

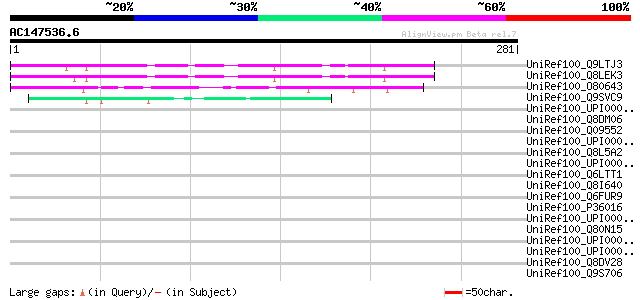
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147536.6 + phase: 0
(281 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9LTJ3 Arabidopsis thaliana genomic DNA, chromosome 5,... 152 9e-36
UniRef100_Q8LEK3 Hypothetical protein [Arabidopsis thaliana] 152 9e-36
UniRef100_O80643 Hypothetical protein At2g39560 [Arabidopsis tha... 135 9e-31
UniRef100_Q9SVC9 Hypothetical protein F22O6_140 [Arabidopsis tha... 47 7e-04
UniRef100_UPI000042D010 UPI000042D010 UniRef100 entry 38 0.34
UniRef100_Q8DM06 Tlr0320 protein [Synechococcus elongatus] 36 0.98
UniRef100_Q09552 Hypothetical protein F27E5.3 [Caenorhabditis el... 36 0.98
UniRef100_UPI00002DA952 UPI00002DA952 UniRef100 entry 35 1.7
UniRef100_Q8L5A2 HV80H14.14B [Hordeum vulgare] 35 1.7
UniRef100_UPI0000363E15 UPI0000363E15 UniRef100 entry 34 3.7
UniRef100_Q6LTT1 Putative DNA methylase HsdM [Photobacterium pro... 34 3.7
UniRef100_Q8I640 Erythrocyte membrane protein 1 SD105A [Plasmodi... 34 3.7
UniRef100_Q6FUR9 Similar to tr|Q08206 Saccharomyces cerevisiae Y... 34 3.7
UniRef100_P36016 Heat shock protein 70 homolog LHS1 precursor [S... 34 3.7
UniRef100_UPI0000363B2F UPI0000363B2F UniRef100 entry 33 6.3
UniRef100_Q80N15 Structural glycoprotein [Yellow head virus] 33 6.3
UniRef100_UPI0000435D5D UPI0000435D5D UniRef100 entry 33 8.3
UniRef100_UPI00003A9BC4 UPI00003A9BC4 UniRef100 entry 33 8.3
UniRef100_Q8DV28 Hypothetical protein SMU.689 [Streptococcus mut... 33 8.3
UniRef100_Q9S706 Hypothetical protein [Oryza sativa] 33 8.3
>UniRef100_Q9LTJ3 Arabidopsis thaliana genomic DNA, chromosome 5, BAC clone:F2O15
[Arabidopsis thaliana]
Length = 287
Score = 152 bits (384), Expect = 9e-36
Identities = 106/249 (42%), Positives = 146/249 (58%), Gaps = 33/249 (13%)
Query: 1 MKSLSSVGLGLSIVFGCLLLALVAEVYYLL---WWKKKIIKREI----ENDYGNGNPLKE 53
MK++S +G+GLS++FG LLLALV EVYYLL KK++I +E E + KE
Sbjct: 1 MKTISGLGIGLSLMFGFLLLALVGEVYYLLRCKKHKKRVISQESEEEKEEEQQQNGYAKE 60
Query: 54 LFYMFCWKRPSTSLRQTGFTPPELCSSMRINDNFVHDPQVQTSKEFLFKPYGEDVIDADY 113
L +FC+K+P + + G E + +N++ D ++ K G+ +A+
Sbjct: 61 LIQLFCFKKPQSLQQNNGGREGE----VSMNEDGNPDLELGLMKHL---NGGDLGFEAEL 113
Query: 114 MMQHHHDGLLGHPRFLFTIVEESKEDLESEDG---KCGKDSRGRSLGDL-LDVETPYLTP 169
M H+ RFLFTI+EE+K DLESEDG + G SR RSL D+ D TP TP
Sbjct: 114 MKLHNQ-------RFLFTIMEETKADLESEDGGKSRLGSRSRRRSLSDVPNDCNTPGFTP 166
Query: 170 IASPHFFTPMNCSPYCSPYNQHGFNPLFESTTDAEFN---RLKSSPPPKFKFLQEAEEKL 226
+ASP + SP S Y HGFNPLFES + EFN R SSPPPKFKF+++AEEKL
Sbjct: 167 LASP----KKSSSPLES-YPHHGFNPLFESDGELEFNKFFRSSSSPPPKFKFMRDAEEKL 221
Query: 227 RRKMQDDNK 235
R+++ ++ K
Sbjct: 222 RKRLIEEAK 230
>UniRef100_Q8LEK3 Hypothetical protein [Arabidopsis thaliana]
Length = 287
Score = 152 bits (384), Expect = 9e-36
Identities = 106/249 (42%), Positives = 146/249 (58%), Gaps = 33/249 (13%)
Query: 1 MKSLSSVGLGLSIVFGCLLLALVAEVYYLLWWKK---KIIKREI----ENDYGNGNPLKE 53
MK++S +G+GLS++FG LLLALV EVYYLL KK ++I +E E + KE
Sbjct: 1 MKTISGLGIGLSLMFGFLLLALVGEVYYLLRCKKHKNRVISQESEEEKEEEQQQNGYAKE 60
Query: 54 LFYMFCWKRPSTSLRQTGFTPPELCSSMRINDNFVHDPQVQTSKEFLFKPYGEDVIDADY 113
L +FC+K+P + + G E + +N++ D ++ K G+ +A+
Sbjct: 61 LIQLFCFKKPQSLQQNNGGREGE----VSMNEDGNPDLELGLMKHL---NGGDLGFEAEL 113
Query: 114 MMQHHHDGLLGHPRFLFTIVEESKEDLESEDG---KCGKDSRGRSLGDL-LDVETPYLTP 169
M H+ RFLFTI+EE+K DLESEDG + G SR RSL D+ D TP TP
Sbjct: 114 MKLHNQ-------RFLFTIMEETKADLESEDGGKSRLGSRSRRRSLSDVPNDCNTPGFTP 166
Query: 170 IASPHFFTPMNCSPYCSPYNQHGFNPLFESTTDAEFN---RLKSSPPPKFKFLQEAEEKL 226
+ASP + SP S Y HGFNPLFES + EFN R SSPPPKFKF+++AEEKL
Sbjct: 167 LASP----KKSSSPLES-YPHHGFNPLFESDGELEFNKFFRSSSSPPPKFKFMRDAEEKL 221
Query: 227 RRKMQDDNK 235
R+++ ++ K
Sbjct: 222 RKRLIEEAK 230
>UniRef100_O80643 Hypothetical protein At2g39560 [Arabidopsis thaliana]
Length = 233
Score = 135 bits (341), Expect = 9e-31
Identities = 106/251 (42%), Positives = 139/251 (55%), Gaps = 48/251 (19%)
Query: 1 MKSLSSVGLGLSIVFGCLLLALVAEVYYLLWWKKKIIKR--EIENDYGNGNPLKELFYMF 58
M+SLSSVGL LSIVFGCLLLAL+AE+YYLLW KK+ R + NDY +EL ++F
Sbjct: 1 MRSLSSVGLALSIVFGCLLLALLAELYYLLWCKKRSTTRRPDFRNDYSTPG-TRELLFIF 59
Query: 59 CWKRPSTSLRQTGFTPPELCSSMRINDNFVHDPQVQTSKEFLFKPYGEDVIDADYMMQHH 118
C S+S + +P SS ++ D Q Q F+ G
Sbjct: 60 CC---SSSTNPSSSSP----SSSSFSNPKPIDTQQQCPLNNGFENVG------------- 99
Query: 119 HDGLLGHPRFLFTIVEESKEDLESEDGKCGKDSRGRSLGDL-LDVET-----PYLTPIAS 172
GL+ PRFLFTI+EE+ E++ESED ++G+SL DL L++E+ PYLTP AS
Sbjct: 100 GPGLV--PRFLFTIMEETVEEMESED---VVSTKGKSLNDLFLNMESGVITPPYLTPRAS 154
Query: 173 PHFFTPMNCSPYCSPYN--QHGFNPLFESTTDAEFNRL------------KSSPPPKFKF 218
P FTP N + + LFES++DAEFNRL SSP +FKF
Sbjct: 155 PSLFTPPLTPLLMESCNGRKEEISSLFESSSDAEFNRLVRSSPLSSSHSPSSSPLSRFKF 214
Query: 219 LQEAEEKLRRK 229
L++AEEKL +K
Sbjct: 215 LRDAEEKLYKK 225
>UniRef100_Q9SVC9 Hypothetical protein F22O6_140 [Arabidopsis thaliana]
Length = 209
Score = 46.6 bits (109), Expect = 7e-04
Identities = 54/193 (27%), Positives = 78/193 (39%), Gaps = 38/193 (19%)
Query: 11 LSIVFGCLLLALVAEVYYLLWWKKKIIKREI---ENDYGNGN---------PLKELFYMF 58
L VF L+A+ A+ Y+LWWK++ +R I E D + P KEL Y F
Sbjct: 10 LVAVFAFCLVAVTAQFAYVLWWKRRFRRRSIAGSERDAFSSRGGDLTATPPPSKELLYFF 69
Query: 59 CWKRPSTSLRQTGFTPP----------ELCSSMRIN-DNFVHDPQVQTSKEFLFKPYGED 107
+ + R T P ++ S IN +N + P E LF
Sbjct: 70 LFCLENKQFRIGSATAPPLPAAAPPVNDVASKWSINGENLLCGP-----SETLF------ 118
Query: 108 VIDADYMMQ-HHHDGLLGHPRFLFTIVEESKEDLESEDGKCGKDSRGRSLG-DLLDVETP 165
I DY + H G + +FT E+ +D E E+ D + + TP
Sbjct: 119 TIAEDYTSESDHRTGEIDPRGSIFT--EDHVKDDEVEEEVVATDISDDEVDFSHYNQTTP 176
Query: 166 YLTPIASPHFFTP 178
+ TP ASP F+TP
Sbjct: 177 FSTPCASPPFYTP 189
>UniRef100_UPI000042D010 UPI000042D010 UniRef100 entry
Length = 363
Score = 37.7 bits (86), Expect = 0.34
Identities = 34/106 (32%), Positives = 43/106 (40%), Gaps = 7/106 (6%)
Query: 113 YMMQHHHDGLLGHPRFLFTIVEESKEDL-ESEDGKCGKDSRGRSLGDLLDVETPYLTPIA 171
YM+ H LG V + DL SED D+ G G +LDV P P
Sbjct: 251 YMLTESHLHTLGSLPEGAVFVNIGRGDLIRSEDILAALDAEGGLFGAVLDVTDPEPLPSG 310
Query: 172 SPHFFTPMNCSPYCSPYNQHGFNPLFESTTD---AEFNRLKSSPPP 214
P F P S +P+ F F+S D A+ +RLK PP
Sbjct: 311 HPLFTHP---SVIVTPHTSGSFEGYFDSGADVLLAQGDRLKQDLPP 353
>UniRef100_Q8DM06 Tlr0320 protein [Synechococcus elongatus]
Length = 895
Score = 36.2 bits (82), Expect = 0.98
Identities = 23/71 (32%), Positives = 37/71 (51%), Gaps = 1/71 (1%)
Query: 208 LKSSPPPKFKFLQEAEEKLRRKMQDDNK-GINGNEVDNSLITIIVDKKYEREVNHHQCHL 266
LK PPP K++QE EEKLR ++ N N +V +TI + + + ++ H Q +
Sbjct: 601 LKQLPPPDQKYVQELEEKLREFIEFINAIDCNCADVAEQGMTIRQEPQKDTKIIHFQSII 660
Query: 267 QQYHSSTSQVL 277
Q S+V+
Sbjct: 661 QDIQKRLSEVV 671
>UniRef100_Q09552 Hypothetical protein F27E5.3 [Caenorhabditis elegans]
Length = 490
Score = 36.2 bits (82), Expect = 0.98
Identities = 33/98 (33%), Positives = 47/98 (47%), Gaps = 16/98 (16%)
Query: 136 SKEDLESEDGKCGKDSRGRSLGDLLDVETPYLTPIASPHFFTPMNCSPYCSPYNQHGFNP 195
S+ DL+ D + + + + D ET LTPI P PY P +Q G P
Sbjct: 193 SQSDLKKHDSRFAYLPKNQRPRQVSDDETQKLTPIFLP--------QPY-KPSSQQGPPP 243
Query: 196 LFESTTDAEFNRLKSSPPPKFKFLQEAEEKLRRKMQDD 233
+ T F+ K+SPP K L+E+ EK +KM+DD
Sbjct: 244 ML---TRQPFHAQKTSPPK--KPLEESPEK--KKMKDD 274
>UniRef100_UPI00002DA952 UPI00002DA952 UniRef100 entry
Length = 603
Score = 35.4 bits (80), Expect = 1.7
Identities = 17/74 (22%), Positives = 42/74 (55%), Gaps = 6/74 (8%)
Query: 194 NPLFESTTDAEFNR-LKSSPPPKFKFLQEAEEKLRRKMQDDNKGINGNEVDNSLITIIVD 252
N +F + DA+F ++S+P PK ++ +++ R + D K +E+ + ++ +++
Sbjct: 133 NCVFAAGEDAQFTCVIQSAPSPKIRWFKDS-----RLLTDQEKYQTYSELRSGVLVLVIK 187
Query: 253 KKYEREVNHHQCHL 266
ER++ H++C +
Sbjct: 188 NLTERDLGHYECEV 201
>UniRef100_Q8L5A2 HV80H14.14B [Hordeum vulgare]
Length = 254
Score = 35.4 bits (80), Expect = 1.7
Identities = 18/65 (27%), Positives = 31/65 (47%), Gaps = 2/65 (3%)
Query: 74 PPELCSSMRINDNFVHDPQVQTSKEFLFKPYGED--VIDADYMMQHHHDGLLGHPRFLFT 131
P E C+ M + ++ HDP + LF E +++ + H GLL ++LF
Sbjct: 107 PMESCADMPLTPSYAHDPSFAAASATLFLTLEERKVILEEKKLAMEEHTGLLKWEKYLFF 166
Query: 132 IVEES 136
I++ S
Sbjct: 167 IMDTS 171
>UniRef100_UPI0000363E15 UPI0000363E15 UniRef100 entry
Length = 416
Score = 34.3 bits (77), Expect = 3.7
Identities = 30/112 (26%), Positives = 46/112 (40%), Gaps = 17/112 (15%)
Query: 167 LTPIASPHFFTPMNCSPYCSPYNQHGFNPLFESTTDAEFNRLKSSPPPKFKFLQEAEE-K 225
LTP + P+N S P N LF S +AE + +P + AEE
Sbjct: 195 LTP--DDRYIQPVNVSVLTQPIFHPRPNCLFHSIKEAEIQGQRDAP------ISHAEEPS 246
Query: 226 LRRKMQDDNKGING--NEVDNSLITIIVDKKYEREVNHHQCHLQQYHSSTSQ 275
+R+ ++ + + E+ N ++ E E+ Q HLQQ HS Q
Sbjct: 247 VRQHVEVQQRYVENLRQEIQNE------QRRAESELQREQAHLQQQHSEIQQ 292
>UniRef100_Q6LTT1 Putative DNA methylase HsdM [Photobacterium profundum)]
Length = 793
Score = 34.3 bits (77), Expect = 3.7
Identities = 41/185 (22%), Positives = 80/185 (43%), Gaps = 40/185 (21%)
Query: 79 SSMRINDNFVHDPQVQTSKEFLFKPYGEDVIDADYMMQHHHDGLLGHPRFLFTIVEESKE 138
SS RI + D ++ E+++ YGED A+ + + + + EE +
Sbjct: 519 SSERI-ETLRFDKALREPMEYIYNTYGEDAYKAEILAK--------ESKAILAWCEEKEI 569
Query: 139 DLESED-GKCGKDSRGRSLGDLLDVETPYLTPIASPHFFTPMNCSPYCSPYNQHGFNPLF 197
L +++ K + LGDL+D+ + I + + + YNQ F
Sbjct: 570 SLNTKNRNKLLDVATWTRLGDLIDIANTLMKAIGTDIY----------NDYNQ------F 613
Query: 198 ESTTDAEF--NRLKSSPPPKFKFL-------QEAEEKLRRKMQDDNKGINGNEVDNSLIT 248
++T DAE ++K S P K L + AE+ +++K++ + G+++D L +
Sbjct: 614 KATVDAELKSRKIKLSAPEKNAILNAVSWYHENAEKVIKKKLK-----LTGSKLDELLTS 668
Query: 249 IIVDK 253
D+
Sbjct: 669 CDCDE 673
>UniRef100_Q8I640 Erythrocyte membrane protein 1 SD105A [Plasmodium falciparum]
Length = 2860
Score = 34.3 bits (77), Expect = 3.7
Identities = 30/118 (25%), Positives = 50/118 (41%), Gaps = 6/118 (5%)
Query: 133 VEESKEDLESEDGKCGKDSRGRSLGDLLDVETPYLTPIASPHFFTPMNC---SPYCSPYN 189
+++ +E +S K K S + D L TPY T H MNC + +C
Sbjct: 1136 LQKLQEANKSSASKRSKRSTDGTTTDTLTPTTPYSTAEGYVHQEATMNCDTQTQFCEK-K 1194
Query: 190 QHGFNPLFESTTDAEFNRLKSSPPPKFKFLQEAEEKLRRKMQDDNKGINGNEVDNSLI 247
G P + TDA + PPP++K E + K + + + + + + NSL+
Sbjct: 1195 HGGTTPTGTNDTDAPYT--FKQPPPEYKDACECDGKSPQAPKKEEEKKDACTIVNSLL 1250
>UniRef100_Q6FUR9 Similar to tr|Q08206 Saccharomyces cerevisiae YOL036w [Candida
glabrata]
Length = 818
Score = 34.3 bits (77), Expect = 3.7
Identities = 19/69 (27%), Positives = 35/69 (50%), Gaps = 9/69 (13%)
Query: 200 TTDAEFNRLKSSPPPKFKFLQEAEEKLRRKMQDDNK---------GINGNEVDNSLITII 250
+T+ R+ + PP + + +++ LR + D+N G NG+ DNS++ +
Sbjct: 576 STNGSSARISNKPPAEVVPSRHSQDFLRHTVPDNNSTIDNYTLRNGSNGDNSDNSVLYMA 635
Query: 251 VDKKYEREV 259
VDK+Y V
Sbjct: 636 VDKRYNGAV 644
>UniRef100_P36016 Heat shock protein 70 homolog LHS1 precursor [Saccharomyces
cerevisiae]
Length = 881
Score = 34.3 bits (77), Expect = 3.7
Identities = 18/58 (31%), Positives = 32/58 (55%), Gaps = 1/58 (1%)
Query: 196 LFESTTDAEFNRLKSSPPPKFKFLQEAEEKLRRKMQDDNKGINGNEVDNSLITIIVDK 253
L +S ++ N +K F+ + EEKL+RK++ + K N NE ++++I DK
Sbjct: 801 LIKSGDESRLNEIKKLHLRNFRLQKRKEEKLKRKLEQE-KSRNNNETESTVINSADDK 857
>UniRef100_UPI0000363B2F UPI0000363B2F UniRef100 entry
Length = 987
Score = 33.5 bits (75), Expect = 6.3
Identities = 16/72 (22%), Positives = 41/72 (56%), Gaps = 6/72 (8%)
Query: 194 NPLFESTTDAEFNR-LKSSPPPKFKFLQEAEEKLRRKMQDDNKGINGNEVDNSLITIIVD 252
N +F + DA+F ++S+P PK ++ +++ R + + K +E+ + ++ +++
Sbjct: 514 NCIFTAGEDAQFTCVIQSAPSPKIRWFKDS-----RLLTEKEKYQTYSELRSGVLVLVIK 568
Query: 253 KKYEREVNHHQC 264
ER++ H++C
Sbjct: 569 NLTERDLGHYEC 580
>UniRef100_Q80N15 Structural glycoprotein [Yellow head virus]
Length = 1666
Score = 33.5 bits (75), Expect = 6.3
Identities = 17/42 (40%), Positives = 27/42 (63%), Gaps = 2/42 (4%)
Query: 74 PPELCSSMRINDNFVHDPQVQTSKEFLFKPYGEDVIDADYMM 115
PP+LC + RIN + +H + S E+LFKP G+ + DY++
Sbjct: 1211 PPKLCDA-RINWSCLHTGSCKNSSEYLFKPLGQHTSN-DYIV 1250
>UniRef100_UPI0000435D5D UPI0000435D5D UniRef100 entry
Length = 369
Score = 33.1 bits (74), Expect = 8.3
Identities = 19/61 (31%), Positives = 34/61 (55%), Gaps = 3/61 (4%)
Query: 217 KFLQEAEEKLRRKMQDDNKGINGNEVDNSLITIIVDKKYEREVN-HHQCHLQQYHSSTSQ 275
+F EA + + R+M DD G G EV+ S+ II D + +++ N H + H + H + +
Sbjct: 2 RFSLEALQHIHREMDDDQDG--GIEVEESVEFIIEDMQQQQQANKHSKLHHEDQHITLEE 59
Query: 276 V 276
+
Sbjct: 60 L 60
>UniRef100_UPI00003A9BC4 UPI00003A9BC4 UniRef100 entry
Length = 588
Score = 33.1 bits (74), Expect = 8.3
Identities = 20/45 (44%), Positives = 27/45 (59%), Gaps = 1/45 (2%)
Query: 11 LSIVFGCLLLALVAEVYYLLWWKK-KIIKREIENDYGNGNPLKEL 54
+ IV G LL+ALVA V Y L+ KK K + ++ D GN K+L
Sbjct: 535 VGIVVGLLLVALVAGVVYWLYVKKSKTASKHVDKDLGNIEENKKL 579
>UniRef100_Q8DV28 Hypothetical protein SMU.689 [Streptococcus mutans]
Length = 979
Score = 33.1 bits (74), Expect = 8.3
Identities = 14/51 (27%), Positives = 28/51 (54%), Gaps = 1/51 (1%)
Query: 231 QDDNKGINGNEVDNSLITIIVD-KKYEREVNHHQCHLQQYHSSTSQVLPLA 280
QDD K N + D+ +++D K ++ ++ H++ H+ Y + T + LA
Sbjct: 310 QDDIKWYNARKADDGSYKVLIDTKNHKNDLGHYEAHIYGYSTVTQSQIGLA 360
>UniRef100_Q9S706 Hypothetical protein [Oryza sativa]
Length = 733
Score = 33.1 bits (74), Expect = 8.3
Identities = 36/144 (25%), Positives = 57/144 (39%), Gaps = 19/144 (13%)
Query: 134 EESKEDLESEDGKCGKDSRGRSLGDLLDVETPYLTPIASPHFFTPMNCSPYCSPYNQ--- 190
+ +ED ES + K KD + + V+ ++ P+ H F P+ C+P P NQ
Sbjct: 247 DPKREDPESLEFKIMKDRMYKIFSSGIVVDYSHILPMVPYHAFNPLLCNP--KPTNQTGS 304
Query: 191 ----------HGFNPLFESTTDAEFNRLKSSPPPKFKFLQEAEEKLR---RKMQD-DNKG 236
G + + +A + K + PPK K K+R RK D D G
Sbjct: 305 RKRKLVLSNDEGDDAEKAGSKEAAGKQPKPANPPKKKTSSRPIPKIRKSSRKPSDIDPSG 364
Query: 237 INGNEVDNSLITIIVDKKYEREVN 260
+ N + + I D+ E N
Sbjct: 365 KDPNPTNKETGSAIADEAGPTEEN 388
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.319 0.137 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 520,659,227
Number of Sequences: 2790947
Number of extensions: 23593289
Number of successful extensions: 55216
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 5
Number of HSP's successfully gapped in prelim test: 19
Number of HSP's that attempted gapping in prelim test: 55188
Number of HSP's gapped (non-prelim): 27
length of query: 281
length of database: 848,049,833
effective HSP length: 126
effective length of query: 155
effective length of database: 496,390,511
effective search space: 76940529205
effective search space used: 76940529205
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 74 (33.1 bits)
Medicago: description of AC147536.6


