

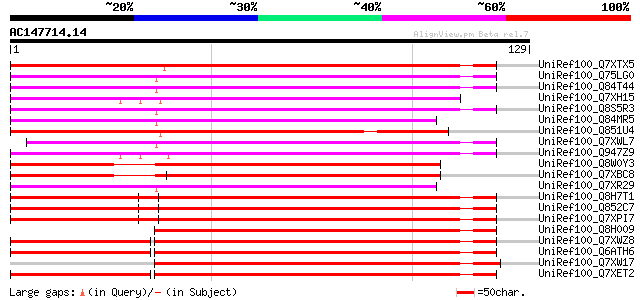
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147714.14 + phase: 0
(129 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q7XTX5 OSJNBa0019K04.23 protein [Oryza sativa] 112 1e-24
UniRef100_Q75LG0 Putative integrase [Oryza sativa] 109 2e-23
UniRef100_Q84T44 Putative copia-type pol polyprotein [Oryza sativa] 108 3e-23
UniRef100_Q7XH15 Putative copia-type pol polyprotein [Oryza sativa] 107 8e-23
UniRef100_Q8S5R3 Putative copia-type pol polyprotein [Oryza sativa] 106 1e-22
UniRef100_Q84MR5 Putative copia-type pol polyprotein [Oryza sativa] 102 1e-21
UniRef100_Q851U4 Putative copia-type retrotransposon protein [Or... 102 3e-21
UniRef100_Q7XWL7 OSJNBb0052B05.12 protein [Oryza sativa] 100 1e-20
UniRef100_Q947Z9 Putative retroelement [Oryza sativa] 99 3e-20
UniRef100_Q8W0Y3 Putative gag protein [Zea mays] 92 3e-18
UniRef100_Q7XBC8 Putative copia-type pol polyprotein [Zea mays] 92 3e-18
UniRef100_Q7XR29 OSJNBa0014F04.18 protein [Oryza sativa] 90 1e-17
UniRef100_Q8H7T1 Putative Zea mays retrotransposon Opie-2 [Oryza... 83 2e-15
UniRef100_Q852C7 Putative gag-pol polyprotein [Oryza sativa] 83 2e-15
UniRef100_Q7XPI7 OSJNBb0004A17.2 protein [Oryza sativa] 83 2e-15
UniRef100_Q8H009 Putative retroelement [Oryza sativa] 82 2e-15
UniRef100_Q7XWZ8 OSJNBb0072N21.1 protein [Oryza sativa] 82 3e-15
UniRef100_Q6ATH6 Putative polyprotein [Oryza sativa] 82 3e-15
UniRef100_Q7XW17 OSJNBb0013O03.1 protein [Oryza sativa] 81 5e-15
UniRef100_Q7XET2 Hypothetical protein [Oryza sativa] 81 5e-15
>UniRef100_Q7XTX5 OSJNBa0019K04.23 protein [Oryza sativa]
Length = 507
Score = 112 bits (281), Expect = 1e-24
Identities = 62/134 (46%), Positives = 84/134 (62%), Gaps = 16/134 (11%)
Query: 1 MKVKSAFLNGAISEEVYVKQPPGFEDLKHPDYVYKDE-------------IEMSMMGELK 47
M VKSAFLNG ISE VYV+QPPGFED K P++VYK EMSMMGEL
Sbjct: 264 MDVKSAFLNGPISELVYVEQPPGFEDPKLPNHVYKLHKALYGLKQAPRAWFEMSMMGELT 323
Query: 48 FFLRIQINQCKDEVYVHQSKYTKDLLMKFKVDDCNEMNTPMHPTCTLSEEDADAKVDQKL 107
FFL +Q+ Q ++ +++ Q+KY KD+L KF ++D + TP+ L +D VDQK+
Sbjct: 324 FFLGLQVKQAQEGIFISQTKYVKDILKKFGMEDAKPIKTPIPTNGHLDLDDNGKSVDQKV 383
Query: 108 FRGIILFDDSILFL 121
+R +I S+L+L
Sbjct: 384 YRSMI---GSLLYL 394
>UniRef100_Q75LG0 Putative integrase [Oryza sativa]
Length = 1507
Score = 109 bits (272), Expect = 2e-23
Identities = 66/170 (38%), Positives = 86/170 (49%), Gaps = 52/170 (30%)
Query: 1 MKVKSAFLNGAISEEVYVKQPPGFEDLKHPDYVYK------------------------- 35
M VKSAFLNG I EEVYVKQPPGFE+ P+YV+K
Sbjct: 1130 MDVKSAFLNGFIQEEVYVKQPPGFENPDFPNYVFKLSKALYGLKQAPRAWYDRLKNFLLA 1189
Query: 36 ------------------------DEIEMSMMGELKFFLRIQINQCKDEVYVHQSKYTKD 71
E EMSMMGEL +FL +QI Q +VHQ+KYTKD
Sbjct: 1190 KGFTMGKVDKTLFVLKHDFAETMRREFEMSMMGELSYFLGLQIKQTPQGTFVHQTKYTKD 1249
Query: 72 LLMKFKVDDCNEMNTPMHPTCTLSEEDADAKVDQKLFRGIILFDDSILFL 121
LL +FK+++C ++TP+ T L ++ VDQK +R +I S+L+L
Sbjct: 1250 LLRRFKMENCKPISTPIDSTAVLDPDEDGEAVDQKEYRSMI---GSLLYL 1296
>UniRef100_Q84T44 Putative copia-type pol polyprotein [Oryza sativa]
Length = 1187
Score = 108 bits (270), Expect = 3e-23
Identities = 63/144 (43%), Positives = 84/144 (57%), Gaps = 26/144 (18%)
Query: 1 MKVKSAFLNGAISEEVYVKQPPGFEDLKHPDYVYK-----------------------DE 37
M VKSAFLNG I+E VYV+QPPGFED +P++VYK
Sbjct: 58 MDVKSAFLNGPINELVYVEQPPGFEDPNYPNHVYKLHKALYGLKQAPRACEEFSRMMTIR 117
Query: 38 IEMSMMGELKFFLRIQINQCKDEVYVHQSKYTKDLLMKFKVDDCNEMNTPMHPTCTLSEE 97
EMSMMGELKFFL +QI Q K+ ++ Q+KY KD+L KF +++ ++TPM L
Sbjct: 118 FEMSMMGELKFFLDLQIKQLKEGTFICQTKYLKDMLKKFGMENAKPIHTPMPSNGHLDLN 177
Query: 98 DADAKVDQKLFRGIILFDDSILFL 121
+ VDQK++R II S+L+L
Sbjct: 178 EQGKDVDQKVYRSII---GSLLYL 198
>UniRef100_Q7XH15 Putative copia-type pol polyprotein [Oryza sativa]
Length = 963
Score = 107 bits (266), Expect = 8e-23
Identities = 64/147 (43%), Positives = 80/147 (53%), Gaps = 35/147 (23%)
Query: 1 MKVKSAFLNGAISEEVYVKQPPGFED---------LKHPD-----YVYKD---------- 36
M VKSAFLNG+I EEVYVKQPPGF LKH D +Y D
Sbjct: 664 MDVKSAFLNGSIQEEVYVKQPPGFTKGKVDKTLFVLKHGDNQLFVQIYVDNIIFGCSTHA 723
Query: 37 -----------EIEMSMMGELKFFLRIQINQCKDEVYVHQSKYTKDLLMKFKVDDCNEMN 85
E EMSMMGEL +FL +QI Q +VHQ+KYTKDLL +FK++ C ++
Sbjct: 724 LVVDFAETMRREFEMSMMGELLYFLGLQIKQTPQGTFVHQTKYTKDLLRRFKMEKCKPIS 783
Query: 86 TPMHPTCTLSEEDADAKVDQKLFRGII 112
TP+ T L ++ VDQK +R +I
Sbjct: 784 TPIGSTAVLDPDEDGEAVDQKEYRSMI 810
>UniRef100_Q8S5R3 Putative copia-type pol polyprotein [Oryza sativa]
Length = 1866
Score = 106 bits (265), Expect = 1e-22
Identities = 62/144 (43%), Positives = 83/144 (57%), Gaps = 26/144 (18%)
Query: 1 MKVKSAFLNGAISEEVYVKQPPGFEDLKHPDYVYK-----------------------DE 37
M VKS FLNG ++E VYV+QPPGFED K+P+ VYK
Sbjct: 1025 MDVKSTFLNGPMNELVYVEQPPGFEDPKYPNRVYKLHKALYGLKQAPRACEEFSRMMTKR 1084
Query: 38 IEMSMMGELKFFLRIQINQCKDEVYVHQSKYTKDLLMKFKVDDCNEMNTPMHPTCTLSEE 97
EMSMMGELKFFL +QI Q K+ ++ Q+KY KD+L KF +++ ++TPM L
Sbjct: 1085 FEMSMMGELKFFLVLQIKQLKEGTFICQTKYLKDMLKKFGMENAKPIHTPMPSNGHLDFN 1144
Query: 98 DADAKVDQKLFRGIILFDDSILFL 121
+ VDQK++R II S+L+L
Sbjct: 1145 EQGKDVDQKVYRSII---GSLLYL 1165
>UniRef100_Q84MR5 Putative copia-type pol polyprotein [Oryza sativa]
Length = 1896
Score = 102 bits (255), Expect = 1e-21
Identities = 57/124 (45%), Positives = 72/124 (57%), Gaps = 18/124 (14%)
Query: 1 MKVKSAFLNGAISEEVYVKQPPGFEDLKHPDYVYK------------------DEIEMSM 42
M VKSAFLNG ISE VYV+QPPGFED K P++VYK EMSM
Sbjct: 1167 MDVKSAFLNGPISELVYVEQPPGFEDPKLPNHVYKLHKALYGLKQAPRACIMTKRFEMSM 1226
Query: 43 MGELKFFLRIQINQCKDEVYVHQSKYTKDLLMKFKVDDCNEMNTPMHPTCTLSEEDADAK 102
MGEL FFL +Q+ Q ++ ++ Q+KY KD++ KF + D + TPM L +D
Sbjct: 1227 MGELTFFLGLQVKQAQEGTFISQTKYVKDIIKKFGMKDVKPIKTPMPTNGHLDLDDNGKC 1286
Query: 103 VDQK 106
VDQK
Sbjct: 1287 VDQK 1290
>UniRef100_Q851U4 Putative copia-type retrotransposon protein [Oryza sativa]
Length = 1183
Score = 102 bits (253), Expect = 3e-21
Identities = 58/116 (50%), Positives = 74/116 (63%), Gaps = 10/116 (8%)
Query: 1 MKVKSAFLNGAISEEVYVKQPPGFEDLKHPDYVYKD-------EIEMSMMGELKFFLRIQ 53
M VKSAFLNG I+E V+V+QPPGFED K+P++VYK E EMSM+GEL FFL +Q
Sbjct: 702 MDVKSAFLNGEIAELVFVEQPPGFEDPKNPNHVYKLSKSPRAWEFEMSMIGELSFFLGLQ 761
Query: 54 INQCKDEVYVHQSKYTKDLLMKFKVDDCNEMNTPMHPTCTLSEEDADAKVDQKLFR 109
I Q KD +V Q+KY KDLL +F ++D + T + T D D +V L R
Sbjct: 762 IKQLKDGTFVSQTKYIKDLLKRFSLEDAKPIKTLL---ATNGHLDLDERVMPPLTR 814
>UniRef100_Q7XWL7 OSJNBb0052B05.12 protein [Oryza sativa]
Length = 1549
Score = 99.8 bits (247), Expect = 1e-20
Identities = 58/140 (41%), Positives = 80/140 (56%), Gaps = 26/140 (18%)
Query: 5 SAFLNGAISEEVYVKQPPGFEDLKHPDYVYK-----------------------DEIEMS 41
S FLNG I+E VYV+Q PGFED K+P++VYK E+S
Sbjct: 1202 STFLNGPINELVYVEQTPGFEDPKYPNHVYKLHKALYGLKQAPRACEEFSRMMTKRFEIS 1261
Query: 42 MMGELKFFLRIQINQCKDEVYVHQSKYTKDLLMKFKVDDCNEMNTPMHPTCTLSEEDADA 101
MMGELKFFL +QI Q K+ ++ Q+KY KD+L KF +++ ++TPM L +
Sbjct: 1262 MMGELKFFLGLQIKQLKEGTFICQTKYLKDMLKKFGMENAKPIHTPMPSNGHLDLNEQGK 1321
Query: 102 KVDQKLFRGIILFDDSILFL 121
VDQK++R II S+L+L
Sbjct: 1322 GVDQKVYRSII---GSLLYL 1338
>UniRef100_Q947Z9 Putative retroelement [Oryza sativa]
Length = 1660
Score = 98.6 bits (244), Expect = 3e-20
Identities = 63/156 (40%), Positives = 82/156 (52%), Gaps = 38/156 (24%)
Query: 1 MKVKSAFLNGAISEEVYVKQPPGFED---------LKHPD-----YVYKDEI-------- 38
M VKS FLNG I EEVYVKQPPGF LKH D +Y D+I
Sbjct: 1229 MDVKSDFLNGFIQEEVYVKQPPGFTMGKVDKTLFVLKHGDNQLFVQIYVDDIIFGCSTHA 1288
Query: 39 -------------EMSMMGELKFFLRIQINQCKDEVYVHQSKYTKDLLMKFKVDDCNEMN 85
EM MM EL + L +QI Q +VHQ+KYTKDLL +FK+++C ++
Sbjct: 1289 LVVDFTETMRREFEMGMMCELSYILGLQIKQTSQGTFVHQTKYTKDLLRRFKMENCKPIS 1348
Query: 86 TPMHPTCTLSEEDADAKVDQKLFRGIILFDDSILFL 121
TP+ T L ++ VDQK +R +I S+L+L
Sbjct: 1349 TPIGSTAVLDPDEDGEAVDQKEYRSMI---GSLLYL 1381
>UniRef100_Q8W0Y3 Putative gag protein [Zea mays]
Length = 1016
Score = 92.0 bits (227), Expect = 3e-18
Identities = 49/107 (45%), Positives = 66/107 (60%), Gaps = 10/107 (9%)
Query: 1 MKVKSAFLNGAISEEVYVKQPPGFEDLKHPDYVYKDEIEMSMMGELKFFLRIQINQCKDE 60
M VKSAFLNG I EVYV+QP GFED + EMSMMG+LK+FL QINQ ++
Sbjct: 825 MDVKSAFLNGPIKGEVYVEQPSGFED----------KFEMSMMGDLKYFLGFQINQLQEG 874
Query: 61 VYVHQSKYTKDLLMKFKVDDCNEMNTPMHPTCTLSEEDADAKVDQKL 107
+++ Q+KY +D+L KF + + + TPM L + VDQK+
Sbjct: 875 IFICQTKYNRDILKKFGMKNVKPIKTPMGTNGHLDLDTRGKSVDQKM 921
Score = 35.8 bits (81), Expect = 0.22
Identities = 16/53 (30%), Positives = 30/53 (56%)
Query: 55 NQCKDEVYVHQSKYTKDLLMKFKVDDCNEMNTPMHPTCTLSEEDADAKVDQKL 107
+Q ++ +++ Q+KY +D+L KF + + + TPM L + VDQK+
Sbjct: 563 SQVEEGIFICQTKYNRDILKKFGMKNVKPIKTPMGTNGHLDLDTRGKSVDQKV 615
>UniRef100_Q7XBC8 Putative copia-type pol polyprotein [Zea mays]
Length = 469
Score = 92.0 bits (227), Expect = 3e-18
Identities = 49/107 (45%), Positives = 66/107 (60%), Gaps = 10/107 (9%)
Query: 1 MKVKSAFLNGAISEEVYVKQPPGFEDLKHPDYVYKDEIEMSMMGELKFFLRIQINQCKDE 60
M VKSAFLNG I EVYV+QP GFED + EMSMMG+LK+FL QINQ ++
Sbjct: 278 MDVKSAFLNGPIKGEVYVEQPSGFED----------KFEMSMMGDLKYFLGFQINQLQEG 327
Query: 61 VYVHQSKYTKDLLMKFKVDDCNEMNTPMHPTCTLSEEDADAKVDQKL 107
+++ Q+KY +D+L KF + + + TPM L + VDQK+
Sbjct: 328 IFICQTKYNRDILKKFGMKNVKPIKTPMGTNGHLDLDTRGKSVDQKM 374
Score = 59.3 bits (142), Expect = 2e-08
Identities = 28/68 (41%), Positives = 43/68 (63%)
Query: 40 MSMMGELKFFLRIQINQCKDEVYVHQSKYTKDLLMKFKVDDCNEMNTPMHPTCTLSEEDA 99
MSMMG+LK+FL QINQ ++ +++ Q+KY +D+L KF + + + TPM L +
Sbjct: 1 MSMMGDLKYFLGFQINQLQEGIFICQTKYNRDILKKFGMKNVKPIKTPMGTNGHLDLDTR 60
Query: 100 DAKVDQKL 107
VDQK+
Sbjct: 61 GKSVDQKV 68
>UniRef100_Q7XR29 OSJNBa0014F04.18 protein [Oryza sativa]
Length = 958
Score = 90.1 bits (222), Expect = 1e-17
Identities = 53/129 (41%), Positives = 69/129 (53%), Gaps = 23/129 (17%)
Query: 1 MKVKSAFLNGAISEEVYVKQPPGFEDLKHPDYVYK-----------------------DE 37
M VKSAFLN I E VY++QP G ED K+P++VYK
Sbjct: 336 MDVKSAFLNEPIHELVYMEQPLGLEDPKYPNHVYKLHKALYGLKQTPRACEEFSRMMTKR 395
Query: 38 IEMSMMGELKFFLRIQINQCKDEVYVHQSKYTKDLLMKFKVDDCNEMNTPMHPTCTLSEE 97
EMSMMGELK FL +QI Q K+ ++ Q+KY KD+L KF +++ + TPM L
Sbjct: 396 FEMSMMGELKLFLGLQIKQLKEGTFICQTKYLKDMLKKFGMENAKPIQTPMPSNGHLDLN 455
Query: 98 DADAKVDQK 106
+ VDQK
Sbjct: 456 EQGKDVDQK 464
>UniRef100_Q8H7T1 Putative Zea mays retrotransposon Opie-2 [Oryza sativa]
Length = 2145
Score = 82.8 bits (203), Expect = 2e-15
Identities = 42/89 (47%), Positives = 60/89 (67%), Gaps = 3/89 (3%)
Query: 33 VYKDEIEMSMMGELKFFLRIQINQCKDEVYVHQSKYTKDLLMKFKVDDCNEMNTPMHPTC 92
V E EMSMMGEL FFL +QI Q K+ ++VHQ+KY+K+LL KF + DC + TPM T
Sbjct: 1499 VMSREFEMSMMGELTFFLGLQIKQTKEGIFVHQTKYSKELLKKFDMADCKPIATPMATTS 1558
Query: 93 TLSEEDADAKVDQKLFRGIILFDDSILFL 121
+L ++ +VDQ+ +R +I S+L+L
Sbjct: 1559 SLGPDEDGEEVDQREYRSMI---GSLLYL 1584
Score = 57.8 bits (138), Expect = 5e-08
Identities = 27/37 (72%), Positives = 31/37 (82%)
Query: 1 MKVKSAFLNGAISEEVYVKQPPGFEDLKHPDYVYKDE 37
M VKSAFLNG I EEVYVKQPPGFE+ K P++V+K E
Sbjct: 1393 MDVKSAFLNGVIEEEVYVKQPPGFENPKFPNHVFKLE 1429
>UniRef100_Q852C7 Putative gag-pol polyprotein [Oryza sativa]
Length = 1969
Score = 82.8 bits (203), Expect = 2e-15
Identities = 42/89 (47%), Positives = 60/89 (67%), Gaps = 3/89 (3%)
Query: 33 VYKDEIEMSMMGELKFFLRIQINQCKDEVYVHQSKYTKDLLMKFKVDDCNEMNTPMHPTC 92
V E EMSMMGEL FFL +QI Q K+ ++VHQ+KY+K+LL KF + DC + TPM T
Sbjct: 1499 VMSREFEMSMMGELTFFLGLQIKQTKEGIFVHQTKYSKELLKKFDMADCKPIATPMATTS 1558
Query: 93 TLSEEDADAKVDQKLFRGIILFDDSILFL 121
+L ++ +VDQ+ +R +I S+L+L
Sbjct: 1559 SLGPDEDGEEVDQREYRSMI---GSLLYL 1584
Score = 57.8 bits (138), Expect = 5e-08
Identities = 27/37 (72%), Positives = 31/37 (82%)
Query: 1 MKVKSAFLNGAISEEVYVKQPPGFEDLKHPDYVYKDE 37
M VKSAFLNG I EEVYVKQPPGFE+ K P++V+K E
Sbjct: 1393 MDVKSAFLNGVIEEEVYVKQPPGFENPKFPNHVFKLE 1429
>UniRef100_Q7XPI7 OSJNBb0004A17.2 protein [Oryza sativa]
Length = 1877
Score = 82.8 bits (203), Expect = 2e-15
Identities = 42/89 (47%), Positives = 60/89 (67%), Gaps = 3/89 (3%)
Query: 33 VYKDEIEMSMMGELKFFLRIQINQCKDEVYVHQSKYTKDLLMKFKVDDCNEMNTPMHPTC 92
V E EMSMMGEL FFL +QI Q K+ ++VHQ+KY+K+LL KF + DC + TPM T
Sbjct: 1581 VMSREFEMSMMGELTFFLGLQIKQTKEGIFVHQTKYSKELLKKFDMADCKPIATPMATTS 1640
Query: 93 TLSEEDADAKVDQKLFRGIILFDDSILFL 121
+L ++ +VDQ+ +R +I S+L+L
Sbjct: 1641 SLGPDEDGEEVDQREYRSMI---GSLLYL 1666
Score = 57.8 bits (138), Expect = 5e-08
Identities = 27/37 (72%), Positives = 31/37 (82%)
Query: 1 MKVKSAFLNGAISEEVYVKQPPGFEDLKHPDYVYKDE 37
M VKSAFLNG I EEVYVKQPPGFE+ K P++V+K E
Sbjct: 1475 MDVKSAFLNGVIEEEVYVKQPPGFENPKFPNHVFKLE 1511
>UniRef100_Q8H009 Putative retroelement [Oryza sativa]
Length = 689
Score = 82.4 bits (202), Expect = 2e-15
Identities = 41/85 (48%), Positives = 57/85 (66%), Gaps = 3/85 (3%)
Query: 37 EIEMSMMGELKFFLRIQINQCKDEVYVHQSKYTKDLLMKFKVDDCNEMNTPMHPTCTLSE 96
E EMSMMGEL +FL +QI Q +VHQ+KYTKDLL +FKV++C ++TP+ T L
Sbjct: 471 EFEMSMMGELSYFLGLQIKQTPQGAFVHQTKYTKDLLRRFKVENCKPISTPIGSTAVLDP 530
Query: 97 EDADAKVDQKLFRGIILFDDSILFL 121
++ VDQK +R +I S+L+L
Sbjct: 531 DEDGEAVDQKEYRSMI---GSLLYL 552
>UniRef100_Q7XWZ8 OSJNBb0072N21.1 protein [Oryza sativa]
Length = 420
Score = 82.0 bits (201), Expect = 3e-15
Identities = 40/85 (47%), Positives = 57/85 (67%), Gaps = 3/85 (3%)
Query: 37 EIEMSMMGELKFFLRIQINQCKDEVYVHQSKYTKDLLMKFKVDDCNEMNTPMHPTCTLSE 96
E EMSMMGEL +FL +QI Q +VHQ+KYTKDLL +FK+++C ++TP+ T L
Sbjct: 202 EFEMSMMGELSYFLGLQIKQTPQRTFVHQTKYTKDLLRRFKMENCKPISTPIGSTAVLDP 261
Query: 97 EDADAKVDQKLFRGIILFDDSILFL 121
++ VDQK +R +I S+L+L
Sbjct: 262 DEDGEAVDQKEYRSMI---GSLLYL 283
Score = 50.8 bits (120), Expect = 7e-06
Identities = 24/35 (68%), Positives = 28/35 (79%)
Query: 1 MKVKSAFLNGAISEEVYVKQPPGFEDLKHPDYVYK 35
M VKSAFLNG I EEVYVKQ PGFE+ P++V+K
Sbjct: 92 MDVKSAFLNGFIQEEVYVKQLPGFENSDFPNHVFK 126
>UniRef100_Q6ATH6 Putative polyprotein [Oryza sativa]
Length = 1326
Score = 82.0 bits (201), Expect = 3e-15
Identities = 41/85 (48%), Positives = 56/85 (65%), Gaps = 3/85 (3%)
Query: 37 EIEMSMMGELKFFLRIQINQCKDEVYVHQSKYTKDLLMKFKVDDCNEMNTPMHPTCTLSE 96
E EMSMMGEL +FL +QI Q +VHQ+KYT DLL +FKV +C ++TP+ T L
Sbjct: 1126 EFEMSMMGELSYFLGLQIKQTPQGTFVHQTKYTNDLLRRFKVKNCKPISTPIGSTAVLDP 1185
Query: 97 EDADAKVDQKLFRGIILFDDSILFL 121
++ D VDQK +R +I S+L+L
Sbjct: 1186 DEDDEAVDQKEYRSMI---GSLLYL 1207
Score = 53.1 bits (126), Expect = 1e-06
Identities = 25/35 (71%), Positives = 28/35 (79%)
Query: 1 MKVKSAFLNGAISEEVYVKQPPGFEDLKHPDYVYK 35
M VKSAFLNG I EEVYVKQPPGFE+ P+ V+K
Sbjct: 1016 MDVKSAFLNGFIQEEVYVKQPPGFENPDFPNLVFK 1050
>UniRef100_Q7XW17 OSJNBb0013O03.1 protein [Oryza sativa]
Length = 832
Score = 81.3 bits (199), Expect = 5e-15
Identities = 41/86 (47%), Positives = 57/86 (65%), Gaps = 3/86 (3%)
Query: 37 EIEMSMMGELKFFLRIQINQCKDEVYVHQSKYTKDLLMKFKVDDCNEMNTPMHPTCTLSE 96
E EMSMMGEL +FL +QI Q +VHQ+KYTKDLL FK+++C ++TP+ T L
Sbjct: 612 EFEMSMMGELSYFLGLQIKQTPQGTFVHQTKYTKDLLRWFKMENCKPISTPIGSTAVLDP 671
Query: 97 EDADAKVDQKLFRGIILFDDSILFLI 122
++ VDQK +R +I S+L+LI
Sbjct: 672 DEDSEAVDQKEYRSMI---GSLLYLI 694
>UniRef100_Q7XET2 Hypothetical protein [Oryza sativa]
Length = 1419
Score = 81.3 bits (199), Expect = 5e-15
Identities = 40/85 (47%), Positives = 57/85 (67%), Gaps = 3/85 (3%)
Query: 37 EIEMSMMGELKFFLRIQINQCKDEVYVHQSKYTKDLLMKFKVDDCNEMNTPMHPTCTLSE 96
E EMSMMGEL +FL +QI Q +VHQ+KYTKDLL +FK+++C ++TP+ T L
Sbjct: 1127 EFEMSMMGELSYFLGLQIKQTPQGTFVHQTKYTKDLLRRFKMENCKPISTPIGSTAVLDP 1186
Query: 97 EDADAKVDQKLFRGIILFDDSILFL 121
++ VDQK +R +I S+L+L
Sbjct: 1187 DEDGEAVDQKEYRSMI---GSLLYL 1208
Score = 54.3 bits (129), Expect = 6e-07
Identities = 25/35 (71%), Positives = 29/35 (82%)
Query: 1 MKVKSAFLNGAISEEVYVKQPPGFEDLKHPDYVYK 35
M VKSAFLNG I EEVYVKQPPGFE+ P++V+K
Sbjct: 1017 MDVKSAFLNGFIQEEVYVKQPPGFENPDFPNHVFK 1051
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.323 0.141 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 215,862,238
Number of Sequences: 2790947
Number of extensions: 8220583
Number of successful extensions: 22767
Number of sequences better than 10.0: 997
Number of HSP's better than 10.0 without gapping: 973
Number of HSP's successfully gapped in prelim test: 24
Number of HSP's that attempted gapping in prelim test: 21397
Number of HSP's gapped (non-prelim): 1365
length of query: 129
length of database: 848,049,833
effective HSP length: 105
effective length of query: 24
effective length of database: 555,000,398
effective search space: 13320009552
effective search space used: 13320009552
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 67 (30.4 bits)
Medicago: description of AC147714.14


