

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147484.13 + phase: 0 /pseudo
(575 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
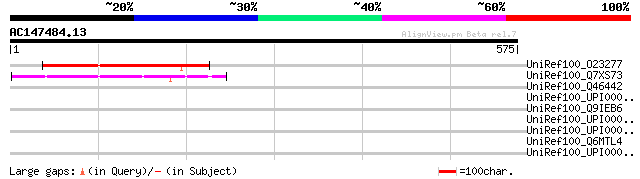
UniRef100_O23277 Hypothetical protein dl3130w [Arabidopsis thali... 169 2e-40
UniRef100_Q7XS73 OSJNBa0020I02.9 protein [Oryza sativa] 135 2e-30
UniRef100_Q46442 Virulence plasmid protein pGP6-D [Chlamydia mur... 39 0.32
UniRef100_UPI0000310A62 UPI0000310A62 UniRef100 entry 37 1.2
UniRef100_Q9IEB6 Gp41 [Human immunodeficiency virus 1] 37 1.6
UniRef100_UPI00002CAE5B UPI00002CAE5B UniRef100 entry 35 6.0
UniRef100_UPI000028760B UPI000028760B UniRef100 entry 35 6.0
UniRef100_Q6MTL4 Hypothetical transmembrane protein [Mycoplasma ... 35 6.0
UniRef100_UPI0000329C30 UPI0000329C30 UniRef100 entry 35 7.8
>UniRef100_O23277 Hypothetical protein dl3130w [Arabidopsis thaliana]
Length = 1268
Score = 169 bits (429), Expect = 2e-40
Identities = 98/197 (49%), Positives = 129/197 (64%), Gaps = 9/197 (4%)
Query: 38 RGLGNMNYQIHYSREAEEIIFQLINNGEWDLLSARIHTVSLKWLFQQENIINSLCHQILK 97
R L N YQI YS EAE IIF L+N EWDL S IH SLKWLFQQE+I SL +QI K
Sbjct: 661 RSLQNERYQISYSLEAERIIFHLLNEYEWDLGSINIHLESLKWLFQQESISKSLIYQIQK 720
Query: 98 FCRNNNLEGDDM--IIGNSNQTVNVQTLAELVSSEDNYGARLFICLLAQLAEEEGQEHDI 155
R NNL G+++ + G+ Q A+L+S DNY A L + LL QLAE+E QE+D+
Sbjct: 721 ISR-NNLIGNEVHNVYGDGRQRSLTYWFAKLISEGDNYAATLLVNLLTQLAEKEEQENDV 779
Query: 156 ISVLNLMATMIHICPAASDQLSLHGIGTTIRTCC--FSTA----TFMSILVLVFNTLSSV 209
IS+LNLM T++ I P AS+ LS++GIG+ I FS + +F ++L+LVFN L+SV
Sbjct: 780 ISILNLMNTIVSIFPTASNNLSMNGIGSVIHRLVSGFSNSSLGTSFRTLLLLVFNILTSV 839
Query: 210 HPKTLSTDQSWVAVTME 226
P L D+SW AV+++
Sbjct: 840 QPAVLMIDESWYAVSIK 856
Score = 37.7 bits (86), Expect = 0.92
Identities = 23/83 (27%), Positives = 44/83 (52%), Gaps = 2/83 (2%)
Query: 484 LTS*VLSKLQL*VKCCRKQMYTNNAQDGLPSEPIKKVLATSYDLGDVCWFLIDLMSPDTL 543
L S ++KL + CRKQM+ N +D E +++ + + +D + C +L+ LM ++
Sbjct: 1159 LNSQQVTKLDRAFQECRKQMHRNGTRDETVEEQVQRKIPSIHDHSEFCNYLVHLMVSNSF 1218
Query: 544 --DMDSWGIHMDGKRLLEEIELF 564
+S K++L+E+E F
Sbjct: 1219 GHPSESETYTQKKKQILDEMEQF 1241
>UniRef100_Q7XS73 OSJNBa0020I02.9 protein [Oryza sativa]
Length = 1176
Score = 135 bits (341), Expect = 2e-30
Identities = 89/248 (35%), Positives = 144/248 (57%), Gaps = 12/248 (4%)
Query: 3 LASLEQYVLLNSSDSHNWTTDSLTVTRLVNLYSLLRGLGNMNYQIHYSREAEEIIFQLIN 62
LAS+EQY+LLN + + SL +T LV+LY+ +RG+ + + I +S EAE+ +F +
Sbjct: 557 LASVEQYILLNGAKFPHEIPGSLMLTLLVHLYAFVRGI-SFRFGIPHSPEAEKTLFHAMT 615
Query: 63 NGEWDLLSARIHTVSLKWLFQQENIINSLCHQILKFCRNNNLEGDDMIIGNSNQTVNVQT 122
+ EWDLL R+H ++LKWLFQ E ++ L +L FC+ E +++ +S Q V++Q
Sbjct: 616 HKEWDLLLIRVHLIALKWLFQNEELMEPLSFHLLNFCK-FFCEDRTVMLSSSTQLVDIQL 674
Query: 123 LAELVSSEDNYGARLFICLLAQLAEEEGQEHDIISVLNLMATMIHICPAASDQLSLHGI- 181
+AELV S + + L + LL+Q+ +E E +++SV+N++ ++ P SDQ GI
Sbjct: 675 IAELVYSGETCISSLLVSLLSQMIKESA-EDEVLSVVNVITEILVSFPCTSDQFVSCGIV 733
Query: 182 ---GTTIRTCCFSTATFMSILVLVFNTLSSVHPKTLS-TDQSWVAVTMESRKSLRHWHSF 237
G+ + C S + L L+FN L S T + D +W+A+TM K L ++S
Sbjct: 734 DALGSIYLSLCSSRIKSVCSL-LIFNILHSASAMTFTCDDDAWLALTM---KLLDCFNSS 789
Query: 238 LDFTSFHQ 245
L +TS Q
Sbjct: 790 LAYTSSEQ 797
>UniRef100_Q46442 Virulence plasmid protein pGP6-D [Chlamydia muridarum]
Length = 246
Score = 39.3 bits (90), Expect = 0.32
Identities = 28/96 (29%), Positives = 48/96 (49%), Gaps = 5/96 (5%)
Query: 119 NVQTLAELVSSEDNYGA-RLFICLLAQLAEEEGQEHDIISVLNLMATMIHICPAASDQL- 176
+++ + ++SE+N RLF+ L++EE + D++SV L+ + I L
Sbjct: 29 SIEMFSTSLNSEENQSLDRLFLSETQNLSDEESYQEDVLSV-KLLTSQIKAIQKQHVLLL 87
Query: 177 --SLHGIGTTIRTCCFSTATFMSILVLVFNTLSSVH 210
++ + CFS+ TF S L LVF T SS +
Sbjct: 88 GEKIYNARKILSKSCFSSTTFSSWLDLVFRTKSSAY 123
>UniRef100_UPI0000310A62 UPI0000310A62 UniRef100 entry
Length = 265
Score = 37.4 bits (85), Expect = 1.2
Identities = 35/124 (28%), Positives = 56/124 (44%), Gaps = 18/124 (14%)
Query: 12 LNSSDSHNWTTDSLTVTRLVNLYSLLR--------GLGNMNYQIHYSREAEEIIFQLINN 63
L S N+ D+ T+ ++V+L SL G GN+ I R F LI
Sbjct: 4 LKKSLGQNFLRDNNTLNKIVSLESLSNQNILEIGPGSGNLTNLIFKQRPKS---FYLIEK 60
Query: 64 GEWDLLSARIHTVSLKWLFQQENIINSLCHQILKFCRNNNLEGDDMIIGNSNQTVNVQTL 123
+ R + + LK +++EN +N L IL F N+ E D ++ GN ++ Q L
Sbjct: 61 DK------RFYEI-LKEKYKRENSLNILNKDILDFNLNDLKEKDVIVFGNLPYNISTQIL 113
Query: 124 AELV 127
A+ +
Sbjct: 114 AKFI 117
>UniRef100_Q9IEB6 Gp41 [Human immunodeficiency virus 1]
Length = 219
Score = 37.0 bits (84), Expect = 1.6
Identities = 30/135 (22%), Positives = 62/135 (45%), Gaps = 19/135 (14%)
Query: 31 VNLYSLLRGLGNMNYQIHYSREAEEIIFQLINNGEWDLLSARIHTVSLKWLFQQENIINS 90
V ++L++G+ + + +A++ QL+ W + R ++L+ L Q + ++NS
Sbjct: 4 VRTHTLIKGIVQQQDNLLRAIQAQQ---QLLRLSVWGIRQLRARLLALETLIQNQQLLNS 60
Query: 91 -------LCHQILKFCRNNNLEGDDMIIGNSNQTVNVQTLAELVSSEDNYGARLFICLLA 143
+C+ +K+ NN +GD+ I GN T E DN + +F +L
Sbjct: 61 WGCKGRLVCYTSVKW--NNTWKGDEYIWGN-------LTWQEWDQQIDNVSSIIFDEILR 111
Query: 144 QLAEEEGQEHDIISV 158
++E E ++ +
Sbjct: 112 AQVQQEENERKLLEL 126
>UniRef100_UPI00002CAE5B UPI00002CAE5B UniRef100 entry
Length = 265
Score = 35.0 bits (79), Expect = 6.0
Identities = 45/163 (27%), Positives = 65/163 (39%), Gaps = 35/163 (21%)
Query: 3 LASLEQYVLLNSSDSHNWTTDSLTVTRLVNLYSLLRGLGNMNYQIHYSREAEEIIFQLIN 62
+A LE VL + S + + D L V R+ S L G + E +E+I ++N
Sbjct: 6 IADLENRVLRHVSVA--FRDDPLRVLRVARFMSKLNSFG-----FRIADETKELISDIVN 58
Query: 63 NGEWDLL---------SARIHTVSLKWLFQ---QENIINSLCHQILKFCRNNNLEGDDMI 110
+GE D L +HT S F+ + N IN + +L G D +
Sbjct: 59 SGELDYLVPERVWLETEKALHTESFDLYFKTLCEFNAINGI---------YPDLNGIDQV 109
Query: 111 IGNSNQTVNVQTLAELVSSEDNYGARLFICLLAQLAEEEGQEH 153
I NS+ T EL A LF C + L +E EH
Sbjct: 110 IANSSLTAYKNVSPELKL------ASLF-CAMDTLTDENLVEH 145
>UniRef100_UPI000028760B UPI000028760B UniRef100 entry
Length = 311
Score = 35.0 bits (79), Expect = 6.0
Identities = 45/165 (27%), Positives = 65/165 (39%), Gaps = 39/165 (23%)
Query: 3 LASLEQYVLLNSSDSHNWTTDSLTVTRLVNLYSLLRGLGNMNYQIHYSREAEEIIFQLIN 62
+A LE VL + S + + D L V R+ S L G + E +E+I ++N
Sbjct: 52 IADLENRVLRHVSVA--FRDDPLRVLRVARFMSKLNSFG-----FRIADETKELISDIVN 104
Query: 63 NGEWDLL---------SARIHTVSLKWLFQ---QENIINSLCHQILKFCRNNNLEGDDMI 110
+GE D L +HT S F+ N IN + +L G D I
Sbjct: 105 SGELDYLVPERVWLETEKALHTESFDLYFKTLCDFNAINDI---------YPDLNGIDQI 155
Query: 111 IGNSNQTV--NVQTLAELVSSEDNYGARLFICLLAQLAEEEGQEH 153
I NS+ T NV +L S +C + L +E EH
Sbjct: 156 IANSSLTAYKNVSPELKLAS---------LLCAMDTLTDENLVEH 191
>UniRef100_Q6MTL4 Hypothetical transmembrane protein [Mycoplasma mycoides]
Length = 275
Score = 35.0 bits (79), Expect = 6.0
Identities = 16/64 (25%), Positives = 34/64 (53%), Gaps = 2/64 (3%)
Query: 19 NWTTDSLTVTRLVNLYSLLRGLGNMNYQIHYSREAEEIIFQLINNGEWDLLSARIHTVSL 78
N+ + SL + VN Y + + ++HY+ E +++ + E+D +ARI+ + +
Sbjct: 191 NYLSFSLIICLFVNYYISI--IIKNKIELHYNDEVSKLVSSALEMYEYDFKAARIYALGI 248
Query: 79 KWLF 82
KW +
Sbjct: 249 KWTY 252
>UniRef100_UPI0000329C30 UPI0000329C30 UniRef100 entry
Length = 169
Score = 34.7 bits (78), Expect = 7.8
Identities = 20/81 (24%), Positives = 41/81 (49%), Gaps = 1/81 (1%)
Query: 108 DMIIGNSNQTVNV-QTLAELVSSEDNYGARLFICLLAQLAEEEGQEHDIISVLNLMATMI 166
++ + N ++T+ + QT+ + N + C++ + +E D +S N+ T +
Sbjct: 83 EISVSNISKTIEIFQTIPLIKKISTNQNIKSINCVIEKASENIPHIIDKLSKSNIEITKL 142
Query: 167 HICPAASDQLSLHGIGTTIRT 187
+I PA D + LH G +I+T
Sbjct: 143 NISPANLDDVFLHLTGNSIKT 163
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.349 0.152 0.538
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 827,897,991
Number of Sequences: 2790947
Number of extensions: 30092312
Number of successful extensions: 135652
Number of sequences better than 10.0: 9
Number of HSP's better than 10.0 without gapping: 2
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 135643
Number of HSP's gapped (non-prelim): 11
length of query: 575
length of database: 848,049,833
effective HSP length: 133
effective length of query: 442
effective length of database: 476,853,882
effective search space: 210769415844
effective search space used: 210769415844
T: 11
A: 40
X1: 14 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.8 bits)
S2: 78 (34.7 bits)
Medicago: description of AC147484.13


