

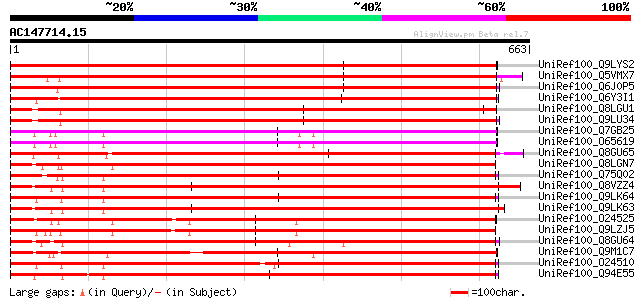
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147714.15 + phase: 0
(663 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9LYS2 ABC transporter-like protein [Arabidopsis thali... 878 0.0
UniRef100_Q5VMX7 Putative multidrug-resistance associated protei... 837 0.0
UniRef100_Q6J0P5 Multidrug-resistance associated protein 3 [Zea ... 811 0.0
UniRef100_Q6Y3I1 Multidrug resistance associated protein 1 [Zea ... 586 e-166
UniRef100_Q8LGU1 Multidrug-resistance related protein [Arabidops... 581 e-164
UniRef100_Q9LU34 Multidrug resistance-associated protein (MRP)-l... 579 e-163
UniRef100_Q7GB25 F20D22.11 protein [Arabidopsis thaliana] 537 e-151
UniRef100_O65619 Multi resistance protein [Arabidopsis thaliana] 537 e-151
UniRef100_Q8GU65 MRP-like ABC transporter [Oryza sativa] 528 e-148
UniRef100_Q8LGN7 Multidrug resistance-associated protein MRP1 [T... 523 e-147
UniRef100_Q75Q02 Multidrug resistance-associated protein [Thlasp... 521 e-146
UniRef100_Q8VZZ4 ATP-binding cassette transporter MRP6 [Arabidop... 520 e-146
UniRef100_Q9LK64 Multidrug resistance-associated protein (MRP); ... 520 e-146
UniRef100_Q9LK63 Multidrug resistance-associated protein (MRP); ... 520 e-146
UniRef100_O24525 Glutathione-conjugate transporter AtMRP4 [Arabi... 515 e-144
UniRef100_Q9LZJ5 ABC transporter-like protein [Arabidopsis thali... 514 e-144
UniRef100_Q8GU64 MRP-like ABC transporter [Oryza sativa] 514 e-144
UniRef100_Q9M1C7 Multi resistance protein homolog [Arabidopsis t... 513 e-144
UniRef100_O24510 MRP-like ABC transporter [Arabidopsis thaliana] 511 e-143
UniRef100_Q94E55 MRP-like ABC transporter [Oryza sativa] 510 e-143
>UniRef100_Q9LYS2 ABC transporter-like protein [Arabidopsis thaliana]
Length = 1389
Score = 878 bits (2268), Expect = 0.0
Identities = 432/622 (69%), Positives = 520/622 (83%)
Query: 1 MSEGVIQQEGPYQQLLATSKEFQDLVNAHKVTDGSNQLVNVTFSRASIKITQTLVENKGK 60
MS+G I + YQ+LLA S++FQDLVNAH+ T GS ++V V +K ++ ++ K
Sbjct: 734 MSDGEITEADTYQELLARSRDFQDLVNAHRETAGSERVVAVENPTKPVKEINRVISSQSK 793
Query: 61 EANGNQLIKQEEREKGDKGLKPYLQYLNQMKGYIFFFVASLGHFIFLVCQILQNLWMAAN 120
++LIKQEEREKGD GL+PY+QY+NQ KGYIFFF+ASL F V QILQN WMAAN
Sbjct: 794 VLKPSRLIKQEEREKGDTGLRPYIQYMNQNKGYIFFFIASLAQVTFAVGQILQNSWMAAN 853
Query: 121 VDNPRVSTFQLIFVYFLLGASSAFFMLTRSLFVIALGLQSSKYLFLQLMNSLFRAPMPFY 180
VDNP+VST +LI VY L+G S ++ RS+ V+ + ++SS LF QL+NSLFRAPM FY
Sbjct: 854 VDNPQVSTLKLILVYLLIGLCSVLCLMVRSVCVVIMCMKSSASLFSQLLNSLFRAPMSFY 913
Query: 181 DCTPLGRILSRVSSELSIMDLDIPFSLTFAVGTTMNFYSTLTVFSVVTWQVLIVAIPMVY 240
D TPLGRILSRVSS+LSI+DLD+PF L F V +++N +L V ++VTWQVL V++PMVY
Sbjct: 914 DSTPLGRILSRVSSDLSIVDLDVPFGLIFVVASSVNTGCSLGVLAIVTWQVLFVSVPMVY 973
Query: 241 ITIRLQRYYFASAKEVMRITGTTKSYVANHVAETVSGAVTIRTFEEEDRFFQKNLDLIDI 300
+ RLQ+YYF +AKE+MRI GTT+SYVANH+AE+V+GA+TIR F+EE+RFF+K+L LID
Sbjct: 974 LAFRLQKYYFQTAKELMRINGTTRSYVANHLAESVAGAITIRAFDEEERFFKKSLTLIDT 1033
Query: 301 NASSFFHNFASNEWLIQRLETISAGVLASAALCMVILPPGTFTSGFIGMALSYGLALNSF 360
NAS FFH+FA+NEWLIQRLET+SA VLAS A CM++LP GTF+SGFIGMALSYGL+LN
Sbjct: 1034 NASPFFHSFAANEWLIQRLETVSAIVLASTAFCMILLPTGTFSSGFIGMALSYGLSLNMG 1093
Query: 361 LVNSIQSQCTLANQIISVERLNQYMHIQSEAKEIVEGNRPPLNWPIAGKVEINDLKIRYR 420
LV S+Q+QC LAN IISVERLNQY H+ EA E++E RPP+NWP+ G+VEI+DL+IRYR
Sbjct: 1094 LVYSVQNQCYLANWIISVERLNQYTHLTPEAPEVIEETRPPVNWPVTGRVEISDLQIRYR 1153
Query: 421 PDGPLVLHGITCTFEVGHKIGIVGRTGSGKSTLISALFRLVEPSGGNIIIDGVDISSIGL 480
+ PLVL GI+CTFE GHKIGIVGRTGSGK+TLISALFRLVEP GG I++DGVDIS IG+
Sbjct: 1154 RESPLVLKGISCTFEGGHKIGIVGRTGSGKTTLISALFRLVEPVGGKIVVDGVDISKIGV 1213
Query: 481 HDLRSRFGIIPQDPTLFTGTVRYNLDPLSQYSDQEIWEVLGKCQLREVVQEKDEGLNSSV 540
HDLRSRFGIIPQDPTLF GTVR+NLDPL Q+SD EIWEVLGKCQL+EVVQEK+ GL+S V
Sbjct: 1214 HDLRSRFGIIPQDPTLFNGTVRFNLDPLCQHSDAEIWEVLGKCQLKEVVQEKENGLDSLV 1273
Query: 541 VEDGSNWSMGQRQLFCLGRALLRRSRILVLDEATASVDNSTDYILLKTIRTEFADCTVIT 600
VEDGSNWSMGQRQLFCLGRA+LRRSR+LVLDEATAS+DN+TD IL KTIR EFADCTVIT
Sbjct: 1274 VEDGSNWSMGQRQLFCLGRAVLRRSRVLVLDEATASIDNATDLILQKTIRREFADCTVIT 1333
Query: 601 VAHRIPTVMDCTMVLSINDGNL 622
VAHRIPTVMDCTMVLSI+DG +
Sbjct: 1334 VAHRIPTVMDCTMVLSISDGRI 1355
Score = 65.5 bits (158), Expect = 5e-09
Identities = 51/198 (25%), Positives = 88/198 (43%), Gaps = 14/198 (7%)
Query: 427 LHGITCTFEVGHKIGIVGRTGSGKSTLISALFRLVEPSGGNIIIDGVDISSIGLHDLRSR 486
L ++ + G K+ + G GSGKSTL++A+ G I D
Sbjct: 556 LRNVSLEVKFGEKVAVCGEVGSGKSTLLAAILGETPCVSGTI-------------DFYGT 602
Query: 487 FGIIPQDPTLFTGTVRYNLDPLSQYSDQEIWEVLGKCQLREVVQEKDEGLNSSVVEDGSN 546
+ Q + TGT+R N+ + E + K L + ++ +G + + E G N
Sbjct: 603 IAYVSQTAWIQTGTIRDNILFGGVMDEHRYRETIQKSSLDKDLELLPDGDQTEIGERGVN 662
Query: 547 WSMGQRQLFCLGRALLRRSRILVLDEATASVDNSTDYILLKT-IRTEFADCTVITVAHRI 605
S GQ+Q L RAL + + I +LD+ ++VD T L + + A V+ V H++
Sbjct: 663 LSGGQKQRIQLARALYQDADIYLLDDPFSAVDAHTASSLFQEYVMDALAGKAVLLVTHQV 722
Query: 606 PTVMDCTMVLSINDGNLS 623
+ VL ++DG ++
Sbjct: 723 DFLPAFDSVLLMSDGEIT 740
>UniRef100_Q5VMX7 Putative multidrug-resistance associated protein [Oryza sativa]
Length = 1474
Score = 837 bits (2162), Expect = 0.0
Identities = 419/629 (66%), Positives = 507/629 (79%), Gaps = 7/629 (1%)
Query: 1 MSEGVIQQEGPYQQLLATSKEFQDLVNAHKVTDGSNQLVNVTFSRA---SIKITQTLVEN 57
MS+G I + PYQ LL +EFQDLVNAHK T G + L N+ R S++ T + +
Sbjct: 813 MSDGKIIRSAPYQDLLEYCQEFQDLVNAHKDTIGISDLNNMPLHREKEISMEETDDIHGS 872
Query: 58 KGKEA----NGNQLIKQEEREKGDKGLKPYLQYLNQMKGYIFFFVASLGHFIFLVCQILQ 113
+ +E+ +QLIK+EERE GD GLKPY+ YL Q KG+++ + + H IF+ QI Q
Sbjct: 873 RYRESVKPSPADQLIKKEEREIGDTGLKPYILYLRQNKGFLYLSICVISHIIFISGQISQ 932
Query: 114 NLWMAANVDNPRVSTFQLIFVYFLLGASSAFFMLTRSLFVIALGLQSSKYLFLQLMNSLF 173
N WMAANV NP VST +LI VY +G + FF+L+RSL ++ LG+Q+S+ LF QL+NSLF
Sbjct: 933 NSWMAANVQNPSVSTLKLIVVYIAIGVCTLFFLLSRSLSIVVLGMQTSRSLFSQLLNSLF 992
Query: 174 RAPMPFYDCTPLGRILSRVSSELSIMDLDIPFSLTFAVGTTMNFYSTLTVFSVVTWQVLI 233
RAPM F+D TPLGR+LSRVSS+LSI+DLD+PF F++ ++N YS L V +V+TWQVL
Sbjct: 993 RAPMSFFDSTPLGRVLSRVSSDLSIVDLDVPFFFMFSISASLNAYSNLGVLAVITWQVLF 1052
Query: 234 VAIPMVYITIRLQRYYFASAKEVMRITGTTKSYVANHVAETVSGAVTIRTFEEEDRFFQK 293
+++PM+ + IRLQRYY ASAKE+MRI GTTKS +ANH+ E++SGA+TIR FEEEDRFF K
Sbjct: 1053 ISVPMIVLVIRLQRYYLASAKELMRINGTTKSSLANHLGESISGAITIRAFEEEDRFFAK 1112
Query: 294 NLDLIDINASSFFHNFASNEWLIQRLETISAGVLASAALCMVILPPGTFTSGFIGMALSY 353
NL+L+D NA F+NFA+ EWLIQRLE +SA VL+ +AL MVILPPGTF+ GF+GMALSY
Sbjct: 1113 NLELVDKNAGPCFYNFAATEWLIQRLELMSAAVLSFSALVMVILPPGTFSPGFVGMALSY 1172
Query: 354 GLALNSFLVNSIQSQCTLANQIISVERLNQYMHIQSEAKEIVEGNRPPLNWPIAGKVEIN 413
GL+LN LV SIQ+QC LANQIISVER+NQYM I SEA E+++ NRP +WP GKVE+
Sbjct: 1173 GLSLNMSLVFSIQNQCNLANQIISVERVNQYMDITSEAAEVIKENRPAPDWPQVGKVELR 1232
Query: 414 DLKIRYRPDGPLVLHGITCTFEVGHKIGIVGRTGSGKSTLISALFRLVEPSGGNIIIDGV 473
DLKI+YR D PLVLHGITCTFE GHKIGIVGRTGSGK+TLI LFRLVEP+GG IIID V
Sbjct: 1233 DLKIKYRQDAPLVLHGITCTFEGGHKIGIVGRTGSGKTTLIGGLFRLVEPAGGKIIIDSV 1292
Query: 474 DISSIGLHDLRSRFGIIPQDPTLFTGTVRYNLDPLSQYSDQEIWEVLGKCQLREVVQEKD 533
DI++IGLHDLRSR GIIPQDPTLF GT+RYNLDPL Q+SDQ+IWEVL KCQL E VQEK+
Sbjct: 1293 DITTIGLHDLRSRLGIIPQDPTLFQGTLRYNLDPLGQFSDQQIWEVLDKCQLLETVQEKE 1352
Query: 534 EGLNSSVVEDGSNWSMGQRQLFCLGRALLRRSRILVLDEATASVDNSTDYILLKTIRTEF 593
+GL+S VVEDGSNWSMGQRQLFCLGRALLRR RILVLDEATAS+DN+TD IL KTIRTEF
Sbjct: 1353 QGLDSLVVEDGSNWSMGQRQLFCLGRALLRRCRILVLDEATASIDNATDAILQKTIRTEF 1412
Query: 594 ADCTVITVAHRIPTVMDCTMVLSINDGNL 622
DCTVITVAHRIPTVMDCTMVL+++DG +
Sbjct: 1413 KDCTVITVAHRIPTVMDCTMVLAMSDGKV 1441
Score = 80.5 bits (197), Expect = 1e-13
Identities = 67/234 (28%), Positives = 111/234 (46%), Gaps = 18/234 (7%)
Query: 427 LHGITCTFEVGHKIGIVGRTGSGKSTLISALFRLVEPSGGNIIIDGVDISSIGLHDLRSR 486
L I + G K+ I G GSGKSTL++++ V + G I + G +
Sbjct: 635 LRNINLVVKSGEKVAICGEVGSGKSTLLASVLGEVPKTEGTIQVCG-------------K 681
Query: 487 FGIIPQDPTLFTGTVRYNLDPLSQYSDQEIWEVLGKCQLREVVQEKDEGLNSSVVEDGSN 546
+ Q+ + TGTV+ N+ S +Q E L KC L + + G ++ + E G N
Sbjct: 682 IAYVSQNAWIQTGTVQENILFGSLMDEQRYKETLEKCSLEKDLAMLPHGDSTQIGERGVN 741
Query: 547 WSMGQRQLFCLGRALLRRSRILVLDEATASVDNSTDYILL-KTIRTEFADCTVITVAHRI 605
S GQ+Q L RAL + + I +LD+ ++VD T L + + +D TV+ V H++
Sbjct: 742 LSGGQKQRVQLARALYQNADIYLLDDPFSAVDAHTASSLFNEYVMGALSDKTVLLVTHQV 801
Query: 606 PTVMDCTMVLSINDGNLSRNT---GLIFSLQSHIDVNNYFLYTVSILSL-SLPL 655
+ +L ++DG + R+ L+ Q D+ N T+ I L ++PL
Sbjct: 802 DFLPVFDSILLMSDGKIIRSAPYQDLLEYCQEFQDLVNAHKDTIGISDLNNMPL 855
>UniRef100_Q6J0P5 Multidrug-resistance associated protein 3 [Zea mays]
Length = 1480
Score = 811 bits (2096), Expect = 0.0
Identities = 409/629 (65%), Positives = 497/629 (78%), Gaps = 7/629 (1%)
Query: 1 MSEGVIQQEGPYQQLLATSKEFQDLVNAHKVTDGSNQLVNVTFSRASIKITQTLVENKG- 59
MS+G I + Y LLA +EFQ+LVNAHK T G + L V R + + + ++ G
Sbjct: 822 MSDGQIIRSASYHDLLAYCQEFQNLVNAHKDTIGVSDLNRVPPHRENEILIKETIDVHGS 881
Query: 60 ------KEANGNQLIKQEEREKGDKGLKPYLQYLNQMKGYIFFFVASLGHFIFLVCQILQ 113
K + +QLIK EERE GD GLKPY+ YL Q KG+ + + + H +F+ QI Q
Sbjct: 882 RYKESLKPSPTDQLIKTEEREMGDTGLKPYILYLRQNKGFFYASLGIISHIVFVCGQISQ 941
Query: 114 NLWMAANVDNPRVSTFQLIFVYFLLGASSAFFMLTRSLFVIALGLQSSKYLFLQLMNSLF 173
N WMA NV+NP VST +L VY +G S FF+L RSL V+ LG+++S+ LF QL+NSLF
Sbjct: 942 NSWMATNVENPDVSTLKLTSVYIAIGIFSVFFLLFRSLAVVVLGVKTSRSLFSQLLNSLF 1001
Query: 174 RAPMPFYDCTPLGRILSRVSSELSIMDLDIPFSLTFAVGTTMNFYSTLTVFSVVTWQVLI 233
RAPM FYD TPLGRILSRVSS+LSI+DLDIPF F++G +N YS L V +VVTWQVL
Sbjct: 1002 RAPMSFYDSTPLGRILSRVSSDLSIVDLDIPFGFMFSIGAGINAYSNLGVLAVVTWQVLF 1061
Query: 234 VAIPMVYITIRLQRYYFASAKEVMRITGTTKSYVANHVAETVSGAVTIRTFEEEDRFFQK 293
V++PM+ + IRLQRYY AS+KE+MRI GTTKS +ANH+ ++++GA+TIR F+EEDRFF+K
Sbjct: 1062 VSLPMIVLAIRLQRYYLASSKELMRINGTTKSALANHLGKSIAGAITIRAFQEEDRFFEK 1121
Query: 294 NLDLIDINASSFFHNFASNEWLIQRLETISAGVLASAALCMVILPPGTFTSGFIGMALSY 353
NL+L+D NA +F+NFA+ EWLIQRLET+SA VL+ +AL M +LP GTF GF+GMALSY
Sbjct: 1122 NLELVDKNAGPYFYNFAATEWLIQRLETMSAAVLSFSALIMALLPQGTFNPGFVGMALSY 1181
Query: 354 GLALNSFLVNSIQSQCTLANQIISVERLNQYMHIQSEAKEIVEGNRPPLNWPIAGKVEIN 413
GL+LN V SIQ+QC LA+QIISVER++QYM I SEA EI+E NRP +WP G+V++
Sbjct: 1182 GLSLNISFVFSIQNQCQLASQIISVERVHQYMDIPSEAAEIIEENRPAPDWPQVGRVDLK 1241
Query: 414 DLKIRYRPDGPLVLHGITCTFEVGHKIGIVGRTGSGKSTLISALFRLVEPSGGNIIIDGV 473
DLKIRYR D PLVLHGITC+F G KIGIVGRTGSGK+TLI ALFRLVEP+GG IIID +
Sbjct: 1242 DLKIRYRQDAPLVLHGITCSFHGGDKIGIVGRTGSGKTTLIGALFRLVEPTGGKIIIDSI 1301
Query: 474 DISSIGLHDLRSRFGIIPQDPTLFTGTVRYNLDPLSQYSDQEIWEVLGKCQLREVVQEKD 533
DI++IGLHDLRSR GIIPQDPTLF GT+RYNLDPL Q+SDQ+IWEVLGKCQL E VQEK+
Sbjct: 1302 DITTIGLHDLRSRLGIIPQDPTLFQGTIRYNLDPLGQFSDQQIWEVLGKCQLLEAVQEKE 1361
Query: 534 EGLNSSVVEDGSNWSMGQRQLFCLGRALLRRSRILVLDEATASVDNSTDYILLKTIRTEF 593
+GL+S VVEDGSNWSMGQRQLFCLGRALLRR RILVLDEATAS+DN+TD IL KTIRTEF
Sbjct: 1362 QGLDSLVVEDGSNWSMGQRQLFCLGRALLRRCRILVLDEATASIDNATDAILQKTIRTEF 1421
Query: 594 ADCTVITVAHRIPTVMDCTMVLSINDGNL 622
DCTVITVAHRIPTVMDC MVL+++DG +
Sbjct: 1422 RDCTVITVAHRIPTVMDCDMVLAMSDGKV 1450
Score = 75.9 bits (185), Expect = 4e-12
Identities = 57/200 (28%), Positives = 95/200 (47%), Gaps = 14/200 (7%)
Query: 427 LHGITCTFEVGHKIGIVGRTGSGKSTLISALFRLVEPSGGNIIIDGVDISSIGLHDLRSR 486
L + + G K+ I G GSGKSTL++A+ V + G I + G +
Sbjct: 644 LKNVNLVVKTGQKVAICGEVGSGKSTLLAAVLGEVPKTEGTIQVCG-------------K 690
Query: 487 FGIIPQDPTLFTGTVRYNLDPLSQYSDQEIWEVLGKCQLREVVQEKDEGLNSSVVEDGSN 546
+ Q+ + TGTV+ N+ S Q E L +C L + ++ G + + E G N
Sbjct: 691 TAYVSQNAWIQTGTVQDNILFGSSMDRQRYQETLERCSLVKDLEMLPYGDRTQIGERGIN 750
Query: 547 WSMGQRQLFCLGRALLRRSRILVLDEATASVDNSTDYILLK-TIRTEFADCTVITVAHRI 605
S GQ+Q L RAL + + I +LD+ ++VD T L + +D TV+ V H++
Sbjct: 751 LSGGQKQRVQLARALYQNADIYLLDDPFSAVDAHTATSLFSGYVMGALSDKTVLLVTHQV 810
Query: 606 PTVMDCTMVLSINDGNLSRN 625
+ +L ++DG + R+
Sbjct: 811 DFLPVFDSILLMSDGQIIRS 830
>UniRef100_Q6Y3I1 Multidrug resistance associated protein 1 [Zea mays]
Length = 1477
Score = 586 bits (1511), Expect = e-166
Identities = 305/639 (47%), Positives = 416/639 (64%), Gaps = 18/639 (2%)
Query: 1 MSEGVIQQEGPYQQLLATSKEFQDLVNAHKVT-----------------DGSNQLVNVTF 43
M G + Q+G Y +LL + F+ LV+AH+ + S++ + +
Sbjct: 807 MEGGQVSQQGKYSELLGSGTAFEKLVSAHQSSITALDTSASQQNQVQGQQESDEYIVPSA 866
Query: 44 SRASIKITQTLVENKGKEANGNQLIKQEEREKGDKGLKPYLQYLNQMKGYIFFFVASLGH 103
+ + + V KG A QL ++EE+ GD G KPY +Y+N KG F +
Sbjct: 867 LQVIRQASDIDVTAKGPSA-AIQLTEEEEKGIGDLGWKPYKEYINVSKGAFQFSGMCIAQ 925
Query: 104 FIFLVCQILQNLWMAANVDNPRVSTFQLIFVYFLLGASSAFFMLTRSLFVIALGLQSSKY 163
+F QI W+A V VS L+ Y L S FF RS F LGL++SK
Sbjct: 926 VLFTCFQIASTYWLAVAVQMGNVSAALLVGAYSGLSIFSCFFAYFRSCFAAILGLKASKA 985
Query: 164 LFLQLMNSLFRAPMPFYDCTPLGRILSRVSSELSIMDLDIPFSLTFAVGTTMNFYSTLTV 223
F LM+S+F+APM F+D TP+GRIL+R SS+LSI+D DIP+S+ F + +T+ V
Sbjct: 986 FFGGLMDSVFKAPMSFFDSTPVGRILTRASSDLSILDFDIPYSMAFVATGGIEVVTTVLV 1045
Query: 224 FSVVTWQVLIVAIPMVYITIRLQRYYFASAKEVMRITGTTKSYVANHVAETVSGAVTIRT 283
VTWQVL+VAIP+ I +QR+Y +SA+E++R+ GTTK+ V N+ +E++ G VTIR
Sbjct: 1046 MGTVTWQVLVVAIPVAVTMIYVQRHYVSSARELVRLNGTTKAPVMNYASESILGVVTIRA 1105
Query: 284 FEEEDRFFQKNLDLIDINASSFFHNFASNEWLIQRLETISAGVLASAALCMVILPPGTFT 343
F +RF N+ LID +A+ FFH A+ EW++ R+E + + + +AAL +V++PPG +
Sbjct: 1106 FAATERFIYSNMQLIDTDATLFFHTIAAQEWVLIRVEALQSLTIITAALFLVLVPPGAIS 1165
Query: 344 SGFIGMALSYGLALNSFLVNSIQSQCTLANQIISVERLNQYMHIQSEAKEIVEGNRPPLN 403
GF G+ LSY L L S + + L N IISVER+ QYMH+ E I+ +RPP +
Sbjct: 1166 PGFAGLCLSYALTLTSAQIFLTRFYSYLENYIISVERIKQYMHLPVEPPAIIPDSRPPTS 1225
Query: 404 WPIAGKVEINDLKIRYRPDGPLVLHGITCTFEVGHKIGIVGRTGSGKSTLISALFRLVEP 463
WP G++++ DLKIRYRP+ PLVL GITCTF G+KIG+VGRTGSGKSTLIS+LFRLV+P
Sbjct: 1226 WPQEGRIDLQDLKIRYRPNAPLVLKGITCTFAAGNKIGVVGRTGSGKSTLISSLFRLVDP 1285
Query: 464 SGGNIIIDGVDISSIGLHDLRSRFGIIPQDPTLFTGTVRYNLDPLSQYSDQEIWEVLGKC 523
+GG I+ID +DI SIGL DLR++ IIPQ+PTLF GTVR NLDPL Q+SD+EIWE L KC
Sbjct: 1286 AGGRILIDKLDICSIGLKDLRTKLSIIPQEPTLFRGTVRNNLDPLGQHSDEEIWEALEKC 1345
Query: 524 QLREVVQEKDEGLNSSVVEDGSNWSMGQRQLFCLGRALLRRSRILVLDEATASVDNSTDY 583
QL+ + L++ V +DG NWS GQRQLFCLGR LLRR++ILVLDEATAS+D++TD
Sbjct: 1346 QLKTAISTTSALLDTVVSDDGDNWSAGQRQLFCLGRVLLRRNKILVLDEATASIDSATDA 1405
Query: 584 ILLKTIRTEFADCTVITVAHRIPTVMDCTMVLSINDGNL 622
IL K IR +F+ CTVIT+AHR+PTV D V+ ++ G L
Sbjct: 1406 ILQKVIRQQFSSCTVITIAHRVPTVTDSDKVMVLSYGKL 1444
Score = 69.3 bits (168), Expect = 3e-10
Identities = 49/201 (24%), Positives = 94/201 (46%), Gaps = 14/201 (6%)
Query: 425 LVLHGITCTFEVGHKIGIVGRTGSGKSTLISALFRLVEPSGGNIIIDGVDISSIGLHDLR 484
L L + G K+ + G GSGKS+L+ AL + G++ + G
Sbjct: 627 LSLRNVNLRVNRGEKVAVCGPVGSGKSSLLYALLGEIPRLSGSVEVFG------------ 674
Query: 485 SRFGIIPQDPTLFTGTVRYNLDPLSQYSDQEIWEVLGKCQLREVVQEKDEGLNSSVVEDG 544
+ Q + +GTVR N+ ++ + + + C L + ++ D G + + + G
Sbjct: 675 -SVAYVSQSSWIQSGTVRDNILFGKPFNKELYDKAIKSCALDKDIENFDHGDLTEIGQRG 733
Query: 545 SNWSMGQRQLFCLGRALLRRSRILVLDEATASVDNSTDYILL-KTIRTEFADCTVITVAH 603
N S GQ+Q L RA+ + + +LD+ ++VD T +L + + T A+ TV+ V H
Sbjct: 734 LNMSGGQKQRIQLARAVYSDADVYLLDDPFSAVDAHTAAVLFYECVMTALAEKTVVLVTH 793
Query: 604 RIPTVMDCTMVLSINDGNLSR 624
++ + + +L + G +S+
Sbjct: 794 QVEFLTETDRILVMEGGQVSQ 814
>UniRef100_Q8LGU1 Multidrug-resistance related protein [Arabidopsis thaliana]
Length = 1294
Score = 581 bits (1498), Expect = e-164
Identities = 300/640 (46%), Positives = 415/640 (63%), Gaps = 24/640 (3%)
Query: 1 MSEGVIQQEGPYQQLLATSKEFQDLVNAHKVTDGSNQLVNVTFSRASIKITQTLVENKGK 60
M EG I Q G Y++LL FQ LVNAH N V V ++ + E K +
Sbjct: 624 MEEGTITQSGKYEELLMMGTAFQQLVNAH------NDAVTVLPLASNESLGDLRKEGKDR 677
Query: 61 EAN------------------GNQLIKQEEREKGDKGLKPYLQYLNQMKGYIFFFVASLG 102
E G QL ++EE+E G G+KP+L Y+ +G+ + + LG
Sbjct: 678 EIRNMTVVEKIEEEIEKTDIPGVQLTQEEEKESGYVGMKPFLDYIGVSRGWCLLWSSVLG 737
Query: 103 HFIFLVCQILQNLWMAANVDNPRVSTFQLIFVYFLLGASSAFFMLTRSLFVIALGLQSSK 162
F+V Q W+A + P+++ LI VY ++ SA F+ R++ LGL++SK
Sbjct: 738 QVGFVVFQAASTYWLAFAIGIPKITNTMLIGVYSIISTLSAGFVYARAITTAHLGLKASK 797
Query: 163 YLFLQLMNSLFRAPMPFYDCTPLGRILSRVSSELSIMDLDIPFSLTFAVGTTMNFYSTLT 222
F N++F+APM F+D TP+GRIL+R SS+L+++D D+PF+ F V + + L
Sbjct: 798 AFFSGFTNAVFKAPMLFFDSTPVGRILTRASSDLNVLDYDVPFAFIFVVAPAVELTAALL 857
Query: 223 VFSVVTWQVLIVAIPMVYITIRLQRYYFASAKEVMRITGTTKSYVANHVAETVSGAVTIR 282
+ + VTWQV+I+A+ + T +Q YY ASA+E++RI GTTK+ V N+ AET G VTIR
Sbjct: 858 IMTYVTWQVIIIALLALAATKVVQDYYLASARELIRINGTTKAPVMNYAAETSLGVVTIR 917
Query: 283 TFEEEDRFFQKNLDLIDINASSFFHNFASNEWLIQRLETISAGVLASAALCMVILPPGTF 342
F +RFF+ L+L+D +A FF + A+ EW+I R+ET+ L + AL ++++P G
Sbjct: 918 AFGTAERFFKNYLNLVDADAVLFFLSNAAMEWVILRIETLQNVTLFTCALLLILIPKGYI 977
Query: 343 TSGFIGMALSYGLALNSFLVNSIQSQCTLANQIISVERLNQYMHIQSEAKEIVEGNRPPL 402
G +G++LSY L L V + CTL+N IISVER+ QYM+I E I++ RPP
Sbjct: 978 APGLVGLSLSYALTLTQTQVFLTRWYCTLSNSIISVERIKQYMNIPEEPPAIIDDKRPPS 1037
Query: 403 NWPIAGKVEINDLKIRYRPDGPLVLHGITCTFEVGHKIGIVGRTGSGKSTLISALFRLVE 462
+WP G + + +LKIRYRP+ PLVL GI+CTF G ++G+VGRTGSGKSTLISALFRLVE
Sbjct: 1038 SWPSNGTIHLQELKIRYRPNAPLVLKGISCTFREGTRVGVVGRTGSGKSTLISALFRLVE 1097
Query: 463 PSGGNIIIDGVDISSIGLHDLRSRFGIIPQDPTLFTGTVRYNLDPLSQYSDQEIWEVLGK 522
P+ G I+IDG+DIS IGL DLR + IIPQ+PT F G +R NLDPL YSD EIW+ L K
Sbjct: 1098 PASGCILIDGIDISKIGLKDLRMKLSIIPQEPTFFRGCIRTNLDPLGVYSDDEIWKALEK 1157
Query: 523 CQLREVVQEKDEGLNSSVVEDGSNWSMGQRQLFCLGRALLRRSRILVLDEATASVDNSTD 582
CQL+ + L+SSV ++G NWS+GQRQLFCLGR LL+R++ILVLDEATAS+D++TD
Sbjct: 1158 CQLKTTISNLPNKLDSSVSDEGENWSVGQRQLFCLGRVLLKRNKILVLDEATASIDSATD 1217
Query: 583 YILLKTIRTEFADCTVITVAHRIPTVMDCTMVLSINDGNL 622
I+ + IR EFADCTVITVAHR+PTV+D MV+ ++ G+L
Sbjct: 1218 AIIQRIIREEFADCTVITVAHRVPTVIDSDMVMVLSFGDL 1257
Score = 53.5 bits (127), Expect = 2e-05
Identities = 54/232 (23%), Positives = 100/232 (42%), Gaps = 18/232 (7%)
Query: 376 ISVERLNQYMHIQSEAKEIVEGNRPPLNWPIAGKVEINDLKIRYRPDGPL-VLHGITCTF 434
+S +RLN ++ + E K + E R L+ V+I + P+ + L I
Sbjct: 408 VSFQRLNNFL-LDDELK-MDEIERSGLD-ASGTAVDIQVGNFGWEPETKIPTLRNIHLEI 464
Query: 435 EVGHKIGIVGRTGSGKSTLISALFRLVEPSGGNIIIDGVDISSIGLHDLRSRFGIIPQDP 494
+ G K+ + G G+GKS+L+ A+ + G + + G + Q
Sbjct: 465 KHGQKVAVCGPVGAGKSSLLHAVLGEIPKVSGTVKVFG-------------SIAYVSQTS 511
Query: 495 TLFTGTVRYNLDPLSQYSDQEIWEVLGKCQLREVVQEKDEGLNSSVVEDGSNWSMGQRQL 554
+ +GT+R N+ + + C L + + G + + + G N S GQ+Q
Sbjct: 512 WIQSGTIRDNILYGKPMESRRYNAAIKACALDKDMNGFGHGDLTEIGQRGINLSGGQKQR 571
Query: 555 FCLGRALLRRSRILVLDEATASVDNSTDYILL-KTIRTEFADCTVITVAHRI 605
L RA+ + + +LD+ ++VD T +L K + + TVI V H++
Sbjct: 572 IQLARAVYADADVYLLDDPFSAVDAHTAGVLFHKCVEDSLKEKTVILVTHQV 623
>UniRef100_Q9LU34 Multidrug resistance-associated protein (MRP)-like; ABC-transporter-
like protein [Arabidopsis thaliana]
Length = 1306
Score = 579 bits (1492), Expect = e-163
Identities = 302/641 (47%), Positives = 415/641 (64%), Gaps = 25/641 (3%)
Query: 1 MSEGVIQQEGPYQQLLATSKEFQDLVNAHKVTDGSNQLVNVTFSRASIKITQTLVENKGK 60
M EG I Q G Y++LL FQ LVNAH N V V ++ + E K +
Sbjct: 635 MEEGTITQSGKYEELLMMGTAFQQLVNAH------NDAVTVLPLASNESLGDLRKEGKDR 688
Query: 61 EAN------------------GNQLIKQEEREKGDKGLKPYLQYLNQMKGYIFFFVASLG 102
E G QL ++EE+E G G+KP+L Y+ +G+ + + LG
Sbjct: 689 EIRNMTVVEKIEEEIEKTDIPGVQLTQEEEKESGYVGMKPFLDYIGVSRGWCLLWSSVLG 748
Query: 103 HFIFLVCQILQNLWMAANVDNPRVSTFQLIFVYFLLGASSAFFMLTRSLFVIALGLQSSK 162
F+V Q W+A + P+++ LI VY ++ SA F+ R++ LGL++SK
Sbjct: 749 QVGFVVFQAASTYWLAFAIGIPKITNTMLIGVYSIISTLSAGFVYARAITTAHLGLKASK 808
Query: 163 YLFLQLMNSLFRAPMPFYDCTPLGRILSRVSSELSIMDLDIPFSLTFAVGTTMNFYSTLT 222
F N++F+APM F+D TP+GRIL+R SS+L+++D D+PF+ F V + + L
Sbjct: 809 AFFSGFTNAVFKAPMLFFDSTPVGRILTRASSDLNVLDYDVPFAFIFVVAPAVELTAALL 868
Query: 223 VFSVVTWQVLIVAIPMVYITIRLQRYYFASAKEVMRITGTTKSYVANHVAETVSGAVTIR 282
+ + VTWQV+I+A+ + T +Q YY ASA+E++RI GTTK+ V N+ AET G VTIR
Sbjct: 869 IMTYVTWQVIIIALLALAATKVVQDYYLASARELIRINGTTKAPVMNYAAETSLGVVTIR 928
Query: 283 TFEEEDRFFQKNLDLIDINASSFFHNFASNEWLIQRLETISAGVLASAALCMVILPPGTF 342
F +RFF+ L+L+D +A FF + A+ EW+I R+ET+ L + AL ++++P G
Sbjct: 929 AFGTAERFFKNYLNLVDADAVLFFLSNAAMEWVILRIETLQNVTLFTCALLLILIPKGYI 988
Query: 343 TSGFIGMALSYGLALNSFLVNSIQSQCTLANQIISVERLNQYMHIQSEAKEIVEGNRPPL 402
G +G++LSY L L V + CTL+N IISVER+ QYM+I E I++ RPP
Sbjct: 989 APGLVGLSLSYALTLTQTQVFLTRWYCTLSNSIISVERIKQYMNIPEEPPAIIDDKRPPS 1048
Query: 403 NWPIAGKVEINDLKIRYRPDGPLVLHGITCTFEVGHKIGIVGRTGSGKSTLISALFRLVE 462
+WP G + + +LKIRYRP+ PLVL GI+CTF G ++G+VGRTGSGKSTLISALFRLVE
Sbjct: 1049 SWPSNGTIHLQELKIRYRPNAPLVLKGISCTFREGTRVGVVGRTGSGKSTLISALFRLVE 1108
Query: 463 PSGGNIIIDGVDISSIGLHDLRSRFGIIPQDPTLFTGTVRYNLDPLSQYSDQEIWEVLGK 522
P+ G I+IDG+DIS IGL DLR + IIPQ+PTLF G +R NLDPL YSD EIW+ L K
Sbjct: 1109 PASGCILIDGIDISKIGLKDLRMKLSIIPQEPTLFRGCIRTNLDPLGVYSDDEIWKALEK 1168
Query: 523 CQLREVVQEKDEGLNSSVVED-GSNWSMGQRQLFCLGRALLRRSRILVLDEATASVDNST 581
CQL+ + L+SS V D G NWS+GQRQLFCLGR LL+R++ILVLDEATAS+D++T
Sbjct: 1169 CQLKTTISNLPNKLDSSEVSDEGENWSVGQRQLFCLGRVLLKRNKILVLDEATASIDSAT 1228
Query: 582 DYILLKTIRTEFADCTVITVAHRIPTVMDCTMVLSINDGNL 622
D I+ + IR EFADCTVITVAHR+PTV+D MV+ ++ G+L
Sbjct: 1229 DAIIQRIIREEFADCTVITVAHRVPTVIDSDMVMVLSFGDL 1269
Score = 59.7 bits (143), Expect = 3e-07
Identities = 56/252 (22%), Positives = 111/252 (43%), Gaps = 18/252 (7%)
Query: 376 ISVERLNQYMHIQSEAKEIVEGNRPPLNWPIAGKVEINDLKIRYRPDGPL-VLHGITCTF 434
+S +RLN ++ + E K + E R L+ V+I + P+ + L I
Sbjct: 408 VSFQRLNNFL-LDDELK-MDEIERSGLD-ASGTAVDIQVGNFGWEPETKIPTLRNIHLEI 464
Query: 435 EVGHKIGIVGRTGSGKSTLISALFRLVEPSGGNIIIDGVDISSIGLHDLRSRFGIIPQDP 494
+ G K+ + G G+GKS+L+ A+ + G + + G + Q
Sbjct: 465 KHGQKVAVCGPVGAGKSSLLHAVLGEIPKVSGTVKVFG-------------SIAYVSQTS 511
Query: 495 TLFTGTVRYNLDPLSQYSDQEIWEVLGKCQLREVVQEKDEGLNSSVVEDGSNWSMGQRQL 554
+ +GT+R N+ + + C L + + G + + + G N S GQ+Q
Sbjct: 512 WIQSGTIRDNILYGKPMESRRYNAAIKACALDKDMNGFGHGDLTEIGQRGINLSGGQKQR 571
Query: 555 FCLGRALLRRSRILVLDEATASVDNSTDYILL-KTIRTEFADCTVITVAHRIPTVMDCTM 613
L RA+ + + +LD+ ++VD T +L K + + TVI V H++ + +
Sbjct: 572 IQLARAVYADADVYLLDDPFSAVDAHTAGVLFHKCVEDSLKEKTVILVTHQVEFLSEVDQ 631
Query: 614 VLSINDGNLSRN 625
+L + +G ++++
Sbjct: 632 ILVMEEGTITQS 643
>UniRef100_Q7GB25 F20D22.11 protein [Arabidopsis thaliana]
Length = 1355
Score = 537 bits (1383), Expect = e-151
Identities = 297/664 (44%), Positives = 402/664 (59%), Gaps = 41/664 (6%)
Query: 1 MSEGVIQQEGPYQQLLATSKEFQDLVNAHK-------VTDGSNQLVNVTFSRASIKI--- 50
+ EG I Q G Y LL +F+ LV+AH + S++ + R S+ +
Sbjct: 659 LKEGRIIQSGKYDDLLQAGTDFKALVSAHHEAIEAMDIPSPSSEDSDENPIRDSLVLHNP 718
Query: 51 --------TQTLVEN------------------KGKEANGNQLIKQEEREKGDKGLKPYL 84
+TL + K K + QL+++EER KG +K YL
Sbjct: 719 KSDVFENDIETLAKEVQEGGSASDLKAIKEKKKKAKRSRKKQLVQEEERVKGKVSMKVYL 778
Query: 85 QYLNQMKGYIFFFVASLGHFIFLVCQILQNLWMA-----ANVDNPRVSTFQLIFVYFLLG 139
Y+ + L F QI N WMA D +V L+ VY L
Sbjct: 779 SYMGAAYKGALIPLIILAQAAFQFLQIASNWWMAWANPQTEGDESKVDPTLLLIVYTALA 838
Query: 140 ASSAFFMLTRSLFVIALGLQSSKYLFLQLMNSLFRAPMPFYDCTPLGRILSRVSSELSIM 199
S+ F+ R+ V GL +++ LFL ++ S+FRAPM F+D TP GRIL+RVS + S++
Sbjct: 839 FGSSVFIFVRAALVATFGLAAAQKLFLNMLRSVFRAPMSFFDSTPAGRILNRVSIDQSVV 898
Query: 200 DLDIPFSLTFAVGTTMNFYSTLTVFSVVTWQVLIVAIPMVYITIRLQRYYFASAKEVMRI 259
DLDIPF L TT+ + V + VTWQV ++ +P+ +Q+YY AS++E++RI
Sbjct: 899 DLDIPFRLGGFASTTIQLCGIVAVMTNVTWQVFLLVVPVAVACFWMQKYYMASSRELVRI 958
Query: 260 TGTTKSYVANHVAETVSGAVTIRTFEEEDRFFQKNLDLIDINASSFFHNFASNEWLIQRL 319
KS + + E+++GA TIR F +E RF ++NL L+D FF + A+ EWL R+
Sbjct: 959 VSIQKSPIIHLFGESIAGAATIRGFGQEKRFIKRNLYLLDCFVRPFFCSIAAIEWLCLRM 1018
Query: 320 ETISAGVLASAALCMVILPPGTFTSGFIGMALSYGLALNSFLVNSIQSQCTLANQIISVE 379
E +S V A + +V P GT G+A++YGL LN L I S C L N+IIS+E
Sbjct: 1019 ELLSTLVFAFCMVLLVSFPHGTIDPSMAGLAVTYGLNLNGRLSRWILSFCKLENKIISIE 1078
Query: 380 RLNQYMHIQSEAKEIVEGNRPPLNWPIAGKVEINDLKIRYRPDGPLVLHGITCTFEVGHK 439
R+ QY I EA I+E RPP +WP G +E+ D+K+RY + P VLHG++C F G K
Sbjct: 1079 RIYQYSQIVGEAPAIIEDFRPPSSWPATGTIELVDVKVRYAENLPTVLHGVSCVFPGGKK 1138
Query: 440 IGIVGRTGSGKSTLISALFRLVEPSGGNIIIDGVDISSIGLHDLRSRFGIIPQDPTLFTG 499
IGIVGRTGSGKSTLI ALFRL+EP+ G I ID +DIS IGLHDLRSR GIIPQDPTLF G
Sbjct: 1139 IGIVGRTGSGKSTLIQALFRLIEPTAGKITIDNIDISQIGLHDLRSRLGIIPQDPTLFEG 1198
Query: 500 TVRYNLDPLSQYSDQEIWEVLGKCQLREVVQEKDEGLNSSVVEDGSNWSMGQRQLFCLGR 559
T+R NLDPL ++SD +IWE L K QL +VV+ KD L+S V+E+G NWS+GQRQL LGR
Sbjct: 1199 TIRANLDPLEEHSDDKIWEALDKSQLGDVVRGKDLKLDSPVLENGDNWSVGQRQLVSLGR 1258
Query: 560 ALLRRSRILVLDEATASVDNSTDYILLKTIRTEFADCTVITVAHRIPTVMDCTMVLSIND 619
ALL++++ILVLDEATASVD +TD ++ K IRTEF DCTV T+AHRIPTV+D +VL ++D
Sbjct: 1259 ALLKQAKILVLDEATASVDTATDNLIQKIIRTEFEDCTVCTIAHRIPTVIDSDLVLVLSD 1318
Query: 620 GNLS 623
G ++
Sbjct: 1319 GRVA 1322
Score = 69.3 bits (168), Expect = 3e-10
Identities = 67/294 (22%), Positives = 127/294 (42%), Gaps = 33/294 (11%)
Query: 343 TSGFIGMALSYGLALNSFLVNSIQSQ---------CTLANQIISVERLNQYMH---IQSE 390
TS F+G L+ G L++ I + +A +S++R++ ++ +Q +
Sbjct: 390 TSIFLGTQLTAGGVLSALATFRILQEPLRNFPDLVSMMAQTKVSLDRISGFLQEEELQED 449
Query: 391 AKEIVEGNRPPLNWPIAGKVEINDLKIRYRP-DGPLVLHGITCTFEVGHKIGIVGRTGSG 449
A ++ + +EI D + P L GI E G ++ + G GSG
Sbjct: 450 ATVVIPRGLSNI------AIEIKDGVFCWDPFSSRPTLSGIQMKVEKGMRVAVCGTVGSG 503
Query: 450 KSTLISALFRLVEPSGGNIIIDGVDISSIGLHDLRSRFGIIPQDPTLFTGTVRYNLDPLS 509
KS+ IS + + G + I G G + Q + +G + N+ S
Sbjct: 504 KSSFISCILGEIPKISGEVRICGTT-------------GYVSQSAWIQSGNIEENILFGS 550
Query: 510 QYSDQEIWEVLGKCQLREVVQEKDEGLNSSVVEDGSNWSMGQRQLFCLGRALLRRSRILV 569
+ V+ C L++ ++ G + + E G N S GQ+Q L RAL + + I +
Sbjct: 551 PMEKTKYKNVIQACSLKKDIELFSHGDQTIIGERGINLSGGQKQRVQLARALYQDADIYL 610
Query: 570 LDEATASVDNSTDYILLKT-IRTEFADCTVITVAHRIPTVMDCTMVLSINDGNL 622
LD+ +++D T L + I + A+ TV+ V H++ + ++L + +G +
Sbjct: 611 LDDPFSALDAHTGSDLFRDYILSALAEKTVVFVTHQVEFLPAADLILVLKEGRI 664
>UniRef100_O65619 Multi resistance protein [Arabidopsis thaliana]
Length = 1514
Score = 537 bits (1383), Expect = e-151
Identities = 297/664 (44%), Positives = 402/664 (59%), Gaps = 41/664 (6%)
Query: 1 MSEGVIQQEGPYQQLLATSKEFQDLVNAHK-------VTDGSNQLVNVTFSRASIKI--- 50
+ EG I Q G Y LL +F+ LV+AH + S++ + R S+ +
Sbjct: 818 LKEGRIIQSGKYDDLLQAGTDFKALVSAHHEAIEAMDIPSPSSEDSDENPIRDSLVLHNP 877
Query: 51 --------TQTLVEN------------------KGKEANGNQLIKQEEREKGDKGLKPYL 84
+TL + K K + QL+++EER KG +K YL
Sbjct: 878 KSDVFENDIETLAKEVQEGGSASDLKAIKEKKKKAKRSRKKQLVQEEERVKGKVSMKVYL 937
Query: 85 QYLNQMKGYIFFFVASLGHFIFLVCQILQNLWMA-----ANVDNPRVSTFQLIFVYFLLG 139
Y+ + L F QI N WMA D +V L+ VY L
Sbjct: 938 SYMGAAYKGALIPLIILAQAAFQFLQIASNWWMAWANPQTEGDESKVDPTLLLIVYTALA 997
Query: 140 ASSAFFMLTRSLFVIALGLQSSKYLFLQLMNSLFRAPMPFYDCTPLGRILSRVSSELSIM 199
S+ F+ R+ V GL +++ LFL ++ S+FRAPM F+D TP GRIL+RVS + S++
Sbjct: 998 FGSSVFIFVRAALVATFGLAAAQKLFLNMLRSVFRAPMSFFDSTPAGRILNRVSIDQSVV 1057
Query: 200 DLDIPFSLTFAVGTTMNFYSTLTVFSVVTWQVLIVAIPMVYITIRLQRYYFASAKEVMRI 259
DLDIPF L TT+ + V + VTWQV ++ +P+ +Q+YY AS++E++RI
Sbjct: 1058 DLDIPFRLGGFASTTIQLCGIVAVMTNVTWQVFLLVVPVAVACFWMQKYYMASSRELVRI 1117
Query: 260 TGTTKSYVANHVAETVSGAVTIRTFEEEDRFFQKNLDLIDINASSFFHNFASNEWLIQRL 319
KS + + E+++GA TIR F +E RF ++NL L+D FF + A+ EWL R+
Sbjct: 1118 VSIQKSPIIHLFGESIAGAATIRGFGQEKRFIKRNLYLLDCFVRPFFCSIAAIEWLCLRM 1177
Query: 320 ETISAGVLASAALCMVILPPGTFTSGFIGMALSYGLALNSFLVNSIQSQCTLANQIISVE 379
E +S V A + +V P GT G+A++YGL LN L I S C L N+IIS+E
Sbjct: 1178 ELLSTLVFAFCMVLLVSFPHGTIDPSMAGLAVTYGLNLNGRLSRWILSFCKLENKIISIE 1237
Query: 380 RLNQYMHIQSEAKEIVEGNRPPLNWPIAGKVEINDLKIRYRPDGPLVLHGITCTFEVGHK 439
R+ QY I EA I+E RPP +WP G +E+ D+K+RY + P VLHG++C F G K
Sbjct: 1238 RIYQYSQIVGEAPAIIEDFRPPSSWPATGTIELVDVKVRYAENLPTVLHGVSCVFPGGKK 1297
Query: 440 IGIVGRTGSGKSTLISALFRLVEPSGGNIIIDGVDISSIGLHDLRSRFGIIPQDPTLFTG 499
IGIVGRTGSGKSTLI ALFRL+EP+ G I ID +DIS IGLHDLRSR GIIPQDPTLF G
Sbjct: 1298 IGIVGRTGSGKSTLIQALFRLIEPTAGKITIDNIDISQIGLHDLRSRLGIIPQDPTLFEG 1357
Query: 500 TVRYNLDPLSQYSDQEIWEVLGKCQLREVVQEKDEGLNSSVVEDGSNWSMGQRQLFCLGR 559
T+R NLDPL ++SD +IWE L K QL +VV+ KD L+S V+E+G NWS+GQRQL LGR
Sbjct: 1358 TIRANLDPLEEHSDDKIWEALDKSQLGDVVRGKDLKLDSPVLENGDNWSVGQRQLVSLGR 1417
Query: 560 ALLRRSRILVLDEATASVDNSTDYILLKTIRTEFADCTVITVAHRIPTVMDCTMVLSIND 619
ALL++++ILVLDEATASVD +TD ++ K IRTEF DCTV T+AHRIPTV+D +VL ++D
Sbjct: 1418 ALLKQAKILVLDEATASVDTATDNLIQKIIRTEFEDCTVCTIAHRIPTVIDSDLVLVLSD 1477
Query: 620 GNLS 623
G ++
Sbjct: 1478 GRVA 1481
Score = 69.3 bits (168), Expect = 3e-10
Identities = 67/294 (22%), Positives = 127/294 (42%), Gaps = 33/294 (11%)
Query: 343 TSGFIGMALSYGLALNSFLVNSIQSQ---------CTLANQIISVERLNQYMH---IQSE 390
TS F+G L+ G L++ I + +A +S++R++ ++ +Q +
Sbjct: 549 TSIFLGTQLTAGGVLSALATFRILQEPLRNFPDLVSMMAQTKVSLDRISGFLQEEELQED 608
Query: 391 AKEIVEGNRPPLNWPIAGKVEINDLKIRYRP-DGPLVLHGITCTFEVGHKIGIVGRTGSG 449
A ++ + +EI D + P L GI E G ++ + G GSG
Sbjct: 609 ATVVIPRGLSNI------AIEIKDGVFCWDPFSSRPTLSGIQMKVEKGMRVAVCGTVGSG 662
Query: 450 KSTLISALFRLVEPSGGNIIIDGVDISSIGLHDLRSRFGIIPQDPTLFTGTVRYNLDPLS 509
KS+ IS + + G + I G G + Q + +G + N+ S
Sbjct: 663 KSSFISCILGEIPKISGEVRICGTT-------------GYVSQSAWIQSGNIEENILFGS 709
Query: 510 QYSDQEIWEVLGKCQLREVVQEKDEGLNSSVVEDGSNWSMGQRQLFCLGRALLRRSRILV 569
+ V+ C L++ ++ G + + E G N S GQ+Q L RAL + + I +
Sbjct: 710 PMEKTKYKNVIQACSLKKDIELFSHGDQTIIGERGINLSGGQKQRVQLARALYQDADIYL 769
Query: 570 LDEATASVDNSTDYILLKT-IRTEFADCTVITVAHRIPTVMDCTMVLSINDGNL 622
LD+ +++D T L + I + A+ TV+ V H++ + ++L + +G +
Sbjct: 770 LDDPFSALDAHTGSDLFRDYILSALAEKTVVFVTHQVEFLPAADLILVLKEGRI 823
>UniRef100_Q8GU65 MRP-like ABC transporter [Oryza sativa]
Length = 1545
Score = 528 bits (1360), Expect = e-148
Identities = 282/641 (43%), Positives = 401/641 (61%), Gaps = 25/641 (3%)
Query: 1 MSEGVIQQEGPYQQLLATSKEFQDLVNAH----KVTDGSNQLVNVTFS--RASIKITQTL 54
M +G+I Q G Y +LL +F LV AH ++ D S Q+V +S +A +I
Sbjct: 877 MRDGMIVQSGKYDELLDAGSDFLALVAAHDSSMELVDQSRQVVKTEYSQPKAVARIPSLR 936
Query: 55 VENKGK----------EANGNQLIKQEEREKGDKGLKPYLQYLNQMKGYIFFFVASLGHF 104
+ GK EA +++I++EERE G + Y Y+ + G+
Sbjct: 937 SRSIGKGEKVLVAPDIEAATSKIIREEERESGQVSWRVYKLYMTEAWGWWGVVGMLAFAI 996
Query: 105 IFLVCQILQNLWMAANVD-----NPRVSTFQLIFVYFLLGASSAFFMLTRSLFVIALGLQ 159
++ V ++ + W++ NP + I VY + A S + +SL LGLQ
Sbjct: 997 VWQVTEMASDYWLSYETSGSIPFNPSL----FIGVYVAIAAVSIILQVIKSLLETILGLQ 1052
Query: 160 SSKYLFLQLMNSLFRAPMPFYDCTPLGRILSRVSSELSIMDLDIPFSLTFAVGTTMNFYS 219
+++ F ++ +S+ APM F+D TP GRILSR SS+ + +D+ + F + + ++ S
Sbjct: 1053 TAQIFFKKMFDSILHAPMSFFDTTPSGRILSRASSDQTTIDIVLSFFVGLTISMYISVLS 1112
Query: 220 TLTVFSVVTWQVLIVAIPMVYITIRLQRYYFASAKEVMRITGTTKSYVANHVAETVSGAV 279
T+ V V W +I IP+V + I + Y A+++E+ R+ G TK+ V +H +ETV GA
Sbjct: 1113 TIIVTCQVAWPSVIAVIPLVLLNIWYRNRYLATSRELTRLEGVTKAPVIDHFSETVLGAT 1172
Query: 280 TIRTFEEEDRFFQKNLDLIDINASSFFHNFASNEWLIQRLETISAGVLASAALCMVILPP 339
TIR F+++ FFQ+NLD I+ + +FHN+A+NEWL RLE I VLA A M+ LP
Sbjct: 1173 TIRCFKKDKEFFQENLDRINSSLRMYFHNYAANEWLGFRLELIGTLVLAITAFLMISLPS 1232
Query: 340 GTFTSGFIGMALSYGLALNSFLVNSIQSQCTLANQIISVERLNQYMHIQSEAKEIVEGNR 399
F+GM+LSYGL+LNS + +I C L N +++VER+NQ+ + SEA +E +
Sbjct: 1233 NFIKKEFVGMSLSYGLSLNSLVYFAISISCMLENDMVAVERVNQFSTLPSEAVWKIEDHL 1292
Query: 400 PPLNWPIAGKVEINDLKIRYRPDGPLVLHGITCTFEVGHKIGIVGRTGSGKSTLISALFR 459
P NWP G ++I+DLK+RYRP+ PL+L GIT + G KIG+VGRTGSGKSTLI ALFR
Sbjct: 1293 PSPNWPTHGDIDIDDLKVRYRPNTPLILKGITVSISGGEKIGVVGRTGSGKSTLIQALFR 1352
Query: 460 LVEPSGGNIIIDGVDISSIGLHDLRSRFGIIPQDPTLFTGTVRYNLDPLSQYSDQEIWEV 519
LVEP G +IIDG+DI ++GLHDLRSRFGIIPQ+P LF GT+R N+DP+ QYSD EIW
Sbjct: 1353 LVEPVQGTMIIDGIDICTLGLHDLRSRFGIIPQEPVLFEGTIRSNIDPIGQYSDAEIWRA 1412
Query: 520 LGKCQLREVVQEKDEGLNSSVVEDGSNWSMGQRQLFCLGRALLRRSRILVLDEATASVDN 579
L CQL++VV K + L++ V + G NWS+GQRQL CLGR +L+R+RIL +DEATASVD+
Sbjct: 1413 LEGCQLKDVVASKPQKLDALVADSGENWSVGQRQLLCLGRVILKRTRILFMDEATASVDS 1472
Query: 580 STDYILLKTIRTEFADCTVITVAHRIPTVMDCTMVLSINDG 620
TD + K R EF+ CT+I++AHRIPTVMDC VL ++ G
Sbjct: 1473 QTDATIQKITRQEFSSCTIISIAHRIPTVMDCDRVLVLDAG 1513
Score = 67.0 bits (162), Expect = 2e-09
Identities = 62/251 (24%), Positives = 109/251 (42%), Gaps = 20/251 (7%)
Query: 408 GKVEINDLKIRYRPDGPLVLHGITCTFEVGHKIGIVGRTGSGKSTLISALFRLVEPSGGN 467
G+ E + + P VL GI G +VG GSGKS+L+S + ++ G
Sbjct: 680 GEEEEEEKDVEETPVLETVLKGINIEVRRGELAAVVGTVGSGKSSLLSCIMGEMDKVSGK 739
Query: 468 IIIDGVDISSIGLHDLRSRFGIIPQDPTLFTGTVRYNLDPLSQYSDQEIW-EVLGKCQLR 526
+ I G + Q + GT++ N+ Q D E + EVL C L
Sbjct: 740 VRICGST-------------AYVAQTAWIQNGTIQENI-LFGQPMDAERYKEVLRSCSLE 785
Query: 527 EVVQEKDEGLNSSVVEDGSNWSMGQRQLFCLGRALLRRSRILVLDEATASVDNSTDYILL 586
+ ++ + G + + E G N S GQ+Q L RA+ + I +LD+ ++VD T +
Sbjct: 786 KDLEMMEFGDQTEIGERGINLSGGQKQRIQLARAVYQNCDIYLLDDVFSAVDAHTGSSIF 845
Query: 587 K-TIRTEFADCTVITVAHRIPTVMDCTMVLSINDGNLSRNTGLIFSLQSHIDVNNYFLYT 645
K +R T++ V H++ + + + + DG + ++ +D + FL
Sbjct: 846 KECLRGMLKGKTILLVTHQVDFLHNVDNIFVMRDGMIVQSG----KYDELLDAGSDFLAL 901
Query: 646 VSILSLSLPLL 656
V+ S+ L+
Sbjct: 902 VAAHDSSMELV 912
>UniRef100_Q8LGN7 Multidrug resistance-associated protein MRP1 [Triticum aestivum]
Length = 764
Score = 523 bits (1348), Expect = e-147
Identities = 283/641 (44%), Positives = 398/641 (61%), Gaps = 24/641 (3%)
Query: 1 MSEGVIQQEGPYQQLLATSKEFQDLVNAHKVTDGSNQLVN----VTFSRASIKITQTLVE 56
M EG I Q G Y +L+ +F LV AH + S +LV V+ +
Sbjct: 95 MKEGTIVQSGKYDELIQRGSDFAALVAAH---NSSMELVEGAAPVSDEKGETPAISRQPS 151
Query: 57 NKGK-------EANG-------NQLIKQEEREKGDKGLKPYLQYLNQMKGYIFFFVASLG 102
KG EA+G +LIK+EER G L Y QY+ + G+ +
Sbjct: 152 RKGSGRRPSSGEAHGVVAEKASARLIKEEERASGHVSLAVYKQYMTEAWGWWGVALVVAV 211
Query: 103 HFIFLVCQILQNLWMAANVDNPRVSTFQ---LIFVYFLLGASSAFFMLTRSLFVIALGLQ 159
+ + + W+A D ++F+ I VY ++ +S + RS V +GLQ
Sbjct: 212 SVAWQGSVLASDYWLAYETDAENAASFRPALFIEVYAIIAVASVVLVSGRSFLVAFIGLQ 271
Query: 160 SSKYLFLQLMNSLFRAPMPFYDCTPLGRILSRVSSELSIMDLDIPFSLTFAVGTTMNFYS 219
++ F Q++NS+ APM F+D TP GRILSR SS+ + +DL +PF + +V + S
Sbjct: 272 TANSFFKQILNSILHAPMSFFDTTPSGRILSRASSDQTNVDLFLPFFVWLSVSMYITVIS 331
Query: 220 TLTVFSVVTWQVLIVAIPMVYITIRLQRYYFASAKEVMRITGTTKSYVANHVAETVSGAV 279
L V V W +I IP++ + + + YY A+++E+ R+ TK+ V +H +ETV G +
Sbjct: 332 VLVVTCQVAWPSVIAIIPLLILNLWYRGYYLATSRELTRLESITKAPVIHHFSETVQGVM 391
Query: 280 TIRTFEEEDRFFQKNLDLIDINASSFFHNFASNEWLIQRLETISAGVLASAALCMVILPP 339
TIR F + D FFQ+NL+ ++ + FHN +NEWL RLE + VL AL MV LP
Sbjct: 392 TIRCFRKGDGFFQENLNRVNSSLRMDFHNNGANEWLGFRLELAGSFVLCFTALLMVTLPK 451
Query: 340 GTFTSGFIGMALSYGLALNSFLVNSIQSQCTLANQIISVERLNQYMHIQSEAKEIVEGNR 399
F+G++LSYGL+LNS L ++ C + N+++SVER+ Q+++I EA+ ++
Sbjct: 452 SFIQPEFVGLSLSYGLSLNSVLFWAVWMSCFIENKMVSVERIKQFVNIPCEAEWRIKDCL 511
Query: 400 PPLNWPIAGKVEINDLKIRYRPDGPLVLHGITCTFEVGHKIGIVGRTGSGKSTLISALFR 459
P NWP G +E+ DLK+RYR + PLVL GIT + G KIG+VGRTGSGKSTLI ALFR
Sbjct: 512 PVANWPTRGDIEVIDLKVRYRHNTPLVLKGITLSIHGGEKIGVVGRTGSGKSTLIQALFR 571
Query: 460 LVEPSGGNIIIDGVDISSIGLHDLRSRFGIIPQDPTLFTGTVRYNLDPLSQYSDQEIWEV 519
+VEPS G IIIDGVDI ++GLHDLRSRFGIIPQ+P LF GT+R N+DPL +YSD EIW+
Sbjct: 572 IVEPSEGKIIIDGVDICTLGLHDLRSRFGIIPQEPVLFEGTIRSNIDPLEEYSDVEIWQA 631
Query: 520 LGKCQLREVVQEKDEGLNSSVVEDGSNWSMGQRQLFCLGRALLRRSRILVLDEATASVDN 579
L +CQL+E V K E L++SVV++G NWS+GQRQL CLGR +L+ S+IL +DEATASVD+
Sbjct: 632 LDRCQLKEAVTSKPEKLDASVVDNGENWSVGQRQLLCLGRVMLKHSKILFMDEATASVDS 691
Query: 580 STDYILLKTIRTEFADCTVITVAHRIPTVMDCTMVLSINDG 620
TD ++ + IR +FA+CT+I++AHRIPTVMDC VL ++ G
Sbjct: 692 QTDAVIQRIIREDFAECTIISIAHRIPTVMDCDRVLVVDAG 732
Score = 39.7 bits (91), Expect = 0.29
Identities = 25/99 (25%), Positives = 51/99 (51%), Gaps = 1/99 (1%)
Query: 525 LREVVQEKDEGLNSSVVEDGSNWSMGQRQLFCLGRALLRRSRILVLDEATASVDNST-DY 583
L + ++ + G + + E G N S GQ+Q L RA+ + I +LD+ ++VD T
Sbjct: 2 LEKDMEMMEFGDQTEIGERGINLSGGQKQRIQLARAVYQDCDIYLLDDVFSAVDAHTGSE 61
Query: 584 ILLKTIRTEFADCTVITVAHRIPTVMDCTMVLSINDGNL 622
I + +R + TV+ V H++ + + ++ + +G +
Sbjct: 62 IFKECVRGALKNKTVVLVTHQVDFLHNADIIYVMKEGTI 100
>UniRef100_Q75Q02 Multidrug resistance-associated protein [Thlaspi caerulescens]
Length = 1514
Score = 521 bits (1343), Expect = e-146
Identities = 284/649 (43%), Positives = 396/649 (60%), Gaps = 33/649 (5%)
Query: 1 MSEGVIQQEGPYQQLLATSKEFQDLVNAHKVTDGSNQLVNVTFSRASIKITQTLVENKG- 59
M +G I Q G Y +L + +F +L+ AH+ V+ S+ T L E G
Sbjct: 835 MKDGRISQAGKYNDILNSGTDFMELIGAHQEALAVVNSVDTN----SVSETSALGEENGV 890
Query: 60 --------------------KEANGN---QLIKQEEREKGDKGLKPYLQYLNQMKGYIFF 96
K +G QL+++EEREKG L Y +Y+ G
Sbjct: 891 VRDDAIGFDGKQEGQDLKNDKPDSGEPQRQLVQEEEREKGSVALSVYWKYITLAYGGALV 950
Query: 97 FVASLGHFIFLVCQILQNLWMA-----ANVDNPRVSTFQLIFVYFLLGASSAFFMLTRSL 151
L +F + QI N WMA + V+ L+ VY L S+ +L R+
Sbjct: 951 PFILLAQVLFQLLQIGSNYWMAWATPVSKDVEATVNLSTLMIVYVALAVGSSLCILFRAT 1010
Query: 152 FVIALGLQSSKYLFLQLMNSLFRAPMPFYDCTPLGRILSRVSSELSIMDLDIPFSLTFAV 211
++ G +++ LF ++ + +FR+PM F+D TP GRI++R S++ S +DLDIP+
Sbjct: 1011 LLVTAGYKTATELFHRMHHCIFRSPMSFFDSTPSGRIMNRASTDQSAVDLDIPYQFGSVA 1070
Query: 212 GTTMNFYSTLTVFSVVTWQVLIVAIPMVYITIRLQRYYFASAKEVMRITGTTKSYVANHV 271
T + + V S V+W V +V IP+V +I QRYY A+A+E+ R+ G K+ + H
Sbjct: 1071 ITVIQLIGIIGVMSQVSWLVFLVFIPVVAASIWYQRYYIAAARELSRLVGVCKAPLIQHF 1130
Query: 272 AETVSGAVTIRTFEEEDRFFQKNLDLIDINASSFFHNFASNEWLIQRLETISAGVLASAA 331
AET+SG+ TIR+F +E RF N+ L D + F++ + EWL RL+ +S+ A +
Sbjct: 1131 AETISGSTTIRSFSQESRFRSDNMRLSDGYSRPKFYSAGAMEWLCFRLDVLSSLTFAFSL 1190
Query: 332 LCMVILPPGTFTSGFIGMALSYGLALNSFLVNSIQSQCTLANQIISVERLNQYMHIQSEA 391
+ ++ +P G G+A++YGL+LN+ I + C L N+IISVER+ QY + E
Sbjct: 1191 VFLISIPTGVIDPSLAGLAVTYGLSLNTMQAWLIWTLCNLENKIISVERILQYASVPGEP 1250
Query: 392 KEIVEGNRPPLNWPIAGKVEINDLKIRYRPDGPLVLHGITCTFEVGHKIGIVGRTGSGKS 451
++E NRP +WP G+V+I DL++RY P PLVL GITCTF+ G + GIVGRTGSGKS
Sbjct: 1251 PLVIESNRPEQSWPSRGEVDIRDLQVRYAPHMPLVLRGITCTFKGGLRTGIVGRTGSGKS 1310
Query: 452 TLISALFRLVEPSGGNIIIDGVDISSIGLHDLRSRFGIIPQDPTLFTGTVRYNLDPLSQY 511
TLI LFR+VEPS G I IDGV+I +IGLHDLR R IIPQDPT+F GTVR NLDPL +Y
Sbjct: 1311 TLIQTLFRIVEPSAGEIRIDGVNILTIGLHDLRLRLSIIPQDPTMFEGTVRSNLDPLEEY 1370
Query: 512 SDQEIWEVLGKCQLREVVQEKDEGLNSSVVEDGSNWSMGQRQLFCLGRALLRRSRILVLD 571
+D +IWE L KCQL + V++K++ L+SSV E+G NWSMGQRQL CLGR LL+RS+ILV D
Sbjct: 1371 TDDQIWEALDKCQLGDEVRKKEQKLDSSVSENGENWSMGQRQLVCLGRVLLKRSKILVND 1430
Query: 572 EATASVDNSTDYILLKTIRTEFADCTVITVAHRIPTVMDCTMVLSINDG 620
EATASVD +TDY++ KT+R FADCTVIT+AHRI +V+D MVL + +G
Sbjct: 1431 EATASVDTATDYLIQKTLRDHFADCTVITIAHRISSVIDSDMVLLLGNG 1479
Score = 67.4 bits (163), Expect = 1e-09
Identities = 68/282 (24%), Positives = 123/282 (43%), Gaps = 16/282 (5%)
Query: 344 SGFIGMALSYGLALNSFLVNSIQSQCTLANQIISVERLNQYMHIQSEAKEIVEGNRPPLN 403
SG I AL+ L + N + L +S++R+ Y+ + + ++VE P +
Sbjct: 576 SGKILSALATFRILQEPIYNLPDTISMLVQTKVSLDRIASYLCLDNLQPDVVE-RLPQGS 634
Query: 404 WPIAGKVEINDLKIRYRPDGPLVLHGITCTFEVGHKIGIVGRTGSGKSTLISALFRLVEP 463
IA +V + L P L I G K+ + G GSGKS+L+S++ V
Sbjct: 635 SDIAVEVTNSTLSWDVSSANP-TLKDINFKVFNGMKVAVCGTVGSGKSSLLSSILGEVPK 693
Query: 464 SGGNIIIDGVDISSIGLHDLRSRFGIIPQDPTLFTGTVRYNLDPLSQYSDQEIWEVLGKC 523
G++ + G + Q P + +G + N+ + +VL C
Sbjct: 694 ISGSLKVCGTK-------------AYVAQSPWIQSGKIEDNILFGKPMERERYEKVLEAC 740
Query: 524 QLREVVQEKDEGLNSSVVEDGSNWSMGQRQLFCLGRALLRRSRILVLDEATASVDNSTDY 583
L++ ++ G + + E G N S GQ+Q + RAL + + I + D+ ++VD T
Sbjct: 741 SLKKDLEILSFGDQTVIGERGINLSGGQKQRIQIARALYQDADIYLFDDPFSAVDAHTGS 800
Query: 584 ILLKTIRTE-FADCTVITVAHRIPTVMDCTMVLSINDGNLSR 624
L K + +VI V H++ + ++L + DG +S+
Sbjct: 801 HLFKEVLLGLLCSKSVIYVTHQVEFLPAADLILFMKDGRISQ 842
>UniRef100_Q8VZZ4 ATP-binding cassette transporter MRP6 [Arabidopsis thaliana]
Length = 1466
Score = 520 bits (1339), Expect = e-146
Identities = 288/669 (43%), Positives = 409/669 (61%), Gaps = 19/669 (2%)
Query: 1 MSEGVIQQEGPYQQLLATSKEFQDLVNAHKVTDGSNQLVNVTFSRASIKIT----QTLVE 56
M +G I Q G Y ++L + +F +LV AH T+ + + AS K T ++
Sbjct: 797 MKDGKITQAGKYNEILDSGTDFMELVGAH--TEALATIDSCETGYASEKSTTDKENEVIH 854
Query: 57 NKGKEANGN------QLIKQEEREKGDKGLKPYLQYLNQMKGYIFFFVASLGHFIFLVCQ 110
+K K+ NG+ QL+++EEREKG G Y +Y+ G + + +F +
Sbjct: 855 HKEKQENGSDNKPSGQLVQEEEREKGKVGFTVYKKYMALAYGGAVIPLILVVQVLFQLLS 914
Query: 111 ILQNLWMA-----ANVDNPRVSTFQLIFVYFLLGASSAFFMLTRSLFVIALGLQSSKYLF 165
I N WM + P VS F LI VY LL +S+F +L R+L V G + + LF
Sbjct: 915 IGSNYWMTWVTPVSKDVEPPVSGFTLILVYVLLAVASSFCILIRALLVAMTGFKMATELF 974
Query: 166 LQLMNSLFRAPMPFYDCTPLGRILSRVSSELSIMDLDIPFSLTFAVGTTMNFYSTLTVFS 225
Q+ +FRA M F+D TP+GRIL+R S++ S+ DL +P + +N + V
Sbjct: 975 TQMHLRIFRASMSFFDATPMGRILNRASTDQSVADLRLPGQFAYVAIAAINILGIIGVIV 1034
Query: 226 VVTWQVLIVAIPMVYITIRLQRYYFASAKEVMRITGTTKSYVANHVAETVSGAVTIRTFE 285
V WQVLIV IP+V ++YY ++A+E+ R+ G ++S V +H +ET+SG TIR+F+
Sbjct: 1035 QVAWQVLIVFIPVVAACAWYRQYYISAARELARLAGISRSPVVHHFSETLSGITTIRSFD 1094
Query: 286 EEDRFFQKNLDLIDINASSFFHNFASNEWLIQRLETISAGVLASAALCMVILPPGTFTSG 345
+E RF + L D + FH+ + EWL RLE +S AS+ + +V P G
Sbjct: 1095 QEPRFRGDIMRLSDCYSRLKFHSTGAMEWLCFRLELLSTFAFASSLVILVSAPEGVINPS 1154
Query: 346 FIGMALSYGLALNSFLVNSIQSQCTLANQIISVERLNQYMHIQSEAKEIVEGNRPPLNWP 405
G+A++Y L LN+ I + C L N++ISVER+ QY +I SE ++E RP +WP
Sbjct: 1155 LAGLAITYALNLNTLQATLIWTLCDLENKMISVERMLQYTNIPSEPPLVIETTRPEKSWP 1214
Query: 406 IAGKVEINDLKIRYRPDGPLVLHGITCTFEVGHKIGIVGRTGSGKSTLISALFRLVEPSG 465
G++ I +L++RY P P+VLHG+TCTF G K GIVGRTG GKSTLI LFR+VEP+
Sbjct: 1215 SRGEITICNLQVRYGPHLPMVLHGLTCTFPGGLKTGIVGRTGCGKSTLIQTLFRIVEPAA 1274
Query: 466 GNIIIDGVDISSIGLHDLRSRFGIIPQDPTLFTGTVRYNLDPLSQYSDQEIWEVLGKCQL 525
G I IDG++I SIGLHDLRSR IIPQDPT+F GT+R NLDPL +Y+D +IWE L CQL
Sbjct: 1275 GEIRIDGINILSIGLHDLRSRLSIIPQDPTMFEGTIRSNLDPLEEYTDDQIWEALDNCQL 1334
Query: 526 REVVQEKDEGLNSSVVEDGSNWSMGQRQLFCLGRALLRRSRILVLDEATASVDNSTDYIL 585
+ V++K+ L+S V E+G NWS+GQRQL CLGR LL+RS++LVLDEATAS+D +TD ++
Sbjct: 1335 GDEVRKKELKLDSPVSENGQNWSVGQRQLVCLGRVLLKRSKLLVLDEATASIDTATDNLI 1394
Query: 586 LKTIRTEFADCTVITVAHRIPTVMDCTMVLSINDGNLSRNTGLIFSLQSHIDVNNYFL-- 643
+T+R FADCTVIT+AHRI +V+D MVL ++ G + + L+ + + F+
Sbjct: 1395 QETLRHHFADCTVITIAHRISSVIDSDMVLLLDQGLIKEHDSPARLLEDRSSLFSKFVAE 1454
Query: 644 YTVSILSLS 652
YT S S S
Sbjct: 1455 YTTSSESKS 1463
Score = 71.6 bits (174), Expect = 7e-11
Identities = 91/396 (22%), Positives = 169/396 (41%), Gaps = 23/396 (5%)
Query: 233 IVAIPMVYITIRLQRYYFASAKEVMRIT-GTTKSYVANHVAETVSGAVTIRTFEEEDRFF 291
I A P I + L Y FA +E + + +K +E + ++ E +F
Sbjct: 428 IAAFPAT-ILVMLANYPFAKLEEKFQSSLMKSKDNRMKKTSEVLLNMKILKLQGWEMKFL 486
Query: 292 QKNLDLIDINASSFFHNFASNEWLIQRLETISAGVLASAALCMVILPPGTFTSGFIGMAL 351
K L+L I A + F N I + + +++ A +L SG I AL
Sbjct: 487 SKILELRHIEAG-WLKKFVYNSSAINSVLWAAPSFISATAFGACLLLKIPLESGKILAAL 545
Query: 352 SYGLALNSFLVNSIQSQCTLANQIISVERLNQYMHIQSEAKEIVEGNRPPLNWPIAGKVE 411
+ L + ++ + +S+ R+ ++ + +++V G P + +A VE
Sbjct: 546 ATFRILQGPIYKLPETISMIVQTKVSLNRIASFLCLDDLQQDVV-GRLPSGSSEMA--VE 602
Query: 412 INDLKIRYRPDGPL-VLHGITCTFEVGHKIGIVGRTGSGKSTLISALFRLVEPSGGNIII 470
I++ + P+ L + G + I G GSGKS+L+S++ V GN+ +
Sbjct: 603 ISNGTFSWDESSPIPTLRDMNFKVSQGMNVAICGTVGSGKSSLLSSILGEVPKISGNLKV 662
Query: 471 DGVDISSIGLHDLRSRFGIIPQDPTLFTGTVRYNLDPLSQYSDQEIWE-VLGKCQLREVV 529
G R I Q P + +G V N+ + ++E ++ VL C L + +
Sbjct: 663 CG-------------RKAYIAQSPWIQSGKVEENI-LFGKPMEREWYDRVLEACSLNKDL 708
Query: 530 QEKDEGLNSSVVEDGSNWSMGQRQLFCLGRALLRRSRILVLDEATASVDNSTDYILLKTI 589
+ + + E G N S GQ+Q + RAL + + I + D+ ++VD T L K +
Sbjct: 709 EILPFHDQTVIGERGINLSGGQKQRIQIARALYQDADIYLFDDPFSAVDAHTGSHLFKEV 768
Query: 590 RTE-FADCTVITVAHRIPTVMDCTMVLSINDGNLSR 624
TVI V H++ + + ++L + DG +++
Sbjct: 769 LLGLLRHKTVIYVTHQVEFLPEADLILVMKDGKITQ 804
>UniRef100_Q9LK64 Multidrug resistance-associated protein (MRP); ABC-transoprter
[Arabidopsis thaliana]
Length = 1514
Score = 520 bits (1339), Expect = e-146
Identities = 280/645 (43%), Positives = 399/645 (61%), Gaps = 26/645 (4%)
Query: 1 MSEGVIQQEGPYQQLLATSKEFQDLVNAHKVT-------DGSNQLVNVTFSRASIKITQT 53
M +G I Q G Y +L + +F +L+ AH+ D ++ + ++ +
Sbjct: 836 MKDGRISQAGKYNDILNSGTDFMELIGAHQEALAVVDSVDANSVSEKSALGQENVIVKDA 895
Query: 54 LVENKGKEANG------------NQLIKQEEREKGDKGLKPYLQYLNQMKGYIFFFVASL 101
+ ++ E+ Q+I++EEREKG L Y +Y+ G L
Sbjct: 896 IAVDEKLESQDLKNDKLESVEPQRQIIQEEEREKGSVALDVYWKYITLAYGGALVPFILL 955
Query: 102 GHFIFLVCQILQNLWMA------ANVDNPRVSTFQLIFVYFLLGASSAFFMLTRSLFVIA 155
G +F + QI N WMA +V P V L+ VY L S+ +L R+ ++
Sbjct: 956 GQVLFQLLQIGSNYWMAWATPVSEDVQAP-VKLSTLMIVYVALAFGSSLCILLRATLLVT 1014
Query: 156 LGLQSSKYLFLQLMNSLFRAPMPFYDCTPLGRILSRVSSELSIMDLDIPFSLTFAVGTTM 215
G +++ LF ++ + +FR+PM F+D TP GRI+SR S++ S +DL++P+ T +
Sbjct: 1015 AGYKTATELFHKMHHCIFRSPMSFFDSTPSGRIMSRASTDQSAVDLELPYQFGSVAITVI 1074
Query: 216 NFYSTLTVFSVVTWQVLIVAIPMVYITIRLQRYYFASAKEVMRITGTTKSYVANHVAETV 275
+ V S V+W V +V IP+V +I QRYY A+A+E+ R+ G K+ + H +ET+
Sbjct: 1075 QLIGIIGVMSQVSWLVFLVFIPVVAASIWYQRYYIAAARELSRLVGVCKAPLIQHFSETI 1134
Query: 276 SGAVTIRTFEEEDRFFQKNLDLIDINASSFFHNFASNEWLIQRLETISAGVLASAALCMV 335
SGA TIR+F +E RF N+ L D + F+ + EWL RL+ +S+ + + +V
Sbjct: 1135 SGATTIRSFSQEFRFRSDNMRLSDGYSRPKFYTAGAMEWLCFRLDMLSSLTFVFSLVFLV 1194
Query: 336 ILPPGTFTSGFIGMALSYGLALNSFLVNSIQSQCTLANQIISVERLNQYMHIQSEAKEIV 395
+P G G+A++YGL+LN+ I + C L N+IISVER+ QY + SE ++
Sbjct: 1195 SIPTGVIDPSLAGLAVTYGLSLNTLQAWLIWTLCNLENKIISVERILQYASVPSEPPLVI 1254
Query: 396 EGNRPPLNWPIAGKVEINDLKIRYRPDGPLVLHGITCTFEVGHKIGIVGRTGSGKSTLIS 455
E NRP +WP G+VEI DL++RY P PLVL GITCTF+ G + GIVGRTGSGKSTLI
Sbjct: 1255 ESNRPEQSWPSRGEVEIRDLQVRYAPHMPLVLRGITCTFKGGLRTGIVGRTGSGKSTLIQ 1314
Query: 456 ALFRLVEPSGGNIIIDGVDISSIGLHDLRSRFGIIPQDPTLFTGTVRYNLDPLSQYSDQE 515
LFR+VEPS G I IDGV+I +IGLHDLR R IIPQDPT+F GT+R NLDPL +Y+D +
Sbjct: 1315 TLFRIVEPSAGEIRIDGVNILTIGLHDLRLRLSIIPQDPTMFEGTMRSNLDPLEEYTDDQ 1374
Query: 516 IWEVLGKCQLREVVQEKDEGLNSSVVEDGSNWSMGQRQLFCLGRALLRRSRILVLDEATA 575
IWE L KCQL + V++K++ L+SSV E+G NWSMGQRQL CLGR LL+RS+ILVLDEATA
Sbjct: 1375 IWEALDKCQLGDEVRKKEQKLDSSVSENGDNWSMGQRQLVCLGRVLLKRSKILVLDEATA 1434
Query: 576 SVDNSTDYILLKTIRTEFADCTVITVAHRIPTVMDCTMVLSINDG 620
SVD +TD ++ KT+R F+DCTVIT+AHRI +V+D MVL +++G
Sbjct: 1435 SVDTATDNLIQKTLREHFSDCTVITIAHRISSVIDSDMVLLLSNG 1479
Score = 67.0 bits (162), Expect = 2e-09
Identities = 69/282 (24%), Positives = 122/282 (42%), Gaps = 16/282 (5%)
Query: 344 SGFIGMALSYGLALNSFLVNSIQSQCTLANQIISVERLNQYMHIQSEAKEIVEGNRPPLN 403
SG I AL+ L + N + + +S++RL Y+ + + +IVE P +
Sbjct: 577 SGKILSALATFRILQEPIYNLPDTISMIVQTKVSLDRLASYLCLDNLQPDIVE-RLPKGS 635
Query: 404 WPIAGKVEINDLKIRYRPDGPLVLHGITCTFEVGHKIGIVGRTGSGKSTLISALFRLVEP 463
+A +V + L P L I G K+ + G GSGKS+L+S+L V
Sbjct: 636 SDVAVEVINSTLSWDVSSSNP-TLKDINFKVFPGMKVAVCGTVGSGKSSLLSSLLGEVPK 694
Query: 464 SGGNIIIDGVDISSIGLHDLRSRFGIIPQDPTLFTGTVRYNLDPLSQYSDQEIWEVLGKC 523
G++ + G + Q P + +G + N+ + +VL C
Sbjct: 695 VSGSLKVCGTK-------------AYVAQSPWIQSGKIEDNILFGKPMERERYDKVLEAC 741
Query: 524 QLREVVQEKDEGLNSSVVEDGSNWSMGQRQLFCLGRALLRRSRILVLDEATASVDNSTDY 583
L + ++ G + + E G N S GQ+Q + RAL + + I + D+ ++VD T
Sbjct: 742 SLSKDLEILSFGDQTVIGERGINLSGGQKQRIQIARALYQDADIYLFDDPFSAVDAHTGS 801
Query: 584 ILLKTIRTE-FADCTVITVAHRIPTVMDCTMVLSINDGNLSR 624
L K + +VI V H++ + ++L + DG +S+
Sbjct: 802 HLFKEVLLGLLCSKSVIYVTHQVEFLPAADLILVMKDGRISQ 843
>UniRef100_Q9LK63 Multidrug resistance-associated protein (MRP); ABC-transoprter
[Arabidopsis thaliana]
Length = 1441
Score = 520 bits (1339), Expect = e-146
Identities = 286/647 (44%), Positives = 403/647 (62%), Gaps = 18/647 (2%)
Query: 1 MSEGVIQQEGPYQQLLATSKEFQDLVNAHKVTDGSNQLVNVTFSRASIKIT----QTLVE 56
M +G I Q G Y ++L + +F +LV AH T+ + + AS K T ++
Sbjct: 797 MKDGKITQAGKYHEILDSGTDFMELVGAH--TEALATIDSCETGYASEKSTTDKENEVLH 854
Query: 57 NKGKEANGN------QLIKQEEREKGDKGLKPYLQYLNQMKGYIFFFVASLGHFIFLVCQ 110
+K K+ NG+ QL+++EEREKG G Y +Y+ G + + +F +
Sbjct: 855 HKEKQENGSDNKPSGQLVQEEEREKGKVGFTVYKKYMALAYGGAVIPLILVVQVLFQLLS 914
Query: 111 ILQNLWMA-----ANVDNPRVSTFQLIFVYFLLGASSAFFMLTRSLFVIALGLQSSKYLF 165
I N WM + P VS F LI VY LL +S+F +L R+L V G + + LF
Sbjct: 915 IGSNYWMTWVTPVSKDVEPPVSGFTLILVYVLLAVASSFCILIRALLVAMTGFKMATELF 974
Query: 166 LQLMNSLFRAPMPFYDCTPLGRILSRVSSELSIMDLDIPFSLTFAVGTTMNFYSTLTVFS 225
Q+ +FRA M F+D TP+GRIL+R S++ S+ DL +P + +N + V
Sbjct: 975 TQMHLRIFRASMSFFDATPMGRILNRASTDQSVADLRLPGQFAYVAIAAINILGIIGVIV 1034
Query: 226 VVTWQVLIVAIPMVYITIRLQRYYFASAKEVMRITGTTKSYVANHVAETVSGAVTIRTFE 285
V WQVLIV IP+V ++YY ++A+E+ R+ G ++S V +H +ET+SG TIR+F+
Sbjct: 1035 QVAWQVLIVFIPVVAACAWYRQYYISAARELARLAGISRSPVVHHFSETLSGITTIRSFD 1094
Query: 286 EEDRFFQKNLDLIDINASSFFHNFASNEWLIQRLETISAGVLASAALCMVILPPGTFTSG 345
+E RF + L D + FH+ + EWL RLE +S AS+ + +V P G
Sbjct: 1095 QEPRFRGDIMRLSDCYSRLKFHSTGAMEWLCFRLELLSTFAFASSLVILVSAPEGVINPS 1154
Query: 346 FIGMALSYGLALNSFLVNSIQSQCTLANQIISVERLNQYMHIQSEAKEIVEGNRPPLNWP 405
G+A++Y L LN+ I + C L N++ISVER+ QY +I SE ++E RP +WP
Sbjct: 1155 LAGLAITYALNLNTLQATLIWTLCDLENKMISVERMLQYTNIPSEPPLVIETTRPEKSWP 1214
Query: 406 IAGKVEINDLKIRYRPDGPLVLHGITCTFEVGHKIGIVGRTGSGKSTLISALFRLVEPSG 465
G++ I +L++RY P P+VLHG+TCTF G K GIVGRTG GKSTLI LFR+VEP+
Sbjct: 1215 SRGEITICNLQVRYGPHLPMVLHGLTCTFPGGLKTGIVGRTGCGKSTLIQTLFRIVEPAA 1274
Query: 466 GNIIIDGVDISSIGLHDLRSRFGIIPQDPTLFTGTVRYNLDPLSQYSDQEIWEVLGKCQL 525
G I IDG++I SIGLHDLRSR IIPQDPT+F GT+R NLDPL +Y+D +IWE L CQL
Sbjct: 1275 GEIRIDGINILSIGLHDLRSRLSIIPQDPTMFEGTIRSNLDPLEEYTDDQIWEALDNCQL 1334
Query: 526 REVVQEKDEGLNSSVVEDGSNWSMGQRQLFCLGRALLRRSRILVLDEATASVDNSTDYIL 585
+ V++K+ L+S V E+G NWS+GQRQL CLGR LL+RS++LVLDEATAS+D +TD ++
Sbjct: 1335 GDEVRKKELKLDSPVSENGQNWSVGQRQLVCLGRVLLKRSKLLVLDEATASIDTATDNLI 1394
Query: 586 LKTIRTEFADCTVITVAHRIPTVMDCTMVLSINDGNLSRNTGLIFSL 632
+T+R FADCTVIT+AHRI +V+D MVL ++ G S T L+F+L
Sbjct: 1395 QETLRHHFADCTVITIAHRISSVIDSDMVLLLDQGCESDYT-LLFTL 1440
Score = 71.6 bits (174), Expect = 7e-11
Identities = 91/396 (22%), Positives = 169/396 (41%), Gaps = 23/396 (5%)
Query: 233 IVAIPMVYITIRLQRYYFASAKEVMRIT-GTTKSYVANHVAETVSGAVTIRTFEEEDRFF 291
I A P I + L Y FA +E + + +K +E + ++ E +F
Sbjct: 428 IAAFPAT-ILVMLANYPFAKLEEKFQSSLMKSKDNRMKKTSEVLLNMKILKLQGWEMKFL 486
Query: 292 QKNLDLIDINASSFFHNFASNEWLIQRLETISAGVLASAALCMVILPPGTFTSGFIGMAL 351
K L+L I A + F N I + + +++ A +L SG I AL
Sbjct: 487 SKILELRHIEAG-WLKKFVYNSSAINSVLWAAPSFISATAFGACLLLKIPLESGKILAAL 545
Query: 352 SYGLALNSFLVNSIQSQCTLANQIISVERLNQYMHIQSEAKEIVEGNRPPLNWPIAGKVE 411
+ L + ++ + +S+ R+ ++ + +++V G P + +A VE
Sbjct: 546 ATFRILQGPIYKLPETISMIVQTKVSLNRIASFLCLDDLQQDVV-GRLPSGSSEMA--VE 602
Query: 412 INDLKIRYRPDGPL-VLHGITCTFEVGHKIGIVGRTGSGKSTLISALFRLVEPSGGNIII 470
I++ + P+ L + G + I G GSGKS+L+S++ V GN+ +
Sbjct: 603 ISNGTFSWDDSSPIPTLRDMNFKVSQGMNVAICGTVGSGKSSLLSSILGEVPKISGNLKV 662
Query: 471 DGVDISSIGLHDLRSRFGIIPQDPTLFTGTVRYNLDPLSQYSDQEIWE-VLGKCQLREVV 529
G R I Q P + +G V N+ + ++E ++ VL C L + +
Sbjct: 663 CG-------------RKAYIAQSPWIQSGKVEENI-LFGKPMEREWYDRVLEACSLNKDL 708
Query: 530 QEKDEGLNSSVVEDGSNWSMGQRQLFCLGRALLRRSRILVLDEATASVDNSTDYILLKTI 589
+ + + E G N S GQ+Q + RAL + + I + D+ ++VD T L K +
Sbjct: 709 EILPFHDQTVIGERGINLSGGQKQRIQIARALYQDADIYLFDDPFSAVDAHTGSHLFKEV 768
Query: 590 RTE-FADCTVITVAHRIPTVMDCTMVLSINDGNLSR 624
TVI V H++ + + ++L + DG +++
Sbjct: 769 LLGLLRHKTVIYVTHQVEFLPEADLILVMKDGKITQ 804
>UniRef100_O24525 Glutathione-conjugate transporter AtMRP4 [Arabidopsis thaliana]
Length = 1516
Score = 515 bits (1327), Expect = e-144
Identities = 282/655 (43%), Positives = 404/655 (61%), Gaps = 42/655 (6%)
Query: 1 MSEGVIQQEGPYQQLLATSKEFQDLVNAHKVTDGSNQLVNVTFSRASIKIT--------- 51
M +G I + G Y +L+++ +F +LV AH+ S +LV A++ +
Sbjct: 837 MRDGKIVESGKYDELVSSGLDFGELVAAHET---SMELVEAGADSAAVATSPRTPTSPHA 893
Query: 52 ---QTLVENKGKE----------------ANGNQLIKQEEREKGDKGLKPYLQYLNQMKG 92
+T +E+ +G++LIK+EERE G L Y QY + G
Sbjct: 894 SSPRTSMESPHLSDLNDEHIKSFLGSHIVEDGSKLIKEEERETGQVSLGVYKQYCTEAYG 953
Query: 93 YIFFFVASLGHFIFLVCQILQNLWMAANVDNPRVSTFQ---LIFVYFLLGASSAFFMLTR 149
+ + + + + W+A +F I Y ++ S + R
Sbjct: 954 WWGIVLVLFFSLTWQGSLMASDYWLAYETSAKNAISFDASVFILGYVIIALVSIVLVSIR 1013
Query: 150 SLFVIALGLQSSKYLFLQLMNSLFRAPMPFYDCTPLGRILSRVSSELSIMDLDIPFSLTF 209
S +V LGL++++ F Q++NS+ APM F+D TP GRILSR S++ + +D+ IPF L
Sbjct: 1014 SYYVTHLGLKTAQIFFRQILNSILHAPMSFFDTTPSGRILSRASTDQTNVDILIPFML-- 1071
Query: 210 AVGTTMNFYSTLTVFSVVT----WQVLIVAIPMVYITIRLQRYYFASAKEVMRITGTTKS 265
G ++ Y+TL +VT W IP+ ++ I + YY AS++E+ R+ TK+
Sbjct: 1072 --GLVVSMYTTLLSIFIVTCQYAWPTAFFVIPLGWLNIWYRNYYLASSRELTRMDSITKA 1129
Query: 266 YVANHVAETVSGAVTIRTFEEEDRFFQKNLDLIDINASSFFHNFASNEWLIQRLETISAG 325
+ +H +E+++G +TIR+F +++ F Q+N+ ++ N FHN SNEWL RLE + +
Sbjct: 1130 PIIHHFSESIAGVMTIRSFRKQELFRQENVKRVNDNLRMDFHNNGSNEWLGFRLELVGSW 1189
Query: 326 VLASAALCMVILPPGTFTSGFIGMALSYGLALNSFLVNSIQSQCTLANQIISVERLNQYM 385
VL +AL MV+LP +G++LSYGL+LNS L +I C + N+++SVER+ Q+
Sbjct: 1190 VLCISALFMVLLPSNVIRPENVGLSLSYGLSLNSVLFFAIYMSCFVENKMVSVERIKQFT 1249
Query: 386 HIQSEAKEIVEGNRPPLNWPIAGKVEINDLKIRYRPDGPLVLHGITCTFEVGHKIGIVGR 445
I SE++ + PP NWP G V + DLK+RYRP+ PLVL GIT + G K+G+VGR
Sbjct: 1250 DIPSESEWERKETLPPSNWPFHGNVHLEDLKVRYRPNTPLVLKGITLDIKGGEKVGVVGR 1309
Query: 446 TGSGKSTLISALFRLVEPSGGNIIIDGVDISSIGLHDLRSRFGIIPQDPTLFTGTVRYNL 505
TGSGKSTLI LFRLVEPSGG IIIDG+DIS++GLHDLRSRFGIIPQ+P LF GTVR N+
Sbjct: 1310 TGSGKSTLIQVLFRLVEPSGGKIIIDGIDISTLGLHDLRSRFGIIPQEPVLFEGTVRSNI 1369
Query: 506 DPLSQYSDQEIWEVLGKCQLREVVQEKDEGLNSSVVEDGSNWSMGQRQLFCLGRALLRRS 565
DP QYSD+EIW+ L +CQL++VV K E L+S VV++G NWS+GQRQL CLGR +L+RS
Sbjct: 1370 DPTEQYSDEEIWKSLERCQLKDVVATKPEKLDSLVVDNGENWSVGQRQLLCLGRVMLKRS 1429
Query: 566 RILVLDEATASVDNSTDYILLKTIRTEFADCTVITVAHRIPTVMDCTMVLSINDG 620
R+L LDEATASVD+ TD ++ K IR +FA CT+I++AHRIPTVMD VL I+ G
Sbjct: 1430 RLLFLDEATASVDSQTDAVIQKIIREDFASCTIISIAHRIPTVMDGDRVLVIDAG 1484
Score = 75.9 bits (185), Expect = 4e-12
Identities = 77/320 (24%), Positives = 139/320 (43%), Gaps = 27/320 (8%)
Query: 314 WLIQRLETISAGVLASAALCMVILPPGTFTSGFIGMALSYGLALNSFLVNSI-------- 365
WL + L +I+ ++ + ++I T+ +G+ L G + + I
Sbjct: 539 WLSKFLYSIAGNIIVLWSTPVLISALTFATALALGVKLDAGTVFTTTTIFKILQEPIRTF 598
Query: 366 -QSQCTLANQIISVERLNQYMHIQSEAKEIVEGNRPPLNWPIAGKVEINDLKIRYRP-DG 423
QS +L+ +IS+ RL+ YM + +++ VE L VE+ D + D
Sbjct: 599 PQSMISLSQAMISLGRLDSYMMSKELSEDAVER---ALGCDGNTAVEVRDGSFSWDDEDN 655
Query: 424 PLVLHGITCTFEVGHKIGIVGRTGSGKSTLISALFRLVEPSGGNIIIDGVDISSIGLHDL 483
L I + G IVG GSGKS+L++++ + G + + G
Sbjct: 656 EPALSDINFKVKKGELTAIVGTVGSGKSSLLASVLGEMHRISGQVRVCGST--------- 706
Query: 484 RSRFGIIPQDPTLFTGTVRYNLDPLSQYSDQEIWEVLGKCQLREVVQEKDEGLNSSVVED 543
G + Q + GTV+ N+ ++ +VL C L + +Q + G + + E
Sbjct: 707 ----GYVAQTSWIENGTVQDNILFGLPMVREKYNKVLNVCSLEKDLQMMEFGDKTEIGER 762
Query: 544 GSNWSMGQRQLFCLGRALLRRSRILVLDEATASVDNST-DYILLKTIRTEFADCTVITVA 602
G N S GQ+Q L RA+ + + +LD+ ++VD T I K +R TV+ V
Sbjct: 763 GINLSGGQKQRIQLARAVYQECDVYLLDDVFSAVDAHTGSDIFKKCVRGALKGKTVLLVT 822
Query: 603 HRIPTVMDCTMVLSINDGNL 622
H++ + + +L + DG +
Sbjct: 823 HQVDFLHNVDCILVMRDGKI 842
>UniRef100_Q9LZJ5 ABC transporter-like protein [Arabidopsis thaliana]
Length = 1539
Score = 514 bits (1324), Expect = e-144
Identities = 285/677 (42%), Positives = 407/677 (60%), Gaps = 61/677 (9%)
Query: 1 MSEGVIQQEGPYQQLLATSKEFQDLVNAHKVT-------DGSNQLVNVTFS----RASIK 49
M +G+I Q G Y +L+++ +F +LV AH+ + S NV + + SI
Sbjct: 835 MRDGMIVQSGKYDELVSSGLDFGELVAAHETSMELVEAGSASATAANVPMASPITQRSIS 894
Query: 50 I------------------------TQTLVENKGKEAN---------------GNQLIKQ 70
I T ++ + E N G++LIK+
Sbjct: 895 IESPRQPKSPKVHRTTSMESPRVLRTTSMESPRLSELNDESIKSFLGSNIPEDGSRLIKE 954
Query: 71 EEREKGDKGLKPYLQYLNQMKGYIFFFVASLGHFIFLVCQILQNLWMAANVDNPRVSTFQ 130
EERE G + Y Y + G+ + + + + W+A +F
Sbjct: 955 EEREVGQVSFQVYKLYSTEAYGWWGMILVVFFSVAWQASLMASDYWLAYETSAKNEVSFD 1014
Query: 131 ---LIFVYFLLGASSAFFMLTRSLFVIALGLQSSKYLFLQLMNSLFRAPMPFYDCTPLGR 187
I VY ++ A S + R+ +V LGL++++ F Q++NSL APM F+D TP GR
Sbjct: 1015 ATVFIRVYVIIAAVSIVLVCLRAFYVTHLGLKTAQIFFKQILNSLVHAPMSFFDTTPSGR 1074
Query: 188 ILSRVSSELSIMDLDIPFSLTFAVGTTMNFYSTLTVFSVVT----WQVLIVAIPMVYITI 243
ILSR S++ + +D+ IPF +G Y+TL +VT W + IP+ ++ I
Sbjct: 1075 ILSRASTDQTNVDIFIPFM----IGLVATMYTTLLSIFIVTCQYAWPTVFFIIPLGWLNI 1130
Query: 244 RLQRYYFASAKEVMRITGTTKSYVANHVAETVSGAVTIRTFEEEDRFFQKNLDLIDINAS 303
+ YY AS++E+ R+ TK+ V +H +E+++G +TIR F+++ F Q+N+ ++ N
Sbjct: 1131 WYRGYYLASSRELTRLDSITKAPVIHHFSESIAGVMTIRAFKKQPMFRQENVKRVNANLR 1190
Query: 304 SFFHNFASNEWLIQRLETISAGVLASAALCMVILPPGTFTSGFIGMALSYGLALNSFLVN 363
FHN SNEWL RLE I + VL +AL MV+LP +G++LSYGL+LN L
Sbjct: 1191 MDFHNNGSNEWLGFRLELIGSWVLCISALFMVMLPSNIIKPENVGLSLSYGLSLNGVLFW 1250
Query: 364 SIQSQCTLANQIISVERLNQYMHIQSEAKEIVEGNRPPLNWPIAGKVEINDLKIRYRPDG 423
+I C + N+++SVER+ Q+ I +EAK ++ +RPP NWP G + + D+K+RYRP+
Sbjct: 1251 AIYLSCFIENKMVSVERIKQFTDIPAEAKWEIKESRPPPNWPYKGNIRLEDVKVRYRPNT 1310
Query: 424 PLVLHGITCTFEVGHKIGIVGRTGSGKSTLISALFRLVEPSGGNIIIDGVDISSIGLHDL 483
PLVL G+T + G KIG+VGRTGSGKSTLI LFRLVEPSGG IIIDG+DI ++GLHDL
Sbjct: 1311 PLVLKGLTIDIKGGEKIGVVGRTGSGKSTLIQVLFRLVEPSGGKIIIDGIDICTLGLHDL 1370
Query: 484 RSRFGIIPQDPTLFTGTVRYNLDPLSQYSDQEIWEVLGKCQLREVVQEKDEGLNSSVVED 543
RSRFGIIPQ+P LF GTVR N+DP +YSD+EIW+ L +CQL++VV K E L+S V ++
Sbjct: 1371 RSRFGIIPQEPVLFEGTVRSNIDPTEKYSDEEIWKSLERCQLKDVVASKPEKLDSLVADN 1430
Query: 544 GSNWSMGQRQLFCLGRALLRRSRILVLDEATASVDNSTDYILLKTIRTEFADCTVITVAH 603
G NWS+GQRQL CLGR +L+RSRIL LDEATASVD+ TD ++ K IR +F+DCT+I++AH
Sbjct: 1431 GENWSVGQRQLLCLGRVMLKRSRILFLDEATASVDSQTDAMIQKIIREDFSDCTIISIAH 1490
Query: 604 RIPTVMDCTMVLSINDG 620
RIPTVMDC VL I+ G
Sbjct: 1491 RIPTVMDCDRVLVIDAG 1507
Score = 79.7 bits (195), Expect = 3e-13
Identities = 76/317 (23%), Positives = 142/317 (43%), Gaps = 25/317 (7%)
Query: 314 WLIQRLETISAGVLASAALCMVILPPGTFTSGFIGMALSYGLALNSFLVNSI-------- 365
WL + L +I+ ++ + ++I T+ F+G+ L G + + I
Sbjct: 537 WLSKFLYSIAGNIIVLWSTPVLISALTFTTAVFLGVKLDAGTVFTTTTIFKILQEPIRTF 596
Query: 366 -QSQCTLANQIISVERLNQYMHIQSEAKEIVEGNRPPLNWPIAGKVEINDLKIRYRPDGP 424
QS +L+ +IS+ RL+ YM + ++E VE ++ + +A +++ D P
Sbjct: 597 PQSMISLSQAMISLGRLDAYMMSRELSEETVERSQG-CDGNVAVEIKDGSFSWDDEDDEP 655
Query: 425 LVLHGITCTFEVGHKIGIVGRTGSGKSTLISALFRLVEPSGGNIIIDGVDISSIGLHDLR 484
+ I + G IVG GSGKS+L++++ + G + + G
Sbjct: 656 AI-ENINFEVKKGELAAIVGTVGSGKSSLLASVLGEMHKLSGKVRVCGTT---------- 704
Query: 485 SRFGIIPQDPTLFTGTVRYNLDPLSQYSDQEIWEVLGKCQLREVVQEKDEGLNSSVVEDG 544
+ Q + GTV+ N+ + + EVL C L + +Q + G + + E G
Sbjct: 705 ---AYVAQTSWIQNGTVQDNILFGLPMNRSKYNEVLKVCCLEKDMQIMEFGDQTEIGERG 761
Query: 545 SNWSMGQRQLFCLGRALLRRSRILVLDEATASVDNST-DYILLKTIRTEFADCTVITVAH 603
N S GQ+Q L RA+ + S + +LD+ ++VD T I K +R T++ V H
Sbjct: 762 INLSGGQKQRIQLARAVYQESDVYLLDDVFSAVDAHTGSDIFKKCVRGALKGKTILLVTH 821
Query: 604 RIPTVMDCTMVLSINDG 620
++ + + +L + DG
Sbjct: 822 QVDFLHNVDRILVMRDG 838
>UniRef100_Q8GU64 MRP-like ABC transporter [Oryza sativa]
Length = 1527
Score = 514 bits (1323), Expect = e-144
Identities = 273/641 (42%), Positives = 392/641 (60%), Gaps = 27/641 (4%)
Query: 1 MSEGVIQQEGPYQQLLATSKEFQDLVNAHKVTDGSNQLV-------------NVTFSRAS 47
M +G + Q G Y LL T +F LV AH + S +LV N+ SR
Sbjct: 861 MRDGAVAQSGRYHDLLRTGTDFAALVAAH---ESSMELVESAAPGPSPSPAGNLPLSRQP 917
Query: 48 IKITQTLVENKGKEANGN--------QLIKQEEREKGDKGLKPYLQYLNQMKGYIFFFVA 99
+ E + +NG+ +LIK EER G Y QY+ + G+ +
Sbjct: 918 SSAPK---ERESASSNGDIKTAKASSRLIKAEERASGHVSFTVYRQYMTEAWGWWGLMLV 974
Query: 100 SLGHFIFLVCQILQNLWMAANVDNPRVSTFQLIFVYFLLGASSAFFMLTRSLFVIALGLQ 159
+ + + W+A I VY ++ A S + RSL V +GL
Sbjct: 975 LAVSVAWQGSTMAADYWLAYQTSGDAFRPALFIKVYAIIAAVSVVIVTVRSLLVATIGLD 1034
Query: 160 SSKYLFLQLMNSLFRAPMPFYDCTPLGRILSRVSSELSIMDLDIPFSLTFAVGTTMNFYS 219
++ F Q+++++ APM F+D TP GRIL+R SS+ + +DL +PF + +V +
Sbjct: 1035 TANIFFRQVLSTILHAPMSFFDTTPSGRILTRASSDQTNVDLLLPFFVWMSVSMYITVIG 1094
Query: 220 TLTVFSVVTWQVLIVAIPMVYITIRLQRYYFASAKEVMRITGTTKSYVANHVAETVSGAV 279
+ + V W +++ +P++ + + ++YY ++++E+ R+ TK+ V +H +ETV G +
Sbjct: 1095 VVIMTCQVAWPSVVLVVPLLMLNLWFRKYYISTSRELTRLESITKAPVIHHFSETVQGVM 1154
Query: 280 TIRTFEEEDRFFQKNLDLIDINASSFFHNFASNEWLIQRLETISAGVLASAALCMVILPP 339
IR F+++D FF +NL ++ + FHN A+NEWL RLE I + VL AL MV LP
Sbjct: 1155 VIRCFQKQDNFFHENLSRLNASLKMDFHNNAANEWLGLRLELIGSLVLCVTALLMVTLPS 1214
Query: 340 GTFTSGFIGMALSYGLALNSFLVNSIQSQCTLANQIISVERLNQYMHIQSEAKEIVEGNR 399
++G++LSYGL+LNS + +I C + N+++SVER+ Q+ +I SEA+ ++
Sbjct: 1215 NIVLPEYVGLSLSYGLSLNSVMFWAIWLSCNIENKMVSVERIKQFTNIPSEAEWRIKETA 1274
Query: 400 PPLNWPIAGKVEINDLKIRYRPDGPLVLHGITCTFEVGHKIGIVGRTGSGKSTLISALFR 459
P NWP G ++I DLK RYR + PLVL GIT + G KIG+VGRTGSGKSTLI ALFR
Sbjct: 1275 PSANWPHKGDIDIIDLKFRYRHNTPLVLKGITLSIHGGEKIGVVGRTGSGKSTLIQALFR 1334
Query: 460 LVEPSGGNIIIDGVDISSIGLHDLRSRFGIIPQDPTLFTGTVRYNLDPLSQYSDQEIWEV 519
+VEPS G IIIDG+DI ++GLHDLRSRFGIIPQ+P LF GT+R N+DPL YSD EIW+
Sbjct: 1335 IVEPSEGKIIIDGIDICTLGLHDLRSRFGIIPQEPVLFEGTIRSNIDPLQLYSDDEIWQA 1394
Query: 520 LGKCQLREVVQEKDEGLNSSVVEDGSNWSMGQRQLFCLGRALLRRSRILVLDEATASVDN 579
L +CQL++ V K E L++SVV++G NWS+GQRQL CLGR +L+ SRIL +DEATASVD+
Sbjct: 1395 LERCQLKDAVTSKPEKLDASVVDNGENWSVGQRQLLCLGRVMLKHSRILFMDEATASVDS 1454
Query: 580 STDYILLKTIRTEFADCTVITVAHRIPTVMDCTMVLSINDG 620
TD ++ K IR EF+ CT+I++AHRIPTVMDC VL I+ G
Sbjct: 1455 RTDAVIQKIIREEFSACTIISIAHRIPTVMDCDRVLVIDAG 1495
Score = 73.6 bits (179), Expect = 2e-11
Identities = 75/329 (22%), Positives = 137/329 (40%), Gaps = 34/329 (10%)
Query: 314 WLIQRLETISAGVLASAALCMVILPPGTFTSGFIGMALSYGLA---------LNSFLVNS 364
WL + + +IS ++A + + I TS +G+ L GL L + N
Sbjct: 558 WLTRFMYSISGNIIALWSAPIAIAALVFATSVLLGVRLDAGLVFTATSFFKILQEPMRNF 617
Query: 365 IQSQCTLANQIISVERLNQYMHIQSEAKEIVEG--NRPPLNWPIAGKVEINDLKIRYRPD 422
QS ++ ++S+ RL+ YM + E+ EG R P V + + + +
Sbjct: 618 PQSIIQVSQAMVSLGRLDSYM----TSAELDEGAVERGPAVGAGMTAVRVRGGEFAWEEE 673
Query: 423 GPL-----VLHGITCTFEVGHKIGIVGRTGSGKSTLISALFRLVEPSGGNIIIDGVDISS 477
VL GI G +VG GSGKS+L+ + + G + +
Sbjct: 674 EEAAGQQAVLRGIDIDVRAGTLAAVVGMVGSGKSSLLGCILGEMRKISGEVTV------- 726
Query: 478 IGLHDLRSRFGIIPQDPTLFTGTVRYNLDPLSQYSDQEIWEVLGKCQLREVVQEKDEGLN 537
R +PQ + GT+ N+ + E + C L + ++ + G
Sbjct: 727 ------RGSMAYVPQTAWIQNGTIEENILFGRGMQRERYREAIRVCSLDKDLEMMEFGDQ 780
Query: 538 SSVVEDGSNWSMGQRQLFCLGRALLRRSRILVLDEATASVDNST-DYILLKTIRTEFADC 596
+ + E G N S GQ+Q L RA+ + + + +LD+ ++VD T I +R D
Sbjct: 781 TEIGERGINLSGGQKQRIQLARAVYQDADVYLLDDVFSAVDAHTGSDIFRDCVRGALRDK 840
Query: 597 TVITVAHRIPTVMDCTMVLSINDGNLSRN 625
TV+ V H++ + + + + DG ++++
Sbjct: 841 TVLLVTHQLDFLRNAHAIYVMRDGAVAQS 869
>UniRef100_Q9M1C7 Multi resistance protein homolog [Arabidopsis thaliana]
Length = 1490
Score = 513 bits (1322), Expect = e-144
Identities = 281/648 (43%), Positives = 400/648 (61%), Gaps = 46/648 (7%)
Query: 1 MSEGVIQQEGPYQQLLATSKEFQDLVNAHKVTDGSNQLVNVTFSRASIK---------IT 51
M G + Q G +++LL + F+ LV AH + + ++++ S + K I
Sbjct: 826 MQNGRVMQAGKFEELLKQNIGFEVLVGAHN--EALDSILSIEKSSRNFKEGSKDDTASIA 883
Query: 52 QTL-----------VENKGKEANGNQLIKQEEREKGDKGLKPYLQYLNQMKGYIFFFVAS 100
++L ENK KEA +L++ EE EKG G + YL YL +KG +
Sbjct: 884 ESLQTHCDSEHNISTENKKKEA---KLVQDEETEKGVIGKEVYLAYLTTVKGGLLVPFII 940
Query: 101 LGHFIFLVCQILQNLWMAANVDN-----PRVSTFQLIFVYFLLGASSAFFMLTRSLFVIA 155
L F + QI N WMA P++ +++ VY LL A S+ +L R++ V
Sbjct: 941 LAQSCFQMLQIASNYWMAWTAPPTAESIPKLGMGRILLVYALLAAGSSLCVLARTILVAI 1000
Query: 156 LGLQSSKYLFLQLMNSLFRAPMPFYDCTPLGRILSRVSSELSIMDLDIPFSLTFAVGTTM 215
GL +++ F +++ S+FRAPM F+D TP GRIL+R S++ S++DL++ L + + +
Sbjct: 1001 GGLSTAETFFSRMLCSIFRAPMSFFDSTPTGRILNRASTDQSVLDLEMAVKLGWCAFSII 1060
Query: 216 NFYSTLTVFSVVTWQVLIVAIPMVYITIRLQRYYFASAKEVMRITGTTKSYVANHVAETV 275
T+ V S V WQ RYY +A+E+ R++G ++ + +H AE++
Sbjct: 1061 QIVGTIFVMSQVAWQ----------------RYYTPTARELSRMSGVERAPILHHFAESL 1104
Query: 276 SGAVTIRTFEEEDRFFQKNLDLIDINASSFFHNFASNEWLIQRLETISAGVLASAALCMV 335
+GA TIR F++ DRF NL LID ++ +FH ++ EWL RL +S V A + + +V
Sbjct: 1105 AGATTIRAFDQRDRFISSNLVLIDSHSRPWFHVASAMEWLSFRLNLLSHFVFAFSLVLLV 1164
Query: 336 ILPPGTFTSGFIGMALSYGLALNSFLVNSIQSQCTLANQIISVERLNQYMHIQSEAKEIV 395
LP G G+ ++YGL+LN I + C N++ISVER+ QY I SEA ++
Sbjct: 1165 TLPEGVINPSIAGLGVTYGLSLNVLQATVIWNICNAENKMISVERILQYSKIPSEAPLVI 1224
Query: 396 EGNRPPLNWPIAGKVEINDLKIRYRPDGPLVLHGITCTFEVGHKIGIVGRTGSGKSTLIS 455
+G+RP NWP G + DL++RY P VL ITC F G KIG+VGRTGSGKSTLI
Sbjct: 1225 DGHRPLDNWPNVGSIVFRDLQVRYAEHFPAVLKNITCEFPGGKKIGVVGRTGSGKSTLIQ 1284
Query: 456 ALFRLVEPSGGNIIIDGVDISSIGLHDLRSRFGIIPQDPTLFTGTVRYNLDPLSQYSDQE 515
ALFR+VEPS G I+ID VDI+ IGLHDLRSR GIIPQDP LF GT+R NLDPL+QY+D E
Sbjct: 1285 ALFRIVEPSQGTIVIDNVDITKIGLHDLRSRLGIIPQDPALFDGTIRLNLDPLAQYTDHE 1344
Query: 516 IWEVLGKCQLREVVQEKDEGLNSSVVEDGSNWSMGQRQLFCLGRALLRRSRILVLDEATA 575
IWE + KCQL +V++ KDE L+++VVE+G NWS+GQRQL CLGR LL++S ILVLDEATA
Sbjct: 1345 IWEAIDKCQLGDVIRAKDERLDATVVENGENWSVGQRQLVCLGRVLLKKSNILVLDEATA 1404
Query: 576 SVDNSTDYILLKTIRTEFADCTVITVAHRIPTVMDCTMVLSINDGNLS 623
SVD++TD ++ K I EF D TV+T+AHRI TV++ +VL ++DG ++
Sbjct: 1405 SVDSATDGVIQKIINQEFKDRTVVTIAHRIHTVIESDLVLVLSDGRIA 1452
Score = 75.5 bits (184), Expect = 5e-12
Identities = 67/285 (23%), Positives = 125/285 (43%), Gaps = 24/285 (8%)
Query: 343 TSGFIGMALSYGLALNSFLVNSIQSQCTLANQIISVERLNQYMH---IQSEAKEIVEGNR 399
T+G + AL+ L S + L +S +R+ Y+ Q +A E +
Sbjct: 566 TAGAVLSALATFQMLQSPIFGLPDLLSALVQSKVSADRIASYLQQSETQKDAVEYCSKDH 625
Query: 400 PPLNWPIAGKVEINDLKIRYRPDGPL-VLHGITCTFEVGHKIGIVGRTGSGKSTLISALF 458
L+ VEI + + P+ L I + G K+ + G GSGKS+L+S++
Sbjct: 626 TELS------VEIENGAFSWEPESSRPTLDDIELKVKSGMKVAVCGAVGSGKSSLLSSIL 679
Query: 459 RLVEPSGGNIIIDGVDISSIGLHDLRSRFGIIPQDPTLFTGTVRYNLDPLSQYSDQEIWE 518
++ G + + G + +PQ P + +GT+R N+ S Y ++
Sbjct: 680 GEIQKLKGTVRVSG-------------KQAYVPQSPWILSGTIRDNILFGSMYESEKYER 726
Query: 519 VLGKCQLREVVQEKDEGLNSSVVEDGSNWSMGQRQLFCLGRALLRRSRILVLDEATASVD 578
+ C L + + G + + E G N S GQ+Q + RA+ + + I +LD+ ++VD
Sbjct: 727 TVKACALIKDFELFSNGDLTEIGERGINMSGGQKQRIQIARAVYQNADIYLLDDPFSAVD 786
Query: 579 NSTDYILLK-TIRTEFADCTVITVAHRIPTVMDCTMVLSINDGNL 622
T L + + D TV+ V H++ + ++L + +G +
Sbjct: 787 AHTGRELFEDCLMGILKDKTVLYVTHQVEFLPAADLILVMQNGRV 831
>UniRef100_O24510 MRP-like ABC transporter [Arabidopsis thaliana]
Length = 1515
Score = 511 bits (1316), Expect = e-143
Identities = 280/651 (43%), Positives = 396/651 (60%), Gaps = 37/651 (5%)
Query: 1 MSEGVIQQEGPYQQLLATSKEFQDLVNAHKVT-------DGSNQLVNVTFSRASIKITQT 53
M +G I Q G Y +L + +F +L+ AH+ D ++ + ++ +
Sbjct: 836 MKDGRISQAGKYNDILNSGTDFMELIGAHQEALAVVDSVDANSVSEKSALGQENVIVKDA 895
Query: 54 LVENKGKEANG------------NQLIKQEEREKGDKGLKPYLQYLNQMKGYIFFFVASL 101
+ ++ E+ Q+I++EEREKG L Y +Y+ G L
Sbjct: 896 IAVDEKLESQDLKNDKLESVEPQRQIIQEEEREKGSVALDVYWKYITLAYGGALVPFILL 955
Query: 102 GHFIFLVCQILQNLWMA------ANVDNPRVSTFQLIFVYFLLGASSAFFMLTRSLFVIA 155
G +F + QI N WMA +V P V L+ VY L S+ +L R+ ++
Sbjct: 956 GQVLFQLLQIGSNYWMAWATPVSEDVQAP-VKLSTLMIVYVALAFGSSLCILLRATLLVT 1014
Query: 156 LGLQSSKYLFLQLMNSLFRAPMPFYDCTPLGRILSRVSSELSIMDLDIPFSLTFAVGTTM 215
G +++ LF ++ + +FR+PM F+D TP GRI+SR S++ S +DL++P+ T +
Sbjct: 1015 AGYKTATELFHKMHHCIFRSPMSFFDSTPSGRIMSRASTDQSAVDLELPYQFGSVAITVI 1074
Query: 216 NFYSTLTVFSVVTWQVLIVAIPMVYITIRLQRYYFASAKEVMRITGTTKSYVANHVAETV 275
+ V S V+W V +V IP+V +I QRYY A+A+E+ R+ G K+ + H +ET+
Sbjct: 1075 QLIGIIGVMSQVSWLVFLVFIPVVAASIWYQRYYIAAARELSRLVGVCKAPLIQHFSETI 1134
Query: 276 SGAVTIRTFEEEDRFFQKNLDLIDINASSFFHNFASNEWLIQRLETISAGVLASAALCMV 335
SGA TIR+F +E RF N+ L D+ + EWL RL+ +L+S C+
Sbjct: 1135 SGATTIRSFSQEFRFRSDNMRLSDVTLGPNSIQLGAMEWLCFRLD-----MLSSLTFCLF 1189
Query: 336 I------LPPGTFTSGFIGMALSYGLALNSFLVNSIQSQCTLANQIISVERLNQYMHIQS 389
+P G G+A++YGL+LN+ I + C L N+IISVER+ QY + S
Sbjct: 1190 NWFSWSPIPTGVIDPSLAGLAVTYGLSLNTLQAWLIWTLCNLENKIISVERILQYASVPS 1249
Query: 390 EAKEIVEGNRPPLNWPIAGKVEINDLKIRYRPDGPLVLHGITCTFEVGHKIGIVGRTGSG 449
E ++E NRP +WP G+VEI DL++RY P PLVL GITCTF+ G + GIVGRTGSG
Sbjct: 1250 EPPLVIESNRPEQSWPSRGEVEIRDLQVRYAPHMPLVLRGITCTFKGGLRTGIVGRTGSG 1309
Query: 450 KSTLISALFRLVEPSGGNIIIDGVDISSIGLHDLRSRFGIIPQDPTLFTGTVRYNLDPLS 509
KSTLI LFR+VEPS G I IDGV+I +IGLHDLR R IIPQDPT+F GT+R NLDPL
Sbjct: 1310 KSTLIQTLFRIVEPSAGEIRIDGVNILTIGLHDLRLRLSIIPQDPTMFEGTMRSNLDPLE 1369
Query: 510 QYSDQEIWEVLGKCQLREVVQEKDEGLNSSVVEDGSNWSMGQRQLFCLGRALLRRSRILV 569
+Y+D +IWE L KCQL + V++K++ L+SSV E+G NWSMGQRQL CLGR LL+RS+ILV
Sbjct: 1370 EYTDDQIWEALDKCQLGDEVRKKEQKLDSSVSENGDNWSMGQRQLVCLGRVLLKRSKILV 1429
Query: 570 LDEATASVDNSTDYILLKTIRTEFADCTVITVAHRIPTVMDCTMVLSINDG 620
LDEATASVD +TD ++ KT+R F+DCTVIT+AHRI +V+D MVL +++G
Sbjct: 1430 LDEATASVDTATDNLIQKTLREHFSDCTVITIAHRISSVIDSDMVLLLSNG 1480
Score = 67.0 bits (162), Expect = 2e-09
Identities = 69/282 (24%), Positives = 122/282 (42%), Gaps = 16/282 (5%)
Query: 344 SGFIGMALSYGLALNSFLVNSIQSQCTLANQIISVERLNQYMHIQSEAKEIVEGNRPPLN 403
SG I AL+ L + N + + +S++RL Y+ + + +IVE P +
Sbjct: 577 SGKILSALATFRILQEPIYNLPDTISMIVQTKVSLDRLASYLCLDNLQPDIVE-RLPKGS 635
Query: 404 WPIAGKVEINDLKIRYRPDGPLVLHGITCTFEVGHKIGIVGRTGSGKSTLISALFRLVEP 463
+A +V + L P L I G K+ + G GSGKS+L+S+L V
Sbjct: 636 SDVAVEVINSTLSWDVSSSNP-TLKDINFKVFPGMKVAVCGTVGSGKSSLLSSLLGEVPK 694
Query: 464 SGGNIIIDGVDISSIGLHDLRSRFGIIPQDPTLFTGTVRYNLDPLSQYSDQEIWEVLGKC 523
G++ + G + Q P + +G + N+ + +VL C
Sbjct: 695 VSGSLKVCGTK-------------AYVAQSPWIQSGKIEDNILFGKPMERERYDKVLEAC 741
Query: 524 QLREVVQEKDEGLNSSVVEDGSNWSMGQRQLFCLGRALLRRSRILVLDEATASVDNSTDY 583
L + ++ G + + E G N S GQ+Q + RAL + + I + D+ ++VD T
Sbjct: 742 SLSKDLEILSFGDQTVIGERGINLSGGQKQRIQIARALYQDADIYLFDDPFSAVDAHTGS 801
Query: 584 ILLKTIRTE-FADCTVITVAHRIPTVMDCTMVLSINDGNLSR 624
L K + +VI V H++ + ++L + DG +S+
Sbjct: 802 HLFKEVLLGLLCSKSVIYVTHQVEFLPAADLILVMKDGRISQ 843
>UniRef100_Q94E55 MRP-like ABC transporter [Oryza sativa]
Length = 1493
Score = 510 bits (1314), Expect = e-143
Identities = 278/642 (43%), Positives = 402/642 (62%), Gaps = 23/642 (3%)
Query: 1 MSEGVIQQEGPYQQLLATSKEFQDLVNAHK----------VTDGSNQLVNV--TFSRASI 48
M G I Q G Y ++L + +EF +LV AHK VT+G N+ + T S A
Sbjct: 820 MKGGRIAQAGKYDEILGSGEEFMELVGAHKDALTALDAIDVTNGGNEASSSSKTASLARS 879
Query: 49 KITQTLVENKGKEANGN----QLIKQEEREKGDKGLKPYLQYLN-QMKGYIFFFVASLGH 103
+ + GKE + N QL+++EEREKG G Y +YL +G + F+ L
Sbjct: 880 VSVEKKDKQNGKEDDANAQSGQLVQEEEREKGRVGFWVYWKYLTLAYRGALVPFIL-LAQ 938
Query: 104 FIFLVCQILQNLWMA-----ANVDNPRVSTFQLIFVYFLLGASSAFFMLTRSLFVIALGL 158
+F V QI N WMA + P VS LI+VY L S+ +L R+L ++
Sbjct: 939 ILFQVLQIASNYWMAWAAPVSKDVEPPVSMSTLIYVYVALAFGSSLCILVRALILVTAAY 998
Query: 159 QSSKYLFLQLMNSLFRAPMPFYDCTPLGRILSRVSSELSIMDLDIPFSLTFAVGTTMNFY 218
+++ LF ++ S+FRAPM F+D TP GRIL+R S++ S +D I + + + +
Sbjct: 999 KTATLLFNKMHMSIFRAPMSFFDSTPSGRILNRASTDQSEVDTSIAYQMGSVAFSIIQLV 1058
Query: 219 STLTVFSVVTWQVLIVAIPMVYITIRLQRYYFASAKEVMRITGTTKSYVANHVAETVSGA 278
+ V S V WQV +V IP++ QRYY +A+E+ R+ G K+ + H AE+++G+
Sbjct: 1059 GIIAVMSQVAWQVFVVFIPVLAACFWYQRYYIDTARELQRLVGVCKAPIIQHFAESITGS 1118
Query: 279 VTIRTFEEEDRFFQKNLDLIDINASSFFHNFASNEWLIQRLETISAGVLASAALCMVILP 338
TIR+F +E++F N L+D + F+N A+ EWL RL+ +S+ A + + +V LP
Sbjct: 1119 TTIRSFGKENQFVSTNSHLMDAFSRPKFYNAAAMEWLCFRLDMLSSLTFAFSLIFLVNLP 1178
Query: 339 PGTFTSGFIGMALSYGLALNSFLVNSIQSQCTLANQIISVERLNQYMHIQSEAKEIVEGN 398
G G G+A++YGL LN + S C L N+IISVER+ QYM I +E V+ +
Sbjct: 1179 TGLIDPGISGLAVTYGLNLNMLQAWVVWSMCNLENKIISVERILQYMSIPAEPPLSVQDD 1238
Query: 399 RPPLNWPIAGKVEINDLKIRYRPDGPLVLHGITCTFEVGHKIGIVGRTGSGKSTLISALF 458
+ +WP G++ +N++ +RY P P VL G+T TF G K GIVGRTGSGKSTLI ALF
Sbjct: 1239 KLTQDWPSEGEIMLNNVHVRYAPHLPFVLKGLTVTFPGGMKTGIVGRTGSGKSTLIQALF 1298
Query: 459 RLVEPSGGNIIIDGVDISSIGLHDLRSRFGIIPQDPTLFTGTVRYNLDPLSQYSDQEIWE 518
R+++P+ G I++D +DI +IGLHDLRSR IIPQ+PT+F GTVR NLDP+ +Y+D +IWE
Sbjct: 1299 RIIDPTVGQILVDSIDICTIGLHDLRSRLSIIPQEPTMFEGTVRTNLDPIGEYTDSQIWE 1358
Query: 519 VLGKCQLREVVQEKDEGLNSSVVEDGSNWSMGQRQLFCLGRALLRRSRILVLDEATASVD 578
L +CQL + V+ K+ L+S V+E+G NWS+GQRQL CLGR +L+RS+ILVLDEATASVD
Sbjct: 1359 ALDRCQLGDEVRRKELRLDSPVIENGENWSVGQRQLVCLGRVILKRSKILVLDEATASVD 1418
Query: 579 NSTDYILLKTIRTEFADCTVITVAHRIPTVMDCTMVLSINDG 620
+TD ++ KT+R +F+D TVIT+AHRI +V+D MVL +++G
Sbjct: 1419 TATDNLIQKTLRQQFSDATVITIAHRITSVLDSDMVLLLDNG 1460
Score = 62.0 bits (149), Expect = 5e-08
Identities = 64/293 (21%), Positives = 128/293 (42%), Gaps = 18/293 (6%)
Query: 333 CMVILPPGTFTSGFIGMALSYGLALNSFLVNSIQSQCTLANQIISVERLNQYMHIQSEAK 392
CM++ P SG + AL+ L + N + L +S++R+ ++ ++
Sbjct: 552 CMLMGIP--LESGKVLSALATFRVLQEPIYNLPDTISMLIQTKVSLDRIASFLCLEELPT 609
Query: 393 EIVEGNRPPLNWPIAGKVEINDLKIRYRPDGPLVLHGITCTFEVGHKIGIVGRTGSGKST 452
+ V P + +A +V P+ P L + + G +I + G GSGKS+
Sbjct: 610 DAVL-KLPSGSSDVAIEVRNGCFSWDASPEVP-TLKDLNFQAQQGMRIAVCGTVGSGKSS 667
Query: 453 LISALFRLVEPSGGNIIIDGVDISSIGLHDLRSRFGIIPQDPTLFTGTVRYNLDPLSQYS 512
L+S + + G + G + Q + +G ++ N+ Q
Sbjct: 668 LLSCILGEIPKLSGEVKTCGT-------------MAYVSQSAWIQSGKIQDNILFGKQMD 714
Query: 513 DQEIWEVLGKCQLREVVQEKDEGLNSSVVEDGSNWSMGQRQLFCLGRALLRRSRILVLDE 572
+++ VL C L++ ++ G + + E G N S GQ+Q + RAL + + I + D+
Sbjct: 715 NEKYDRVLESCSLKKDLEILPFGDQTVIGERGINLSGGQKQRIQIARALYQDADIYLFDD 774
Query: 573 ATASVDNST-DYILLKTIRTEFADCTVITVAHRIPTVMDCTMVLSINDGNLSR 624
++VD T ++ + + E A TV+ V H+I + ++L + G +++
Sbjct: 775 PFSAVDAHTGSHLFKECLLGELASKTVVYVTHQIEFLPAADLILVMKGGRIAQ 827
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.323 0.138 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,037,978,390
Number of Sequences: 2790947
Number of extensions: 42256962
Number of successful extensions: 180077
Number of sequences better than 10.0: 21476
Number of HSP's better than 10.0 without gapping: 14022
Number of HSP's successfully gapped in prelim test: 7455
Number of HSP's that attempted gapping in prelim test: 142426
Number of HSP's gapped (non-prelim): 27891
length of query: 663
length of database: 848,049,833
effective HSP length: 134
effective length of query: 529
effective length of database: 474,062,935
effective search space: 250779292615
effective search space used: 250779292615
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 78 (34.7 bits)
Medicago: description of AC147714.15


