAVTSTLH 897
G LG IDLP+T G P F + F V D++ YN +LGRP + A +
Sbjct: 663 TPG-HTWPLGHIDLPVTFGGPANFRTERVNFDVADLSLPYNAVLGRPALVKFMAAVHYAY 721
Query: 898 QKLKFAKSGKLVTIHGE 914
++K +++HG+
Sbjct: 722 LQMKMPCPDSPISVHGD 738
Score = 47.4 bits (111), Expect = 0.004
Identities = 70/350 (20%), Positives = 138/350 (39%), Gaps = 24/350 (6%)
Query: 73 TPSTAAGTSGTANNVLIP--ATNAVSINATLPQTTAAVTEPLVHAIPQGVNINTQHGSIP 130
T ++ A T+G+A++ A AV +A P +T + E L +I +
Sbjct: 129 TKASGAATTGSASSRRRARRAAAAVRRSAATPSSTPSTREDLRGEQDARASIERRRNDRS 188
Query: 131 VTKTMEEMMEELAKELRHEIKANRGNADSFKTQD-------LCLVSKVDVPKKFKIPDFD 183
E A R + RGN S + + V P +F+ +
Sbjct: 189 AAHATEG-----ASSSRVSPRHGRGNQPSVPPVGGVGCRAFVASLRNVRWPPRFRPTIAE 243
Query: 184 RYNGLTCPQNHIIKYVRKMGNYKDNDSLMIHCFQDSLMEDAAEWYTSLSKNDIHTFDELA 243
+Y+G P + Y + +D +M + F +L A W +L +H++++L
Sbjct: 244 KYDGSVNPAEFLQVYTTGIEAAGGDDRVMANFFPMTLKGQARGWLMNLPPASVHSWEDLC 303
Query: 244 AAFKSHYGFNTRLKPNREF-LRSLSQKKEESFREYAQRWRGAAARITPALDEEEMTQTFL 302
F ++ T +P E L ++ ++ +ES R Y QR+ I P + + F
Sbjct: 304 QQFTMNFQ-GTYPRPGEEADLHAVQRRDDESLRSYIQRFCQVRNTI-PCIPAHAVIYAFR 361
Query: 303 KTLKKD-YVERMIIAAPNNFSEMVTMGTRLEEAVREGIIVFEKAESSVNASKRYGN---- 357
++ + +E++ P +E+ + R+ A +E + + S V AS G+
Sbjct: 362 GGVRHNRMLEKIASKEPQTTAELFQLADRV--ARKEEAWTWNPSGSGVAASAAPGSAAQA 419
Query: 358 GHHKKKETEVGMVSAGAGQSMATVAPINATQMPPSYPYQKQQLPVQQQQQ 407
G ++ + + S G +A AT+ +K++ V +++
Sbjct: 420 GRRDRRRKKRSVRSDDEGHVLAVEGASRATRKGRPASDKKKEASVPSRER 469
>UniRef100_Q7XL20 OSJNBa0079M09.11 protein [Oryza sativa]
Length = 2028
Score = 380 bits (975), Expect = e-103
Identities = 238/738 (32%), Positives = 375/738 (50%), Gaps = 51/738 (6%)
Query: 1205 IYYLSKKLTDCETRYTMLEKTCCALAWAAKRLRHYLVNHTTWLISRMDPIKYIFEKAVVT 1264
+Y++S+ L D +TRY +K A+ A+++LRHY H +++ P+ I T
Sbjct: 1319 VYFVSEALRDAKTRYPQAQKMLYAILMASRKLRHYFQAHRVTVVTSY-PLGQILHNREGT 1377
Query: 1265 GKIARWQVLLSEYDIVFKTQKAIKGSILADHPAYQPLDDYQPIESDFPNEEIVYLKSKDC 1324
G++ +W + LSE+D+ F+ + AIK LAD A P E V +
Sbjct: 1378 GRVVKWAIELSEFDLHFEPRHAIKSQALADFVAEWT-----------PAPETVSIPEAST 1426
Query: 1325 EEPLIGEGPDPNSKWGLVFDGAANAYGKGIGAVIVSPQGHHIPFTAQILFECTNNMVEYE 1384
+ + + + W + FDG+ + G G G ++SP G + + ++ F TNNM EYE
Sbjct: 1427 DPSQLPQ----TAHWVMQFDGSLSLQGAGAGVTLISPNGDVLRYLVRLDFRATNNMAEYE 1482
Query: 1385 ACIFGIEEAIGMRIKHLDIYGDSALIINQIKGEWETHHAKLIPYRDYARRLLTYFTKVEL 1444
+ G+ A G+ I+ L + GDS L++NQ+ E+ ++ Y RR+ +F +EL
Sbjct: 1483 GLLAGLRVAAGLGIRRLLVLGDSQLVVNQVCKEYRCSDPQMDAYVRQVRRMERHFDGIEL 1542
Query: 1445 HHIPRDENQMADALATLSSMFRVNHWNDVPIIKVQRLERPS--------------HVFTI 1490
H+PR +N +AD L+ L+S + P +RL +PS H +
Sbjct: 1543 RHVPRRDNMIADELSRLAS----SRAQTPPGAFEERLTQPSARPDPSGETDAPDRHPRPV 1598
Query: 1491 GDVIDQAGENMVDNK---PWYYDIKQCLLSREYPPGASNKDKKTLRRLASRFLLDGDILY 1547
G E++ + W +I+ L + P ++ ++R++ R++L LY
Sbjct: 1599 GVQASGPEESVPSSLRLIAWISEIQTYLTDKTLPEDREGSER--VQRISKRYVLVEGTLY 1656
Query: 1548 KRNYDMVLLRCVDEHEAEQLMHDVHDGTFGTHATGHTMSRKLLRAGYYWLTMEHDCYQHA 1607
+R + +LL+C+ + + +L+ D+H+G G H+ T+ K R G+YW T +D
Sbjct: 1657 RRAANGILLKCIPQEQGVELLADIHEGECGAHSASRTLVGKAFRQGFYWPTALNDAVDLV 1716
Query: 1608 RKCHKCQIYADKIHVPPHALNVISSPWPFSMVGIDMIGRIEPKASNGHRFILVAIDYFTK 1667
R+C CQ +A +IH P AL +I WPF++ G+D++G + +A G ++ VAID FTK
Sbjct: 1717 RRCRACQFHAKQIHQPAQALQIIPLSWPFAVWGLDILGPFK-RAPGGFEYLYVAIDKFTK 1775
Query: 1668 WVEAASYTNVTKQVVAKFIKNNIICRYGVPSKIITDNGTNLNNNVVQALCEEFKIEHHNS 1727
W EA + K KFIK I R+GVP++IITDNGT + + CE+ I+ +
Sbjct: 1776 WPEAYPVVKIDKHSALKFIK-GITARFGVPNRIITDNGTQFTSELFGDYCEDMGIKLCFA 1834
Query: 1728 SPYRPQINGVVEAAN----KNIKRIVQKMVTTYRD-WHEMLPYALHGYRTTVRSSTGATP 1782
SP P+ NG VE AN K +K ++ + D W E LP L RTT +TG TP
Sbjct: 1835 SPAHPRSNGQVERANAEILKGLKTKTFNILKKHGDSWIEELPAVLWANRTTPSRATGETP 1894
Query: 1783 FSLVYGMEAVLPLEVEIPSLRVIMEAKLSEAEWCQSRYDQLNLVEEKRMDAMARGQSYQA 1842
F LVYG EAVLP E+ + S R M EA+ Q R D L+ +EE+R A R YQ
Sbjct: 1895 FFLVYGAEAVLPSELTLRSPRATM---YCEADQDQLRRDDLDYLEERRRRAALRAARYQQ 1951
Query: 1843 RMKTAFDKKVHPREFKEGELVLKRRISQQPDPRGKWTPNYEGSYVVKKAFSGGALILTHM 1902
++ + V R +LVL+R Q K +P +EG Y V G++ L
Sbjct: 1952 SLRRYHQRHVRARSLCVDDLVLRR--VQTRAGLSKLSPIWEGPYRVIGVPRPGSVRLATG 2009
Query: 1903 DGVELPNPVNADLVKKYF 1920
DG ELPNP N + +++++
Sbjct: 2010 DGTELPNPWNIEHLRRFY 2027
Score = 54.3 bits (129), Expect = 4e-05
Identities = 34/116 (29%), Positives = 59/116 (50%), Gaps = 5/116 (4%)
Query: 803 ISNVLVDTGSSLNVMPKSTLDQLSYRGTPLRRSTFLVKAFDGTRKSVLGEIDLPITIG-P 861
+ VL+D G++LN++ + D + G L+ S ++ G LG IDLP+T G P
Sbjct: 625 LGRVLIDGGAALNILSPAAFDAIKAPGMVLQPSQPIIGVTPG-HTWPLGHIDLPVTFGGP 683
Query: 862 ETFL---ITFQVMDINASYNCLLGRP*IHDAGAVTSTLHQKLKFAKSGKLVTIHGE 914
F ++F V D++ YN +LGRP + A + ++K G +++ G+
Sbjct: 684 ANFRTERVSFDVADLSLPYNAVLGRPALVKFMAAVHYAYLQMKMPGPGGPISVRGD 739
>UniRef100_Q60D74 Putative polyprotein [Oryza sativa]
Length = 1869
Score = 379 bits (973), Expect = e-103
Identities = 245/751 (32%), Positives = 373/751 (49%), Gaps = 59/751 (7%)
Query: 1178 RLLEQEGRAIQPHQEEIEIINLGIPTAIYYLSKKLTDCETRYTMLEKTCCALAWAAKRLR 1237
RL E+ R P +E+ + P +Y++S+ L D +TRY +K A+ A+++LR
Sbjct: 1169 RLGEERPRDAAPSEEDRPHRKVQRP--VYFVSEALRDAKTRYPQAQKMLYAILMASRKLR 1226
Query: 1238 HYLVNHTTWLISRMDPIKYIFEKAVVTGKIARWQVLLSEYDIVFKTQKAIKGSILADHPA 1297
HY H +++ P+ I TG++ +W + LSE+D+ F+ + AIK LAD A
Sbjct: 1227 HYFQAHRVTVVTSY-PLGQILHNREGTGRVVKWAIELSEFDLHFEPRHAIKSQALADFVA 1285
Query: 1298 YQPLDDYQPIESDFPNEEIVYLKSKDCEEPLIGEGPDP---NSKWGLVFDGAANAYGKGI 1354
P E+V + P GP + W + FDG+ + G G
Sbjct: 1286 EWT-----------PAPELVSV-------PEASSGPSQLPHTAHWVMQFDGSLSLQGAGA 1327
Query: 1355 GAVIVSPQGHHIPFTAQILFECTNNMVEYEACIFGIEEAIGMRIKHLDIYGDSALIINQI 1414
G + SP G + + ++ F TNNM EYE + G+ A G+ I+ L + GDS L++NQ+
Sbjct: 1328 GVTLTSPSGDVLRYLVRLDFRATNNMAEYEGLLAGLRVAAGLGIRRLLVLGDSQLVVNQV 1387
Query: 1415 KGEWETHHAKLIPYRDYARRLLTYFTKVELHHIPRDENQMADALATLSSMFRVNHWNDVP 1474
E+ ++ Y RR+ +F +EL H+PR +N MAD L+ L+S + P
Sbjct: 1388 CKEYRCSDPQMDAYVRQVRRMERHFDGIELRHVPRRDNMMADELSRLAS----SRAQTPP 1443
Query: 1475 IIKVQRLERPSHVFTIGDVIDQAGENMVDNKPWYYDIKQCLLSREYPPGASNKDKKTLRR 1534
+RL +PS + W +I+ L + P ++ + R
Sbjct: 1444 GAFEERLAQPSALIA-----------------WIAEIQAYLTDKTLPEDREGSER--VHR 1484
Query: 1535 LASRFLLDGDILYKRNYDMVLLRCVDEHEAEQLMHDVHDGTFGTHATGHTMSRKLLRAGY 1594
++ R++L LY+R + +LL+C+ + L+ D+H+G G H+ T+ K R G+
Sbjct: 1485 ISKRYVLVEGTLYRRAANGILLKCIPREQGVVLLADIHEGECGAHSASRTLVGKAFRQGF 1544
Query: 1595 YWLTMEHDCYQHARKCHKCQIYADKIHVPPHALNVISSPWPFSMVGIDMIGRIEPKASNG 1654
YW T +D R+C CQ +A +IH P AL I WPF++ G+ ++G +A G
Sbjct: 1545 YWPTALNDAVDLVRRCRACQFHAKQIHQPAQALQTIPLSWPFAVWGLHILGPFR-RAPGG 1603
Query: 1655 HRFILVAIDYFTKWVEAASYTNVTKQVVAKFIKNNIICRYGVPSKIITDNGTNLNNNVVQ 1714
++ VAID FTKW EA + K KFIK I R+GVP++IITDNGT + +
Sbjct: 1604 FEYLYVAIDKFTKWPEAYPVVKIDKHSALKFIK-GITARFGVPNRIITDNGTQFTSELFG 1662
Query: 1715 ALCEEFKIEHHNSSPYRPQINGVVEAAN----KNIKRIVQKMVTTYRD-WHEMLPYALHG 1769
CE+ I+ +SP P+ NG VE AN K +K ++ + D W E LP L
Sbjct: 1663 DYCEDMGIKLCFASPAHPRSNGQVERANAEILKGLKTKTFNILKKHGDSWIEELPAVLWA 1722
Query: 1770 YRTTVRSSTGATPFSLVYGMEAVLPLEVEIPSLRVIMEAKLSEAEWCQSRYDQLNLVEEK 1829
RTT +TG TPF LVYG EAVLP E+ + S R M EA+ Q R D L+ +EE+
Sbjct: 1723 NRTTPSRATGETPFFLVYGAEAVLPSELTLKSPRATM---YCEADQDQLRRDDLDYLEER 1779
Query: 1830 RMDAMARGQSYQARMKTAFDKKVHPREFKEGELVLKRRISQQPDPRGKWTPNYEGSYVVK 1889
R A R YQ ++ V R +LVL+R Q K +P +EG Y V
Sbjct: 1780 RRRAALRAARYQQSLRRYHQCHVRARSLCVDDLVLRR--VQTRAGLSKLSPMWEGPYRVV 1837
Query: 1890 KAFSGGALILTHMDGVELPNPVNADLVKKYF 1920
G++ L DG ELPNP N + +++++
Sbjct: 1838 GVPRPGSIRLATGDGTELPNPWNIEHLRRFY 1868
Score = 56.2 bits (134), Expect = 1e-05
Identities = 38/137 (27%), Positives = 65/137 (46%), Gaps = 5/137 (3%)
Query: 782 PEAGKHHNLALHISVNCTPDIISNVLVDTGSSLNVMPKSTLDQLSYRGTPLRRSTFLVKA 841
P + +A+ +S + VL+D G++LN++ + D + G LR S ++
Sbjct: 476 PCVARGGQIAMVVSPTVCNVKLGRVLIDGGAALNILSPAAFDAIKALGMMLRPSQPIIGV 535
Query: 842 FDGTRKSVLGEIDLPITIG-PETFL---ITFQVMDINASYNCLLGRP*IHDAGAVTSTLH 897
G LG IDLP+T G P F + F V D++ YN +LGRP + A +
Sbjct: 536 TPG-HTWPLGHIDLPVTFGGPANFRTERVNFDVADLSPPYNAVLGRPALVKFMAAVHYAY 594
Query: 898 QKLKFAKSGKLVTIHGE 914
++K +++HG+
Sbjct: 595 LQMKMPGPDGPISVHGD 611
Score = 47.0 bits (110), Expect = 0.006
Identities = 51/240 (21%), Positives = 102/240 (42%), Gaps = 12/240 (5%)
Query: 174 PKKFKIPDFDRYNGLTCPQNHIIKYVRKMGNYKDNDSLMIHCFQDSLMEDAAEWYTSLSK 233
P +F+ ++Y+G P + Y + +D +M + F +L A W +L
Sbjct: 107 PPRFRPTIAEKYDGSVNPAEFLQVYTTGIEAAGGDDRVMANFFPMALKGQARGWLMNLPP 166
Query: 234 NDIHTFDELAAAFKSHYGFNTRLKPNREF-LRSLSQKKEESFREYAQRWRGAAARITPAL 292
+H++++L F ++ T +P E L ++ ++ +ES R Y QR+ I P +
Sbjct: 167 ASVHSWEDLCQQFTMNFQ-GTYPRPGEEADLHAVQRRDDESLRSYIQRFCQVRNTI-PCI 224
Query: 293 DEEEMTQTFLKTLKKD-YVERMIIAAPNNFSEMVTMGTRLEEAVREGIIVFEKAESSVNA 351
+ F ++ + +E++ P +E+ + R+ A +E + + S V A
Sbjct: 225 PAHAVIYAFRGGVRHNRMLEKIASKEPQTTAELFQLADRV--ARKEEAWTWNPSGSGVAA 282
Query: 352 SKRYGN----GHHKKKETEVGMVSAGAGQSMATVAPINATQ--MPPSYPYQKQQLPVQQQ 405
S G+ G ++ + + S G +A ATQ P S +K P +++
Sbjct: 283 SAAPGSTAQTGRRDRRRKKRSVHSDDEGHVLAVEGASRATQKGRPASDKKKKAGAPSRER 342
>UniRef100_Q84T73 Putative polyprotein [Oryza sativa]
Length = 2026
Score = 379 bits (972), Expect = e-103
Identities = 240/744 (32%), Positives = 372/744 (49%), Gaps = 63/744 (8%)
Query: 1205 IYYLSKKLTDCETRYTMLEKTCCALAWAAKRLRHYLVNHTTWLISRMDPIKYIFEKAVVT 1264
+Y++S+ L D +TRY +K A+ A+++LRHY H +++ P+ I T
Sbjct: 1317 VYFVSEALRDAKTRYPQAQKMLYAILMASRKLRHYFQAHRVTVVTSY-PLGQILHNREGT 1375
Query: 1265 GKIARWQVLLSEYDIVFKTQKAIKGSILADHPAYQPLDDYQPIESDFPNEEIVYLKSKDC 1324
G++ +W + LSE+D+ F+ + AIK LAD A P E V +
Sbjct: 1376 GRVVKWAIELSEFDLHFEPRHAIKSQALADFVAEWT-----------PAPETVSIPEASA 1424
Query: 1325 EEPLIGEGPDPNSKWGLVFDGAANAYGKGIGAVIVSPQGHHIPFTAQILFECTNNMVEYE 1384
+ + + W + FDG+ + G G G + SP G + + ++ F TNNM EYE
Sbjct: 1425 DPSQLPHA----AHWVMQFDGSLSLQGAGAGVTLTSPNGDVLRYLVRLDFRATNNMAEYE 1480
Query: 1385 ACIFGIEEAIGMRIKHLDIYGDSALIINQIKGEWETHHAKLIPYRDYARRLLTYFTKVEL 1444
+ G+ A G+ I+ L + GDS L++NQ+ E+ ++ Y RR+ +F +EL
Sbjct: 1481 GLLAGLRVAAGLGIRRLLVLGDSQLVVNQVCKEYRCSDPQMDAYVRQVRRMERHFDGIEL 1540
Query: 1445 HHIPRDENQMADALATLSSMFRVNHWNDVPIIKVQRLERPSHVFTIGDVIDQAGENMVDN 1504
H+PR +N +AD L+ L+S + P +RL +PS D GE +
Sbjct: 1541 RHVPRRDNMIADELSRLAS----SRTQTPPGAFEERLTQPSAR------PDPLGETDAPD 1590
Query: 1505 KP-----------------------WYYDIKQCLLSREYPPGASNKDKKTLRRLASRFLL 1541
+P W +I+ L + P ++ ++R++ R++L
Sbjct: 1591 RPPRPVGVQASGPEGSAPSSLRLIAWISEIQTYLTDKTLPEDREGSER--VQRISKRYVL 1648
Query: 1542 DGDILYKRNYDMVLLRCVDEHEAEQLMHDVHDGTFGTHATGHTMSRKLLRAGYYWLTMEH 1601
LY+R + +LL+C+ + + +L+ D+H+G G H+ T+ K R G+YW T +
Sbjct: 1649 VEGTLYRRAANGILLKCIPQEQGVELLADIHEGECGAHSASRTLVGKAFRQGFYWPTALN 1708
Query: 1602 DCYQHARKCHKCQIYADKIHVPPHALNVISSPWPFSMVGIDMIGRIEPKASNGHRFILVA 1661
D R+C CQ +A +IH P AL VI WPF++ G+D++G + +A G ++ VA
Sbjct: 1709 DAVDLVRRCRACQFHARQIHQPAQALQVIPLSWPFAVWGLDILGPFK-RAPGGFEYLYVA 1767
Query: 1662 IDYFTKWVEAASYTNVTKQVVAKFIKNNIICRYGVPSKIITDNGTNLNNNVVQALCEEFK 1721
ID FTKW EA + K KFIK I R+GVP++IITDNGT + + CE+
Sbjct: 1768 IDKFTKWPEAYPVVKIDKHSALKFIK-GITARFGVPNRIITDNGTQFTSELFGDYCEDMG 1826
Query: 1722 IEHHNSSPYRPQINGVVEAAN----KNIKRIVQKMVTTYRD-WHEMLPYALHGYRTTVRS 1776
I+ +SP P+ NG VE AN K +K ++ + D W E LP L RTT
Sbjct: 1827 IKLCFASPAHPRSNGQVERANAEILKGLKTKTFNILKKHGDSWIEELPVVLWANRTTPSR 1886
Query: 1777 STGATPFSLVYGMEAVLPLEVEIPSLRVIMEAKLSEAEWCQSRYDQLNLVEEKRMDAMAR 1836
+TG TPF LVYG EAVLP E+ + S R M EA+ Q R D L+ +EE+R A R
Sbjct: 1887 ATGETPFFLVYGAEAVLPSELTLRSPRATM---YCEADQDQLRRDDLDYLEERRRRAALR 1943
Query: 1837 GQSYQARMKTAFDKKVHPREFKEGELVLKRRISQQPDPRGKWTPNYEGSYVVKKAFSGGA 1896
YQ ++ + V R +LVL+R Q K +P +EG Y V G+
Sbjct: 1944 AARYQQSLRRYHQRHVRARSLCVDDLVLRR--VQTRAGLSKLSPMWEGPYRVIGVPRPGS 2001
Query: 1897 LILTHMDGVELPNPVNADLVKKYF 1920
+ L DG ELPNP N + +++++
Sbjct: 2002 VRLATGDGTELPNPWNIEHLRRFY 2025
Score = 57.8 bits (138), Expect = 3e-06
Identities = 36/116 (31%), Positives = 59/116 (50%), Gaps = 5/116 (4%)
Query: 803 ISNVLVDTGSSLNVMPKSTLDQLSYRGTPLRRSTFLVKAFDGTRKSVLGEIDLPITIG-P 861
+ VL+D G++LN++ D + G LR S ++ G LG IDLP+T G P
Sbjct: 623 LGRVLIDGGAALNILSPVAFDAIKAPGMVLRPSQPIIGVTPG-HTWPLGHIDLPVTFGGP 681
Query: 862 ETFL---ITFQVMDINASYNCLLGRP*IHDAGAVTSTLHQKLKFAKSGKLVTIHGE 914
F ++F V D++ YN +LGRP + A + ++K G +++HG+
Sbjct: 682 ANFRTERVSFDVADLSLPYNAVLGRPALVKFMAAVHYAYLQMKMPGPGGPISVHGD 737
>UniRef100_Q6L5D6 Putative polyprotein [Oryza sativa]
Length = 1514
Score = 378 bits (970), Expect = e-102
Identities = 237/724 (32%), Positives = 366/724 (49%), Gaps = 62/724 (8%)
Query: 1205 IYYLSKKLTDCETRYTMLEKTCCALAWAAKRLRHYLVNHTTWLISRMDPIKYIFEKAVVT 1264
+Y++S+ L D +TRY +K A+ A+++LRHY H +++ P+ I T
Sbjct: 844 VYFVSEALRDAKTRYPQAQKMLYAILMASRKLRHYFQAHRVTVVTSY-PLGQILHNREGT 902
Query: 1265 GKIARWQVLLSEYDIVFKTQKAIKGSILADHPAYQPLDDYQPIESDFPNEEIVYLKSKDC 1324
G++ RW + LSE+D+ F+ + AIK LAD A ++ P +
Sbjct: 903 GRVVRWAIELSEFDLRFEPRHAIKSQALADFVA-----EWTPAP-------------EPV 944
Query: 1325 EEPLIGEGPDP---NSKWGLVFDGAANAYGKGIGAVIVSPQGHHIPFTAQILFECTNNMV 1381
P GP + W + FDG+ + G G G + SP G + + ++ F TNNM
Sbjct: 945 SAPEASSGPTQLPHTAYWVMQFDGSLSLQGAGAGVTLTSPSGDVLRYLVRLDFRATNNMA 1004
Query: 1382 EYEACIFGIEEAIGMRIKHLDIYGDSALIINQIKGEWETHHAKLIPYRDYARRLLTYFTK 1441
EYE + G+ A G+ I+ L + GDS L++NQ+ E++ ++ Y RR+ +F +
Sbjct: 1005 EYEGLLAGLRVAAGLGIRRLLVLGDSQLVVNQVCKEYQCSDPQMDAYVRQVRRMERHFDR 1064
Query: 1442 VELHHIPRDENQMADALATLSSMFRVNHWNDVPIIKVQRLERPSHVFTIGDVIDQAGENM 1501
+EL H+PR +N +AD L+ L+S R + P F ++ + +
Sbjct: 1065 IELRHVPRRDNAVADELSRLAS---------------SRAQTPPGAFE-----ERLAQPL 1104
Query: 1502 VDNKPWYYDIKQCLLSREYPPGASNKDKKTLRRLASRFLLDGDILYKRNYDMVLLRCVDE 1561
+ W +I+ L + P ++ +RR++ R++L LY+R + VLL+C+
Sbjct: 1105 I---AWITEIQAYLADKTLPEDREGSER--VRRISKRYVLVEGTLYRRAANGVLLKCIPR 1159
Query: 1562 HEAEQLMHDVHDGTFGTHATGHTMSRKLLRAGYYWLTMEHDCYQHARKCHKCQIYADKIH 1621
+ +L+ DVH+G G H+ T+ K R G+YW T +D R+C CQ +A + H
Sbjct: 1160 EQGVELLADVHEGECGAHSVSRTLVGKAFRQGFYWPTALNDAVDLVRRCRACQFHARQTH 1219
Query: 1622 VPPHALNVISSPWPFSMVGIDMIGRIEPKASNGHRFILVAIDYFTKWVEAASYTNVTKQV 1681
P AL I WPF++ G+D++GR A G ++ VA+D FTKW EA + K
Sbjct: 1220 QPAQALQTIPLSWPFAVWGLDILGR----APGGFEYLYVAVDKFTKWPEAYPVIKIDKHS 1275
Query: 1682 VAKFIKNNIICRYGVPSKIITDNGTNLNNNVVQALCEEFKIEHHNSSPYRPQINGVVEAA 1741
KFI+ I R+GVP++IITDNGT + + CE+ I+ +SP P+ NG VE A
Sbjct: 1276 ALKFIR-GITARFGVPNRIITDNGTQFTSELFGDYCEDMGIKLCFASPTHPRSNGQVERA 1334
Query: 1742 N----KNIKRIVQKMVTTYRD-WHEMLPYALHGYRTTVRSSTGATPFSLVYGMEAVLPLE 1796
N K +K ++ + D W E LP L RTT +TG TPF LVYG EAVLP E
Sbjct: 1335 NAEILKGLKTKTFNILKKHGDSWIEELPAVLWANRTTPSRATGETPFFLVYGAEAVLPSE 1394
Query: 1797 VEIPSLRVIMEAKLSEAEWCQSRYDQLNLVEEKRMDAMARGQSYQARMKTAFDKKVHPRE 1856
+ + S RV M EA+ Q R D L+ +EE+R A R YQ ++ + V R
Sbjct: 1395 LTLRSPRVTM---YCEADQDQLRRDDLDYLEERRRRAALRAARYQQSLRRYHQRHVRARS 1451
Query: 1857 FKEGELVLKRRISQQPDPRGKWTPNYEGSYVVKKAFSGGALILTHMDGVELPNPVNADLV 1916
+LVL+R Q K +P +EG Y V G++ L DG ELPNP N + +
Sbjct: 1452 LCVDDLVLRR--VQTRAGLSKLSPMWEGPYRVIGVPRPGSVRLATGDGTELPNPWNIEHL 1509
Query: 1917 KKYF 1920
+++
Sbjct: 1510 CRFY 1513
Score = 60.1 bits (144), Expect = 7e-07
Identities = 41/146 (28%), Positives = 69/146 (47%), Gaps = 6/146 (4%)
Query: 774 LWFSENELPEA-GKHHNLALHISVNCTPDIISNVLVDTGSSLNVMPKSTLDQLSYRGTPL 832
L F + + P + +A+ +S + VL+D G++LN++ + D + G L
Sbjct: 580 LTFDQTDHPSCVARGGQIAMVVSPTVCNVKLGRVLIDGGAALNILSPAAFDAIKAPGMVL 639
Query: 833 RRSTFLVKAFDGTRKSVLGEIDLPITIGPE----TFLITFQVMDINASYNCLLGRP*IHD 888
R S ++ G LG IDLP+T G T + F V D++ YN +LGRP +
Sbjct: 640 RPSQPIIGVTPG-HTWPLGHIDLPVTFGGSANFRTERVDFDVADLSLPYNAVLGRPALVK 698
Query: 889 AGAVTSTLHQKLKFAKSGKLVTIHGE 914
A + ++K G +T+HG+
Sbjct: 699 FMAAVHYAYLQMKMPGPGGPITVHGD 724
>UniRef100_Q8S0W5 Putative polyprotein [Oryza sativa]
Length = 2030
Score = 377 bits (968), Expect = e-102
Identities = 237/738 (32%), Positives = 372/738 (50%), Gaps = 51/738 (6%)
Query: 1205 IYYLSKKLTDCETRYTMLEKTCCALAWAAKRLRHYLVNHTTWLISRMDPIKYIFEKAVVT 1264
+Y++S+ L D +TRY +K A+ A+++LRHY H +++ P+ I T
Sbjct: 1321 VYFISETLRDAKTRYPQAQKMLYAILMASRKLRHYFQAHRVTVVTSY-PLGQILHNREGT 1379
Query: 1265 GKIARWQVLLSEYDIVFKTQKAIKGSILADHPAYQPLDDYQPIESDFPNEEIVYLKSKDC 1324
G++ +W + LSE+D+ F+ + AIK LAD A P E V +
Sbjct: 1380 GRVVKWAIELSEFDLHFEPRHAIKSQALADFVAEWT-----------PAPETVSMPEAST 1428
Query: 1325 EEPLIGEGPDPNSKWGLVFDGAANAYGKGIGAVIVSPQGHHIPFTAQILFECTNNMVEYE 1384
+ + + W + FDG+ + G G G + SP G + + ++ F TNNM EYE
Sbjct: 1429 NLSQLPQ----TAHWVMQFDGSLSLQGAGAGVTLTSPNGDVLRYLVRLDFRATNNMAEYE 1484
Query: 1385 ACIFGIEEAIGMRIKHLDIYGDSALIINQIKGEWETHHAKLIPYRDYARRLLTYFTKVEL 1444
+ G+ A G+ I+ L + GDS L++NQ+ E+ ++ Y RR+ +F +EL
Sbjct: 1485 GLLAGLRVAAGLGIRRLLVLGDSQLVVNQVCKEYRCSDPQMNAYVRQVRRMERHFDGIEL 1544
Query: 1445 HHIPRDENQMADALATLSSMFRVNHWNDVPIIKVQRLERPSH-----------------V 1487
H+PR +N +AD L+ L+S + P +RL +PS V
Sbjct: 1545 RHVPRRDNMIADELSRLAS----SRAQTPPGAFEERLTQPSARPNPLGETDAPDRPPRAV 1600
Query: 1488 FTIGDVIDQAGENMVDNKPWYYDIKQCLLSREYPPGASNKDKKTLRRLASRFLLDGDILY 1547
++ + N + W +I+ L + P ++ ++R++ R++L LY
Sbjct: 1601 GVQASGLEGSAPNSLRLIAWISEIQTYLTDKTLPEDRQGSER--VQRISKRYVLVEGTLY 1658
Query: 1548 KRNYDMVLLRCVDEHEAEQLMHDVHDGTFGTHATGHTMSRKLLRAGYYWLTMEHDCYQHA 1607
+R + +LL+C+ + + +L+ D+H+G G H+ T+ K R G+YW T +D
Sbjct: 1659 RRAANGILLKCIPQEQGVELLADIHEGECGAHSASRTLVGKAFRQGFYWPTALNDAVDLV 1718
Query: 1608 RKCHKCQIYADKIHVPPHALNVISSPWPFSMVGIDMIGRIEPKASNGHRFILVAIDYFTK 1667
R+C CQ +A +IH P AL +I WPF++ G+D++G + +A G ++ VAID FTK
Sbjct: 1719 RRCRACQFHAKQIHQPAQALQIIPLSWPFAVWGLDILGPFK-RALGGFEYLYVAIDKFTK 1777
Query: 1668 WVEAASYTNVTKQVVAKFIKNNIICRYGVPSKIITDNGTNLNNNVVQALCEEFKIEHHNS 1727
W EA + K KFIK I R+GVP++IITDNGT + + CE+ I+ +
Sbjct: 1778 WPEAYPVVKIDKHSALKFIK-GITARFGVPNRIITDNGTQFTSELFGDYCEDMGIKLCFA 1836
Query: 1728 SPYRPQINGVVEAAN----KNIKRIVQKMVTTYRD-WHEMLPYALHGYRTTVRSSTGATP 1782
SP P+ NG VE AN K +K ++ + D W E LP L RTT +TG TP
Sbjct: 1837 SPAHPRSNGQVERANAEILKGLKTKTFNILKKHGDSWIEELPAVLWANRTTPSRATGETP 1896
Query: 1783 FSLVYGMEAVLPLEVEIPSLRVIMEAKLSEAEWCQSRYDQLNLVEEKRMDAMARGQSYQA 1842
F LVYG EAVLP E+ + S R M EA+ Q R D L+ +EE+R A R YQ
Sbjct: 1897 FFLVYGAEAVLPSELTLRSPRATM---YCEADQDQLRRDDLDYLEERRRRAALRAARYQQ 1953
Query: 1843 RMKTAFDKKVHPREFKEGELVLKRRISQQPDPRGKWTPNYEGSYVVKKAFSGGALILTHM 1902
++ + V R +LVL+R Q K +P +EG Y V G++ L
Sbjct: 1954 SLRRYHQRHVRARSLCVDDLVLRR--VQTRAGLSKLSPIWEGPYRVIGVPRPGSVRLATG 2011
Query: 1903 DGVELPNPVNADLVKKYF 1920
DG ELPNP N + +++++
Sbjct: 2012 DGTELPNPWNIEHLRRFY 2029
Score = 58.9 bits (141), Expect = 1e-06
Identities = 36/116 (31%), Positives = 60/116 (51%), Gaps = 5/116 (4%)
Query: 803 ISNVLVDTGSSLNVMPKSTLDQLSYRGTPLRRSTFLVKAFDGTRKSVLGEIDLPITIG-P 861
+ VL+D G++LN++ + D + G LR S ++ G LG IDLP+T G P
Sbjct: 624 LGRVLIDGGAALNILSPAAFDAIKAPGMVLRPSQPIIGVTPG-HTWPLGHIDLPVTFGGP 682
Query: 862 ETFL---ITFQVMDINASYNCLLGRP*IHDAGAVTSTLHQKLKFAKSGKLVTIHGE 914
F ++F V D++ YN +LGRP + A + ++K G +++HG+
Sbjct: 683 ANFRTERVSFDVADLSLPYNAVLGRPALVKFMAAVHYAYLQMKMPGPGGPISVHGD 738
>UniRef100_Q84JT1 Putative polyprotein [Oryza sativa]
Length = 1911
Score = 372 bits (955), Expect = e-101
Identities = 239/741 (32%), Positives = 374/741 (50%), Gaps = 57/741 (7%)
Query: 1205 IYYLSKKLTDCETRYTMLEKTCCALAWAAKRLRHYLVNHTTWLISRMDPIKYIFEKAVVT 1264
+Y++S+ L D +TRY +K A+ A+++LRHY H +++ P+ I T
Sbjct: 1202 VYFVSEALRDAKTRYPQAQKMLYAILMASRKLRHYFQAHRVTVVTSY-PLGQILHNREGT 1260
Query: 1265 GKIARWQVLLSEYDIVFKTQKAIKGSILADHPAYQPLDDYQPIESDFPNEEIVYLKSKDC 1324
G++ RW + LSE+D+ F+++ AIK LAD A ++ P +
Sbjct: 1261 GRVVRWTIELSEFDLRFESRHAIKSQALADFVA-----EWTPAP-------------EPV 1302
Query: 1325 EEPLIGEGPDP---NSKWGLVFDGAANAYGKGIGAVIVSPQGHHIPFTAQILFECTNNMV 1381
P GP + W + FDG+ + G G G + SP G + + ++ F TNNM
Sbjct: 1303 SAPEASSGPSQLPHTAYWVMQFDGSLSLQGAGAGVTMTSPSGDVLRYLVRLDFRATNNMA 1362
Query: 1382 EYEACIFGIEEAIGMRIKHLDIYGDSALIINQIKGEWETHHAKLIPYRDYARRLLTYFTK 1441
EYE + G+ A G+ I+ L + GDS L++NQ+ E++ ++ Y RR+ +F
Sbjct: 1363 EYEGLLAGLRVAAGLGIRRLLVLGDSQLVVNQVCKEYQCSDPQMDAYVRQVRRMERHFDG 1422
Query: 1442 VELHHIPRDENQMADALATLSSMFRVNHWNDVPIIKVQRLERPS-HVFTIGDV------- 1493
+EL H+PR +N +AD L+ L+S + P + +RL +PS +G+
Sbjct: 1423 IELRHVPRRDNAVADELSRLAS----SRAQTPPGVFEERLAQPSARPDPLGETDAPERPP 1478
Query: 1494 ----IDQAG-ENMVDNKP----WYYDIKQCLLSREYPPGASNKDKKTLRRLASRFLLDGD 1544
+ +G E + P W +I+ L+ + P ++ +RR++ R++L
Sbjct: 1479 RPVGVQASGPEGSAPSSPRLIAWITEIQAYLVDKTLPEDREGSER--VRRISKRYVLVEG 1536
Query: 1545 ILYKRNYDMVLLRCVDEHEAEQLMHDVHDGTFGTHATGHTMSRKLLRAGYYWLTMEHDCY 1604
LY+R + VLL+C+ + +L+ DVH+G G H+ T+ K R G+YW T +D
Sbjct: 1537 TLYRRAANGVLLKCIPREQGVELLADVHEGECGAHSASRTLVGKAFRQGFYWPTALNDAV 1596
Query: 1605 QHARKCHKCQIYADKIHVPPHALNVISSPWPFSMVGIDMIGRIEPKASNGHRFILVAIDY 1664
R+C CQ +A + H AL I WPF++ G D++G +A G ++ V +D
Sbjct: 1597 DLVRRCKACQFHARQTHQLAQALQTIPLSWPFAVWGFDILGPFR-RAPGGFEYLYVTVDK 1655
Query: 1665 FTKWVEAASYTNVTKQVVAKFIKNNIICRYGVPSKIITDNGTNLNNNVVQALCEEFKIEH 1724
FTKW +A + K KFI+ I R+GVP++IITDNGT + + CE+ I+
Sbjct: 1656 FTKWPQAYPVIKIDKHSALKFIR-GITARFGVPNRIITDNGTQFTSELFGDYCEDMGIKL 1714
Query: 1725 HNSSPYRPQINGVVEAAN----KNIKRIVQKMVTTYRD-WHEMLPYALHGYRTTVRSSTG 1779
+SP P+ NG VE AN K +K ++ + D W E LP L RTT +TG
Sbjct: 1715 CFASPAHPRSNGQVERANAEILKGLKTKTFNILKKHGDSWIEELPAVLWANRTTPSRATG 1774
Query: 1780 ATPFSLVYGMEAVLPLEVEIPSLRVIMEAKLSEAEWCQSRYDQLNLVEEKRMDAMARGQS 1839
TPF LVYG EAVLP E+ + S RV M EA+ Q R D L+ +EE+R A R
Sbjct: 1775 ETPFFLVYGAEAVLPSELTLRSPRVTM---YCEADQDQLRRDDLDYLEERRRRAALRAAR 1831
Query: 1840 YQARMKTAFDKKVHPREFKEGELVLKRRISQQPDPRGKWTPNYEGSYVVKKAFSGGALIL 1899
YQ ++ + V R +LVL+R Q K +P +EG Y V G++ L
Sbjct: 1832 YQQSLRRYHQRHVRARSLCVDDLVLRR--VQTRAGLSKLSPMWEGPYRVIGVPRPGSVRL 1889
Query: 1900 THMDGVELPNPVNADLVKKYF 1920
DG ELPNP N + +++++
Sbjct: 1890 ATGDGTELPNPWNIEHLRRFY 1910
Score = 52.8 bits (125), Expect = 1e-04
Identities = 36/137 (26%), Positives = 62/137 (44%), Gaps = 5/137 (3%)
Query: 782 PEAGKHHNLALHISVNCTPDIISNVLVDTGSSLNVMPKSTLDQLSYRGTPLRRSTFLVKA 841
P + +A+ +S + VL+D G++LN++ + + G L S ++
Sbjct: 603 PCVARGGQIAMVVSPTVCNVKLGRVLIDGGAALNILSPAAFGAIKAPGMVLWPSQPIIGV 662
Query: 842 FDGTRKSVLGEIDLPITIGPETFL----ITFQVMDINASYNCLLGRP*IHDAGAVTSTLH 897
G LG IDLP+T G + F V D++ YN +LGRP + A +
Sbjct: 663 TPG-HTWPLGHIDLPVTFGGSANFRMERVDFDVADLSLPYNAVLGRPALVKFMAAVHYAY 721
Query: 898 QKLKFAKSGKLVTIHGE 914
++K G +T+HG+
Sbjct: 722 LQMKMPGPGGPITVHGD 738
Score = 42.0 bits (97), Expect = 0.19
Identities = 26/109 (23%), Positives = 53/109 (47%), Gaps = 2/109 (1%)
Query: 174 PKKFKIPDFDRYNGLTCPQNHIIKYVRKMGNYKDNDSLMIHCFQDSLMEDAAEWYTSLSK 233
P +F+ ++Y+G P + Y + +D +M + F +L A W +L
Sbjct: 234 PPRFRPTITEKYDGNVNPAEFLQIYTTGIEATGGDDRVMANFFPMALKGQARGWLMNLPP 293
Query: 234 NDIHTFDELAAAFKSHYGFNTRLKPNREF-LRSLSQKKEESFREYAQRW 281
+H++++L F +++ T +P E L ++ ++ +ES R Y QR+
Sbjct: 294 ASVHSWEDLCQQFTTNFQ-GTYPRPGEEADLHAVQRRDDESLRSYIQRF 341
>UniRef100_Q5W669 Putatve polyprotein [Oryza sativa]
Length = 1905
Score = 372 bits (955), Expect = e-101
Identities = 248/784 (31%), Positives = 380/784 (47%), Gaps = 88/784 (11%)
Query: 1165 EDEEGDDIPYEITRLLEQEGRAIQPHQEEIEIINLGIPTAIYYLSKKLTDCETRYTMLEK 1224
E+ GD P R +PH++ + +Y++S+ L D +TRY +K
Sbjct: 1181 EERPGDTAP----------SRGDRPHRK--------VQWPVYFVSEALRDAKTRYPQAQK 1222
Query: 1225 TCCALAWAAKRLRHYLVNHTTWLISRMDPIKYIFEKAVVTGKIARWQVLLSEYDIVFKTQ 1284
A+ A+++LRHY H +++ P+ I TG++ +W + LSE+D+ F+ +
Sbjct: 1223 MLYAILMASRKLRHYFQAHRVTVVTSY-PLGQILHNREGTGRVVKWAIELSEFDLHFEPR 1281
Query: 1285 KAIKGSILADHPAYQPLDDYQPIESDFPNEEIVYLKSKDCEEPLIGEGPDPNSKWGLVFD 1344
AIK LAD A P E V + + + W + FD
Sbjct: 1282 HAIKSQALADFVAEWT-----------PAPETVSIPEASTNPSQLPH----TTHWVMQFD 1326
Query: 1345 GAANAYGKGIGAVIVSPQGHHIPFTAQILFECTNNMVEYEACIFGIEEAIGMRIKHLDIY 1404
G+ + G G G + SP G + + ++ F TNNM EYE + G+ A G+ I+ L +
Sbjct: 1327 GSLSLQGAGAGVTLTSPTGDVLRYLVRLDFRATNNMAEYEGLLAGLRVAAGLGIRRLLVL 1386
Query: 1405 GDSALIINQIKGEWETHHAKLIPYRDYARRLLTYFTKVELHHIPRDENQMADALATLSSM 1464
GDS L++NQ+ E+ ++ Y RR+ +F +EL H+PR +N +AD L+ L+S
Sbjct: 1387 GDSQLVVNQVCKEYRCSDPQMDAYVRQVRRMERHFDGIELRHVPRRDNMIADELSRLAS- 1445
Query: 1465 FRVNHWNDVPIIKVQRLERPSHVFTIGDVIDQAGENMVDNKP------------------ 1506
+ P +RL RPS D GE ++P
Sbjct: 1446 ---SRAQTPPGAFEERLTRPSAR------PDPLGETDAPDRPPRPVGVQASGPEGSAPSS 1496
Query: 1507 -----WYYDIKQCLLSREYPPGASNKDKKTLRRLASRFLLDGDILYKRNYDMVLLRCVDE 1561
W +I+ L + P ++ ++R++ R++L LY+R + +LL+C+ +
Sbjct: 1497 LRLIAWISEIQTYLTDKTLPEDREGSER--VQRISKRYVLVEGTLYRRAANGILLKCIPQ 1554
Query: 1562 HEAEQLMHDVHDGTFGTHATGHTMSRKLLRAGYYWLTMEHDCYQHARKCHKCQIYADKIH 1621
+ +L+ D+H+G G H+ T K R G+YW T +D R+C CQ +A +IH
Sbjct: 1555 EQGVELLADIHEGECGAHSASRTQVGKAFRQGFYWPTALNDAVDLVRRCRACQFHAKQIH 1614
Query: 1622 VPPHALNVISSPWPFSMVGIDMIGRIEPKASNGHRFILVAIDYFTKWVEAASYTNVTKQV 1681
P AL +I WPF++ G+D++G I+ +A G ++ VAID FTKW EA + K
Sbjct: 1615 QPAQALQIIPLSWPFAVWGLDILGPIK-RAPGGFEYLYVAIDKFTKWPEAYPVVKIDKHS 1673
Query: 1682 VAKFIKNNIICRYGVPSKIITDNGTNLNNNVVQALCEEFKIEHHNSSPYRPQINGVVEAA 1741
KFIK I R+GVP++IITDNGT + + CE+ I+ +SP P+ NG VE A
Sbjct: 1674 ALKFIK-GITARFGVPNRIITDNGTQFTSELFGDYCEDMGIKLCFASPAHPRSNGQVERA 1732
Query: 1742 N----KNIKRIVQKMVTTYRD-WHEMLPYALHGYRTTVRSSTGATPFSLVYGMEAVLPLE 1796
N K +K ++ + D W E LP L RTT +TG TPF LVYG EAVLP E
Sbjct: 1733 NAEILKGLKTKTFNILKKHGDSWIEELPAVLWANRTTPSRATGETPFFLVYGAEAVLPSE 1792
Query: 1797 VEIPSLRVIMEAKLSEAEWCQSRYDQLNLVEEKRMDAMARGQSYQARMKTAFDKKVHPRE 1856
+ + S R M EA+ Q R D L+ +EE+R A R Y R V R
Sbjct: 1793 LTLRSPRATM---YCEADQDQLRRDDLDYLEERRRRAALRAARYHQR-------HVRARS 1842
Query: 1857 FKEGELVLKRRISQQPDPRGKWTPNYEGSYVVKKAFSGGALILTHMDGVELPNPVNADLV 1916
+LVL+R Q K +P +EG Y V G++ L DG ELPNP N + +
Sbjct: 1843 LCVDDLVLRR--VQTRAGLSKLSPMWEGPYRVIGVPRPGSVRLATGDGTELPNPWNIEHL 1900
Query: 1917 KKYF 1920
++++
Sbjct: 1901 RRFY 1904
Score = 53.5 bits (127), Expect = 6e-05
Identities = 36/137 (26%), Positives = 64/137 (46%), Gaps = 5/137 (3%)
Query: 782 PEAGKHHNLALHISVNCTPDIISNVLVDTGSSLNVMPKSTLDQLSYRGTPLRRSTFLVKA 841
P + +A+ +S + VL+D G++LN++ + D + G L+ S ++
Sbjct: 603 PCVARGGQIAMVVSPTICNVKLGRVLIDGGAALNIISPAAFDAIKAPGMVLQPSQPIIGV 662
Query: 842 FDGTRKSVLGEIDLPITIGPE----TFLITFQVMDINASYNCLLGRP*IHDAGAVTSTLH 897
G LG IDLP+T G T + F V D++ YN +LGRP + A +
Sbjct: 663 TPG-HTWPLGHIDLPVTFGGSANFRTERVNFDVADLSLPYNAVLGRPALVKFMAAVHYAY 721
Query: 898 QKLKFAKSGKLVTIHGE 914
++K G +++ G+
Sbjct: 722 LQMKMPGPGGPISVRGD 738
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.321 0.136 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,231,494,515
Number of Sequences: 2790947
Number of extensions: 142846912
Number of successful extensions: 428650
Number of sequences better than 10.0: 4561
Number of HSP's better than 10.0 without gapping: 3707
Number of HSP's successfully gapped in prelim test: 856
Number of HSP's that attempted gapping in prelim test: 413425
Number of HSP's gapped (non-prelim): 9835
length of query: 1920
length of database: 848,049,833
effective HSP length: 142
effective length of query: 1778
effective length of database: 451,735,359
effective search space: 803185468302
effective search space used: 803185468302
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 83 (36.6 bits)
Medicago: description of AC147714.4


