

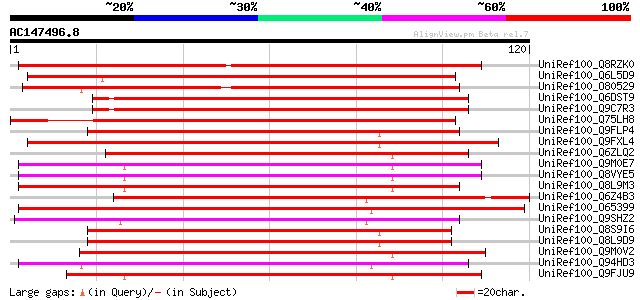
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147496.8 + phase: 1 /partial
(120 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8RZK0 Beta 1,3-glucanase-like [Oryza sativa] 142 1e-33
UniRef100_Q6L5D9 Hypothetical protein OJ1651_G11.6 [Oryza sativa] 139 1e-32
UniRef100_O80529 F14J9.12 protein [Arabidopsis thaliana] 134 5e-31
UniRef100_Q6DST9 Hypothetical protein [Arabidopsis thaliana] 127 4e-29
UniRef100_Q9C7R3 Beta-1,3 glucanase, putative; 26636-27432 [Arab... 127 4e-29
UniRef100_Q75LH8 Putative glucanase [Oryza sativa] 117 6e-26
UniRef100_Q9FLP4 Beta-1,3-glucanase-like protein [Arabidopsis th... 102 3e-21
UniRef100_Q9FXL4 Elicitor inducible beta-1,3-glucanase NtEIG-E76... 101 4e-21
UniRef100_Q6ZLQ2 Putative glycosyl hydrolase [Oryza sativa] 101 4e-21
UniRef100_Q9M0E7 Beta-1, 3-glucanase-like protein [Arabidopsis t... 100 1e-20
UniRef100_Q8VYE5 Putative beta-1,3-glucanase [Arabidopsis thaliana] 100 1e-20
UniRef100_Q8L9M3 Beta-1,3-glucanase-like protein [Arabidopsis th... 99 2e-20
UniRef100_Q6Z4B3 Putative beta-1,3-glucanase [Oryza sativa] 99 2e-20
UniRef100_O65399 Putative glucan endo-1,3-beta-glucosidase 1 pre... 99 2e-20
UniRef100_Q9SHZ2 Putative beta-1,3-glucanase [Arabidopsis thaliana] 99 3e-20
UniRef100_Q8S9I6 AT5g55180/MCO15_13 [Arabidopsis thaliana] 99 3e-20
UniRef100_Q8L9D9 Beta-1,3-glucanase-like protein [Arabidopsis th... 99 3e-20
UniRef100_Q9M0V2 Hypothetical protein AT4g05430 [Arabidopsis tha... 99 3e-20
UniRef100_Q94HD3 Hypothetical protein OSJNBa0045C13.18 [Oryza sa... 99 3e-20
UniRef100_Q9FJU9 Beta-1,3-glucanase-like protein [Arabidopsis th... 98 4e-20
>UniRef100_Q8RZK0 Beta 1,3-glucanase-like [Oryza sativa]
Length = 279
Score = 142 bits (359), Expect = 1e-33
Identities = 67/107 (62%), Positives = 77/107 (71%), Gaps = 1/107 (0%)
Query: 3 SPSSTVPITAPSTSNSPVSSGASWCIASPSASQRSLQVALDYACGYGGTDCSAIQPGGSC 62
S S P A + G SWC+AS SAS +LQVALDYACGYG DCSAIQPGGSC
Sbjct: 69 SSPSAFPSLATAGGGGGGGGGGSWCVASQSASPTALQVALDYACGYGA-DCSAIQPGGSC 127
Query: 63 YNPNSVHDHASFAFNKYYQKNPVPNSCNFGGNAVLTNTNPSKASTIY 109
+NP++VHDHAS+AFN YYQKNPV SC+FGG A +TNT+PS S Y
Sbjct: 128 FNPDTVHDHASYAFNSYYQKNPVATSCDFGGTATITNTDPSSGSCQY 174
>UniRef100_Q6L5D9 Hypothetical protein OJ1651_G11.6 [Oryza sativa]
Length = 281
Score = 139 bits (350), Expect = 1e-32
Identities = 65/111 (58%), Positives = 76/111 (67%), Gaps = 12/111 (10%)
Query: 5 SSTVPITAPSTSNSPV------------SSGASWCIASPSASQRSLQVALDYACGYGGTD 52
S+T P P T +P G SWC+ASPSAS +LQVALDYACG GG D
Sbjct: 76 STTNPAAMPGTQTTPSLANPVAAGGGGGGGGGSWCVASPSASTAALQVALDYACGQGGVD 135
Query: 53 CSAIQPGGSCYNPNSVHDHASFAFNKYYQKNPVPNSCNFGGNAVLTNTNPS 103
CSAIQ GG C+NPN+V DHASFAFN YYQKNPV SC+F G A+LT+T+P+
Sbjct: 136 CSAIQSGGGCFNPNTVRDHASFAFNSYYQKNPVQTSCDFAGTAILTSTDPN 186
>UniRef100_O80529 F14J9.12 protein [Arabidopsis thaliana]
Length = 213
Score = 134 bits (337), Expect = 5e-31
Identities = 66/106 (62%), Positives = 74/106 (69%), Gaps = 7/106 (6%)
Query: 4 PSSTVPITAPST-----SNSPVSSGASWCIASPSASQRSLQVALDYACGYGGTDCSAIQP 58
PS TVP+ P SNSP SG SWC+A P ASQ SLQ ALDYACG DCS +Q
Sbjct: 110 PSGTVPVPVPVVAPPVVSNSPSVSGQSWCVAKPGASQVSLQQALDYACGIA--DCSQLQQ 167
Query: 59 GGSCYNPNSVHDHASFAFNKYYQKNPVPNSCNFGGNAVLTNTNPSK 104
GG+CY+P S+ HASFAFN YYQKNP P SC+FGG A L NTNPS+
Sbjct: 168 GGNCYSPISLQSHASFAFNSYYQKNPSPQSCDFGGAASLVNTNPSE 213
>UniRef100_Q6DST9 Hypothetical protein [Arabidopsis thaliana]
Length = 315
Score = 127 bits (320), Expect = 4e-29
Identities = 59/87 (67%), Positives = 67/87 (76%), Gaps = 1/87 (1%)
Query: 20 VSSGASWCIASPSASQRSLQVALDYACGYGGTDCSAIQPGGSCYNPNSVHDHASFAFNKY 79
V SG WCIA +AS SLQVALDYACGYGG DC IQ G +CY PN++ DHASFAFN Y
Sbjct: 143 VGSG-QWCIAKANASPTSLQVALDYACGYGGADCGQIQQGAACYEPNTIRDHASFAFNSY 201
Query: 80 YQKNPVPNSCNFGGNAVLTNTNPSKAS 106
YQK+P +SCNFGG A LT+T+PSK S
Sbjct: 202 YQKHPGSDSCNFGGAAQLTSTDPSKGS 228
>UniRef100_Q9C7R3 Beta-1,3 glucanase, putative; 26636-27432 [Arabidopsis thaliana]
Length = 228
Score = 127 bits (320), Expect = 4e-29
Identities = 59/87 (67%), Positives = 67/87 (76%), Gaps = 1/87 (1%)
Query: 20 VSSGASWCIASPSASQRSLQVALDYACGYGGTDCSAIQPGGSCYNPNSVHDHASFAFNKY 79
V SG WCIA +AS SLQVALDYACGYGG DC IQ G +CY PN++ DHASFAFN Y
Sbjct: 143 VGSG-QWCIAKANASPTSLQVALDYACGYGGADCGQIQQGAACYEPNTIRDHASFAFNSY 201
Query: 80 YQKNPVPNSCNFGGNAVLTNTNPSKAS 106
YQK+P +SCNFGG A LT+T+PSK S
Sbjct: 202 YQKHPGSDSCNFGGAAQLTSTDPSKTS 228
>UniRef100_Q75LH8 Putative glucanase [Oryza sativa]
Length = 594
Score = 117 bits (293), Expect = 6e-26
Identities = 51/103 (49%), Positives = 67/103 (64%), Gaps = 10/103 (9%)
Query: 1 TYSPSSTVPITAPSTSNSPVSSGASWCIASPSASQRSLQVALDYACGYGGTDCSAIQPGG 60
TY P +T+P + G +WC+A +LQ LDYACG GG DC+AIQP G
Sbjct: 362 TYQPPATMP----------AAGGQTWCVAKAGLMDAALQSGLDYACGMGGADCTAIQPMG 411
Query: 61 SCYNPNSVHDHASFAFNKYYQKNPVPNSCNFGGNAVLTNTNPS 103
+CYNPN++ HAS+AFN Y+Q+NP P SC+FGG +L N NP+
Sbjct: 412 ACYNPNTLQAHASYAFNSYFQRNPSPASCDFGGAGMLVNINPT 454
>UniRef100_Q9FLP4 Beta-1,3-glucanase-like protein [Arabidopsis thaliana]
Length = 471
Score = 102 bits (253), Expect = 3e-21
Identities = 46/87 (52%), Positives = 57/87 (64%), Gaps = 1/87 (1%)
Query: 19 PVSSGASWCIASPSASQRSLQVALDYACGYGGTDCSAIQPGGSCYNPNSVHDHASFAFNK 78
P G +WC+A+ ++ LQ LDYACG GG DC IQPG +CYNP S+ HAS+AFN
Sbjct: 365 PSLVGQTWCVANGKTTKEKLQEGLDYACGEGGADCRPIQPGATCYNPESLEAHASYAFNS 424
Query: 79 YYQKNP-VPNSCNFGGNAVLTNTNPSK 104
YYQKN +CNFGG A + + PSK
Sbjct: 425 YYQKNARGVGTCNFGGAAYVVSQPPSK 451
>UniRef100_Q9FXL4 Elicitor inducible beta-1,3-glucanase NtEIG-E76 [Nicotiana tabacum]
Length = 467
Score = 101 bits (251), Expect = 4e-21
Identities = 46/110 (41%), Positives = 67/110 (60%), Gaps = 1/110 (0%)
Query: 5 SSTVPITAPSTSNSPVSSGASWCIASPSASQRSLQVALDYACGYGGTDCSAIQPGGSCYN 64
S + +TAP + G +WC+A+ A++ LQ ALDYACG GG DC IQ G +CY+
Sbjct: 358 SKSTVVTAPEPALEASKVGNTWCVANEKAAREKLQAALDYACGEGGADCRPIQQGATCYD 417
Query: 65 PNSVHDHASFAFNKYYQKNP-VPNSCNFGGNAVLTNTNPSKASTIYDHGF 113
P+++ HAS+AFN YYQKN ++C+F G A + +P S + G+
Sbjct: 418 PDTLEAHASYAFNSYYQKNTRGVSTCDFSGAAYVVTQHPKYGSCKFPTGY 467
>UniRef100_Q6ZLQ2 Putative glycosyl hydrolase [Oryza sativa]
Length = 129
Score = 101 bits (251), Expect = 4e-21
Identities = 44/85 (51%), Positives = 58/85 (67%), Gaps = 1/85 (1%)
Query: 23 GASWCIASPSASQRSLQVALDYACGYGGTDCSAIQPGGSCYNPNSVHDHASFAFNKYYQK 82
G +WC+A P SQ LQ ALD+ACG GG DC+ +QPGG CY P+++ HAS+AFN +YQ+
Sbjct: 6 GVTWCVARPGVSQEDLQNALDWACGQGGADCTPLQPGGRCYQPDTLLSHASYAFNIFYQQ 65
Query: 83 NPVPN-SCNFGGNAVLTNTNPSKAS 106
N + +CNFGG + NPS S
Sbjct: 66 NGNSDIACNFGGAGTIIKRNPSFGS 90
>UniRef100_Q9M0E7 Beta-1, 3-glucanase-like protein [Arabidopsis thaliana]
Length = 512
Score = 99.8 bits (247), Expect = 1e-20
Identities = 51/122 (41%), Positives = 73/122 (59%), Gaps = 15/122 (12%)
Query: 3 SPSSTVPITAPSTSNSPVSSGAS--------------WCIASPSASQRSLQVALDYACGY 48
SP+++ T+PS S+SP+ +G S WCIAS AS LQ ALD+ACG
Sbjct: 332 SPTNSTTGTSPSPSSSPIINGNSTVTIGGGGGGGTKKWCIASSQASVTELQTALDWACGP 391
Query: 49 GGTDCSAIQPGGSCYNPNSVHDHASFAFNKYYQKNPVPN-SCNFGGNAVLTNTNPSKAST 107
G DCSA+QP C+ P++V HAS+AFN YYQ++ + C+F G +V + +PS +
Sbjct: 392 GNVDCSAVQPDQPCFEPDTVLSHASYAFNTYYQQSGASSIDCSFNGASVEVDKDPSYGNC 451
Query: 108 IY 109
+Y
Sbjct: 452 LY 453
>UniRef100_Q8VYE5 Putative beta-1,3-glucanase [Arabidopsis thaliana]
Length = 534
Score = 99.8 bits (247), Expect = 1e-20
Identities = 51/122 (41%), Positives = 73/122 (59%), Gaps = 15/122 (12%)
Query: 3 SPSSTVPITAPSTSNSPVSSGAS--------------WCIASPSASQRSLQVALDYACGY 48
SP+++ T+PS S+SP+ +G S WCIAS AS LQ ALD+ACG
Sbjct: 354 SPTNSTTGTSPSPSSSPIINGNSTVTIGGGGGGGTKKWCIASSQASVTELQTALDWACGP 413
Query: 49 GGTDCSAIQPGGSCYNPNSVHDHASFAFNKYYQKNPVPN-SCNFGGNAVLTNTNPSKAST 107
G DCSA+QP C+ P++V HAS+AFN YYQ++ + C+F G +V + +PS +
Sbjct: 414 GNVDCSAVQPDQPCFEPDTVLSHASYAFNTYYQQSGASSIDCSFNGASVEVDKDPSYGNC 473
Query: 108 IY 109
+Y
Sbjct: 474 LY 475
>UniRef100_Q8L9M3 Beta-1,3-glucanase-like protein [Arabidopsis thaliana]
Length = 488
Score = 99.0 bits (245), Expect = 2e-20
Identities = 51/117 (43%), Positives = 71/117 (60%), Gaps = 15/117 (12%)
Query: 3 SPSSTVPITAPSTSNSPVSSGAS--------------WCIASPSASQRSLQVALDYACGY 48
SP+++ T+PS S+SP+ +G S WCIAS AS LQ ALD+ACG
Sbjct: 354 SPTNSTTGTSPSPSSSPIINGNSTVTIGGGGGGGTKKWCIASSQASVTELQTALDWACGP 413
Query: 49 GGTDCSAIQPGGSCYNPNSVHDHASFAFNKYYQKNPVPN-SCNFGGNAVLTNTNPSK 104
G DCSA+QP C+ P++V HAS+AFN YYQ++ + C+F G +V + +PSK
Sbjct: 414 GNVDCSAVQPDQPCFEPDTVLSHASYAFNTYYQQSGASSIDCSFNGASVEVDKDPSK 470
>UniRef100_Q6Z4B3 Putative beta-1,3-glucanase [Oryza sativa]
Length = 540
Score = 99.0 bits (245), Expect = 2e-20
Identities = 49/97 (50%), Positives = 65/97 (66%), Gaps = 2/97 (2%)
Query: 25 SWCIASPSASQRSLQVALDYACGYGGTDCSAIQPGGSCYNPNSVHDHASFAFNKYYQ-KN 83
++CIAS A ++++Q A+D+ACG G TDC+AIQPG CY PN V HASFAF+ YYQ +
Sbjct: 391 TFCIASDDADEKAVQAAMDWACGPGRTDCTAIQPGQGCYEPNDVRSHASFAFDSYYQSQG 450
Query: 84 PVPNSCNFGGNAVLTNTNPSKASTIYDHGFKLQSDTT 120
SC F G ++T T+PS S I+ G KL S+ T
Sbjct: 451 KAAGSCYFQGVGMVTTTDPSHDSCIFP-GSKLLSNVT 486
>UniRef100_O65399 Putative glucan endo-1,3-beta-glucosidase 1 precursor (EC 3.2.1.39)
((1->3)-beta-glucan endohydrolase)
((1->3)-beta-glucanase) [Arabidopsis thaliana]
Length = 402
Score = 99.0 bits (245), Expect = 2e-20
Identities = 50/118 (42%), Positives = 72/118 (60%), Gaps = 1/118 (0%)
Query: 3 SPSSTVPITAPSTSNSPVSSGASWCIASPSASQRSLQVALDYACGYGGTDCSAIQPGGSC 62
+P + ++ T + ++ ++CIA ++LQ ALD+ACG G ++CS IQPG SC
Sbjct: 249 TPVYLLHVSGSGTFLANDTTNQTYCIAMDGVDAKTLQAALDWACGPGRSNCSEIQPGESC 308
Query: 63 YNPNSVHDHASFAFNKYYQK-NPVPNSCNFGGNAVLTNTNPSKASTIYDHGFKLQSDT 119
Y PN+V HASFAFN YYQK SC+F G A++T T+PS S I+ K+ + T
Sbjct: 309 YQPNNVKGHASFAFNSYYQKEGRASGSCDFKGVAMITTTDPSHGSCIFPGSKKVGNRT 366
>UniRef100_Q9SHZ2 Putative beta-1,3-glucanase [Arabidopsis thaliana]
Length = 473
Score = 98.6 bits (244), Expect = 3e-20
Identities = 49/112 (43%), Positives = 61/112 (53%), Gaps = 9/112 (8%)
Query: 2 YSPSSTVPITAPSTSNSPVSSGA--------SWCIASPSASQRSLQVALDYACGYGGTDC 53
Y P+T P SG +WC+A+ A + LQ LDYACG GG DC
Sbjct: 352 YRDGGHTPVTGGDQVTKPPMSGGVSKSLNGYTWCVANGDAGEERLQGGLDYACGEGGADC 411
Query: 54 SAIQPGGSCYNPNSVHDHASFAFNKYYQ-KNPVPNSCNFGGNAVLTNTNPSK 104
IQPG +CY+P+++ HASFAFN YYQ K SC FGG A + + PSK
Sbjct: 412 RPIQPGANCYSPDTLEAHASFAFNSYYQKKGRAGGSCYFGGAAYVVSQPPSK 463
>UniRef100_Q8S9I6 AT5g55180/MCO15_13 [Arabidopsis thaliana]
Length = 460
Score = 98.6 bits (244), Expect = 3e-20
Identities = 44/85 (51%), Positives = 55/85 (63%), Gaps = 1/85 (1%)
Query: 19 PVSSGASWCIASPSASQRSLQVALDYACGYGGTDCSAIQPGGSCYNPNSVHDHASFAFNK 78
P G +WC+A+ ++ LQ LDYACG GG DC IQPG +CYNP S+ HAS+AFN
Sbjct: 365 PSLVGQTWCVANGKTTKEKLQEGLDYACGEGGADCRPIQPGATCYNPESLEAHASYAFNS 424
Query: 79 YYQKNP-VPNSCNFGGNAVLTNTNP 102
YYQKN +CNFGG A + + P
Sbjct: 425 YYQKNARGVGTCNFGGAAYVVSQPP 449
>UniRef100_Q8L9D9 Beta-1,3-glucanase-like protein [Arabidopsis thaliana]
Length = 460
Score = 98.6 bits (244), Expect = 3e-20
Identities = 44/85 (51%), Positives = 55/85 (63%), Gaps = 1/85 (1%)
Query: 19 PVSSGASWCIASPSASQRSLQVALDYACGYGGTDCSAIQPGGSCYNPNSVHDHASFAFNK 78
P G +WC+A+ ++ LQ LDYACG GG DC IQPG +CYNP S+ HAS+AFN
Sbjct: 365 PSLVGQTWCVANGKTTKEKLQEGLDYACGEGGADCRPIQPGATCYNPESLEAHASYAFNS 424
Query: 79 YYQKNP-VPNSCNFGGNAVLTNTNP 102
YYQKN +CNFGG A + + P
Sbjct: 425 YYQKNARGVGTCNFGGAAYVVSQPP 449
>UniRef100_Q9M0V2 Hypothetical protein AT4g05430 [Arabidopsis thaliana]
Length = 119
Score = 98.6 bits (244), Expect = 3e-20
Identities = 46/95 (48%), Positives = 58/95 (60%), Gaps = 1/95 (1%)
Query: 17 NSPVSSGASWCIASPSASQRSLQVALDYACGYGGTDCSAIQPGGSCYNPNSVHDHASFAF 76
N+ G +WC+A P A+Q LQ ALD+ACG G DCS I+ G CY P+++ HASFAF
Sbjct: 14 NTTFLEGTTWCVARPGATQAELQRALDWACGIGRVDCSVIERHGDCYEPDTIVSHASFAF 73
Query: 77 NKYYQKNPVPN-SCNFGGNAVLTNTNPSKASTIYD 110
N YYQ N +C FGG A T NP++ S D
Sbjct: 74 NAYYQTNGNNRIACYFGGTATFTKINPNRKSPPQD 108
>UniRef100_Q94HD3 Hypothetical protein OSJNBa0045C13.18 [Oryza sativa]
Length = 345
Score = 98.6 bits (244), Expect = 3e-20
Identities = 49/108 (45%), Positives = 64/108 (58%), Gaps = 4/108 (3%)
Query: 3 SPSSTVPITAPST---SNSPVSSGASWCIASPSASQRSLQVALDYACGYGGTDCSAIQPG 59
+P S P A S S + G WC+A P+ LQ A+DYAC G DC I G
Sbjct: 231 APCSAPPTAAMSPQPCSGEGGNGGGQWCVAKPTVPLDRLQEAMDYACSQDGVDCQEISGG 290
Query: 60 GSCYNPNSVHDHASFAFNKYYQK-NPVPNSCNFGGNAVLTNTNPSKAS 106
GSC+ P+++ HAS+AFN Y+QK + SC+FGG AVL N++PS AS
Sbjct: 291 GSCFYPDNIAAHASYAFNSYWQKMKHIGGSCSFGGTAVLINSDPSMAS 338
>UniRef100_Q9FJU9 Beta-1,3-glucanase-like protein [Arabidopsis thaliana]
Length = 506
Score = 98.2 bits (243), Expect = 4e-20
Identities = 49/99 (49%), Positives = 65/99 (65%), Gaps = 3/99 (3%)
Query: 14 STSNSPVSSGAS--WCIASPSASQRSLQVALDYACGYGGTDCSAIQPGGSCYNPNSVHDH 71
S S+ SSG+S WCIAS AS+R L+ ALD+ACG G DC+AIQP C+ P+++ H
Sbjct: 355 SNSSGTNSSGSSNSWCIASSKASERDLKGALDWACGPGNVDCTAIQPSQPCFQPDTLVSH 414
Query: 72 ASFAFNKYYQKNPVPN-SCNFGGNAVLTNTNPSKASTIY 109
ASF FN Y+Q+N + +C+FGG V N +PS IY
Sbjct: 415 ASFVFNSYFQQNRATDVACSFGGAGVKVNKDPSYDKCIY 453
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.310 0.125 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 233,347,572
Number of Sequences: 2790947
Number of extensions: 9884756
Number of successful extensions: 22272
Number of sequences better than 10.0: 215
Number of HSP's better than 10.0 without gapping: 126
Number of HSP's successfully gapped in prelim test: 89
Number of HSP's that attempted gapping in prelim test: 21905
Number of HSP's gapped (non-prelim): 278
length of query: 120
length of database: 848,049,833
effective HSP length: 96
effective length of query: 24
effective length of database: 580,118,921
effective search space: 13922854104
effective search space used: 13922854104
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 67 (30.4 bits)
Medicago: description of AC147496.8


