

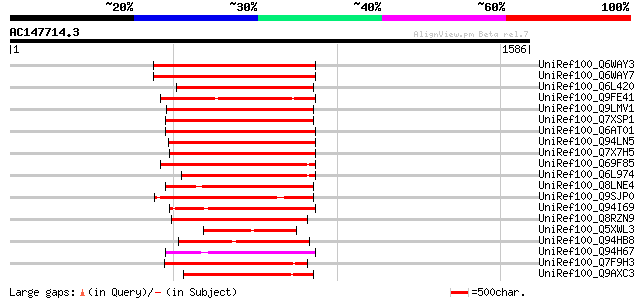
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147714.3 - phase: 0 /pseudo
(1586 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q6WAY3 Gag/pol polyprotein [Pisum sativum] 784 0.0
UniRef100_Q6WAY7 Gag/pol polyprotein [Pisum sativum] 782 0.0
UniRef100_Q6L420 Putative polyprotein [Solanum demissum] 572 e-161
UniRef100_Q9FE41 Oryza sativa (japonica cultivar-group) genomic ... 447 e-123
UniRef100_Q9LMV1 F5M15.26 [Arabidopsis thaliana] 405 e-111
UniRef100_Q7XSP1 OSJNBa0070M12.15 protein [Oryza sativa] 402 e-110
UniRef100_Q6AT01 Putative polyprotein [Oryza sativa] 393 e-107
UniRef100_Q94LN5 Putative retroelement pol polyprotein [Oryza sa... 384 e-104
UniRef100_Q7X7H5 OSJNBa0085C10.2 protein [Oryza sativa] 382 e-104
UniRef100_Q69F85 Gag-pol polyprotein [Phaseolus vulgaris] 382 e-104
UniRef100_Q6L974 GAG-POL [Vitis vinifera] 381 e-104
UniRef100_Q8LNE4 Putative retroelement [Oryza sativa] 373 e-101
UniRef100_Q9SJP0 Putative retroelement pol polyprotein [Arabidop... 367 2e-99
UniRef100_Q94I69 Putative retroelement [Oryza sativa] 363 2e-98
UniRef100_Q8RZN9 Putative polyprotein [Oryza sativa] 363 2e-98
UniRef100_Q5XWL3 Gag-pol polyprotein-like [Solanum tuberosum] 363 2e-98
UniRef100_Q94HB8 Putative retroelement [Oryza sativa] 357 2e-96
UniRef100_Q94H67 Putative gag-pol protein [Oryza sativa] 355 6e-96
UniRef100_Q7F9H3 OSJNBa0032N05.3 protein [Oryza sativa] 355 8e-96
UniRef100_Q9AXC3 Hypothetical protein [Antirrhinum hispanicum] 353 2e-95
>UniRef100_Q6WAY3 Gag/pol polyprotein [Pisum sativum]
Length = 2262
Score = 784 bits (2025), Expect = 0.0
Identities = 372/495 (75%), Positives = 434/495 (87%), Gaps = 3/495 (0%)
Query: 441 NFEFPVYEAEDEEGDD--IPYEITRLLEQERKAIQPHQEEIELINLGTEENKREIKVGAA 498
+F+ P+Y+AE+E +D +P E+ RLL+QE + IQPHQEE+E++NLGTE+ REIK+GAA
Sbjct: 1121 DFDNPIYQAEEESEEDCELPAELVRLLKQEERVIQPHQEELEVVNLGTEDATREIKIGAA 1180
Query: 499 LEEGVKRKIFQLL*EYPDIFAWSYEDMPGLDPKIVEHRIPTKPECPPVRQKLRRTHPDMA 558
LE+ VKR++ ++L EY +IFAWSY+DMPGLD IV HR+P + CP V+QKLRRT PDMA
Sbjct: 1181 LEDSVKRRLIEMLREYVEIFAWSYQDMPGLDTDIVVHRLPLREGCPSVKQKLRRTSPDMA 1240
Query: 559 LKIKSEV*KQIDAGFLMTIEYPEWVANIVPVPKKDGKVRMCVDFRDLNKASPKDNFPLPH 618
KIK EV KQ DAGFL YP W+ANIVPVPKKDGKVRMCVD+RDLN+ASPKD+FPLPH
Sbjct: 1241 TKIKEEVQKQWDAGFLAVTSYPPWMANIVPVPKKDGKVRMCVDYRDLNRASPKDDFPLPH 1300
Query: 619 IDVLVDNTAQSKVFSFMDGFSGYNQIKMSPEDREKTSFITPWGTFCYKVMPFGLINAGAT 678
IDVLVDNTAQS VFSFMDGFSGYNQIKM+PED EKT+FITPWGTFCYKVMPFGL NAGAT
Sbjct: 1301 IDVLVDNTAQSSVFSFMDGFSGYNQIKMAPEDMEKTTFITPWGTFCYKVMPFGLKNAGAT 1360
Query: 679 YQRGMTTLFHDMIHKEVEVYVDDMIVKSKDEEQHVEYLTKMFERLRKYKLRLNPNKCTFG 738
YQR MTTLFHDM+HKE+EVYVDDMI KS+ EE+H+ L K+F+RLRK+KLRLNPNKCTFG
Sbjct: 1361 YQRAMTTLFHDMMHKEIEVYVDDMIAKSQTEEEHLVNLQKLFDRLRKFKLRLNPNKCTFG 1420
Query: 739 VRSGKLLGFIVSQKGIEVDPDKVRAIREMPAPQTEKQVRGFLRRLNYISRFISHMTATCG 798
VRSGKLLGFIVS+KGIEVDP KV+AI+EMP P+TEKQVRGFL RLNYI+RFISH+TATC
Sbjct: 1421 VRSGKLLGFIVSEKGIEVDPAKVKAIQEMPEPKTEKQVRGFLGRLNYIARFISHLTATCE 1480
Query: 799 PIFKLLRKNQPIVWNDECQGAFDSIKNYLLEPPILVPPMEGKPLIMYLSVFDESVGCVLG 858
PIFKLLRKNQ I WND+CQ AFD IK YL +PPIL+PP+ G+PLIMYLSV + S+GCVLG
Sbjct: 1481 PIFKLLRKNQAIKWNDDCQKAFDKIKEYLQKPPILIPPVPGRPLIMYLSVTENSMGCVLG 1540
Query: 859 QQDETGKKEHAIYYLSKKFTDCETRYMMLEKTCCAPAWAAKRLRHYLVNHTTWLISRMDP 918
+ DE+G+KEHAIYYLSKKFTDCETRY +LEKTCCA AWAA+RLR Y++NHTT LIS+MDP
Sbjct: 1541 RHDESGRKEHAIYYLSKKFTDCETRYSLLEKTCCALAWAARRLRQYMLNHTTLLISKMDP 1600
Query: 919 IKYIFEKPAVLLGRL 933
+KYIFEKPA L GR+
Sbjct: 1601 VKYIFEKPA-LTGRV 1614
>UniRef100_Q6WAY7 Gag/pol polyprotein [Pisum sativum]
Length = 1814
Score = 782 bits (2019), Expect = 0.0
Identities = 374/495 (75%), Positives = 431/495 (86%), Gaps = 3/495 (0%)
Query: 441 NFEFPVYEAEDEEGDD--IPYEITRLLEQERKAIQPHQEEIELINLGTEENKREIKVGAA 498
+F P+Y+AE+E +D +P E+ RLL QE + IQPHQEE+E++NLGTE+ KREIK+GAA
Sbjct: 1123 DFGNPIYQAEEESEEDCELPAELVRLLRQEERVIQPHQEELEVVNLGTEDAKREIKIGAA 1182
Query: 499 LEEGVKRKIFQLL*EYPDIFAWSYEDMPGLDPKIVEHRIPTKPECPPVRQKLRRTHPDMA 558
LE+ VKR++ ++L EY +IFAWSY+DMPGLD IV HR+P + CP V+QKLRRT PDMA
Sbjct: 1183 LEDSVKRRLIEMLREYVEIFAWSYQDMPGLDTDIVVHRLPLREGCPSVKQKLRRTSPDMA 1242
Query: 559 LKIKSEV*KQIDAGFLMTIEYPEWVANIVPVPKKDGKVRMCVDFRDLNKASPKDNFPLPH 618
KIK EV KQ DAGFL YP WVANIVPVPKKDGKVRMCVD+RDLN+ASPKD+FPLPH
Sbjct: 1243 TKIKEEVQKQWDAGFLAVTSYPPWVANIVPVPKKDGKVRMCVDYRDLNRASPKDDFPLPH 1302
Query: 619 IDVLVDNTAQSKVFSFMDGFSGYNQIKMSPEDREKTSFITPWGTFCYKVMPFGLINAGAT 678
IDVLVDNTAQS VFSFMDGFSGYNQIKM+PED EKT+FITPWGTF YKVMPFGL NAGAT
Sbjct: 1303 IDVLVDNTAQSSVFSFMDGFSGYNQIKMAPEDMEKTTFITPWGTFYYKVMPFGLKNAGAT 1362
Query: 679 YQRGMTTLFHDMIHKEVEVYVDDMIVKSKDEEQHVEYLTKMFERLRKYKLRLNPNKCTFG 738
YQR MTTLFHDM+HKE+EVYVDDMI KS+ EE+H+ L K+F+RLRK+KLRLNPNKCTFG
Sbjct: 1363 YQRAMTTLFHDMMHKEIEVYVDDMIAKSQTEEEHLVNLQKLFDRLRKFKLRLNPNKCTFG 1422
Query: 739 VRSGKLLGFIVSQKGIEVDPDKVRAIREMPAPQTEKQVRGFLRRLNYISRFISHMTATCG 798
VRSGKLLGFIVS+KGIEVDP KV+AI+EMP P+TEKQVRGFL RLNYI+RFISH+TATC
Sbjct: 1423 VRSGKLLGFIVSEKGIEVDPAKVKAIQEMPEPKTEKQVRGFLGRLNYIARFISHLTATCE 1482
Query: 799 PIFKLLRKNQPIVWNDECQGAFDSIKNYLLEPPILVPPMEGKPLIMYLSVFDESVGCVLG 858
PIFKLLRKNQ I WND+CQ AFD IK YL +PPIL PP+ G+PLIMYLSV + S+GCVLG
Sbjct: 1483 PIFKLLRKNQAIKWNDDCQKAFDKIKEYLQKPPILTPPVPGRPLIMYLSVTENSMGCVLG 1542
Query: 859 QQDETGKKEHAIYYLSKKFTDCETRYMMLEKTCCAPAWAAKRLRHYLVNHTTWLISRMDP 918
Q DE+G+KEHAIYYLSKKFTDCETRY +LEKTCCA AWAA+RLR Y++NHTT LIS+MDP
Sbjct: 1543 QHDESGRKEHAIYYLSKKFTDCETRYSLLEKTCCALAWAARRLRQYMLNHTTLLISKMDP 1602
Query: 919 IKYIFEKPAVLLGRL 933
+KYIFEKPA L GR+
Sbjct: 1603 VKYIFEKPA-LTGRV 1616
>UniRef100_Q6L420 Putative polyprotein [Solanum demissum]
Length = 1483
Score = 572 bits (1474), Expect = e-161
Identities = 262/418 (62%), Positives = 337/418 (79%)
Query: 509 QLL*EYPDIFAWSYEDMPGLDPKIVEHRIPTKPECPPVRQKLRRTHPDMALKIKSEV*KQ 568
+ L +Y D+FA SY+DMPGL +V H++P P+ PPV++KLR+ DM++ IK E+ KQ
Sbjct: 523 EALLDYKDVFASSYDDMPGLSTDMVVHKLPIDPKFPPVKKKLRKLKTDMSVIIKEEITKQ 582
Query: 569 IDAGFLMTIEYPEWVANIVPVPKKDGKVRMCVDFRDLNKASPKDNFPLPHIDVLVDNTAQ 628
++A + +YP W+ANIVPVPKKDGKVRMCVD RDLNKASPKD+FPLP+ +L+DN A+
Sbjct: 583 LEAKVIRVAQYPSWLANIVPVPKKDGKVRMCVDCRDLNKASPKDDFPLPNSHILLDNCAE 642
Query: 629 SKVFSFMDGFSGYNQIKMSPEDREKTSFITPWGTFCYKVMPFGLINAGATYQRGMTTLFH 688
++ SF+D ++GY+QI M ED EK SFITPWGT+CY+VMPFGL NAGATY R MTT+FH
Sbjct: 643 HEIASFVDCYAGYHQIIMDDEDAEKMSFITPWGTYCYRVMPFGLKNAGATYMRAMTTMFH 702
Query: 689 DMIHKEVEVYVDDMIVKSKDEEQHVEYLTKMFERLRKYKLRLNPNKCTFGVRSGKLLGFI 748
DM+HKE+EVYVDD+I+KSK + HV+ L + FERLR+Y L+LNP KC FGV SG+LLGFI
Sbjct: 703 DMMHKEIEVYVDDVIIKSKKQSNHVQDLRRFFERLRRYNLKLNPAKCVFGVLSGELLGFI 762
Query: 749 VSQKGIEVDPDKVRAIREMPAPQTEKQVRGFLRRLNYISRFISHMTATCGPIFKLLRKNQ 808
S++GIE+DP K++AI E+P P+ + +V FL RLNYISRFI+ +T TC PIFKLL+KN
Sbjct: 763 GSRRGIELDPLKIKAIHELPPPKNKTEVMSFLGRLNYISRFIAQLTTTCEPIFKLLKKNA 822
Query: 809 PIVWNDECQGAFDSIKNYLLEPPILVPPMEGKPLIMYLSVFDESVGCVLGQQDETGKKEH 868
+ W +EC+ AF+ IKNYLL PP+LVPP G+PLI+Y+SV D S GCVLGQ D TGKKEH
Sbjct: 823 TVEWTEECREAFERIKNYLLNPPVLVPPEPGRPLILYMSVLDNSFGCVLGQHDGTGKKEH 882
Query: 869 AIYYLSKKFTDCETRYMMLEKTCCAPAWAAKRLRHYLVNHTTWLISRMDPIKYIFEKP 926
AIYYLSKKFT E++Y +LE+TCCA W A++L+HYL ++TT+LISRMDP+KYIF+KP
Sbjct: 883 AIYYLSKKFTVYESKYTLLERTCCALTWVAQKLKHYLSSYTTYLISRMDPLKYIFQKP 940
>UniRef100_Q9FE41 Oryza sativa (japonica cultivar-group) genomic DNA, chromosome 1, PAC
clone:P0433F09 [Oryza sativa]
Length = 2876
Score = 447 bits (1150), Expect = e-123
Identities = 225/474 (47%), Positives = 311/474 (65%), Gaps = 8/474 (1%)
Query: 460 EITRLLEQERKAIQPHQEEIELINLGTEENKREIKVGAALEEGVKRKIFQLL*EYPDIFA 519
++ L+Q QP +E+ +NLGTE++ R I V L E + L E+ D FA
Sbjct: 1759 DVAGALKQLDDGGQPTIDELVEMNLGTEDDPRPIFVSGMLTEEEREDYRSFLMEFRDCFA 1818
Query: 520 WSYEDMPGLDPKIVEHRIPTKPECPPVRQKLRRTHPDMALKIKSEV*KQIDAGFLMTIEY 579
W+Y++MPGLD ++ H++ P+ PV+Q RR P+ ++ +EV + I+ GF+ I+Y
Sbjct: 1819 WTYKEMPGLDSRVATHKLAIDPQFRPVKQPPRRLRPEFQDQVIAEVDRLINVGFIKEIQY 1878
Query: 580 PEWVANIVPVPKKDGKVRMCVDFRDLNKASPKDNFPLPHIDVLVDNTAQSKVFSFMDGFS 639
P W+ANIVPV KK+G+VR+CVDFRDLN+A PKD+FPLP +++VD+T S
Sbjct: 1879 PRWLANIVPVEKKNGQVRVCVDFRDLNRACPKDDFPLPITEMVVDSTTG------YGALS 1932
Query: 640 GYNQIKMSPEDREKTSFITPWGTFCYKVMPFGLINAGATYQRGMTTLFHDMIHKEVEVYV 699
GYNQIKM D T+F TP G F Y VMPFGL NAGATYQR M + D+IH VE YV
Sbjct: 1933 GYNQIKMDLLDAFDTAFRTPKGNFYYTVMPFGLKNAGATYQRAMQFVLDDLIHHSVECYV 1992
Query: 700 DDMIVKSKDEEQHVEYLTKMFERLRKYKLRLNPNKCTFGVRSGKLLGFIVSQKGIEVDPD 759
DDM+VK+KD E H E L +FERLR+++L++NP KC F V+SG LGF++ +GIE++P
Sbjct: 1993 DDMVVKTKDHEHHQEDLRIVFERLRRHQLKMNPLKCAFAVQSGVFLGFVIRHRGIEIEPK 2052
Query: 760 KVRAIREMPAPQTEKQVRGFLRRLNYISRFISHMTATCGPIFKLLRKNQPIVWNDECQGA 819
K++AI MP PQ K +R +L YI RFIS+++ P KL++K P VW++ECQ
Sbjct: 2053 KIKAILNMPPPQELKDLRKLQGKLAYIRRFISNLSGRIQPFSKLMKKGTPFVWDEECQNG 2112
Query: 820 FDSIKNYLLEPPILVPPMEGKPLIMYLSVFDESVGCVLGQQDETGKKEHAIYYLSKKFTD 879
FDSIK YLL PP+L P++G+PLI+Y++ S+G +L Q ++ G KE A YYLS+
Sbjct: 2113 FDSIKRYLLNPPVLAAPVKGRPLILYIATQPASIGALLAQHNDEG-KEVACYYLSRTMVG 2171
Query: 880 CETRYMMLEKTCCAPAWAAKRLRHYLVNHTTWLISRMDPIKYIFEKPAVLLGRL 933
E Y +EK C A +A K+LRHY++ H LI+R DPI+Y+ +P VL GRL
Sbjct: 2172 AEQNYSPIEKLCLALIFALKKLRHYMLAHQIQLIARADPIRYVLSQP-VLTGRL 2224
>UniRef100_Q9LMV1 F5M15.26 [Arabidopsis thaliana]
Length = 1838
Score = 405 bits (1042), Expect = e-111
Identities = 199/448 (44%), Positives = 294/448 (65%), Gaps = 3/448 (0%)
Query: 480 ELINLGTEENKREIKVGAALEEGVKRKIFQLL*EYPDIFAWSYEDMPGLDPKIVEHRIPT 539
E++N+ + R + VGA + ++ ++ LL FAWS EDM G+DP I H +
Sbjct: 759 EMVNIDESDPTRCVGVGAEISPSIRLELIALLKRNSKTFAWSIEDMKGIDPAITAHELNV 818
Query: 540 KPECPPVRQKLRRTHPDMALKIKSEV*KQIDAGFLMTIEYPEWVANIVPVPKKDGKVRMC 599
P PV+QK R+ P+ A + EV K + AG ++ ++YPEW+AN V V KK+GK R+C
Sbjct: 819 DPTFKPVKQKRRKLGPERARAVNEEVEKLLKAGQIIEVKYPEWLANPVVVKKKNGKWRVC 878
Query: 600 VDFRDLNKASPKDNFPLPHIDVLVDNTAQSKVFSFMDGFSGYNQIKMSPEDREKTSFITP 659
VD+ DLNKA PKD++PLPHID LV+ T+ + + SFMD FSGYNQI M +D+EKTSF+T
Sbjct: 879 VDYTDLNKACPKDSYPLPHIDRLVEATSGNGLLSFMDAFSGYNQILMHKDDQEKTSFVTD 938
Query: 660 WGTFCYKVMPFGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSKDEEQHVEYLTKM 719
GT+CYKVM FGL NAGATYQR + + D I + VEVY+DDM+VKS E HVE+L+K
Sbjct: 939 RGTYCYKVMSFGLKNAGATYQRFVNKMLADQIGRTVEVYIDDMLVKSLKPEDHVEHLSKC 998
Query: 720 FERLRKYKLRLNPNKCTFGVRSGKLLGFIVSQKGIEVDPDKVRAIREMPAPQTEKQVRGF 779
F+ L Y ++LNP KCTFGV SG+ LG++V+++GIE +P ++RAI E+P+P+ ++V+
Sbjct: 999 FDVLNTYGMKLNPTKCTFGVTSGEFLGYVVTKRGIEANPKQIRAILELPSPRNAREVQRL 1058
Query: 780 LRRLNYISRFISHMTATCGPIFKLLRKNQPIVWNDECQGAFDSIKNYLLEPPILVPPMEG 839
R+ ++RFIS T C P + LL++ W+ + + AF+ +K+YL PPILV P G
Sbjct: 1059 TGRIAALNRFISRSTDKCLPFYNLLKRRAQFDWDKDSEEAFEKLKDYLSTPPILVKPEVG 1118
Query: 840 KPLIMYLSVFDESVGCVLGQQDETGKKEHAIYYLSKKFTDCETRYMMLEKTCCAPAWAAK 899
+ L +Y++V D +V VL ++D ++ I+Y SK + ETRY ++EK A +A+
Sbjct: 1119 ETLYLYIAVSDHAVSSVLVREDR--GEQRPIFYTSKSLVEAETRYPVIEKAALAVVTSAR 1176
Query: 900 RLRHYLVNHTTWLISRMDPIKYIFEKPA 927
+LR Y +HT +++ P++ P+
Sbjct: 1177 KLRPYFQSHTIAVLTD-QPLRVALHSPS 1203
>UniRef100_Q7XSP1 OSJNBa0070M12.15 protein [Oryza sativa]
Length = 1685
Score = 402 bits (1032), Expect = e-110
Identities = 201/454 (44%), Positives = 294/454 (64%), Gaps = 6/454 (1%)
Query: 477 EEIELINLGTEENKREIKVGAALEEGVKRKIFQLL*EYPDIFAWSYEDMPGLDPKIVEHR 536
++++ +++G + R + L + K+ +LL E+ D FAW Y +MPGL IVEHR
Sbjct: 904 DDLDEVDIGPGDRPRPTFISKNLSSEFRTKLMELLKEFRDCFAWEYYEMPGLSRSIVEHR 963
Query: 537 IPTKPECPPVRQKLRRTHPDMALKIKSEV*KQIDAGFLMTIEYPEWVANIVPVPKKDGKV 596
+P KP P +Q RR DM +K+E+ + DAGF+ Y EWV++IVPV KK+GKV
Sbjct: 964 LPLKPGIRPHQQPPRRCKADMLEPVKAEIKRLYDAGFIHPCRYAEWVSSIVPVIKKNGKV 1023
Query: 597 RMCVDFRDLNKASPKDNFPLPHIDVLVDNTAQSKVFSFMDGFSGYNQIKMSPEDREKTSF 656
R+C+DFRDLNKA+PKD +P+P D LVD + K+ SFMDG +GYNQI M+ ED KT F
Sbjct: 1024 RVCIDFRDLNKATPKDEYPMPVADQLVDAASGHKILSFMDGNAGYNQIFMTEEDIHKTVF 1083
Query: 657 ITPW--GTFCYKVMPFGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSKDEEQHVE 714
P G F + VM FGL +AGATYQR M ++HD+I VEVY+DD++VKS++ E H+
Sbjct: 1084 RCPGAIGLFEWVVMTFGLKSAGATYQRAMNYIYHDLIGWLVEVYIDDVVVKSREIEDHIA 1143
Query: 715 YLTKMFERLRKYKLRLNPNKCTFGVRSGKLLGFIVSQKGIEVDPDKVRAIREMPAPQTEK 774
L K+FER RKY L++NP KC FGV +G+ LGF+V ++GIE+ + AI+++ P +
Sbjct: 1144 DLRKVFERTRKYGLKMNPTKCAFGVSAGQFLGFLVHERGIEIPQRSINAIKKIKPPGNKT 1203
Query: 775 QVRGFLRRLNYISRFISHMTATCGPIFKL--LRKNQPIVWNDECQGAFDSIKNYLLEPPI 832
+++ + ++N++ RFIS+++ P L LR +Q W E Q A D+IK YL PP+
Sbjct: 1204 ELQEMIGKINFVRRFISNLSGKLEPFTPLLRLRADQQFTWGAEQQKALDNIKEYLSSPPV 1263
Query: 833 LVPPMEGKPLIMYLSVFDESVGCVLGQQDETGKKEHAIYYLSKKFTDCETRYMMLEKTCC 892
L+PP +G P +YLS D+S+G VL Q+ E KE ++YLS++ D ETRY +EK C
Sbjct: 1264 LIPPQKGIPFWLYLSAGDKSIGSVLIQELE--GKERVVFYLSRRLLDAETRYSPVEKLCL 1321
Query: 893 APAWAAKRLRHYLVNHTTWLISRMDPIKYIFEKP 926
++ +LRHYL+++ ++ + D IKY+ P
Sbjct: 1322 CLYFSCTKLRHYLLSNECTVVCKADVIKYMLSAP 1355
>UniRef100_Q6AT01 Putative polyprotein [Oryza sativa]
Length = 1739
Score = 393 bits (1009), Expect = e-107
Identities = 204/461 (44%), Positives = 298/461 (64%), Gaps = 7/461 (1%)
Query: 477 EEIELINLGTEENKREIKVGAALEEGVKRKIFQLL*EYPDIFAWSYEDMPGLDPKIVEHR 536
+++E I++G + R V L + K+ +LL E+ D FAW Y +MPGL IVEHR
Sbjct: 859 DDLEEIDIGPGDRPRPTFVSKNLSPEFRTKLIELLKEFRDCFAWEYYEMPGLSRSIVEHR 918
Query: 537 IPTKPECPPVRQKLRRTHPDMALKIKSEV*KQIDAGFLMTIEYPEWVANIVPVPKKDGKV 596
+P KP P +Q LRR DM +K+E+ + DAGF+ Y +WV++IVPV KK+GKV
Sbjct: 919 LPIKPGVRPRQQPLRRCKADMLEPVKAEIKRLYDAGFIRPWRYAKWVSSIVPVIKKNGKV 978
Query: 597 RMCVDFRDLNKASPKDNFPLPHIDVLVDNTAQSKVFSFMDGFSGYNQIKMSPEDREKTSF 656
R+C+DFRDLNKA+PKD +P+P D LVD + +K+ SFMDG +GYNQI M+ ED KT+F
Sbjct: 979 RVCIDFRDLNKATPKDEYPMPVADQLVDAASGNKILSFMDGNAGYNQIFMAEEDIHKTAF 1038
Query: 657 ITPW--GTFCYKVMPFGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSKDEEQHVE 714
P G F + VM FGL +AGATYQR M ++HD+I V VY+DD++VKSK+ E +
Sbjct: 1039 RCPCAIGLFEWVVMTFGLKSAGATYQRAMNYIYHDLIGWLVGVYIDDVVVKSKEIEDQIA 1098
Query: 715 YLTKMFERLRKYKLRLNPNKCTFGVRSGKLLGFIVSQKGIEVDPDKVRAIREMPAPQTEK 774
L K+FER RKY L++NP KC FGV +G+ LGF+V +GIEV V AI+++ P+ +
Sbjct: 1099 DLRKVFERTRKYGLKMNPTKCAFGVSAGQFLGFLVHDRGIEVTQRSVNAIKKIQPPENKT 1158
Query: 775 QVRGFLRRLNYISRFISHMTATCGPIFKLLR--KNQPIVWNDECQGAFDSIKNYLLEPPI 832
+++ + +++++ RFIS+++ P LLR +Q E Q A D IK YL PP+
Sbjct: 1159 ELQEMIGKIHFVRRFISNLSGRLEPFTPLLRLKADQQFTCGAEQQKALDDIKEYLSSPPV 1218
Query: 833 LVPPMEGKPLIMYLSVFDESVGCVLGQQDETGKKEHAIYYLSKKFTDCETRYMMLEKTCC 892
L+PP +G P +YLS ++S+G VL Q+ E KE ++YLS++ + ETRY ++K C
Sbjct: 1219 LIPPHKGIPFRLYLSAGEKSIGSVLIQELE--GKERVVFYLSRRLLNAETRYSPVKKLCL 1276
Query: 893 APAWAAKRLRHYLVNHTTWLISRMDPIKYIFEKPAVLLGRL 933
++ RLRHYL+++ +I + D +KY+ P +L GR+
Sbjct: 1277 CLYFSCTRLRHYLLSNECTVICKADVVKYMLSAP-ILKGRV 1316
>UniRef100_Q94LN5 Putative retroelement pol polyprotein [Oryza sativa]
Length = 1580
Score = 384 bits (987), Expect = e-104
Identities = 200/454 (44%), Positives = 293/454 (64%), Gaps = 8/454 (1%)
Query: 484 LGTEENKREIKVGAALEEGVKRKIFQLL*EYPDIFAWSYEDMPGLDPKIVEHRIPTKPEC 543
L + ++ EI + A+ +G + + +LL E+ D F W Y +MPG IVEHR+P KP
Sbjct: 524 LMSADDLEEIDISPAIGQG-RHLLIELLKEFRDCFTWEYYEMPGHSRSIVEHRLPLKPGV 582
Query: 544 PPVRQKLRRTHPDMALKIKSEV*KQIDAGFLMTIEYPEWVANIVPVPKKDGKVRMCVDFR 603
P +Q RR DM +K+E+ + DAGF+ Y EWV++IVPV KK+GKVR+C+DFR
Sbjct: 583 RPHQQLPRRCKADMLEPVKAEIKRLYDAGFIRPCRYAEWVSSIVPVIKKNGKVRVCIDFR 642
Query: 604 DLNKASPKDNFPLPHIDVLVDNTAQSKVFSFMDGFSGYNQIKMSPEDREKTSFITPW--G 661
LNKA+PKD +P+ D LVD + K+ SFMDG +GYNQI M+ ED KT+F P G
Sbjct: 643 YLNKATPKDEYPMLVADQLVDAASGHKILSFMDGNAGYNQIFMAEEDIHKTTFRCPGAIG 702
Query: 662 TFCYKVMPFGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSKDEEQHVEYLTKMFE 721
F + V+ F L +AGATYQR M ++HD+I VEVY+DD++VKSK+ E H+ L K+FE
Sbjct: 703 LFEWVVITFVLKSAGATYQRAMNYIYHDLIGWLVEVYIDDVVVKSKEIEDHIADLRKVFE 762
Query: 722 RLRKYKLRLNPNKCTFGVRSGKLLGFIVSQKGIEVDPDKVRAIREMPAPQTEKQVRGFLR 781
R RKY L++NP KC FGV +G+ LGF+V ++GIEV + AI+++ P+ + +++ +
Sbjct: 763 RTRKYGLKMNPTKCAFGVSAGQFLGFLVHERGIEVTQRSINAIKKIKPPEDKTELQEMIG 822
Query: 782 RLNYISRFISHMTATCGPIFKLLR--KNQPIVWNDECQGAFDSIKNYLLEPPILVPPMEG 839
++N++ RFIS ++ P LLR +Q W E Q A D+IK YL PP+L+PP +G
Sbjct: 823 KINFVRRFISILSGKLEPFTPLLRLKADQQFAWGAEQQKALDNIKEYLSSPPVLIPPQKG 882
Query: 840 KPLIMYLSVFDESVGCVLGQQDETGKKEHAIYYLSKKFTDCETRYMMLEKTCCAPAWAAK 899
P +YLS D+S+ VL Q+ E +KE ++YLS++ + ETRY +EK C ++
Sbjct: 883 IPFRLYLSAGDKSISSVLIQELE--RKERVVFYLSRRLLNAETRYSPVEKLCLCLYFSCT 940
Query: 900 RLRHYLVNHTTWLISRMDPIKYIFEKPAVLLGRL 933
RLRHYL+++ +I + D +KY+ P +L GR+
Sbjct: 941 RLRHYLLSNECTVICKADVVKYMLSAP-ILKGRI 973
>UniRef100_Q7X7H5 OSJNBa0085C10.2 protein [Oryza sativa]
Length = 861
Score = 382 bits (982), Expect = e-104
Identities = 197/450 (43%), Positives = 287/450 (63%), Gaps = 7/450 (1%)
Query: 488 ENKREIKVGAALEEGVKRKIFQLL*EYPDIFAWSYEDMPGLDPKIVEHRIPTKPECPPVR 547
+ R + L + K+ +LL E+ D FAW Y +MPGL IVEHR+P KP P +
Sbjct: 27 DQPRPTFISKNLSSEFRTKLIELLKEFRDCFAWEYYEMPGLSRSIVEHRLPIKPGVRPRQ 86
Query: 548 QKLRRTHPDMALKIKSEV*KQIDAGFLMTIEYPEWVANIVPVPKKDGKVRMCVDFRDLNK 607
Q RR DM +K+E+ + DAGF+ Y EWV++IVPV KK+GKVR+C+DFRDLNK
Sbjct: 87 QPPRRCKADMLEPVKAEIKRLYDAGFIHPYRYAEWVSSIVPVIKKNGKVRVCIDFRDLNK 146
Query: 608 ASPKDNFPLPHIDVLVDNTAQSKVFSFMDGFSGYNQIKMSPEDREKTSFITP--WGTFCY 665
A+PKD +P+P D L D + +K+ SFMD +GYNQI M+ ED KT+F P G F +
Sbjct: 147 ATPKDEYPMPVADQLFDAASGNKILSFMDRNTGYNQIFMAEEDIHKTAFRCPSAIGLFEW 206
Query: 666 KVMPFGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSKDEEQHVEYLTKMFERLRK 725
VM F L +AGATYQR M ++HD+I VEV +DD++VKSK+ + H+ L K+FER RK
Sbjct: 207 VVMTFDLKSAGATYQRAMNYIYHDLIGSLVEVDIDDVVVKSKEVDDHIADLRKVFERTRK 266
Query: 726 YKLRLNPNKCTFGVRSGKLLGFIVSQKGIEVDPDKVRAIREMPAPQTEKQVRGFLRRLNY 785
Y L++NP KC FGV + + LGF+V ++GIEV V AI+++ P+ + +++ + ++N+
Sbjct: 267 YGLKMNPTKCAFGVSARQFLGFLVHERGIEVTQRSVNAIQKIKPPENKTELQEMIGKINF 326
Query: 786 ISRFISHMTATCGPIFKLLR--KNQPIVWNDECQGAFDSIKNYLLEPPILVPPMEGKPLI 843
+ RFIS+++ P LLR +Q W E Q A D+IK YL PP+L+PP +G P
Sbjct: 327 VRRFISNLSGRLEPFTPLLRLKADQQFTWGAEQQKALDNIKEYLSSPPVLIPPQKGIPFR 386
Query: 844 MYLSVFDESVGCVLGQQDETGKKEHAIYYLSKKFTDCETRYMMLEKTCCAPAWAAKRLRH 903
+YLS ++S G VL Q+ E KE ++Y+S++ D ETRY +EK C + RLRH
Sbjct: 387 LYLSAGEKSNGSVLIQELE--GKERVVFYISRRLLDAETRYSPMEKLCLCLYFLCTRLRH 444
Query: 904 YLVNHTTWLISRMDPIKYIFEKPAVLLGRL 933
YL+++ +I + D ++Y+ P +L G++
Sbjct: 445 YLLSNECTVICKADVVRYMLSAP-ILKGQI 473
>UniRef100_Q69F85 Gag-pol polyprotein [Phaseolus vulgaris]
Length = 1859
Score = 382 bits (981), Expect = e-104
Identities = 201/474 (42%), Positives = 289/474 (60%), Gaps = 5/474 (1%)
Query: 461 ITRLLEQERKAIQPHQEEIE-LINLGTEENKREIKVGAALEEGVKRKIFQLL*EYPDIFA 519
+ L++ + + P E E L + + R+ +G +L+ + I + L + D+FA
Sbjct: 816 LVALVDLDPRLDDPRMEAGEDLQPIFLRDKDRKTYMGTSLKPDDRETIGKTLTKNADLFA 875
Query: 520 WSYEDMPGLDPKIVEHRIPTKPECPPVRQKLRRTHPDMALKIKSEV*KQIDAGFLMTIEY 579
W+ DMPG+ ++ HR+ E P+ QK R+ + + E K I AGF+ Y
Sbjct: 876 WTAADMPGVKSDVITHRLSVYTEARPIAQKKRKLGEERRKAAREETDKLIQAGFIQKAHY 935
Query: 580 PEWVANIVPVPKKDGKVRMCVDFRDLNKASPKDNFPLPHIDVLVDNTAQSKVFSFMDGFS 639
W+AN+V V K +GK RMCVD+ DLNKA PKD++PLP ID LVD A ++ SF+D +S
Sbjct: 936 TTWLANVVMVKKTNGKWRMCVDYTDLNKACPKDSYPLPTIDRLVDGAAGHQILSFLDAYS 995
Query: 640 GYNQIKMSPEDREKTSFITPWGTFCYKVMPFGLINAGATYQRGMTTLFHDMIHKEVEVYV 699
GYNQI+M DREKT+F T F Y+VMPFGL NAGATYQR M +FHDMI + VEVYV
Sbjct: 996 GYNQIQMYHRDREKTAFRTDSDNFFYEVMPFGLKNAGATYQRLMDHVFHDMIGRNVEVYV 1055
Query: 700 DDMIVKSKDEEQHVEYLTKMFERLRKYKLRLNPNKCTFGVRSGKLLGFIVSQKGIEVDPD 759
DD++VKS EQHV L ++F+ LR+Y++RLNP KC FGV GK LGF+++ +GIE +P+
Sbjct: 1056 DDIVVKSDSCEQHVSDLKEVFQALRQYRMRLNPEKCAFGVEGGKFLGFMLTHRGIEANPE 1115
Query: 760 KVRAIREMPAPQTEKQVRGFLRRLNYISRFISHMTATCGPIFKLLRKNQPIVWNDECQGA 819
K +AI EM +P+ K+++ + RL +SRF+ + PI KLL+K W DEC+
Sbjct: 1116 KCKAITEMRSPKGLKEIQRLVSRLTSLSRFVPKLAERTRPIIKLLKKTSKFEWTDECEQN 1175
Query: 820 FDSIKNYLLEPPILVPPMEGKPLIMYLSVFDESVGCVLGQQDETGKKEHAIYYLSKKFTD 879
F +K +L PP++ P +P+++YL+V +E+V L Q E +E +Y++S+ D
Sbjct: 1176 FQQLKAFLASPPVIQKPNAREPIVVYLAVSNEAVSSALVQ--EIKAEERPVYFVSRVLHD 1233
Query: 880 CETRYMMLEKTCCAPAWAAKRLRHYLVNHTTWLISRMDPIKYIFEKPAVLLGRL 933
ETRY M+EK A A+R+R Y NH ++ PI I KP L GR+
Sbjct: 1234 AETRYQMVEKVAFALVITARRMRMYFQNHKV-IVRTNYPIMKILTKPD-LAGRM 1285
>UniRef100_Q6L974 GAG-POL [Vitis vinifera]
Length = 1027
Score = 381 bits (979), Expect = e-104
Identities = 191/409 (46%), Positives = 271/409 (65%), Gaps = 3/409 (0%)
Query: 525 MPGLDPKIVEHRIPTKPECPPVRQKLRRTHPDMALKIKSEV*KQIDAGFLMTIEYPEWVA 584
M G+ P I HR+ PVRQ++RR HPD I++E+ K ++AGF+ + YP+W+A
Sbjct: 1 MKGIHPSITSHRLNVVSTARPVRQRIRRFHPDRQRVIRNEIDKLLEAGFIREVSYPDWLA 60
Query: 585 NIVPVPKKDGKVRMCVDFRDLNKASPKDNFPLPHIDVLVDNTAQSKVFSFMDGFSGYNQI 644
N+V VPKK+GK R+CVD+ +LN A PKD+FPLP ID +VD+T+ + SF+D FSGY+QI
Sbjct: 61 NVVVVPKKEGKWRVCVDYTNLNNACPKDSFPLPRIDQIVDSTSGQGMLSFLDAFSGYHQI 120
Query: 645 KMSPEDREKTSFITPWGTFCYKVMPFGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIV 704
MSP+D EK +FITP +CYKVMPFGL NAGATYQR MT +F +I VEVY+DD++V
Sbjct: 121 PMSPDDEEKIAFITPHDLYCYKVMPFGLKNAGATYQRLMTKIFKPLIGHSVEVYIDDIVV 180
Query: 705 KSKDEEQHVEYLTKMFERLRKYKLRLNPNKCTFGVRSGKLLGFIVSQKGIEVDPDKVRAI 764
KSK EQH+ +L ++F LR+Y ++LNP+KC FGV + K LGF+VSQ+GIEV PD+V+A+
Sbjct: 181 KSKTREQHILHLQEVFYLLRRYGMKLNPSKCAFGVSARKFLGFMVSQRGIEVSPDQVKAV 240
Query: 765 REMPAPQTEKQVRGFLRRLNYISRFISHMTATCGPIFKLLRKNQPIVWNDECQGAFDSIK 824
E P P+ +K+++ +L + RFI+ P F +RK W D CQ A + IK
Sbjct: 241 METPPPRNKKELQRLTGKLVALGRFIARFIDELRPFFLAIRKAGTHGWTDNCQNALERIK 300
Query: 825 NYLLEPPILVPPMEGKPLIMYLSVFDESVGCVLGQQDETGKKEHAIYYLSKKFTDCETRY 884
+YL++PPIL P+ + L MYL+V + ++ VL + + K++ IYY+S+ D ETRY
Sbjct: 301 HYLMQPPILSSPIPKEKLYMYLAVSEWAISAVL-FRCPSPKEQKPIYYVSRALADVETRY 359
Query: 885 MMLEKTCCAPAWAAKRLRHYLVNHTTWLISRMDPIKYIFEKPAVLLGRL 933
+E A AA++LR Y H ++ P++ I KP L GR+
Sbjct: 360 SKMELISLALRSAAQKLRPYFQAHPV-IVLTDQPLRNILHKPD-LTGRM 406
>UniRef100_Q8LNE4 Putative retroelement [Oryza sativa]
Length = 1170
Score = 373 bits (958), Expect = e-101
Identities = 195/454 (42%), Positives = 286/454 (62%), Gaps = 21/454 (4%)
Query: 477 EEIELINLGTEENKREIKVGAALEEGVKRKIFQLL*EYPDIFAWSYEDMPGLDPKIVEHR 536
+E+E I++G + R + L + K+ +LL E+ D FAW Y +MPGL IVEHR
Sbjct: 614 DELEEIDIGPGDRPRPTFIRKNLSPEFRTKLIELLKEFRDCFAWEYYEMPGLSRSIVEHR 673
Query: 537 IPTKPECPPVRQKLRRTHPDMALKIKSEV*KQIDAGFLMTIEYPEWVANIVPVPKKDGKV 596
+P KP P +Q RR +M +K+E+ + DA VPV KK+GKV
Sbjct: 674 LPIKPGVRPCQQPPRRCKANMLEPVKAEIKRLYDA---------------VPVIKKNGKV 718
Query: 597 RMCVDFRDLNKASPKDNFPLPHIDVLVDNTAQSKVFSFMDGFSGYNQIKMSPEDREKTSF 656
R+C+DFRDLNKA+PKD +P+P D LVD + +K+ SF+DG +GYNQI M+ ED K +F
Sbjct: 719 RVCIDFRDLNKATPKDEYPMPVADQLVDAASGNKILSFVDGNAGYNQIFMAEEDIHKMAF 778
Query: 657 ITPW--GTFCYKVMPFGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSKDEEQHVE 714
P G F + VM FGL +AGATYQR M ++HD+I VEVY+D+++VKSK+ E H+
Sbjct: 779 RCPSAIGLFEWVVMTFGLKSAGATYQRAMNYIYHDLIGWLVEVYIDNVVVKSKEVEDHIA 838
Query: 715 YLTKMFERLRKYKLRLNPNKCTFGVRSGKLLGFIVSQKGIEVDPDKVRAIREMPAPQTEK 774
L K+FER RKY L++NP KC FGV +G+ LGF+V ++GIEV V AI+++ P+ +
Sbjct: 839 DLRKVFERTRKYGLKMNPTKCAFGVSAGQFLGFLVHERGIEVTQRSVNAIKKIQPPENKT 898
Query: 775 QVRGFLRRLNYISRFISHMTATCGPIFKLLR--KNQPIVWNDECQGAFDSIKNYLLEPPI 832
+++ + ++N++ RFIS+++ P LLR NQ W E Q A D+IK YL PP+
Sbjct: 899 ELQEMIGKINFVRRFISNLSGRLEPFTPLLRLKANQQFTWGAEQQKALDNIKEYLSSPPV 958
Query: 833 LVPPMEGKPLIMYLSVFDESVGCVLGQQDETGKKEHAIYYLSKKFTDCETRYMMLEKTCC 892
L+PP +G P +YLS ++S+G VL Q+ E KE ++YLS++ D ET+Y +EK C
Sbjct: 959 LIPPQKGIPFRLYLSAGEKSIGSVLIQELE--GKERVVFYLSRRLLDAETQYSPVEKLCL 1016
Query: 893 APAWAAKRLRHYLVNHTTWLISRMDPIKYIFEKP 926
++ RLRHYL+++ +I + D ++Y+ P
Sbjct: 1017 CLYFSCTRLRHYLLSNECTVICKADVVRYMLLAP 1050
>UniRef100_Q9SJP0 Putative retroelement pol polyprotein [Arabidopsis thaliana]
Length = 1787
Score = 367 bits (941), Expect = 2e-99
Identities = 190/486 (39%), Positives = 293/486 (60%), Gaps = 30/486 (6%)
Query: 442 FEFPVYEAEDEEGDDIPYEITRLLEQERKAIQPHQEEIELINLGTEENKREIKVGAALEE 501
FEF V + I Y + + Q+ + P + + +N+ + R + + L
Sbjct: 686 FEFAVVDKP------IIYNVILVPTQDERP-NPQKGTVVQVNIDESDPSRCVGIRIDLPS 738
Query: 502 GVKRKIFQLL*EYPDIFAWSYEDMPGLDPKIVEHRIPTKPECPPVRQKLRRTHPDMALKI 561
++ ++ L + FAWS EDMPG+D + H + P P++QK R+ PD +
Sbjct: 739 ELQNELVNFLRQNAATFAWSVEDMPGIDSAVTCHELNVDPTYKPLKQKRRKLGPDRTKDV 798
Query: 562 KSEV*KQIDAGFLMTIEYPEWVANIVPVPKKDGKVRMCVDFRDLNKASPKDNFPLPHIDV 621
EV K +DAG ++ + YP+W+ N V V KK+GK R+C+DF DLNKA PKD+FPLPHID
Sbjct: 799 NEEVKKLLDAGSIVEVRYPDWLRNPVVVKKKNGKWRVCIDFTDLNKACPKDSFPLPHIDR 858
Query: 622 LVDNTAQSKVFSFMDGFSGYNQIKMSPEDREKTSFITPWGTFCYKVMPFGLINAGATYQR 681
LV+ TA +++ SFMD FSGYNQI M DREKT FIT GT+CYKVMPFGL NAGATY R
Sbjct: 859 LVEATAGNELLSFMDAFSGYNQILMHQNDREKTVFITDQGTYCYKVMPFGLKNAGATYPR 918
Query: 682 GMTTLFHDMIHKEVEVYVDDMIVKSKDEEQHVEYLTKMFERLRKYKLRLNPNKCTFGVRS 741
+ +F D + +EVY+DDM+VKS E+H+ +L + F+ L +Y ++LNP+KCTFGV S
Sbjct: 919 LVNQMFTDQLDHSMEVYIDDMLVKSLRAEEHITHLRQCFQVLNRYNMKLNPSKCTFGVTS 978
Query: 742 GKLLGFIVSQKGIEVDPDKVRAIREMPAPQTEKQVRGFLRRLNYISRFISHMTATCGPIF 801
G+ LG++V+++GIE +P ++ AI ++P+P+ ++V+ + R+ ++RFIS T C P +
Sbjct: 979 GEFLGYLVTRRGIEANPKQISAIIDLPSPRNTREVQRLIGRIAALNRFISRSTDKCLPFY 1038
Query: 802 KLLRKNQPIVWNDECQGAFDSIKNYLLEPPILVPPMEGKPLIMYLSVFDESVGCVLGQQD 861
+LLR N+ W+++C+ EG+ L +Y++V +V VL ++D
Sbjct: 1039 QLLRANKRFEWDEKCE--------------------EGETLYLYIAVSTSAVSGVLVRED 1078
Query: 862 ETGKKEHAIYYLSKKFTDCETRYMMLEKTCCAPAWAAKRLRHYLVNHTTWLISRMDPIKY 921
++H I+Y+SK E RY LEK A +A++LR Y ++T +++ P++
Sbjct: 1079 R--GEQHPIFYVSKTLDGAELRYPTLEKLAFAVVISARKLRPYFKSYTVEVLTN-QPLRT 1135
Query: 922 IFEKPA 927
I P+
Sbjct: 1136 ILHSPS 1141
>UniRef100_Q94I69 Putative retroelement [Oryza sativa]
Length = 2014
Score = 363 bits (933), Expect = 2e-98
Identities = 197/450 (43%), Positives = 278/450 (61%), Gaps = 21/450 (4%)
Query: 488 ENKREIKVGAALEEGVKRKIFQLL*EYPDIFAWSYEDMPGLDPKIVEHRIPTKPECPPVR 547
++ EI +G + K+ +LL E+ D FAW Y +MPGL IVEHR+P KP P +
Sbjct: 1081 DDLEEIDIGPEF----RAKLIELLKEFRDCFAWEYYEMPGLSRSIVEHRLPIKPGVRPHQ 1136
Query: 548 QKLRRTHPDMALKIKSEV*KQIDAGFLMTIEYPEWVANIVPVPKKDGKVRMCVDFRDLNK 607
Q LRR DM +K E+ + DA F+ Y EWV++IVPV KK+ DLNK
Sbjct: 1137 QPLRRCKADMLEPVKVEIKRLYDACFIRPCRYAEWVSSIVPVIKKN----------DLNK 1186
Query: 608 ASPKDNFPLPHIDVLVDNTAQSKVFSFMDGFSGYNQIKMSPEDREKTSFITPW--GTFCY 665
A+PKD +P+P D LVD + +K+ S+MDG GYNQI M+ ED KT+F P G F +
Sbjct: 1187 ATPKDEYPMPVADQLVDAASGNKILSYMDGNVGYNQIFMAEEDIHKTAFRCPGAIGLFEW 1246
Query: 666 KVMPFGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSKDEEQHVEYLTKMFERLRK 725
VM FGL +AGATYQR M ++HD+I VEVY+DD++VKSK+ E H+ L K+FER RK
Sbjct: 1247 VVMTFGLKSAGATYQRAMNYIYHDLIGWLVEVYIDDVVVKSKEIEDHIADLRKVFERTRK 1306
Query: 726 YKLRLNPNKCTFGVRSGKLLGFIVSQKGIEVDPDKVRAIREMPAPQTEKQVRGFLRRLNY 785
Y L++NP KC FGV G+ LGF+V ++GIEV V AI+++ P+ + +++ ++N+
Sbjct: 1307 YGLKMNPTKCAFGVSVGQFLGFLVHERGIEVTQRSVNAIKKIQPPENKTELQEMNGKINF 1366
Query: 786 ISRFISHMTATCGPIFKLLR--KNQPIVWNDECQGAFDSIKNYLLEPPILVPPMEGKPLI 843
+ RFIS+++ P LLR +Q W E Q A D IK YL PP+L+PP +G
Sbjct: 1367 VRRFISYLSGRLEPFTPLLRLKADQQFTWGAEQQKALDDIKEYLSSPPVLIPPQKGISFR 1426
Query: 844 MYLSVFDESVGCVLGQQDETGKKEHAIYYLSKKFTDCETRYMMLEKTCCAPAWAAKRLRH 903
+YLS ++S+G VL Q E KE ++YLS++ D ETRY +EK C ++ RL H
Sbjct: 1427 LYLSAGEKSIGSVLIQ--ELDGKERVVFYLSRRLLDVETRYSPMEKLCLCLYFSCTRLSH 1484
Query: 904 YLVNHTTWLISRMDPIKYIFEKPAVLLGRL 933
YL+++ +I + D IKY+ P +L GR+
Sbjct: 1485 YLLSNECTVICKADVIKYMLSAP-ILKGRV 1513
>UniRef100_Q8RZN9 Putative polyprotein [Oryza sativa]
Length = 2001
Score = 363 bits (933), Expect = 2e-98
Identities = 184/416 (44%), Positives = 267/416 (63%), Gaps = 2/416 (0%)
Query: 493 IKVGAALEEGVKRKIFQLL*EYPDIFAWSYEDMPGLDPKIVEHRIPTKPECPPVRQKLRR 552
I +G LE+ ++ +I +++ E +FAWS +++ G+D ++EH + K P +QKLRR
Sbjct: 956 ILIGENLEKHIEEEILKVVKENMAVFAWSPDELQGVDRSLIEHNLAIKSGYKPKKQKLRR 1015
Query: 553 THPDMALKIKSEV*KQIDAGFLMTIEYPEWVANIVPVPKKDGKVRMCVDFRDLNKASPKD 612
D K E+ K + A + + +PEW+AN V V K +GK RMC+DF DLNKA PKD
Sbjct: 1016 MSTDRQQAAKIELEKLLKAKVIREVMHPEWLANPVLVKKANGKWRMCIDFTDLNKACPKD 1075
Query: 613 NFPLPHIDVLVDNTAQSKVFSFMDGFSGYNQIKMSPEDREKTSFITPWGTFCYKVMPFGL 672
+FPLP ID LVD TA ++ SF+D +SGY+Q+ M ED EKTSFITP+GT+C+ MPFGL
Sbjct: 1076 DFPLPRIDQLVDATAGCELMSFLDAYSGYHQVFMVKEDEEKTSFITPFGTYCFIRMPFGL 1135
Query: 673 INAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSKDEEQHVEYLTKMFERLRKYKLRLNP 732
NAGAT+ R + + + + VE Y+DD++VKSK H + L + FE LRK ++LNP
Sbjct: 1136 KNAGATFARLIGKVLAKQLGRNVEAYIDDIVVKSKQAFTHGKDLQETFENLRKCSVKLNP 1195
Query: 733 NKCTFGVRSGKLLGFIVSQKGIEVDPDKVRAIREMPAPQTEKQVRGFLRRLNYISRFISH 792
KC FGVR+GKLLGF+VS++GIE +PDK+ AI +M P+ ++V+ R+ +SRF+S
Sbjct: 1196 EKCVFGVRAGKLLGFLVSKRGIEANPDKIAAIHQMEPPKNTREVQRLTGRMASLSRFLSK 1255
Query: 793 MTATCGPIFKLLRKNQPIVWNDECQGAFDSIKNYLLEPPILVPPMEGKPLIMYLSVFDES 852
P FK LR W ECQ AFD +K YL E P L P +G+PL+MY++ +
Sbjct: 1256 SAEKGLPFFKTLRGANTFEWTAECQQAFDDLKKYLHEMPTLASPPKGQPLLMYVAATPAT 1315
Query: 853 VGCVLGQQDETGKKEHAIYYLSKKFTDCETRYMMLEKTCCAPAWAAKRLRHYLVNH 908
V VL Q++E ++ +Y++S+ +TRY +EK A A+++LRHY ++H
Sbjct: 1316 VSAVLVQEEE--NRQVPVYFVSEALQGPKTRYSEVEKLIYAIVMASRKLRHYFLSH 1369
>UniRef100_Q5XWL3 Gag-pol polyprotein-like [Solanum tuberosum]
Length = 1716
Score = 363 bits (932), Expect = 2e-98
Identities = 172/286 (60%), Positives = 220/286 (76%), Gaps = 5/286 (1%)
Query: 591 KKDGKVRMCVDFRDLNKASPKDNFPLPHIDVLVDNTAQSKVFSFMDGFSGYNQIKMSPED 650
+K+GK+R+CVD++DLNKA PKDNF LP+I +L+DN A ++ SF+D ++GY+QI M ED
Sbjct: 1003 EKNGKIRICVDYKDLNKAGPKDNFSLPNIHILIDNCATHEMQSFVDCYAGYHQILMDEED 1062
Query: 651 REKTSFITPWGTFCYKVMPFGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSKDEE 710
EK +FITPWG + Y+VM FGL N GATY R MTT+FHDMIHKE+EV VDD+I+KS +
Sbjct: 1063 AEKNAFITPWGVYHYRVMSFGLKNGGATYIRAMTTIFHDMIHKEIEVCVDDVIIKSCESS 1122
Query: 711 QHVEYLTKMFERLRKYKLRLNPNKCTFGVRSGKLLGFIVSQKGIEVDPDKVRAIREMPAP 770
++ +L K F+RLR+Y L LN K FG LLGFIVS++GIE+DP K++AI+E+P P
Sbjct: 1123 DYLTHLRKFFDRLRRYNLNLNLTKSAFG-----LLGFIVSRRGIELDPSKIKAIQELPPP 1177
Query: 771 QTEKQVRGFLRRLNYISRFISHMTATCGPIFKLLRKNQPIVWNDECQGAFDSIKNYLLEP 830
+T+K+V FL RLNYISRFI+ T C PIFKLL+K+ W ECQ AFD+IKNYL P
Sbjct: 1178 KTKKEVMSFLGRLNYISRFIAQSTVVCQPIFKLLKKDVSTKWTGECQTAFDAIKNYLSNP 1237
Query: 831 PILVPPMEGKPLIMYLSVFDESVGCVLGQQDETGKKEHAIYYLSKK 876
P+LVPP EG L++Y+SV + + GCVLGQ DETGKKE AIYYLSKK
Sbjct: 1238 PVLVPPREGSHLLLYMSVSNNAFGCVLGQHDETGKKERAIYYLSKK 1283
>UniRef100_Q94HB8 Putative retroelement [Oryza sativa]
Length = 2079
Score = 357 bits (915), Expect = 2e-96
Identities = 177/399 (44%), Positives = 255/399 (63%), Gaps = 10/399 (2%)
Query: 516 DIFAWSYEDMPGLDPKIVEHRIPTKPECPPVRQKLRRTHPDMALKIKSEV*KQIDAGFLM 575
D+FAW DMPG+ +++EH++ +P+ PV+Q+LRR PD I+ E+ K + AGF+
Sbjct: 1069 DVFAWQPSDMPGVPREVIEHKLMVRPDAKPVKQRLRRFAPDRKQAIREELDKLLKAGFIR 1128
Query: 576 TIEYPEWVANIVPVPKKDGKVRMCVDFRDLNKASPKDNFPLPHIDVLVDNTAQSKVFSFM 635
+ +PEW+AN V V K +GK RMCVDF DLNKA PKD+FPLP ID LV +TA ++ SF+
Sbjct: 1129 EVLHPEWLANPVIVRKANGKWRMCVDFTDLNKACPKDHFPLPRIDQLVYSTAGCELLSFL 1188
Query: 636 DGFSGYNQIKMSPEDREKTSFITPWGTFCYKVMPFGLINAGATYQRGMTTLFHDMIHKEV 695
D +SGY+QI M ED EKT+FITP+G FC+ MPFGLI AG T+QR + V
Sbjct: 1189 DAYSGYHQISMVKEDEEKTAFITPFGVFCHVKMPFGLITAGNTFQR--------TLGNNV 1240
Query: 696 EVYVDDMIVKSKDEEQHVEYLTKMFERLRKYKLRLNPNKCTFGVRSGKLLGFIVSQKGIE 755
E YVD ++VK+K + ++ L + F+ LR+Y+L LNP KCTFGV SGKLLGF+VS +GIE
Sbjct: 1241 EAYVDGIVVKTKTSDSLIDDLRETFDNLRRYRLMLNPEKCTFGVPSGKLLGFLVSGRGIE 1300
Query: 756 VDPDKVRAIREMPAPQTEKQVRGFLRRLNYISRFISHMTATCGPIFKLLRKNQPIVWNDE 815
+P+K++AI M +P K+V+ + +SRF++ + P F LL+K VW E
Sbjct: 1301 ANPEKIKAIENMKSPTRLKEVQKLTGYMAALSRFVARIGEQGQPFFALLKKQDKFVWTQE 1360
Query: 816 CQGAFDSIKNYLLEPPILVPPMEGKPLIMYLSVFDESVGCVLGQQDETGKKEHAIYYLSK 875
+ AF ++K YL PP+LV P + L +Y++ SV V+ + E K + +YY+S+
Sbjct: 1361 TKEAFITLKRYLSNPPVLVAPQPNEELFLYIAATPHSVSIVIVVERE--KVQRPVYYVSE 1418
Query: 876 KFTDCETRYMMLEKTCCAPAWAAKRLRHYLVNHTTWLIS 914
D +TRY ++K A +++LRHY H ++S
Sbjct: 1419 ALHDAKTRYPQIQKLLYAIIMTSRKLRHYFQAHRVTIVS 1457
>UniRef100_Q94H67 Putative gag-pol protein [Oryza sativa]
Length = 2273
Score = 355 bits (911), Expect = 6e-96
Identities = 193/461 (41%), Positives = 276/461 (59%), Gaps = 26/461 (5%)
Query: 477 EEIELINLGTEENKREIKVGAALEEGVKRKIFQLL*EYPDIFAWSYEDMPGLDPKIVEHR 536
+++E I++G + R + L + K+ +LL E+ D FAW Y +MPGL IVEHR
Sbjct: 1334 DDLEEIDIGPGDRPRPTFICKNLSTEFRAKLIELLKEFRDCFAWEYYEMPGLSRSIVEHR 1393
Query: 537 IPTKPECPPVRQKLRRTHPDMALKIKSEV*KQIDAGFLMTIEYPEWVANIVPVPKKDGKV 596
+P KP P +Q LRR DM +K E+ + DA F+ Y EWV+N
Sbjct: 1394 LPIKPGVRPHQQPLRRCKADMLEPVKVEIKRLYDACFIRPCRYAEWVSN----------- 1442
Query: 597 RMCVDFRDLNKASPKDNFPLPHIDVLVDNTAQSKVFSFMDGFSGYNQIKMSPEDREKTSF 656
LNKA+PKD +P+P D LVD + +K+ S+MDG GYNQI M+ ED KT+F
Sbjct: 1443 --------LNKATPKDEYPMPVADQLVDAASGNKILSYMDGNVGYNQIFMAEEDIHKTAF 1494
Query: 657 ITPW--GTFCYKVMPFGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSKDEEQHVE 714
P G F + VM FGL +AGATYQR M ++HD+I VEVY+DD++VKSK+ E H+
Sbjct: 1495 RCPGAIGLFEWVVMTFGLKSAGATYQRAMNYIYHDLIGWLVEVYIDDVVVKSKEIEDHIA 1554
Query: 715 YLTKMFERLRKYKLRLNPNKCTFGVRSGKLLGFIVSQKGIEVDPDKVRAIREMPAPQTEK 774
L K+FER RKY L++NP KC FGV G+ LGF+V ++GIEV V AI+++ P+ +
Sbjct: 1555 DLRKVFERTRKYGLKMNPTKCAFGVSVGQFLGFLVHERGIEVTQRSVNAIKKIQPPENKT 1614
Query: 775 QVRGFLRRLNYISRFISHMTATCGPIFKLLR--KNQPIVWNDECQGAFDSIKNYLLEPPI 832
+++ ++N++ RFIS+++ P LLR +Q W E Q A D IK YL PP+
Sbjct: 1615 ELQEMNGKINFVRRFISYLSGRLEPFTPLLRLKADQQFTWGAEQQKALDDIKEYLSSPPV 1674
Query: 833 LVPPMEGKPLIMYLSVFDESVGCVLGQQDETGKKEHAIYYLSKKFTDCETRYMMLEKTCC 892
L+PP +G +YLS ++S+G VL Q E KE ++YLS++ D ETRY +EK C
Sbjct: 1675 LIPPQKGISFRLYLSAGEKSIGSVLIQ--ELDGKERVVFYLSRRLLDVETRYSPMEKLCL 1732
Query: 893 APAWAAKRLRHYLVNHTTWLISRMDPIKYIFEKPAVLLGRL 933
++ RL HYL+++ +I + D IKY+ P +L GR+
Sbjct: 1733 CLYFSCTRLSHYLLSNECTVICKADVIKYMLSAP-ILKGRV 1772
>UniRef100_Q7F9H3 OSJNBa0032N05.3 protein [Oryza sativa]
Length = 707
Score = 355 bits (910), Expect = 8e-96
Identities = 182/435 (41%), Positives = 269/435 (61%), Gaps = 3/435 (0%)
Query: 474 PHQEEIELINLGTEENKREIKVGAALEEGVKRKIFQLL*EYPDIFAWSYEDMPGLDPKIV 533
PH + I + + + R + +G + E I ++L + DIFAW +++ G+ ++
Sbjct: 6 PHGKVIR-VQVDDADPTRFVSLGGDMGEREAENILEVLKKNIDIFAWGPDEVGGVSADLI 64
Query: 534 EHRIPTKPECPPVRQKLRRTHPDMALKIKSEV*KQIDAGFLMTIEYPEWVANIVPVPKKD 593
H + KP+ P +QKLR+ D K+EV K + G + I++PEW+AN V V K +
Sbjct: 65 MHYLAVKPDAKPRKQKLRKMSVDRQEAAKAEVQKLLKTGVIQEIDHPEWLANPVLVRKSN 124
Query: 594 GKVRMCVDFRDLNKASPKDNFPLPHIDVLVDNTAQSKVFSFMDGFSGYNQIKMSPEDREK 653
GK RMCVDF DLNKA PKD+FPLP ID LVD+TA ++ SF+D +SGY+QI M+P K
Sbjct: 125 GKWRMCVDFTDLNKACPKDDFPLPRIDQLVDSTAGCELMSFLDAYSGYHQIHMNPAHIPK 184
Query: 654 TSFITPWGTFCYKVMPFGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSKDEEQHV 713
T+FITP+GTFC+ MPFGL NAGAT+ R + + + + + VE YVDD++VKS+ HV
Sbjct: 185 TAFITPFGTFCHLRMPFGLRNAGATFARLVYKVLYKQLGRNVEAYVDDIVVKSRKAFDHV 244
Query: 714 EYLTKMFERLRKYKLRLNPNKCTFGVRSGKLLGFIVSQKGIEVDPDKVRAIREMPAPQTE 773
L + F+ LR ++LNP KC FGVR+GKLLGF+VS++GIE +P+K+ AI++M P +
Sbjct: 245 SDLQETFDNLRVAGMKLNPEKCVFGVRAGKLLGFLVSERGIEANPEKIDAIQQMKPPSSV 304
Query: 774 KQVRGFLRRLNYISRFISHMTATCGPIFKLLRKNQPIVWNDECQGAFDSIKNYLLEPPIL 833
++V+ ++ +SRF+S P FK LR W ECQ AFD +K Y PP L
Sbjct: 305 REVQKLAGQIAALSRFLSKAAERGLPFFKTLRGAGKFSWTPECQAAFDEVKQYPQSPPTL 364
Query: 834 VPPMEGKPLIMYLSVFDESVGCVLGQQDETGKKEHAIYYLSKKFTDCETRYMMLEKTCCA 893
V P G L++YL+ +V L Q+ E+G+K +Y++S+ + RY+ +EK A
Sbjct: 365 VSPALGSELLLYLAASPVAVSAALVQETESGQK--PVYFISEALQGAKLRYIEMEKFTYA 422
Query: 894 PAWAAKRLRHYLVNH 908
A+++L+HY H
Sbjct: 423 LVMASRKLKHYFQAH 437
>UniRef100_Q9AXC3 Hypothetical protein [Antirrhinum hispanicum]
Length = 1455
Score = 353 bits (906), Expect = 2e-95
Identities = 182/396 (45%), Positives = 253/396 (62%), Gaps = 3/396 (0%)
Query: 531 KIVEHRIPTKPECPPVRQKLRRTHPDMALKIKSEV*KQIDAGFLMTIEYPEWVANIVPVP 590
K+VEH I +P+ PV+QK + P+ IKSE+ + I + ++YPEW+AN+V VP
Sbjct: 659 KMVEHYIDVRPDAKPVQQKRHKMGPERNKVIKSEIQRLIGLNQIREVQYPEWLANVVLVP 718
Query: 591 KKDGKVRMCVDFRDLNKASPKDNFPLPHIDVLVDNTAQSKVFSFMDGFSGYNQIKMSPED 650
K G+ RM +DF DLN A PKD FPLP ID LVD+TA + S MDG GYNQI M+ D
Sbjct: 719 KSGGRWRMFIDFTDLNDACPKDCFPLPSIDQLVDSTAGCALLSTMDGNQGYNQIPMAERD 778
Query: 651 REKTSFITPWGTFCYKVMPFGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSKDEE 710
+EKTSFIT +GT+CY VMPFGL NAG TYQR M +F MI +EVYVDD++ K K+
Sbjct: 779 QEKTSFITDFGTYCYLVMPFGLKNAGTTYQRLMNAMFSPMIGDNLEVYVDDIVAKCKEAS 838
Query: 711 QHVEYLTKMFERLRKYKLRLNPNKCTFGVRSGKLLGFIVSQKGIEVDPDKVRAIREMPAP 770
QH L + F+ + K+ L++N KC+FG++ GK LGF+V+Q+GI+V+P+K +AI +MP P
Sbjct: 839 QHNADLIEAFQIMEKFGLKVNLRKCSFGIQGGKFLGFLVTQRGIKVNPEKSKAILDMPPP 898
Query: 771 QTEKQVRGFLRRLNYISRFISHMTATCGPIFKLLRKNQPIVWNDECQGAFDSIKNYLLEP 830
QT + V+ R+ + RFIS P FK+L + W+DECQ AFD +K YLL P
Sbjct: 899 QTIRDVQRLTGRMASLIRFISKAAEKGLPFFKVLHNVKNFQWSDECQKAFDDLKGYLLCP 958
Query: 831 PILVPPMEGKPLIMYLSVFDESVGCVLGQQDETGKKEHAIYYLSKKFTDCETRYMMLEKT 890
P LV P +G+ L +YL+V E V VL + + G+ IYY S+ D +TRY ++EK
Sbjct: 959 PPLVKPQQGERLYVYLAVRKEVVSAVLARAE--GRDHLPIYYTSRTLKDAKTRYSLIEKF 1016
Query: 891 CCAPAWAAKRLRHYLVNHTTWLISRMDPIKYIFEKP 926
A A++LR Y + H ++++ P++ I P
Sbjct: 1017 AFALVTTAQKLRSYFLTHPITILTK-HPLRAILSHP 1051
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.346 0.153 0.549
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,425,273,526
Number of Sequences: 2790947
Number of extensions: 97761486
Number of successful extensions: 367997
Number of sequences better than 10.0: 27967
Number of HSP's better than 10.0 without gapping: 2419
Number of HSP's successfully gapped in prelim test: 25548
Number of HSP's that attempted gapping in prelim test: 360039
Number of HSP's gapped (non-prelim): 29545
length of query: 1586
length of database: 848,049,833
effective HSP length: 141
effective length of query: 1445
effective length of database: 454,526,306
effective search space: 656790512170
effective search space used: 656790512170
T: 11
A: 40
X1: 15 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.7 bits)
S2: 82 (36.2 bits)
Medicago: description of AC147714.3


