

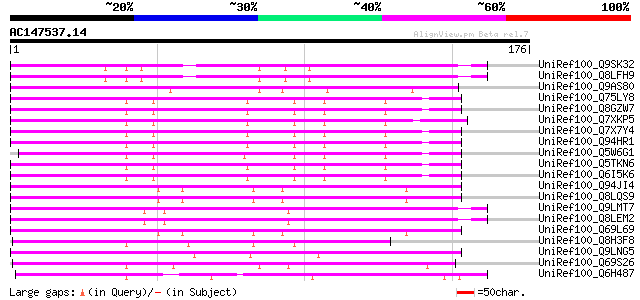
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147537.14 - phase: 0
(176 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9SK32 Expressed protein [Arabidopsis thaliana] 72 6e-12
UniRef100_Q8LFH9 Hypothetical protein [Arabidopsis thaliana] 72 6e-12
UniRef100_Q9AS80 P0028E10.12 protein [Oryza sativa] 66 4e-10
UniRef100_Q75LY8 Putative MuDR family transposase [Oryza sativa] 64 1e-09
UniRef100_Q8GZW7 Putative transposable element [Oryza sativa] 64 2e-09
UniRef100_Q7XKP5 OSJNBb0013O03.11 protein [Oryza sativa] 64 2e-09
UniRef100_Q7X7Y4 OSJNBa0014F04.28 protein [Oryza sativa] 64 2e-09
UniRef100_Q94HR1 Hypothetical protein OSJNBa0065C16.8 [Oryza sat... 63 3e-09
UniRef100_Q5W6G1 Putative polyprotein [Oryza sativa] 63 4e-09
UniRef100_Q5TKN6 Hypothetical protein OJ1058_F05.1 [Oryza sativa] 62 5e-09
UniRef100_Q6I5K6 Hypothetical protein OSJNBb0088F07.13 [Oryza sa... 62 5e-09
UniRef100_Q94JI4 P0638D12.5 protein [Oryza sativa] 60 3e-08
UniRef100_Q8LQS9 P0702H08.15 protein [Oryza sativa] 60 3e-08
UniRef100_Q9LMT7 F2H15.15 protein [Arabidopsis thaliana] 59 4e-08
UniRef100_Q8LEM2 Hypothetical protein [Arabidopsis thaliana] 59 4e-08
UniRef100_Q69L69 Calcineurin-like phosphoesterase-like protein [... 57 2e-07
UniRef100_Q8H3F8 Serine/threonine phosphatase PP7-related-like p... 55 6e-07
UniRef100_Q9LNG5 F21D18.16 [Arabidopsis thaliana] 54 2e-06
UniRef100_Q69S26 Serine/threonine phosphatase PP7-related-like [... 54 2e-06
UniRef100_Q6H487 Hypothetical protein B1040D06.35 [Oryza sativa] 54 2e-06
>UniRef100_Q9SK32 Expressed protein [Arabidopsis thaliana]
Length = 509
Score = 72.0 bits (175), Expect = 6e-12
Identities = 59/182 (32%), Positives = 88/182 (47%), Gaps = 28/182 (15%)
Query: 1 MEAWNSERHAFEIGSGEIGFSLLDVALILGI-----PVVGHRV--ELAEE-------QLF 46
+E W E + F + GE+ +L +VAL+LG+ P+VG +V E+A + +L
Sbjct: 80 VERWRRETNTFHLPLGEMTITLDEVALVLGLEIDGDPIVGSKVGDEVAMDMCGRLLGKLP 139
Query: 47 SELEEEYGASRAKRKVAMTSLEARLDSIGEDVSDDFV----RSFLLFTIG-TFLSSIDG- 100
S +E SR K + L+ ED S D V R++LL+ IG T ++ DG
Sbjct: 140 SAANKEVNCSRVK----LNWLKRTFSECPEDASFDVVKCHTRAYLLYLIGSTIFATTDGD 195
Query: 101 KVDSRYLSFLGNLDDVSGFAWGAAVIEDLCQWLDKRKDNNVQYVGGCLIFLQVSLICLSY 160
KV +YL + D +AWGAA + L + L + + GCL LQ C SY
Sbjct: 196 KVSVKYLPLFEDFDQAGRYAWGAAALACLYRALGNASLKSQSNICGCLTLLQ----CWSY 251
Query: 161 YH 162
+H
Sbjct: 252 FH 253
>UniRef100_Q8LFH9 Hypothetical protein [Arabidopsis thaliana]
Length = 509
Score = 72.0 bits (175), Expect = 6e-12
Identities = 59/182 (32%), Positives = 88/182 (47%), Gaps = 28/182 (15%)
Query: 1 MEAWNSERHAFEIGSGEIGFSLLDVALILGI-----PVVGHRV--ELAEE-------QLF 46
+E W E + F + GE+ +L +VAL+LG+ P+VG +V E+A + +L
Sbjct: 80 VERWRRETNTFHLPLGEMTITLDEVALVLGLEIDGDPIVGSKVDDEVAMDMCGRLLGKLP 139
Query: 47 SELEEEYGASRAKRKVAMTSLEARLDSIGEDVSDDFV----RSFLLFTIG-TFLSSIDG- 100
S +E SR K + L+ ED S D V R++LL+ IG T ++ DG
Sbjct: 140 SAANKEVNCSRVK----LNWLKRTFSECPEDASFDVVKCHTRAYLLYLIGSTIFATTDGD 195
Query: 101 KVDSRYLSFLGNLDDVSGFAWGAAVIEDLCQWLDKRKDNNVQYVGGCLIFLQVSLICLSY 160
KV +YL + D +AWGAA + L + L + + GCL LQ C SY
Sbjct: 196 KVSVKYLPLFEDFDQAGRYAWGAAALACLYRALGNASLKSQSNICGCLTLLQ----CWSY 251
Query: 161 YH 162
+H
Sbjct: 252 FH 253
>UniRef100_Q9AS80 P0028E10.12 protein [Oryza sativa]
Length = 792
Score = 65.9 bits (159), Expect = 4e-10
Identities = 43/165 (26%), Positives = 84/165 (50%), Gaps = 13/165 (7%)
Query: 1 MEAWNSERHAFEIGSGEIGFSLLDVALILGIPVVGHRVELAEEQLFSELEEEY-----GA 55
++ W E H F + GEI +L DVA+ILG+P+ GH V + + +EL E Y
Sbjct: 266 VDRWRPETHTFHMPCGEITITLQDVAMILGLPIAGHAVTVNPTEPQNELVERYLGKAPPP 325
Query: 56 SRAKRKVAMTSLEARLDSIGEDVSDDFV----RSFLLFTI-GTFLSSIDGKVDSRY-LSF 109
R + + ++ + A ++ ED ++ + R+++L I G G + + Y
Sbjct: 326 DRPRPGLRVSWVRAEFNNCPEDADEETIKQHARAYILSLISGLLFPDASGDLYTFYPFPL 385
Query: 110 LGNLDDVSGFAWGAAVIEDLCQWLDK--RKDNNVQYVGGCLIFLQ 152
+ +L+++ ++WG+A + L + + R+ ++ + GCL+ LQ
Sbjct: 386 IADLENIGSYSWGSATLAYLYRAMCDACRRQSDGSNLTGCLLLLQ 430
>UniRef100_Q75LY8 Putative MuDR family transposase [Oryza sativa]
Length = 1403
Score = 64.3 bits (155), Expect = 1e-09
Identities = 44/175 (25%), Positives = 87/175 (49%), Gaps = 24/175 (13%)
Query: 1 MEAWNSERHAFEIGSGEIGFSLLDVALILGIPVVGHRV---------ELAEEQLFS---E 48
++ W E H+F + SGE+ +L DVA+IL +P+ GH V EQLF
Sbjct: 876 VDRWRPETHSFHLPSGEMTITLQDVAMILALPLRGHAVTGRTETPGWRAQVEQLFGITLN 935
Query: 49 LEEEYGASRAKRKVAMTSLEARLDSIGEDVS----DDFVRSFLLFTIGTFL-SSIDGKVD 103
+E+ G + + + ++ L + +D + + R+++L +G L + G +
Sbjct: 936 IEQGQGGKKKQNGIPLSWLSQNFSHLHDDAEPWRVECYARAYILHLLGGVLFPNAGGDIA 995
Query: 104 SR-YLSFLGNLDDVSGFAWGAAVI----EDLCQWLDKRKDNNVQYVGGCLIFLQV 153
S ++ + NL D+ F+WG+AV+ LC+ ++ ++ + GC++ +Q+
Sbjct: 996 SAIWIPLVANLGDLGRFSWGSAVLAWTYRQLCEACCRQAPSS--NMSGCVLLIQL 1048
>UniRef100_Q8GZW7 Putative transposable element [Oryza sativa]
Length = 663
Score = 63.9 bits (154), Expect = 2e-09
Identities = 43/175 (24%), Positives = 86/175 (48%), Gaps = 24/175 (13%)
Query: 1 MEAWNSERHAFEIGSGEIGFSLLDVALILGIPVVGHRV---------ELAEEQLFS---E 48
++ W E H+F + SGE+ +L DVA+IL +P+ GH V +QLF
Sbjct: 80 VDRWRPETHSFHLPSGEMTITLQDVAMILALPLRGHAVTGRTETPGWHAQVQQLFGIPLN 139
Query: 49 LEEEYGASRAKRKVAMTSLEARLDSIGEDVS----DDFVRSFLLFTIGTFL-SSIDGKVD 103
+E+ G + + + ++ L + +D + + R+++L +G L G +
Sbjct: 140 IEQGQGGKKKQNGIPLSWLSQNFSHLDDDAEPWRVECYARAYILHLLGGVLFPDAGGDIA 199
Query: 104 SR-YLSFLGNLDDVSGFAWGAAVI----EDLCQWLDKRKDNNVQYVGGCLIFLQV 153
S ++ + NL D+ F+WG+AV+ LC+ ++ ++ + GC++ +Q+
Sbjct: 200 SAIWIPLVANLGDLGRFSWGSAVLAWTYRQLCEACCRQAPSS--NMSGCVLLIQI 252
>UniRef100_Q7XKP5 OSJNBb0013O03.11 protein [Oryza sativa]
Length = 1495
Score = 63.5 bits (153), Expect = 2e-09
Identities = 44/177 (24%), Positives = 86/177 (47%), Gaps = 24/177 (13%)
Query: 1 MEAWNSERHAFEIGSGEIGFSLLDVALILGIPVVGHRV---------ELAEEQLFS---E 48
++ W E H+F + SGE+ +L DVA+IL +P+ GH V EQLF
Sbjct: 940 VDRWRPETHSFHLPSGEMTITLQDVAMILALPLQGHAVTGRTETPGWRAQVEQLFGIQLN 999
Query: 49 LEEEYGASRAKRKVAMTSLEARLDSIGEDVS----DDFVRSFLLFTIGTFL-SSIDGKVD 103
+E+ G + + + ++ L + +D + + R+++L +G L G +
Sbjct: 1000 IEQGQGGKKMQNGIPLSWLSQNFSHLDDDAEPWRVECYARAYILHLLGGVLFPDAGGDIA 1059
Query: 104 SR-YLSFLGNLDDVSGFAWGAAVI----EDLCQWLDKRKDNNVQYVGGCLIFLQVSL 155
S ++ + NL D+ F+WG+AV+ LC+ + ++ + GC++ +Q+ +
Sbjct: 1060 SAIWIPLVANLGDLGRFSWGSAVLAWTYRQLCEACCCQA--SLSNMSGCVLLIQLCM 1114
>UniRef100_Q7X7Y4 OSJNBa0014F04.28 protein [Oryza sativa]
Length = 1575
Score = 63.5 bits (153), Expect = 2e-09
Identities = 43/175 (24%), Positives = 86/175 (48%), Gaps = 24/175 (13%)
Query: 1 MEAWNSERHAFEIGSGEIGFSLLDVALILGIPVVGHRV---------ELAEEQLFS---E 48
++ W E H+F + SGE+ +L DVA+IL +P+ GH V +QLF
Sbjct: 972 VDRWRPETHSFHLPSGEMTITLQDVAMILALPLRGHAVTGRTDTPGWHAQVQQLFGIPLN 1031
Query: 49 LEEEYGASRAKRKVAMTSLEARLDSIGEDVS----DDFVRSFLLFTIGTFL-SSIDGKVD 103
+E+ G + + + ++ L + +D + + R+++L +G L G +
Sbjct: 1032 IEQGQGGKKKQNGIPLSWLSQNFSHLDDDAEPWRVECYARAYILHLLGEVLFPDAGGDIA 1091
Query: 104 SR-YLSFLGNLDDVSGFAWGAAVI----EDLCQWLDKRKDNNVQYVGGCLIFLQV 153
S ++ + NL D+ F+WG+AV+ LC+ ++ ++ + GC++ +Q+
Sbjct: 1092 SAIWIPLVANLGDLGRFSWGSAVLAWTYRQLCEACCRQAPSS--NMSGCVLLIQM 1144
>UniRef100_Q94HR1 Hypothetical protein OSJNBa0065C16.8 [Oryza sativa]
Length = 681
Score = 63.2 bits (152), Expect = 3e-09
Identities = 44/175 (25%), Positives = 87/175 (49%), Gaps = 24/175 (13%)
Query: 1 MEAWNSERHAFEIGSGEIGFSLLDVALILGIPVVGHRV---------ELAEEQLFS---E 48
++ W E H+F + SGE+ +L DVA+IL +P+ GH V EQLF
Sbjct: 176 VDRWRPETHSFHLPSGEMTITLQDVAMILALPLQGHVVTGRTETPGWRAQVEQLFGIPLN 235
Query: 49 LEEEYGASRAKRKVAMTSLEARLDSIGEDVS----DDFVRSFLLFTIGTFL-SSIDGKVD 103
+E+ G + + + ++ L + +D + + R+++L +G L + G +
Sbjct: 236 IEQGQGGKKKQNGIPLSWLSQNFSHLDDDAEPWRVECYARAYILHLLGGVLFPNAGGDIA 295
Query: 104 SR-YLSFLGNLDDVSGFAWGAAVI----EDLCQWLDKRKDNNVQYVGGCLIFLQV 153
S ++ + NL D+ F+WG+AV+ LC+ ++ ++ + GC++ +Q+
Sbjct: 296 SAIWIPLVANLGDLGRFSWGSAVLAWTYRQLCEACCRQALSS--NMSGCVLLIQL 348
>UniRef100_Q5W6G1 Putative polyprotein [Oryza sativa]
Length = 1567
Score = 62.8 bits (151), Expect = 4e-09
Identities = 43/172 (25%), Positives = 85/172 (49%), Gaps = 24/172 (13%)
Query: 4 WNSERHAFEIGSGEIGFSLLDVALILGIPVVGHRV---------ELAEEQLFS---ELEE 51
W E H+F + SGE+ +L DVA+IL +P+ GH V +QLF +E+
Sbjct: 1033 WRPETHSFHLPSGEMTITLQDVAMILALPLRGHAVTGRTETPGWHAQVQQLFGIPLNIEQ 1092
Query: 52 EYGASRAKRKVAMTSLEARLDSIGEDV----SDDFVRSFLLFTIGTFL-SSIDGKVDSR- 105
G + + + ++ L + +D ++ + R+++L +G L G + S
Sbjct: 1093 GQGGKKKQNGIPLSWLSQNFSHLDDDAEPWRAECYARAYILHLLGGVLFPDAGGDIASAI 1152
Query: 106 YLSFLGNLDDVSGFAWGAAVI----EDLCQWLDKRKDNNVQYVGGCLIFLQV 153
++ + NL D+ F+WG+AV+ LC+ ++ ++ + GC++ +Q+
Sbjct: 1153 WIPLVANLGDLGRFSWGSAVLAWTYRQLCEACCRQAPSS--NMSGCVLLIQM 1202
>UniRef100_Q5TKN6 Hypothetical protein OJ1058_F05.1 [Oryza sativa]
Length = 725
Score = 62.4 bits (150), Expect = 5e-09
Identities = 43/175 (24%), Positives = 86/175 (48%), Gaps = 24/175 (13%)
Query: 1 MEAWNSERHAFEIGSGEIGFSLLDVALILGIPVVGHRV---------ELAEEQLFS---E 48
++ W E H+F + SGE+ +L DVA+IL +P+ GH V +QLF
Sbjct: 128 VDRWRPETHSFHLPSGEMTITLQDVAMILALPLRGHAVTGRTETPGWHAQVQQLFGIPLN 187
Query: 49 LEEEYGASRAKRKVAMTSLEARLDSIGEDVS----DDFVRSFLLFTIGTFL-SSIDGKVD 103
+E+ G + + + ++ L + +D + + R+++L +G L G +
Sbjct: 188 IEQGQGGKKKQNGIPLSWLSQNFSHLDDDAEPWRVECYARAYILHLLGGVLFPDAGGDIA 247
Query: 104 SR-YLSFLGNLDDVSGFAWGAAVI----EDLCQWLDKRKDNNVQYVGGCLIFLQV 153
S ++ + NL D+ F+WG+AV+ LC+ ++ ++ + GC++ +Q+
Sbjct: 248 SAIWIPLVTNLGDLGRFSWGSAVLAWTYRQLCEACCRQAPSS--NMSGCVLLIQM 300
>UniRef100_Q6I5K6 Hypothetical protein OSJNBb0088F07.13 [Oryza sativa]
Length = 1564
Score = 62.4 bits (150), Expect = 5e-09
Identities = 43/175 (24%), Positives = 86/175 (48%), Gaps = 24/175 (13%)
Query: 1 MEAWNSERHAFEIGSGEIGFSLLDVALILGIPVVGHRV---------ELAEEQLFS---E 48
++ W E H+F + SGE+ +L DVA+IL +P+ GH V +QLF
Sbjct: 967 VDRWRPETHSFHLPSGEMTITLQDVAMILALPLRGHAVTGRTETPGWHAQVQQLFGIPLN 1026
Query: 49 LEEEYGASRAKRKVAMTSLEARLDSIGEDVS----DDFVRSFLLFTIGTFL-SSIDGKVD 103
+E+ G + + + ++ L + +D + + R+++L +G L G +
Sbjct: 1027 IEQGQGGKKKQNGIPLSWLSQNFSHLDDDAEPWRVECYARAYILHLLGGVLFPDAGGDIA 1086
Query: 104 SR-YLSFLGNLDDVSGFAWGAAVI----EDLCQWLDKRKDNNVQYVGGCLIFLQV 153
S ++ + NL D+ F+WG+AV+ LC+ ++ ++ + GC++ +Q+
Sbjct: 1087 SAIWIPLVTNLGDLGRFSWGSAVLAWTYRQLCEACCRQAPSS--NMSGCVLLIQM 1139
>UniRef100_Q94JI4 P0638D12.5 protein [Oryza sativa]
Length = 761
Score = 59.7 bits (143), Expect = 3e-08
Identities = 43/166 (25%), Positives = 80/166 (47%), Gaps = 13/166 (7%)
Query: 1 MEAWNSERHAFEIGSGEIGFSLLDVALILGIPVVGHRVELAEEQLFSEL------EEEYG 54
+E W E H+F + SGE+ +L DVA++L +P+ G V + ++++ + G
Sbjct: 155 VERWRPETHSFHLASGEMAVTLQDVAMLLALPIDGRPVCSTTDHDYAQMVIDCLGHDPRG 214
Query: 55 ASR-AKRKVAMTSLEARLDSIGEDVSDD----FVRSFLLFTI-GTFLSSIDGKVDSRYLS 108
S K + L+ + E D VR+++L + G G++ YL
Sbjct: 215 PSMPGKSFLHYKWLKKHFYELPEGADDQTVERHVRAYILSLLCGVLFPDGTGRMSLIYLP 274
Query: 109 FLGNLDDVSGFAWGAAVIEDLCQWL-DKRKDNNVQYVGGCLIFLQV 153
+ +L V ++WG+AV+ L + L +N++ +GG L+ LQ+
Sbjct: 275 LIADLSRVGTYSWGSAVLAFLYRSLCSVASSHNIKNIGGSLLLLQL 320
>UniRef100_Q8LQS9 P0702H08.15 protein [Oryza sativa]
Length = 718
Score = 59.7 bits (143), Expect = 3e-08
Identities = 43/166 (25%), Positives = 80/166 (47%), Gaps = 13/166 (7%)
Query: 1 MEAWNSERHAFEIGSGEIGFSLLDVALILGIPVVGHRVELAEEQLFSEL------EEEYG 54
+E W E H+F + SGE+ +L DVA++L +P+ G V + ++++ + G
Sbjct: 169 VERWRPETHSFHLASGEMAVTLQDVAMLLALPIDGRPVCSTTDHDYAQMVIDCLGHDPRG 228
Query: 55 ASR-AKRKVAMTSLEARLDSIGEDVSDD----FVRSFLLFTI-GTFLSSIDGKVDSRYLS 108
S K + L+ + E D VR+++L + G G++ YL
Sbjct: 229 PSMPGKSFLHYKWLKKHFYELPEGADDQTVERHVRAYILSLLCGVLFPDGTGRMSLIYLP 288
Query: 109 FLGNLDDVSGFAWGAAVIEDLCQWL-DKRKDNNVQYVGGCLIFLQV 153
+ +L V ++WG+AV+ L + L +N++ +GG L+ LQ+
Sbjct: 289 LIADLSRVGTYSWGSAVLAFLYRSLCSVASSHNIKNIGGSLLLLQL 334
>UniRef100_Q9LMT7 F2H15.15 protein [Arabidopsis thaliana]
Length = 478
Score = 59.3 bits (142), Expect = 4e-08
Identities = 45/175 (25%), Positives = 76/175 (42%), Gaps = 17/175 (9%)
Query: 1 MEAWNSERHAFEIGSGEIGFSLLDVALILGIPVVGHRVELAEEQ----------LFSELE 50
+E W E + F GE+ +L +V+LILG+ V G V +E+ L +L
Sbjct: 71 VERWRRETNTFHFPCGEMTITLDEVSLILGLAVDGKPVVGVKEKDEDPSQVCLRLLGKLP 130
Query: 51 E-EYGASRAKRKVAMTSLEARLDSIGEDVSDDFVRSFLLFTIGT--FLSSIDGKVDSRYL 107
+ E +R K S + R++L++ +G+ F ++ K+ YL
Sbjct: 131 KGELSGNRVTAKWLKESFAECPKGATMKEIEYHTRAYLIYIVGSTIFATTDPSKISVDYL 190
Query: 108 SFLGNLDDVSGFAWGAAVIEDLCQWLDKRKDNNVQYVGGCLIFLQVSLICLSYYH 162
+ + +AWGAA + L + + + +GGCL LQ C SY+H
Sbjct: 191 ILFEDFEKAGEYAWGAAALAFLYRQIGNASQRSQSIIGGCLTLLQ----CWSYFH 241
>UniRef100_Q8LEM2 Hypothetical protein [Arabidopsis thaliana]
Length = 478
Score = 59.3 bits (142), Expect = 4e-08
Identities = 45/175 (25%), Positives = 76/175 (42%), Gaps = 17/175 (9%)
Query: 1 MEAWNSERHAFEIGSGEIGFSLLDVALILGIPVVGHRVELAEEQ----------LFSELE 50
+E W E + F GE+ +L +V+LILG+ V G V +E+ L +L
Sbjct: 71 VERWRRETNTFHFPCGEMTITLDEVSLILGLAVDGKPVVGVKEKDEDPSQVCLKLLGKLP 130
Query: 51 E-EYGASRAKRKVAMTSLEARLDSIGEDVSDDFVRSFLLFTIGT--FLSSIDGKVDSRYL 107
+ E +R K S + R++L++ +G+ F ++ K+ YL
Sbjct: 131 KGELSGNRVTAKWLKESFAECPKGATMKEIEYHTRAYLIYIVGSTIFATTDPSKISVDYL 190
Query: 108 SFLGNLDDVSGFAWGAAVIEDLCQWLDKRKDNNVQYVGGCLIFLQVSLICLSYYH 162
+ + +AWGAA + L + + + +GGCL LQ C SY+H
Sbjct: 191 ILFEDFEKAGEYAWGAAALAFLYRQIGNASQRSQSIIGGCLTLLQ----CWSYFH 241
>UniRef100_Q69L69 Calcineurin-like phosphoesterase-like protein [Oryza sativa]
Length = 766
Score = 57.0 bits (136), Expect = 2e-07
Identities = 42/166 (25%), Positives = 80/166 (47%), Gaps = 13/166 (7%)
Query: 1 MEAWNSERHAFEIGSGEIGFSLLDVALILGIPVVGHRVELAEEQLFSEL------EEEYG 54
+E W E H+F + SGE+ +L DV+++L +P+ G V + ++++ + G
Sbjct: 138 VERWRPETHSFHLASGEMTVTLQDVSMLLALPIDGRPVCSTTDHDYAQMVIDCLGHDPRG 197
Query: 55 ASR-AKRKVAMTSLEARLDSIGEDVSDD----FVRSFLLFTI-GTFLSSIDGKVDSRYLS 108
S K + L+ + E D VR+++L + G G++ YL
Sbjct: 198 PSMPGKSFLHYKWLKKHFYELPEGTDDQTVERHVRAYILSLLCGVLFPDGTGRMSLIYLP 257
Query: 109 FLGNLDDVSGFAWGAAVIEDLCQWL-DKRKDNNVQYVGGCLIFLQV 153
+ +L V ++WG+AV+ L + L +N++ +GG L+ LQ+
Sbjct: 258 LIADLSLVGTYSWGSAVLAFLYRSLCSVASSHNIKNIGGSLLLLQL 303
>UniRef100_Q8H3F8 Serine/threonine phosphatase PP7-related-like protein [Oryza
sativa]
Length = 589
Score = 55.5 bits (132), Expect = 6e-07
Identities = 44/142 (30%), Positives = 67/142 (46%), Gaps = 14/142 (9%)
Query: 2 EAWNSERHAFEIGSGEIGFSLLDVALILGIPVVGHRV-------ELAEEQLFSELEEEYG 54
E W+ E + ++ E+ SL D A ILGIPV G V + E+ F L +
Sbjct: 68 ELWSKETNTAQLNGFEMAPSLRDAAYILGIPVTGRVVTTGAVLNKSVEDLCFQYLGQVPD 127
Query: 55 ASRAK-RKVAMTSLEARLDSIGEDVSDD----FVRSFLLFTIGTFL--SSIDGKVDSRYL 107
+ V ++ L+++ I E ++D R++LLF IG+ L G V +YL
Sbjct: 128 CRDCRGSHVKLSWLQSKFSRIPERPTNDQTMYGTRAYLLFLIGSALLPERDRGYVSPKYL 187
Query: 108 SFLGNLDDVSGFAWGAAVIEDL 129
L + D V +AWGAA + L
Sbjct: 188 PLLSDFDKVQEYAWGAAALAHL 209
>UniRef100_Q9LNG5 F21D18.16 [Arabidopsis thaliana]
Length = 1340
Score = 53.9 bits (128), Expect = 2e-06
Identities = 44/165 (26%), Positives = 77/165 (46%), Gaps = 12/165 (7%)
Query: 1 MEAWNSERHAFEIGSGEIGFSLLDVALILGIPVVGHRVELAEEQLFSELEEEYGASRAKR 60
+E W E H F + +GEI +L DV ++LG+ V G V + + +++L E+ R
Sbjct: 89 VERWRPETHTFHLPAGEITVTLQDVNILLGLRVDGPAVTGSTKYNWADLCEDLLGHRPGP 148
Query: 61 K------VAMTSLEARLDSIGEDVSD----DFVRSFLLFTIGTFLSSIDGKVD--SRYLS 108
K V++ L ++ D + R+F+L + FL K D +L
Sbjct: 149 KDLHGSHVSLAWLRENFRNLPADPDEVTLKCHTRAFVLALMSGFLYGDKSKHDVALTFLP 208
Query: 109 FLGNLDDVSGFAWGAAVIEDLCQWLDKRKDNNVQYVGGCLIFLQV 153
L + D+V+ +WG+A + L + L + V + G L+ LQ+
Sbjct: 209 LLRDFDEVAKLSWGSATLALLYRELCRASKRTVSTICGPLVLLQL 253
>UniRef100_Q69S26 Serine/threonine phosphatase PP7-related-like [Oryza sativa]
Length = 619
Score = 53.9 bits (128), Expect = 2e-06
Identities = 45/166 (27%), Positives = 75/166 (45%), Gaps = 16/166 (9%)
Query: 2 EAWNSERHAFEIGSGEIGFSLLDVALILGIPVVGHRV-------ELAEEQLFSELEEEYG 54
E WN+E + E+ SL DV+ ILGIPV GH V E L E G
Sbjct: 81 ERWNNETNTAHFMGFEMAPSLRDVSYILGIPVTGHVVTAEPIGDEAVRRMCLHFLGESPG 140
Query: 55 -ASRAKRKVAMTSLEARLDSIGEDVSDDFV----RSFLLFTIGT--FLSSIDGKVDSRYL 107
+ + +T L + + E+ + + + R++LL+ +G+ F ++ G V RYL
Sbjct: 141 NGEQLCGLIRLTWLYRKFHQLPENPTINEIAYSTRAYLLYLVGSTLFPDTMRGFVSPRYL 200
Query: 108 SFLGNLDDVSGFAWGAAVIEDLCQWLDKRKDNN--VQYVGGCLIFL 151
L + + +AWG+A + L + L N Q++G + +
Sbjct: 201 PLLADFRKIREYAWGSAALAHLYRGLSVAVTPNATTQFLGSATLLM 246
>UniRef100_Q6H487 Hypothetical protein B1040D06.35 [Oryza sativa]
Length = 330
Score = 53.5 bits (127), Expect = 2e-06
Identities = 42/173 (24%), Positives = 86/173 (49%), Gaps = 20/173 (11%)
Query: 3 AWNSERHAFEIGSGEIGFSLLDVALILGIPVVGHRV----ELAEEQLFSELEEEYGASRA 58
+++ E+ AF I + +L DV +LG+P G + ++ + +LF+ ++E
Sbjct: 63 SYDKEKKAFNINGTFVTMTLDDVDCLLGLPSKGDDIFEAPKINKPELFNLYKQE-----G 117
Query: 59 KRKVAMTSL-EARLDSIGEDVSDDFVRSFLLFTIGTFLSSIDGK-VDSRYLSFLGNLDDV 116
+ + + +L EA ++S D D F+R F+LF+IG+F+ + V S YL+ + ++D +
Sbjct: 118 QTTITLQALREAIINSSSYD--DHFIRRFILFSIGSFICPTTQRYVRSEYLNLVDDVDKM 175
Query: 117 SGFAWGAAVIEDLCQWLDKRKDNNVQYVGG-CLIFL------QVSLICLSYYH 162
W + + L + + K ++ G CL+ + Q+ + + YH
Sbjct: 176 RELNWSSLTLNQLLKGIIKFREKTTNIEGNVCLLQIWYWEKVQIDKLAATIYH 228
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.324 0.142 0.425
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 291,093,011
Number of Sequences: 2790947
Number of extensions: 11339736
Number of successful extensions: 32851
Number of sequences better than 10.0: 143
Number of HSP's better than 10.0 without gapping: 97
Number of HSP's successfully gapped in prelim test: 46
Number of HSP's that attempted gapping in prelim test: 32710
Number of HSP's gapped (non-prelim): 148
length of query: 176
length of database: 848,049,833
effective HSP length: 119
effective length of query: 57
effective length of database: 515,927,140
effective search space: 29407846980
effective search space used: 29407846980
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 70 (31.6 bits)
Medicago: description of AC147537.14


