

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147537.12 + phase: 0
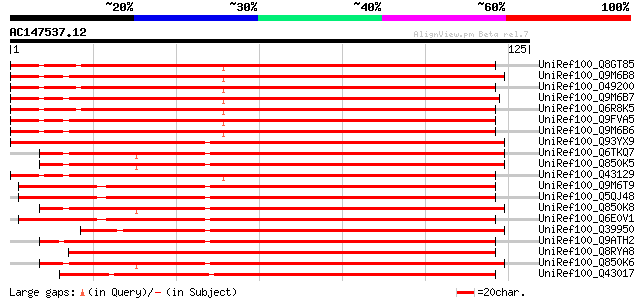
(125 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8GT85 Fiber lipid transfer protein [Gossypium barbade... 126 1e-28
UniRef100_Q9M6B8 Lipid transfer protein precursor [Gossypium hir... 125 2e-28
UniRef100_O49200 Lipid transfer protein precursor [Gossypium hir... 125 2e-28
UniRef100_Q9M6B7 Lipid transfer protein precursor [Gossypium hir... 124 4e-28
UniRef100_Q6R8K5 Lipid transfer protein [Gossypium barbadense] 124 5e-28
UniRef100_Q9FVA5 Lipid transfer protein 3 precursor [Gossypium h... 124 6e-28
UniRef100_Q9M6B6 Lipid transfer protein precursor [Gossypium hir... 122 1e-27
UniRef100_Q93YX9 Lipid transfer protein [Davidia involucrata] 119 2e-26
UniRef100_Q6TKQ7 Lipid transfer protein [Vitis aestivalis] 117 6e-26
UniRef100_Q850K5 Lipid transfer protein isoform 4 [Vitis vinifera] 116 1e-25
UniRef100_Q43129 Nonspecific lipid-transfer protein precursor [G... 116 1e-25
UniRef100_Q9M6T9 Lipid-transfer protein [Nicotiana glauca] 115 2e-25
UniRef100_Q5QJ48 Lipid-transfer protein [Nicotiana attenuata] 114 4e-25
UniRef100_Q850K8 Nonspecific lipid transfer protein 1 [Vitis ber... 114 4e-25
UniRef100_Q6E0V1 Lipid transfer protein [Nicotiana glauca] 113 1e-24
UniRef100_Q39950 Nonspecific lipid-transfer protein precursor [H... 113 1e-24
UniRef100_Q9ATH2 Lipid transfer protein precursor [Corylus avell... 112 1e-24
UniRef100_Q8RYA8 Lipid transfer precursor protein [Hevea brasili... 112 2e-24
UniRef100_Q850K6 Lipid transfer protein isoform 1 [Vitis vinifera] 112 2e-24
UniRef100_Q43017 Nonspecific lipid-transfer protein 1 precursor ... 112 2e-24
>UniRef100_Q8GT85 Fiber lipid transfer protein [Gossypium barbadense]
Length = 120
Score = 126 bits (317), Expect = 1e-28
Identities = 61/118 (51%), Positives = 84/118 (70%), Gaps = 3/118 (2%)
Query: 1 MSSSMVLVKVTCLTMMMCMVLGLPQTLDALSCLQVETKLMPCVPYVTGNG-GYVPQPCCD 59
M+SSM L K+ C+ ++ CMV+G P A++C QV + L PC+ Y+TGNG G VP CC
Sbjct: 1 MASSMSL-KLACVAVL-CMVVGAPLAHGAVTCGQVTSSLAPCIGYLTGNGAGGVPPGCCG 58
Query: 60 GVKAINNQAVTKSDRQAACRCIKTATSAIHGLNMDILAGLPSKCGVHLPYTLGPSTTC 117
G+K++N+ A T DRQAAC+CIK+A + I G+N I +GLP KCGV++PY + PST C
Sbjct: 59 GIKSLNSAAQTTPDRQAACKCIKSAAAGISGINYGIASGLPGKCGVNIPYKISPSTDC 116
>UniRef100_Q9M6B8 Lipid transfer protein precursor [Gossypium hirsutum]
Length = 120
Score = 125 bits (315), Expect = 2e-28
Identities = 60/120 (50%), Positives = 84/120 (70%), Gaps = 3/120 (2%)
Query: 1 MSSSMVLVKVTCLTMMMCMVLGLPQTLDALSCLQVETKLMPCVPYVTGNG-GYVPQPCCD 59
M+SSM L K+ C+ +++CMV+G P ++C QV L PC+ Y+ GNG G VPQ CC
Sbjct: 1 MASSMSL-KLACV-VVLCMVVGAPLAQGTVTCGQVTGSLAPCINYLRGNGAGAVPQGCCS 58
Query: 60 GVKAINNQAVTKSDRQAACRCIKTATSAIHGLNMDILAGLPSKCGVHLPYTLGPSTTCEK 119
G+K++N+ A T DRQAAC+CIK+A + I G+N I +GLP KCGV++PY + PST C +
Sbjct: 59 GIKSLNSAAQTTPDRQAACKCIKSAAAGIPGINYGIASGLPGKCGVNIPYKISPSTDCSR 118
>UniRef100_O49200 Lipid transfer protein precursor [Gossypium hirsutum]
Length = 120
Score = 125 bits (315), Expect = 2e-28
Identities = 61/118 (51%), Positives = 83/118 (69%), Gaps = 3/118 (2%)
Query: 1 MSSSMVLVKVTCLTMMMCMVLGLPQTLDALSCLQVETKLMPCVPYVTGNG-GYVPQPCCD 59
M+SSM L K C+ ++ CMV+G P A++C QV + L PC+ Y+TGNG G VP CC
Sbjct: 1 MASSMSL-KPACVAVL-CMVVGAPLAQGAVTCGQVTSSLAPCIGYLTGNGAGGVPPGCCG 58
Query: 60 GVKAINNQAVTKSDRQAACRCIKTATSAIHGLNMDILAGLPSKCGVHLPYTLGPSTTC 117
G+K++N+ A T DRQAAC+CIK+A + I G+N I +GLP KCGV++PY + PST C
Sbjct: 59 GIKSLNSAAQTTPDRQAACKCIKSAAAGISGINYGIASGLPGKCGVNIPYKISPSTDC 116
>UniRef100_Q9M6B7 Lipid transfer protein precursor [Gossypium hirsutum]
Length = 120
Score = 124 bits (312), Expect = 4e-28
Identities = 58/119 (48%), Positives = 83/119 (69%), Gaps = 3/119 (2%)
Query: 1 MSSSMVLVKVTCLTMMMCMVLGLPQTLDALSCLQVETKLMPCVPYVTGNG-GYVPQPCCD 59
M+SSM L K+TC+ ++ CMV+G P A+SC Q+ + L PC+ Y+ GNG G P CC+
Sbjct: 1 MASSMSL-KLTCV-VVFCMVVGAPLAQGAISCGQITSALAPCIAYLKGNGAGSAPPACCN 58
Query: 60 GVKAINNQAVTKSDRQAACRCIKTATSAIHGLNMDILAGLPSKCGVHLPYTLGPSTTCE 118
G++++N+ A T DRQAAC CIK+A + I G+N AGLP KCG+++PY + PST C+
Sbjct: 59 GIRSLNSAAKTTPDRQAACSCIKSAATGISGINYSTAAGLPGKCGINIPYKISPSTDCK 117
>UniRef100_Q6R8K5 Lipid transfer protein [Gossypium barbadense]
Length = 120
Score = 124 bits (311), Expect = 5e-28
Identities = 60/118 (50%), Positives = 83/118 (69%), Gaps = 3/118 (2%)
Query: 1 MSSSMVLVKVTCLTMMMCMVLGLPQTLDALSCLQVETKLMPCVPYVTGNG-GYVPQPCCD 59
M+SSM L K+ C+ ++ CMV+G P A++C QV + L PC+ Y+TGNG G VP CC
Sbjct: 1 MASSMSL-KLACVAVL-CMVVGAPLAQGAVTCGQVTSSLAPCIGYLTGNGAGGVPPGCCG 58
Query: 60 GVKAINNQAVTKSDRQAACRCIKTATSAIHGLNMDILAGLPSKCGVHLPYTLGPSTTC 117
G+K++N+ A T DRQAAC+CIK+A + I G+N I +G P KCGV++PY + PST C
Sbjct: 59 GIKSLNSAAQTTPDRQAACKCIKSAAAGISGINYGIASGPPGKCGVNIPYKISPSTDC 116
>UniRef100_Q9FVA5 Lipid transfer protein 3 precursor [Gossypium hirsutum]
Length = 120
Score = 124 bits (310), Expect = 6e-28
Identities = 59/118 (50%), Positives = 83/118 (70%), Gaps = 3/118 (2%)
Query: 1 MSSSMVLVKVTCLTMMMCMVLGLPQTLDALSCLQVETKLMPCVPYVTGNG-GYVPQPCCD 59
M+SSM L K+ C+ +++CMV+G P A++C QV L PC+ Y+ G+G G VP CC
Sbjct: 1 MASSMSL-KLACV-VVLCMVVGAPLAQGAVTCGQVTNSLAPCINYLRGSGAGAVPPGCCS 58
Query: 60 GVKAINNQAVTKSDRQAACRCIKTATSAIHGLNMDILAGLPSKCGVHLPYTLGPSTTC 117
G+K++N+ A T DRQAACRCIK+A + I G+N + +GLP KCGV++PY + PST C
Sbjct: 59 GIKSLNSAAQTTPDRQAACRCIKSAAAGITGINFGLASGLPGKCGVNIPYKISPSTDC 116
>UniRef100_Q9M6B6 Lipid transfer protein precursor [Gossypium hirsutum]
Length = 120
Score = 122 bits (307), Expect = 1e-27
Identities = 57/118 (48%), Positives = 83/118 (70%), Gaps = 3/118 (2%)
Query: 1 MSSSMVLVKVTCLTMMMCMVLGLPQTLDALSCLQVETKLMPCVPYVTGNG-GYVPQPCCD 59
M+SSM L K+ C+ +++CMV+G P A++C QV + L PC+ Y+ G G G +P CC
Sbjct: 1 MASSMYL-KLACV-VVLCMVVGAPLAQGAVTCGQVTSSLAPCINYLRGTGAGAIPPGCCS 58
Query: 60 GVKAINNQAVTKSDRQAACRCIKTATSAIHGLNMDILAGLPSKCGVHLPYTLGPSTTC 117
G+K++N+ A T DRQAAC+CIK+A + I G+N + +GLP KCGV++PY + PST C
Sbjct: 59 GIKSLNSAAQTTPDRQAACKCIKSAAAGIPGINFGLASGLPGKCGVNIPYKISPSTDC 116
>UniRef100_Q93YX9 Lipid transfer protein [Davidia involucrata]
Length = 120
Score = 119 bits (298), Expect = 2e-26
Identities = 54/119 (45%), Positives = 79/119 (66%), Gaps = 1/119 (0%)
Query: 1 MSSSMVLVKVTCLTMMMCMVLGLPQTLDALSCLQVETKLMPCVPYVTGNGGYVPQPCCDG 60
M S V++KV + ++MCMV+ P A++C V L PC+ Y+ GG VP CC+G
Sbjct: 1 MGRSGVVIKVGIVAVLMCMVVSAPHAEAAITCGTVTVSLAPCLNYLK-KGGPVPPACCNG 59
Query: 61 VKAINNQAVTKSDRQAACRCIKTATSAIHGLNMDILAGLPSKCGVHLPYTLGPSTTCEK 119
+K++N A T +DRQAAC C+KTA+++I G+N+ + LP KCGV++PY + PST C K
Sbjct: 60 IKSLNAAAKTTADRQAACNCLKTASTSIAGINLSYASSLPGKCGVNVPYKISPSTDCSK 118
>UniRef100_Q6TKQ7 Lipid transfer protein [Vitis aestivalis]
Length = 119
Score = 117 bits (293), Expect = 6e-26
Identities = 52/113 (46%), Positives = 79/113 (69%), Gaps = 3/113 (2%)
Query: 8 VKVTCLTMMMCMVLGLPQTLDA-LSCLQVETKLMPCVPYVTGNGGYVPQPCCDGVKAINN 66
VK+ C+ M++CMV+ P ++A ++C QV + L PC+ Y+ GG VP CC G+K++N+
Sbjct: 7 VKLACV-MVICMVMAAPAAVEAAITCGQVASALSPCISYLQ-KGGAVPPACCSGIKSLNS 64
Query: 67 QAVTKSDRQAACRCIKTATSAIHGLNMDILAGLPSKCGVHLPYTLGPSTTCEK 119
A T +DRQAAC+C+K +S + G+N+ + +GLP KCGV +PY + PST C K
Sbjct: 65 SAKTTADRQAACKCLKNFSSTVSGINLSLASGLPGKCGVSVPYKISPSTDCTK 117
>UniRef100_Q850K5 Lipid transfer protein isoform 4 [Vitis vinifera]
Length = 119
Score = 116 bits (291), Expect = 1e-25
Identities = 53/113 (46%), Positives = 79/113 (69%), Gaps = 3/113 (2%)
Query: 8 VKVTCLTMMMCMVLGLPQTLDA-LSCLQVETKLMPCVPYVTGNGGYVPQPCCDGVKAINN 66
VK+ C+ M++CMV+ P ++A ++C QV + L PC+ Y+ GG VP CC G+K++N+
Sbjct: 7 VKLACV-MVICMVVAAPAVVEATVTCGQVASALSPCISYLQ-KGGAVPAGCCSGIKSLNS 64
Query: 67 QAVTKSDRQAACRCIKTATSAIHGLNMDILAGLPSKCGVHLPYTLGPSTTCEK 119
A T DRQAAC+C+KT +S++ G+N + +GLP KCGV +PY + PST C K
Sbjct: 65 AAKTTGDRQAACKCLKTFSSSVSGINYGLASGLPGKCGVSVPYKISPSTDCSK 117
>UniRef100_Q43129 Nonspecific lipid-transfer protein precursor [Gossypium hirsutum]
Length = 120
Score = 116 bits (290), Expect = 1e-25
Identities = 57/118 (48%), Positives = 81/118 (68%), Gaps = 3/118 (2%)
Query: 1 MSSSMVLVKVTCLTMMMCMVLGLPQTLDALSCLQVETKLMPCVPYVTGNG-GYVPQPCCD 59
M+SSM L K+ C+ +++CMV+G P A++ QV L PC+ Y+ G+G G VP CC
Sbjct: 1 MASSMSL-KLACV-VVLCMVVGAPLAQGAVTSGQVTNSLAPCINYLRGSGAGAVPPGCCT 58
Query: 60 GVKAINNQAVTKSDRQAACRCIKTATSAIHGLNMDILAGLPSKCGVHLPYTLGPSTTC 117
G+K++N+ A T RQAACRCIK+A + I G+N + +GLP KCGV++PY + PST C
Sbjct: 59 GIKSLNSAAQTTPVRQAACRCIKSAAAGITGINFGLASGLPGKCGVNIPYKISPSTDC 116
>UniRef100_Q9M6T9 Lipid-transfer protein [Nicotiana glauca]
Length = 117
Score = 115 bits (289), Expect = 2e-25
Identities = 53/115 (46%), Positives = 74/115 (64%), Gaps = 3/115 (2%)
Query: 3 SSMVLVKVTCLTMMMCMVLGLPQTLDALSCLQVETKLMPCVPYVTGNGGYVPQPCCDGVK 62
+ + L+ V C+ + M+ PQ A+SC QV + L PC+ YVT GG +P PCC G+K
Sbjct: 2 AKVALLVVVCMAAVAVMLT--PQADAAISCGQVVSSLTPCISYVT-KGGAIPAPCCSGIK 58
Query: 63 AINNQAVTKSDRQAACRCIKTATSAIHGLNMDILAGLPSKCGVHLPYTLGPSTTC 117
++NNQA + DRQ AC CIK+A +I G+N + + LP KCGV+LPY + PS C
Sbjct: 59 SLNNQATSTPDRQTACNCIKSAVGSISGINFGLASSLPGKCGVNLPYKISPSIDC 113
>UniRef100_Q5QJ48 Lipid-transfer protein [Nicotiana attenuata]
Length = 117
Score = 114 bits (286), Expect = 4e-25
Identities = 53/115 (46%), Positives = 73/115 (63%), Gaps = 3/115 (2%)
Query: 3 SSMVLVKVTCLTMMMCMVLGLPQTLDALSCLQVETKLMPCVPYVTGNGGYVPQPCCDGVK 62
+ + L+ V C+ + M+ P A+SC QV L PC+ YV GG +P PCC G+K
Sbjct: 2 AKVALLVVVCMAAVAVMLT--PHADAAISCGQVVASLSPCISYVR-QGGAIPAPCCSGIK 58
Query: 63 AINNQAVTKSDRQAACRCIKTATSAIHGLNMDILAGLPSKCGVHLPYTLGPSTTC 117
++NNQA + DRQ AC CIK+A +AI+G+N + LPSKCGV+LPY + PS C
Sbjct: 59 SLNNQATSTPDRQTACNCIKSAAAAINGINYSLAGSLPSKCGVNLPYKISPSIDC 113
>UniRef100_Q850K8 Nonspecific lipid transfer protein 1 [Vitis berlandieri x Vitis
vinifera]
Length = 119
Score = 114 bits (286), Expect = 4e-25
Identities = 53/113 (46%), Positives = 80/113 (69%), Gaps = 3/113 (2%)
Query: 8 VKVTCLTMMMCMVLGLPQTLDA-LSCLQVETKLMPCVPYVTGNGGYVPQPCCDGVKAINN 66
VK+ C+ M++CMV+ P ++A ++C QV + L C+ Y+ NGG VP CC G+K +N+
Sbjct: 7 VKLACV-MVICMVVAAPAAVEAAITCGQVSSALSSCLGYLK-NGGAVPPGCCSGIKNLNS 64
Query: 67 QAVTKSDRQAACRCIKTATSAIHGLNMDILAGLPSKCGVHLPYTLGPSTTCEK 119
A T +DRQAAC+C+KT +++I G+N+ + +GLP KCGV +PY + PST C K
Sbjct: 65 AAQTTADRQAACKCLKTFSNSIPGINLGLASGLPGKCGVSVPYKISPSTDCSK 117
>UniRef100_Q6E0V1 Lipid transfer protein [Nicotiana glauca]
Length = 117
Score = 113 bits (282), Expect = 1e-24
Identities = 51/115 (44%), Positives = 74/115 (64%), Gaps = 3/115 (2%)
Query: 3 SSMVLVKVTCLTMMMCMVLGLPQTLDALSCLQVETKLMPCVPYVTGNGGYVPQPCCDGVK 62
+ + L+ V C+ + M+ P A+SC QV + L PC+ YVT GG +P PCC+G++
Sbjct: 2 AKVALLVVVCMAAVSVMLT--PHADAAISCGQVVSSLTPCISYVT-KGGAIPAPCCNGIE 58
Query: 63 AINNQAVTKSDRQAACRCIKTATSAIHGLNMDILAGLPSKCGVHLPYTLGPSTTC 117
++NNQA + DRQ AC CIK+A ++I G+N + LP KCGV+LPY + PS C
Sbjct: 59 SLNNQATSTPDRQTACNCIKSAAASIKGINFSLAGSLPGKCGVNLPYKISPSIDC 113
>UniRef100_Q39950 Nonspecific lipid-transfer protein precursor [Helianthus annuus]
Length = 116
Score = 113 bits (282), Expect = 1e-24
Identities = 54/102 (52%), Positives = 69/102 (66%), Gaps = 2/102 (1%)
Query: 18 CMVLGLPQTLDALSCLQVETKLMPCVPYVTGNGGYVPQPCCDGVKAINNQAVTKSDRQAA 77
CMV+G P T +ALSC QV + L PC+ Y+T GG VP CC GVK++N+ A T DRQAA
Sbjct: 15 CMVVGAPYT-EALSCGQVSSSLAPCISYLT-KGGAVPPACCSGVKSLNSAAKTTPDRQAA 72
Query: 78 CRCIKTATSAIHGLNMDILAGLPSKCGVHLPYTLGPSTTCEK 119
C C+K+A ++I G+N A P KCGV +PY + PST C K
Sbjct: 73 CGCLKSAYNSISGVNAGNAASFPGKCGVSIPYKISPSTDCSK 114
>UniRef100_Q9ATH2 Lipid transfer protein precursor [Corylus avellana]
Length = 115
Score = 112 bits (281), Expect = 1e-24
Identities = 50/110 (45%), Positives = 74/110 (66%), Gaps = 2/110 (1%)
Query: 8 VKVTCLTMMMCMVLGLPQTLDALSCLQVETKLMPCVPYVTGNGGYVPQPCCDGVKAINNQ 67
+K+ C +++CM++ P +L+C Q++ L PCV Y+ NGG +P CC GV+A+N+
Sbjct: 4 LKLVC-AVLLCMMVAAPVARASLTCPQIKGNLTPCVLYLK-NGGVLPPSCCKGVRAVNDA 61
Query: 68 AVTKSDRQAACRCIKTATSAIHGLNMDILAGLPSKCGVHLPYTLGPSTTC 117
+ T SDRQ+AC C+K I GLN ++ AGLP KCGV++PY + PST C
Sbjct: 62 SRTTSDRQSACNCLKDTAKGIAGLNPNLAAGLPGKCGVNIPYKISPSTNC 111
>UniRef100_Q8RYA8 Lipid transfer precursor protein [Hevea brasiliensis]
Length = 116
Score = 112 bits (279), Expect = 2e-24
Identities = 46/103 (44%), Positives = 71/103 (68%)
Query: 15 MMMCMVLGLPQTLDALSCLQVETKLMPCVPYVTGNGGYVPQPCCDGVKAINNQAVTKSDR 74
+++CM++ P T A++C QV++ L+PC+ Y+ G P CC+GV+ INN A T +DR
Sbjct: 10 LVLCMLVAAPMTAQAITCGQVQSALVPCLSYLKTTGPTPPATCCNGVRTINNAAKTTADR 69
Query: 75 QAACRCIKTATSAIHGLNMDILAGLPSKCGVHLPYTLGPSTTC 117
+ AC+C+K+A ++ GLN +AGLP KCGV++PY + ST C
Sbjct: 70 RTACQCLKSAAGSVKGLNPTTVAGLPGKCGVNIPYKISLSTNC 112
>UniRef100_Q850K6 Lipid transfer protein isoform 1 [Vitis vinifera]
Length = 119
Score = 112 bits (279), Expect = 2e-24
Identities = 52/113 (46%), Positives = 77/113 (68%), Gaps = 3/113 (2%)
Query: 8 VKVTCLTMMMCMVLGLPQTLDA-LSCLQVETKLMPCVPYVTGNGGYVPQPCCDGVKAINN 66
VK+ C+ M++CMV+ P ++A ++C QV + + PC Y+ GG VP CC G+K++N+
Sbjct: 7 VKLACV-MVICMVVAAPAAVEATITCGQVASAVGPCASYLQ-KGGSVPAGCCSGIKSLNS 64
Query: 67 QAVTKSDRQAACRCIKTATSAIHGLNMDILAGLPSKCGVHLPYTLGPSTTCEK 119
A T DRQAAC+C+KT + +I G+N + +GLP KCGV +PY + PST C K
Sbjct: 65 AAKTTVDRQAACKCLKTFSGSIPGINFGLASGLPGKCGVSVPYKISPSTDCSK 117
>UniRef100_Q43017 Nonspecific lipid-transfer protein 1 precursor [Prunus dulcis]
Length = 117
Score = 112 bits (279), Expect = 2e-24
Identities = 50/105 (47%), Positives = 72/105 (67%), Gaps = 2/105 (1%)
Query: 13 LTMMMCMVLGLPQTLDALSCLQVETKLMPCVPYVTGNGGYVPQPCCDGVKAINNQAVTKS 72
L + +CMV+ +P A++C QV + L PC+PYV G GG VP CC+G++ +NN A T
Sbjct: 11 LVVALCMVVSVP-IAQAITCGQVSSNLAPCIPYVRG-GGAVPPACCNGIRNVNNLARTTP 68
Query: 73 DRQAACRCIKTATSAIHGLNMDILAGLPSKCGVHLPYTLGPSTTC 117
DRQAAC C+K ++++ G+N + A LP KCGV++PY + PST C
Sbjct: 69 DRQAACNCLKQLSASVPGVNPNNAAALPGKCGVNIPYQISPSTNC 113
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.322 0.135 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 196,872,973
Number of Sequences: 2790947
Number of extensions: 6766252
Number of successful extensions: 11986
Number of sequences better than 10.0: 259
Number of HSP's better than 10.0 without gapping: 193
Number of HSP's successfully gapped in prelim test: 66
Number of HSP's that attempted gapping in prelim test: 11630
Number of HSP's gapped (non-prelim): 260
length of query: 125
length of database: 848,049,833
effective HSP length: 101
effective length of query: 24
effective length of database: 566,164,186
effective search space: 13587940464
effective search space used: 13587940464
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 67 (30.4 bits)
Medicago: description of AC147537.12


