

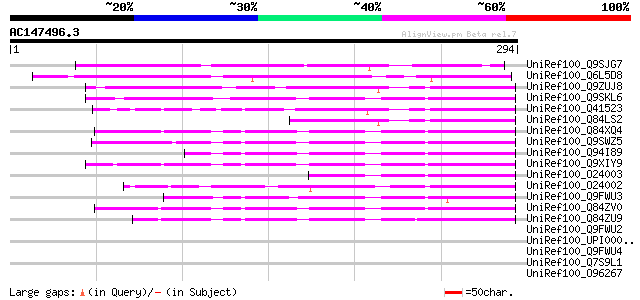
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147496.3 - phase: 0
(294 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9SJG7 Hypothetical protein At2g42900 [Arabidopsis tha... 164 3e-39
UniRef100_Q6L5D8 Hypothetical protein OJ1651_G11.7 [Oryza sativa] 149 7e-35
UniRef100_Q9ZUJ8 Hypothetical protein At2g15130 [Arabidopsis tha... 76 9e-13
UniRef100_Q9SKL6 Expressed protein [Arabidopsis thaliana] 69 1e-10
UniRef100_Q41523 Hypothetical protein [Triticum aestivum] 69 1e-10
UniRef100_Q84LS2 Hypothetical protein [Arabidopsis thaliana] 62 1e-08
UniRef100_Q84XQ4 NtPRp27-like protein [Solanum tuberosum] 61 3e-08
UniRef100_Q9SWZ5 Secretory protein [Triticum aestivum] 59 2e-07
UniRef100_Q94I89 Putative NtPRp27-like protein [Atropa belladonna] 57 6e-07
UniRef100_Q9XIY9 NtPRp27 [Nicotiana tabacum] 56 1e-06
UniRef100_O24003 Hypothetical protein precursor [Hordeum vulgare] 55 2e-06
UniRef100_O24002 Hypothetical protein precursor [Hordeum vulgare] 54 4e-06
UniRef100_Q9FWU3 Putative secretory protein [Oryza sativa] 54 6e-06
UniRef100_Q84ZV0 R 14 protein [Glycine max] 50 5e-05
UniRef100_Q84ZU9 R 13 protein [Glycine max] 50 5e-05
UniRef100_Q9FWU2 Putative secretory protein [Oryza sativa] 46 0.001
UniRef100_UPI000021A836 UPI000021A836 UniRef100 entry 40 0.073
UniRef100_Q9FWU4 Putative basic secretory protein [Oryza sativa] 39 0.13
UniRef100_Q7S9L1 Hypothetical protein [Neurospora crassa] 39 0.13
UniRef100_O96267 Hypothetical protein PFB0875c [Plasmodium falci... 39 0.16
>UniRef100_Q9SJG7 Hypothetical protein At2g42900 [Arabidopsis thaliana]
Length = 276
Score = 164 bits (414), Expect = 3e-39
Identities = 98/252 (38%), Positives = 141/252 (55%), Gaps = 26/252 (10%)
Query: 39 NNNNIIHRILLILFVAIISIWANYEASKTFDIHIVNDTKDSLAGRRFTLFYVSNDKASRI 98
+++ II R+ IL V IS+WAN+EASK F I I+ND KDS +G+RF LF+ S+D A RI
Sbjct: 26 SDSGIILRLFSILLVGAISLWANHEASKGFSISIINDAKDSPSGKRFALFFESDDTAVRI 85
Query: 99 LLNTSSFVEQILYPNGNNDNIDIKNKKIIKSVTLRLARQNLDTITTITADE-KNINSYVI 157
LL+ S FVE+ LY + + + +K + VT++ + D + + + YVI
Sbjct: 86 LLDASFFVERFLY-----EGVPHRLRKPVNHVTVQFCGNSSDRVDRFSVTSGASHGEYVI 140
Query: 158 EISPMLLEDENFNNMAIVGAIQRAMARVWLWDGRSKASPRLLDGMVEYIA--ELAGFHRE 215
+SP L E + F+N A+ A++R+M R+WLW S ASP L+ GMVEY+A H E
Sbjct: 141 RLSPSLTERKKFSN-AVESALRRSMVRIWLWGDESGASPELVAGMVEYLAVESRKRRHFE 199
Query: 216 RLSGGVGESPECEDGRELWWDNKDPTHVARLLHYCEKYDKGFIQRLNEAMKDTWHDRMVG 275
+ G W D + +V LL YCE+ +GFI+RLN M+ W DR V
Sbjct: 200 KFGGN-------------WKDKEKSVYVVSLLDYCERRSEGFIRRLNHGMRLRWDDRTV- 245
Query: 276 DVLGLKATKLCG 287
L ++ CG
Sbjct: 246 ---DLASSGACG 254
>UniRef100_Q6L5D8 Hypothetical protein OJ1651_G11.7 [Oryza sativa]
Length = 297
Score = 149 bits (377), Expect = 7e-35
Identities = 103/284 (36%), Positives = 151/284 (52%), Gaps = 29/284 (10%)
Query: 14 LPHFNPSSYEINQQNDYNNSKFFIPNNNNIIHRILLILFVAIISIWANYEASKTFDIHIV 73
LP SS D ++S P+ +I+ R++ +L VA +S++A +EASK F + IV
Sbjct: 7 LPLATASSSSSGATADESSSS---PSTASIVARVVAVLAVASVSLFARHEASKGFHVDIV 63
Query: 74 NDT-KDSLAGRRFTLFYVSNDKASRILLNTSSFVEQILYPNGNNDNIDIKNKKIIKSVTL 132
N +D++AGRRF LF+VSN KA RIL + VE L+P+ + +K + VT+
Sbjct: 64 NAAPRDTVAGRRFDLFFVSNGKAERILHYANRGVEAALFPDASFP------RKQVTRVTV 117
Query: 133 RLARQNL--DTITTITADEKNINSYVIEISPMLLEDENFNNM-AIVGAIQRAMARVWLWD 189
R+A NL D T+ A YVI +SP L+ + A+ A++RA+AR+WLWD
Sbjct: 118 RMAGHNLTDDDDATVIAAGVAPGEYVISLSPRLMHPASDKAADAVANAVRRAVARMWLWD 177
Query: 190 GRSKASPRLLDGMVEYIAELAGFHRERLSGGVGESPECEDGRELWWDNKDPTHV--ARLL 247
GR A R+ + MV+Y++ A V SP +D D+ D H AR L
Sbjct: 178 GRGAAPARVTEAMVDYLSSGA--------DAVEASPANDD------DDDDAPHCMSARFL 223
Query: 248 HYCEKYDKGFIQRLNEAMKDTWHDRMVGDVLGLKATKLCGLYNA 291
+ E+ GF+ RLN AM+D W D + LG+ A +C Y A
Sbjct: 224 GHLERRRGGFVARLNRAMRDRWSDAAMDAALGVPARPVCAAYRA 267
>UniRef100_Q9ZUJ8 Hypothetical protein At2g15130 [Arabidopsis thaliana]
Length = 225
Score = 76.3 bits (186), Expect = 9e-13
Identities = 61/252 (24%), Positives = 102/252 (40%), Gaps = 35/252 (13%)
Query: 45 HRILLILFVAIISIWANYEASKTFDIHIVNDTKDSLAGRRFTLFYVSNDKASRILLNTSS 104
H+I L+ IS+ D +V++T DS GRRF + L + +
Sbjct: 4 HKIFLV-----ISLMLAVSLVSAVDFSVVDNTGDSPGGRRFRNEIGGVSYGEQSLRDATD 58
Query: 105 FVEQILYPNGNNDNIDIKNKKIIKSVTLRLARQNLDTITTITADEKNINSYVIEISPMLL 164
F ++ +D D+ + L +N + I + DE + N+ L+
Sbjct: 59 FTWRLFQQTNPSDRKDVTK--------ITLFMENSNGIAYSSQDEIHYNA------GSLV 104
Query: 165 EDENFNNMAIVGAIQRAMARVWLWDGRSKASPRLLDGMVEYIAELAGF---HRERLSGGV 221
+D+ + G + + W W+G +A L++G+ +Y+ AG+ H R GG
Sbjct: 105 DDKGYVRRGFTGVVYHEVVHSWQWNGAGRAPGGLIEGIADYVRLKAGYVASHWVRPGGG- 163
Query: 222 GESPECEDGRELWWDNKDPTHVARLLHYCEKYDKGFIQRLNEAMKDTWHDRMVGDVLGLK 281
+ W D T AR L YC GF+ LN+ M+ ++D D+LG
Sbjct: 164 ----------DRWDQGYDVT--ARFLEYCNDLRNGFVAELNKKMRSDYNDGFFVDLLGKD 211
Query: 282 ATKLCGLYNATY 293
+L Y A Y
Sbjct: 212 VNQLWREYKANY 223
>UniRef100_Q9SKL6 Expressed protein [Arabidopsis thaliana]
Length = 225
Score = 69.3 bits (168), Expect = 1e-10
Identities = 61/251 (24%), Positives = 104/251 (41%), Gaps = 33/251 (13%)
Query: 45 HRILLILFVAIISIWANYEASKTFDIHIVNDTKDSLAGRRFTLFYVSNDKASRILLNTSS 104
H+I ++ + ++ N D +V+++ DS GRRF ++ L + +
Sbjct: 4 HKIFFVISLMLVVSLVN-----AVDYSVVDNSGDSTGGRRFRGEIGGISYGTQTLRSATD 58
Query: 105 FVEQILYPNGNNDNIDIKNKKIIKSVT-LRLARQNLDTITTITADEKNIN-SYVIEISPM 162
FV ++ +D KSVT + L +N D + +A+E + N Y+ +S
Sbjct: 59 FVWRLFQQTNPSDR---------KSVTKITLFMENGDGVAYNSANEIHFNVGYLAGVSGD 109
Query: 163 LLEDENFNNMAIVGAIQRAMARVWLWDGRSKASPRLLDGMVEYIAELAGFHRERLSGGVG 222
+ + G + + W W+G +A L++G+ +Y+ RL G
Sbjct: 110 VKRE-------FTGVVYHEVVHSWQWNGAGRAPGGLIEGIADYV---------RLKAGYA 153
Query: 223 ESPECEDGRELWWDNKDPTHVARLLHYCEKYDKGFIQRLNEAMKDTWHDRMVGDVLGLKA 282
S GR WD AR L YC GF+ LN+ M++ + D D+LG
Sbjct: 154 PSHWVGPGRGDRWDQGYDV-TARFLDYCNGLRNGFVAELNKKMRNGYSDGFFVDLLGKDV 212
Query: 283 TKLCGLYNATY 293
+L Y A Y
Sbjct: 213 NQLWREYKAKY 223
>UniRef100_Q41523 Hypothetical protein [Triticum aestivum]
Length = 231
Score = 68.9 bits (167), Expect = 1e-10
Identities = 64/249 (25%), Positives = 109/249 (43%), Gaps = 37/249 (14%)
Query: 49 LILFVAIISIWANYEASKTFDIHIVNDTKDSLAGRRFTLFYVSNDKASRILLNTSSFVEQ 108
L+LF+A+ ++ + TFD N DS G+RF V D A ++L SSF+
Sbjct: 11 LLLFLALAAM----AGAVTFDA--TNTVPDSAGGQRFDQD-VGVDYAKQVLSEASSFI-- 61
Query: 109 ILYPNGNNDNIDIKNKKIIKSVTLRLARQNLDTITTITADEKNINS-YVIEISPMLLEDE 167
+ N N + +++ SVTL + N++ + + + + + YV DE
Sbjct: 62 --WTTFNQPNPE--DRRDYDSVTLAVV-DNIEPVAQTSGNAIQLRAQYVAGF------DE 110
Query: 168 NFNNMAIVGAIQRAMARVWLWDGRSKASPRLLDGMVEYI---AELAGFHRERLSGGVGES 224
+ G + VW W+G+ +A+ L++G+ +Y+ A+LA H G
Sbjct: 111 GDVKKEVKGVLYHEATHVWQWNGQGRANGGLIEGIADYVRLKADLAPTHWRPQGSG---- 166
Query: 225 PECEDGRELWWDNKDPTHVARLLHYCEKYDKGFIQRLNEAMKDTWHDRMVGDVLGLKATK 284
+ W + D T A+ L YC+ GF+ +N +KD + D +LG +
Sbjct: 167 -------DRWDEGYDVT--AKFLDYCDSLKAGFVAEMNSKLKDGYSDDYFVQILGKSVDQ 217
Query: 285 LCGLYNATY 293
L Y A Y
Sbjct: 218 LWNDYKAKY 226
>UniRef100_Q84LS2 Hypothetical protein [Arabidopsis thaliana]
Length = 144
Score = 62.4 bits (150), Expect = 1e-08
Identities = 37/134 (27%), Positives = 59/134 (43%), Gaps = 16/134 (11%)
Query: 163 LLEDENFNNMAIVGAIQRAMARVWLWDGRSKASPRLLDGMVEYIAELAGF---HRERLSG 219
L++D+ + G + + W W+G +A L++G+ +Y+ AG+ H R G
Sbjct: 22 LVDDKGYVRRGFTGVVYHEVVHSWQWNGAGRAPGGLIEGIADYVRLKAGYVASHWVRPGG 81
Query: 220 GVGESPECEDGRELWWDNKDPTHVARLLHYCEKYDKGFIQRLNEAMKDTWHDRMVGDVLG 279
G + W D T AR L YC GF+ LN+ M+ ++D D+LG
Sbjct: 82 G-----------DRWDQGYDVT--ARFLEYCNDLRNGFVAELNKKMRSDYNDGFFVDLLG 128
Query: 280 LKATKLCGLYNATY 293
+L Y A Y
Sbjct: 129 KDVNQLWREYKANY 142
>UniRef100_Q84XQ4 NtPRp27-like protein [Solanum tuberosum]
Length = 226
Score = 61.2 bits (147), Expect = 3e-08
Identities = 54/245 (22%), Positives = 105/245 (42%), Gaps = 26/245 (10%)
Query: 50 ILFVAIISIWANY-EASKTFDIHIVNDTKDSLAGRRFTLFYVSNDKASRILLNTSSFVEQ 108
I F++ + + A + + D + N ++ G RF + + + L+ +SF+
Sbjct: 5 IFFISSLFVLAIFTQKIDAVDYSVTNTAANTPGGARFDRD-IGAQYSQQTLVAATSFIWN 63
Query: 109 ILYPNGNNDNIDIKNKKIIKSVTLRLARQNLDTITTITADEKNINSYVIEISPMLLEDEN 168
I N D +K + V++ + ++D + + +E ++++ I+ + E
Sbjct: 64 IFQQNSPAD------RKNVPKVSMFV--DDMDGVAYASNNEIHVSARYIQGYSGDVRRE- 114
Query: 169 FNNMAIVGAIQRAMARVWLWDGRSKASPRLLDGMVEYIAELAGFHRERLSGGVGESPECE 228
I G + VW W+G A L++G+ +Y+ RL G+G S +
Sbjct: 115 -----ITGVLYHEATHVWQWNGNGGAPGGLIEGIADYV---------RLKAGLGPSHWVK 160
Query: 229 DGRELWWDNKDPTHVARLLHYCEKYDKGFIQRLNEAMKDTWHDRMVGDVLGLKATKLCGL 288
G+ WD A+ L YC GF+ LN+ M++ + D+ D+LG +L
Sbjct: 161 PGQGNRWDQGYDV-TAQFLDYCNSLRNGFVAELNKKMRNGYSDQFFVDLLGKTVDQLWSD 219
Query: 289 YNATY 293
Y A +
Sbjct: 220 YKAKF 224
>UniRef100_Q9SWZ5 Secretory protein [Triticum aestivum]
Length = 224
Score = 58.9 bits (141), Expect = 2e-07
Identities = 56/247 (22%), Positives = 103/247 (41%), Gaps = 28/247 (11%)
Query: 48 LLILFVAIISIWANYEASKTFDIHIVNDTKDSLAGRRFTLFYVSNDKASRILLNTSSFVE 107
L + VA + A A++ N + GRRF V K ++L + S+F+
Sbjct: 3 LQVATVASFVLVALAAAAQAVTFDASNKASGTSGGRRFNQAVVPYSK--KVLSDASAFIW 60
Query: 108 QILYPNGNNDNIDIKNKKIIKSVTLRLARQNLDTITTITADEKNINS-YVIEISPMLLED 166
+ D +K + +VTL + +++ + +A+ ++++ YV IS + ++
Sbjct: 61 KTFNQRAVGD------RKTVDAVTLVV--EDISGVAFTSANGIHLSAQYVGGISGDVKKE 112
Query: 167 ENFNNMAIVGAIQRAMARVWLWDGRSKASPRLLDGMVEYIAELAGFHRERLSGGVGESPE 226
+ G + VW W+G +A+ L++G+ +Y+ RL G
Sbjct: 113 -------VTGVLYHEATHVWQWNGPGQANGGLIEGIADYV---------RLKAGFAPGHW 156
Query: 227 CEDGRELWWDNKDPTHVARLLHYCEKYDKGFIQRLNEAMKDTWHDRMVGDVLGLKATKLC 286
+ G+ WD AR L YC+ GF+ +N MK + D +LG +L
Sbjct: 157 VKPGQGDRWDQGYDV-TARFLDYCDSLKPGFVAHVNAKMKSGYTDDFFAQILGKNVQQLW 215
Query: 287 GLYNATY 293
Y A +
Sbjct: 216 KDYKAKF 222
>UniRef100_Q94I89 Putative NtPRp27-like protein [Atropa belladonna]
Length = 223
Score = 57.0 bits (136), Expect = 6e-07
Identities = 43/192 (22%), Positives = 86/192 (44%), Gaps = 24/192 (12%)
Query: 102 TSSFVEQILYPNGNNDNIDIKNKKIIKSVTLRLARQNLDTITTITADEKNINSYVIEISP 161
++SF+ +I N D +K ++ +++ + ++D + DE ++++ I+
Sbjct: 49 STSFIWRIFEQNSPAD------RKNVQKISMFV--DDMDGVAYAVNDEIHVSARYIQSYS 100
Query: 162 MLLEDENFNNMAIVGAIQRAMARVWLWDGRSKASPRLLDGMVEYIAELAGFHRERLSGGV 221
+ E I G + VW W+G +A L++G+ +Y+ RL G+
Sbjct: 101 GDVRRE------IAGVLYHECTHVWQWNGNGRAPGGLIEGIADYV---------RLKAGL 145
Query: 222 GESPECEDGRELWWDNKDPTHVARLLHYCEKYDKGFIQRLNEAMKDTWHDRMVGDVLGLK 281
S + G+ WD AR L YC GF+ +LN+ M++ + ++ ++LG
Sbjct: 146 APSHWVKPGQGDRWDQGYDV-TARFLDYCNSLKNGFVAQLNKKMRNGYSNQFFVELLGKT 204
Query: 282 ATKLCGLYNATY 293
+L Y A +
Sbjct: 205 VDQLWSDYKAKF 216
>UniRef100_Q9XIY9 NtPRp27 [Nicotiana tabacum]
Length = 242
Score = 56.2 bits (134), Expect = 1e-06
Identities = 53/250 (21%), Positives = 104/250 (41%), Gaps = 29/250 (11%)
Query: 45 HRILLILFVAIISIWANYEASKTFDIHIVNDTKDSLAGRRFTLFYVSNDKASRILLNTSS 104
H+I + +++I+ + D + N ++ G RF + + + L +S
Sbjct: 19 HKIFFFYSLFVLAIFT--QKIHAVDYSVTNTAANTAGGARFNRD-IGAQYSQQTLAAATS 75
Query: 105 FVEQILYPNGNNDNIDIKNKKIIKSVTLRLARQNLDTITTITADEKNIN-SYVIEISPML 163
F+ N D +K ++ V++ + ++D + + +E +++ SY+ S +
Sbjct: 76 FIWNTFQQNFPAD------RKNVQKVSMFV--DDMDGVAYASNNEIHVSASYIQGYSGDV 127
Query: 164 LEDENFNNMAIVGAIQRAMARVWLWDGRSKASPRLLDGMVEYIAELAGFHRERLSGGVGE 223
+ I G + VW W+G A L++G+ +Y+ RL G
Sbjct: 128 RRE-------ITGVLYHESTHVWQWNGNGGAPGGLIEGIADYV---------RLKAGFAP 171
Query: 224 SPECEDGRELWWDNKDPTHVARLLHYCEKYDKGFIQRLNEAMKDTWHDRMVGDVLGLKAT 283
S + G+ WD AR L YC GF+ +LN+ M+ + ++ D+LG
Sbjct: 172 SHWVKPGQGDRWDQGYDV-TARFLDYCNSLRNGFVAQLNKKMRTGYSNQFFIDLLGKTVD 230
Query: 284 KLCGLYNATY 293
+L Y A +
Sbjct: 231 QLWSDYKAKF 240
>UniRef100_O24003 Hypothetical protein precursor [Hordeum vulgare]
Length = 225
Score = 55.1 bits (131), Expect = 2e-06
Identities = 33/120 (27%), Positives = 50/120 (41%), Gaps = 10/120 (8%)
Query: 174 IVGAIQRAMARVWLWDGRSKASPRLLDGMVEYIAELAGFHRERLSGGVGESPECEDGREL 233
+ G + VW W+GR A+ L++G+ +Y+ RL G+ G
Sbjct: 114 VTGVLYHEATHVWQWNGRGTANGGLIEGIADYV---------RLKAGLAPGHWRPQGSGD 164
Query: 234 WWDNKDPTHVARLLHYCEKYDKGFIQRLNEAMKDTWHDRMVGDVLGLKATKLCGLYNATY 293
WD AR L YC+ GF+ +LN MK + D +LG +L Y A +
Sbjct: 165 RWDQGYDI-TARFLDYCDSLMPGFVAQLNAKMKSGYSDDFFAQILGKNVQQLWKDYKAKF 223
>UniRef100_O24002 Hypothetical protein precursor [Hordeum vulgare]
Length = 229
Score = 54.3 bits (129), Expect = 4e-06
Identities = 55/230 (23%), Positives = 97/230 (41%), Gaps = 29/230 (12%)
Query: 67 TFDIHIVNDTKDSLAGRRFTLFYVSNDKASRILLNTSSFVEQILYPNGNNDNIDIKNKKI 126
TFD+ N+ + G+RF Y + A ++L + SSF I + D +++
Sbjct: 23 TFDV--TNEASSTAGGQRFDREYGAA-YAKQVLSDASSFTWGIF------NQPDPSDRRP 73
Query: 127 IKSVTLRLARQNLDTITTITADEKNINSYVIEISPMLLEDENFNNMA--IVGAIQRAMAR 184
T+ LA ++ + I + + IE+S + +N+ + G + +
Sbjct: 74 ADGDTVTLAVRDTNGIASTSGS-------TIELSARSVGGITGDNLKEQVDGVLYHEVVH 126
Query: 185 VWLWDGRSKASPR-LLDGMVEYIAELAGFHRERLSGGVGESPECEDGRELWWDNKDPTHV 243
VW W + + +G+ +Y+ AG+ V + E G W + D T
Sbjct: 127 VWQWGLQDYHEHHGIFEGIADYVRLKAGY--------VAANWVKEGGGSRWDEGYDVT-- 176
Query: 244 ARLLHYCEKYDKGFIQRLNEAMKDTWHDRMVGDVLGLKATKLCGLYNATY 293
AR L YC+ GF+ +N +KD ++D +LG A +L Y A Y
Sbjct: 177 ARFLDYCDSRKPGFVAEMNGKLKDGYNDDYFVQILGTSADQLWNDYKAKY 226
>UniRef100_Q9FWU3 Putative secretory protein [Oryza sativa]
Length = 260
Score = 53.5 bits (127), Expect = 6e-06
Identities = 51/207 (24%), Positives = 87/207 (41%), Gaps = 25/207 (12%)
Query: 90 VSNDKASRILLNTSSFVEQILYPNGNNDNIDIKNKKIIKSVTLRLARQNLDTITTITADE 149
V D A ++L + SSF+ G+ + +K + +VTL + +++D + + D
Sbjct: 74 VGVDYAKQMLADASSFIWDTFEQPGDGGD-----RKPVDAVTLTV--EDIDGVAFTSGDG 126
Query: 150 KNINS-YVIEISPMLLEDENFNNMAIVGAIQRAMARVWLWDGRSKASPRLLDGMVEYIAE 208
++++ YV S + G + VW WDGR A L++G+ +++
Sbjct: 127 IHLSARYVGGYSAA-----GDVRAEVTGVLYHEATHVWQWDGRGGADGGLIEGIADFV-- 179
Query: 209 LAGFHRERLSGGVGESPECEDGRELWWDNKDPTHVARLLHYCEK--YDKGFIQRLNEAMK 266
RL G + G+ WD AR L YC+ +GF+ +LN MK
Sbjct: 180 -------RLRAGYAPPHWVQPGQGDRWDQGYDV-TARFLDYCDSPAVVQGFVAQLNGKMK 231
Query: 267 DTWHDRMVGDVLGLKATKLCGLYNATY 293
D + D + G +L Y A Y
Sbjct: 232 DGYSDDFFVQISGKTIDQLWQDYKAKY 258
>UniRef100_Q84ZV0 R 14 protein [Glycine max]
Length = 641
Score = 50.4 bits (119), Expect = 5e-05
Identities = 54/245 (22%), Positives = 105/245 (42%), Gaps = 26/245 (10%)
Query: 50 ILFVAIISIWANYEASKTFDIHIVNDTKDSLAGRRFTLFYVSNDKASRILLNTSSFVEQI 109
I+ + +S A + ++ + ++N+ + G RF + + A+R L + FV +I
Sbjct: 418 IVDILKVSFDATKQETQGYKFTVINNALTTPGGVRFR-DKIGAEYANRTLELATQFVWRI 476
Query: 110 LYPNGNNDNIDIKNKKIIKSVTLRLARQNLDTITTITADEKNINS-YVIEISPMLLEDEN 168
+D +K ++ V+L + +++ I +E ++++ YV S ++ E
Sbjct: 477 FQQKNPSD------RKNVQKVSLVV--DDMEGIAYTMNNEIHVSARYVNGYSGGDVKRE- 527
Query: 169 FNNMAIVGAIQRAMARVWLWDGRSKASPRLLDGMVEYIAELAGFHRERLSGGVGESPECE 228
I G + + VW W G +A L+ G+ +++ RL S +
Sbjct: 528 -----ITGVLFHQVCYVWQWYGNGEAPGGLIGGIADFV---------RLKANYAASHWRK 573
Query: 229 DGRELWWDNKDPTHVARLLHYCEKYDKGFIQRLNEAMKDTWHDRMVGDVLGLKATKLCGL 288
G+ WD A L YC+ GF+ +LN+ M+ + D+ +LG +L
Sbjct: 574 PGQGQRWDEGYDI-TAHFLDYCDSLKSGFVAQLNQLMRTGYSDQFFVQLLGKPVDQLWRD 632
Query: 289 YNATY 293
Y A Y
Sbjct: 633 YKAQY 637
>UniRef100_Q84ZU9 R 13 protein [Glycine max]
Length = 641
Score = 50.4 bits (119), Expect = 5e-05
Identities = 52/223 (23%), Positives = 95/223 (42%), Gaps = 26/223 (11%)
Query: 72 IVNDTKDSLAGRRFTLFYVSNDKASRILLNTSSFVEQILYPNGNNDNIDIKNKKIIKSVT 131
++N+ +L G RF + + A++ L + + F+ +I N +D +K ++ V+
Sbjct: 440 VINNALTTLGGVRFR-DKIGAEYANQTLDSATQFIWEIFQQNNPSD------RKSVQKVS 492
Query: 132 LRLARQNLDTITTITADEKNINS-YVIEISPMLLEDENFNNMAIVGAIQRAMARVWLWDG 190
L + ++D I +E ++++ YV S ++ E I G + + VW W G
Sbjct: 493 LFV--DDMDGIAYTRKNEIHVSARYVNGYSGGDVKRE------ITGVLFHQVCYVWQWKG 544
Query: 191 RSKASPRLLDGMVEYIAELAGFHRERLSGGVGESPECEDGRELWWDNKDPTHVARLLHYC 250
KA L G+ +++ RL S + G+ W N+ A L YC
Sbjct: 545 NGKAPGGLTGGIADFV---------RLKANYAASHWRKPGQGQKW-NEGYEITAHFLDYC 594
Query: 251 EKYDKGFIQRLNEAMKDTWHDRMVGDVLGLKATKLCGLYNATY 293
+ GF+ +LN+ M+ + D +L L Y A Y
Sbjct: 595 DSLKSGFVGQLNQWMRTDYSDEFFFLLLAKPVNHLWRDYKAMY 637
>UniRef100_Q9FWU2 Putative secretory protein [Oryza sativa]
Length = 233
Score = 46.2 bits (108), Expect = 0.001
Identities = 56/252 (22%), Positives = 101/252 (39%), Gaps = 28/252 (11%)
Query: 45 HRILLILFVAIISIWANYEASKTFDIHIVNDTKDSLAGRRFTLFYVSNDKASRILLNTSS 104
H L +A + A + TFD N + G+RF V D A ++L + SS
Sbjct: 5 HMATLAALMAAAAAAATAAGAVTFDA--TNTASSTAGGQRFDR-EVGVDYAKQVLADASS 61
Query: 105 FVEQILYPNGNNDNIDIKNKKIIKSVTLRLARQNLDTITTITADEKNINS-YVIEISPML 163
F+ G+ + +K + +VTL + +++D + + D ++++ YV S
Sbjct: 62 FIWDAFEQPGDGGD-----RKPVDAVTLTV--EDIDGVAFTSGDGIHLSARYVGGYS--- 111
Query: 164 LEDENFNNMAIVGAIQRAMARVWLWDGRSKASPR-LLDGMVEYIAELAGFHRER-LSGGV 221
+ G + VW W + A+ + +G+ +++ AG+ + G
Sbjct: 112 -SSSGDVRTEVTGVLYHEATHVWQWGLQDYAAHSWVYEGIADFVRLRAGYVAAGWVQPGQ 170
Query: 222 GESPECEDGRELWWDNKDPTHVARLLHYCEKYDKGFIQRLNEAMKDTWHDRMVGDVLGLK 281
G S W D+ T AR YC+ GF+ +N +KD ++ + G
Sbjct: 171 GNS---------WEDSYSVT--ARFFDYCDSVKPGFVADINAKLKDGYNVDYFVQITGKT 219
Query: 282 ATKLCGLYNATY 293
+L Y A Y
Sbjct: 220 VQQLWQDYKAKY 231
>UniRef100_UPI000021A836 UPI000021A836 UniRef100 entry
Length = 313
Score = 40.0 bits (92), Expect = 0.073
Identities = 48/191 (25%), Positives = 83/191 (43%), Gaps = 30/191 (15%)
Query: 89 YVSNDKASRILLNTSSFVEQILYPNGNNDNIDIKNKKIIKSVTLRLARQNLDTITTITAD 148
++S AS L + V++ILY N + +SVTL L +++D + T
Sbjct: 95 FLSRVDASATLPTAVANVQRILY---NTPKDHTTHLPPTRSVTLIL--RDMDGVAYTTGS 149
Query: 149 EKNINSYVIEISPMLLE----------DENFNNMA--IVGAIQRAMARVWLWDGRSKASP 196
E + + I S +E DEN + +A IVG I + + WD +
Sbjct: 150 ELDSDHKEIHFSLHYIESISKREKCSGDEN-DGIAHEIVGVITHELVHCYQWDAKGTCPG 208
Query: 197 RLLDGMVEYIAELAGFHRERLSGGVGESPECEDGRELWWDNKDPTHVARLLHYCE-KYDK 255
L++G+ +++ RL G+ P E E WD + H A L Y E ++
Sbjct: 209 GLIEGIADFV---------RLRCGL-SPPHWEKDLEGSWD-RGYQHTAYFLDYLECRFGD 257
Query: 256 GFIQRLNEAMK 266
G ++++NE ++
Sbjct: 258 GTVRKINEGLR 268
>UniRef100_Q9FWU4 Putative basic secretory protein [Oryza sativa]
Length = 229
Score = 39.3 bits (90), Expect = 0.13
Identities = 54/253 (21%), Positives = 101/253 (39%), Gaps = 31/253 (12%)
Query: 44 IHRILLILFVAIISIWANYEASKTFDIHIVNDTKDSLAGRRFTLFYVSNDKASRILLNTS 103
+H + IL S A+ T+++ N+ + G+RF Y + A ++L S
Sbjct: 3 LHVVAAILLAVAASSSPPPAAAVTYEVS--NEAASTAGGQRFDREYGAG-YAKQVLAAAS 59
Query: 104 SFVEQILYPNGNNDNIDIKNKKIIKSVTLRLARQNLDTITTITADEKNINS-YVIEISPM 162
SF I D ++ + +V L A +++D I + + + + + YV ++
Sbjct: 60 SFTWSIFSQPSAAD------RRPVDAVVL--AVRDVDGIASTSGNTITLGAGYVAGVTG- 110
Query: 163 LLEDENFNNMAIVGAIQRAMARVWLWDGRSK-ASPRLLDGMVEYIAELAGFHRER-LSGG 220
N + G + + VW W + A + +G+ +++ AG+ + G
Sbjct: 111 -----NDFKTQVTGVLYHEVVHVWQWGLQDYGAHSWVYEGIADFVRLRAGYPAAGWVQPG 165
Query: 221 VGESPECEDGRELWWDNKDPTHVARLLHYCEKYDKGFIQRLNEAMKDTWHDRMVGDVLGL 280
G S W D+ T AR YC+ GF+ LN +K+ ++ + G
Sbjct: 166 QGNS---------WEDSYSVT--ARFFDYCDSVKPGFVADLNAKLKNGYNVDYFVQITGK 214
Query: 281 KATKLCGLYNATY 293
+L Y A Y
Sbjct: 215 TVQQLWQDYKAKY 227
>UniRef100_Q7S9L1 Hypothetical protein [Neurospora crassa]
Length = 292
Score = 39.3 bits (90), Expect = 0.13
Identities = 47/210 (22%), Positives = 89/210 (42%), Gaps = 33/210 (15%)
Query: 95 ASRILLNTSSFVEQILYPNGNNDNIDIKNKKIIKSVTLRLARQNLDTITTITADEKNIN- 153
AS +L + V+++LY + + + + + + +A T T + D K I+
Sbjct: 88 ASSLLSDAVKNVQRLLYRSPADKHTTCPLTRSVTVILRDMAGVAYTTGTDLDPDHKEIHF 147
Query: 154 ------SYVIEISPMLLEDENFNNMAIVGAIQRAMARVWLWDGRSKASPRLLDGMVEYI- 206
+++ P L E I G + + + WD A L++G+ +++
Sbjct: 148 SLSYIAAHIPSTPPSRLPAE------ITGVLTHELVHCYQWDAHGSAPGGLIEGIADWVR 201
Query: 207 --AELAGFHRERLSGGVGESPECEDGRELWWDNKDPTHVARLLHYCE-KYDKGFIQRLNE 263
+L+ H ++ + + WD K H A L Y E ++ +G ++RLNE
Sbjct: 202 LNCDLSPPHWKQGDVKIDDP----------WD-KGYQHTAYFLQYLEGRFGEGTVRRLNE 250
Query: 264 AMKDT----WHDRMVGDVLGLKATKLCGLY 289
A++ T + VG VLG + +L G Y
Sbjct: 251 ALRSTREYEAREFWVG-VLGEEVEELWGEY 279
>UniRef100_O96267 Hypothetical protein PFB0875c [Plasmodium falciparum]
Length = 486
Score = 38.9 bits (89), Expect = 0.16
Identities = 36/133 (27%), Positives = 58/133 (43%), Gaps = 8/133 (6%)
Query: 16 HFNPSSYEINQQNDYNNS---KFFIPNNN----NIIHRILLILFVAIISIWANYEASKTF 68
H N I ++D N+ K+ NNN N+I++ + I I I N +K
Sbjct: 75 HMNQYPNNIGLKSDNKNNIVLKYIDENNNVKYDNLINQQIHIYNNEIDKIEPNERINKLR 134
Query: 69 DIHIVNDTKDSLAGRRFTLFYVSNDKASRILLNTSSFVEQILYPNGNNDNI-DIKNKKII 127
I++DTKD + +ND+ + I+ NT +E IL N NI + K +K
Sbjct: 135 KKKILSDTKDREEKYNEPTYKPNNDEENEIIENTKRNIENILNEKLNKSNIVNKKEEKYY 194
Query: 128 KSVTLRLARQNLD 140
+ + NL+
Sbjct: 195 RYIPQNKLNNNLE 207
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.320 0.137 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 516,076,801
Number of Sequences: 2790947
Number of extensions: 22350538
Number of successful extensions: 68623
Number of sequences better than 10.0: 89
Number of HSP's better than 10.0 without gapping: 19
Number of HSP's successfully gapped in prelim test: 72
Number of HSP's that attempted gapping in prelim test: 68400
Number of HSP's gapped (non-prelim): 254
length of query: 294
length of database: 848,049,833
effective HSP length: 126
effective length of query: 168
effective length of database: 496,390,511
effective search space: 83393605848
effective search space used: 83393605848
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 74 (33.1 bits)
Medicago: description of AC147496.3


