

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
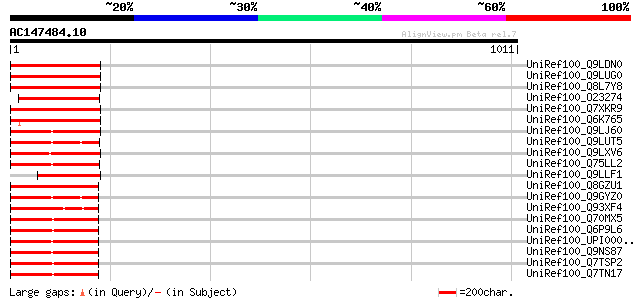
Query= AC147484.10 - phase: 0 /pseudo
(1011 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9LDN0 Phragmoplast-associated kinesin-related protein... 313 2e-83
UniRef100_Q9LUG0 Kinesin-like protein [Arabidopsis thaliana] 308 5e-82
UniRef100_Q8L7Y8 AT3g23670/MDB19_16 [Arabidopsis thaliana] 308 5e-82
UniRef100_O23274 Kinesin like protein [Arabidopsis thaliana] 281 7e-74
UniRef100_Q7XKR9 OSJNBa0038P21.12 protein [Oryza sativa] 273 1e-71
UniRef100_Q6K765 Putative phragmoplast-associated kinesin-relate... 257 1e-66
UniRef100_Q9LJ60 Kinesin (Centromeric protein)-like protein [Ara... 222 4e-56
UniRef100_Q9LUT5 Kinesin-related centromere protein-like [Arabid... 217 1e-54
UniRef100_Q9LXV6 Kinesin-like protein [Arabidopsis thaliana] 211 7e-53
UniRef100_Q75LL2 Kinesin-like protein [Oryza sativa] 208 8e-52
UniRef100_Q9LLF1 Phragmoplast-associated kinesin-related protein... 197 1e-48
UniRef100_Q8GZU1 Kinesin related protein [Lycopersicon esculentum] 191 1e-46
UniRef100_Q9GYZ0 Kinesin-like protein KRP180 [Strongylocentrotus... 189 5e-46
UniRef100_Q93XF4 Kinesin heavy chain [Zea mays] 185 7e-45
UniRef100_Q70MX5 Kinesin family member 15 [Mus musculus] 182 3e-44
UniRef100_Q6P9L6 Kinesin-like 7 [Mus musculus] 182 3e-44
UniRef100_UPI000036B4A8 UPI000036B4A8 UniRef100 entry 182 4e-44
UniRef100_Q9NS87 Kinesin-like protein 2 [Homo sapiens] 182 4e-44
UniRef100_Q7TSP2 Kinesin-like protein KIF15 [Rattus norvegicus] 182 6e-44
UniRef100_Q7TN17 Kinesin-like protein KIF15 [Rattus norvegicus] 182 6e-44
>UniRef100_Q9LDN0 Phragmoplast-associated kinesin-related protein 1 [Arabidopsis
thaliana]
Length = 1292
Score = 313 bits (802), Expect = 2e-83
Identities = 158/180 (87%), Positives = 169/180 (93%)
Query: 1 IYNEQITDLLDPSQRNLQIREDVKSGVYVENLTEEQVSTMKDVTQLLLKGLSNRRIGATS 60
IYNEQITDLLDPSQ+NL IREDVKSGVYVENLTEE V + DV+QLL+KGL NRR GATS
Sbjct: 229 IYNEQITDLLDPSQKNLMIREDVKSGVYVENLTEEYVKNLTDVSQLLIKGLGNRRTGATS 288
Query: 61 INSESSRSHTVFTCVVESRCKSAADGVSRFKTSRINLVDLAGSERQKSTGAAGERLKEAG 120
+N+ESSRSH VFTCVVESRCK+ ADG+S FKTSRINLVDLAGSERQKSTGAAGERLKEAG
Sbjct: 289 VNTESSRSHCVFTCVVESRCKNVADGLSSFKTSRINLVDLAGSERQKSTGAAGERLKEAG 348
Query: 121 NINRSLSQLGNLINILAEVSQTGKQRHIPYRDSRLTFLLQESLGGNAKLAMVCAISPAQS 180
NINRSLSQLGNLINILAE+SQTGK RHIPYRDSRLTFLLQESLGGNAKLAMVCA+SP+QS
Sbjct: 349 NINRSLSQLGNLINILAEISQTGKPRHIPYRDSRLTFLLQESLGGNAKLAMVCAVSPSQS 408
>UniRef100_Q9LUG0 Kinesin-like protein [Arabidopsis thaliana]
Length = 1268
Score = 308 bits (789), Expect = 5e-82
Identities = 155/180 (86%), Positives = 169/180 (93%)
Query: 1 IYNEQITDLLDPSQRNLQIREDVKSGVYVENLTEEQVSTMKDVTQLLLKGLSNRRIGATS 60
IYNEQITDLLDPS +NL IREDVKSGVYVENLTEE V +KD+++LL+KGL+NRR GATS
Sbjct: 189 IYNEQITDLLDPSLKNLMIREDVKSGVYVENLTEEYVKNLKDLSKLLVKGLANRRTGATS 248
Query: 61 INSESSRSHTVFTCVVESRCKSAADGVSRFKTSRINLVDLAGSERQKSTGAAGERLKEAG 120
+N+ESSRSH VFTCVVES CKS ADG+S FKTSRINLVDLAGSERQK TGAAG+RLKEAG
Sbjct: 249 VNAESSRSHCVFTCVVESHCKSVADGLSSFKTSRINLVDLAGSERQKLTGAAGDRLKEAG 308
Query: 121 NINRSLSQLGNLINILAEVSQTGKQRHIPYRDSRLTFLLQESLGGNAKLAMVCAISPAQS 180
NINRSLSQLGNLINILAE+SQTGKQRHIPYRDSRLTFLLQESLGGNAKLAMVCA+SP+QS
Sbjct: 309 NINRSLSQLGNLINILAEISQTGKQRHIPYRDSRLTFLLQESLGGNAKLAMVCAVSPSQS 368
>UniRef100_Q8L7Y8 AT3g23670/MDB19_16 [Arabidopsis thaliana]
Length = 1313
Score = 308 bits (789), Expect = 5e-82
Identities = 155/180 (86%), Positives = 169/180 (93%)
Query: 1 IYNEQITDLLDPSQRNLQIREDVKSGVYVENLTEEQVSTMKDVTQLLLKGLSNRRIGATS 60
IYNEQITDLLDPS +NL IREDVKSGVYVENLTEE V +KD+++LL+KGL+NRR GATS
Sbjct: 234 IYNEQITDLLDPSLKNLMIREDVKSGVYVENLTEEYVKNLKDLSKLLVKGLANRRTGATS 293
Query: 61 INSESSRSHTVFTCVVESRCKSAADGVSRFKTSRINLVDLAGSERQKSTGAAGERLKEAG 120
+N+ESSRSH VFTCVVES CKS ADG+S FKTSRINLVDLAGSERQK TGAAG+RLKEAG
Sbjct: 294 VNAESSRSHCVFTCVVESHCKSVADGLSSFKTSRINLVDLAGSERQKLTGAAGDRLKEAG 353
Query: 121 NINRSLSQLGNLINILAEVSQTGKQRHIPYRDSRLTFLLQESLGGNAKLAMVCAISPAQS 180
NINRSLSQLGNLINILAE+SQTGKQRHIPYRDSRLTFLLQESLGGNAKLAMVCA+SP+QS
Sbjct: 354 NINRSLSQLGNLINILAEISQTGKQRHIPYRDSRLTFLLQESLGGNAKLAMVCAVSPSQS 413
>UniRef100_O23274 Kinesin like protein [Arabidopsis thaliana]
Length = 1662
Score = 281 bits (719), Expect = 7e-74
Identities = 142/162 (87%), Positives = 152/162 (93%)
Query: 18 QIREDVKSGVYVENLTEEQVSTMKDVTQLLLKGLSNRRIGATSINSESSRSHTVFTCVVE 77
QIREDVKSGVYVENLTEE V + DV+QLL+KGL NRR GATS+N+ESSRSH VFTCVVE
Sbjct: 239 QIREDVKSGVYVENLTEEYVKNLTDVSQLLIKGLGNRRTGATSVNTESSRSHCVFTCVVE 298
Query: 78 SRCKSAADGVSRFKTSRINLVDLAGSERQKSTGAAGERLKEAGNINRSLSQLGNLINILA 137
SRCK+ ADG+S FKTSRINLVDLAGSERQKSTGAAGERLKEAGNINRSLSQLGNLINILA
Sbjct: 299 SRCKNVADGLSSFKTSRINLVDLAGSERQKSTGAAGERLKEAGNINRSLSQLGNLINILA 358
Query: 138 EVSQTGKQRHIPYRDSRLTFLLQESLGGNAKLAMVCAISPAQ 179
E+SQTGK RHIPYRDSRLTFLLQESLGGNAKLAMVCA+SP+Q
Sbjct: 359 EISQTGKPRHIPYRDSRLTFLLQESLGGNAKLAMVCAVSPSQ 400
>UniRef100_Q7XKR9 OSJNBa0038P21.12 protein [Oryza sativa]
Length = 1094
Score = 273 bits (699), Expect = 1e-71
Identities = 140/181 (77%), Positives = 162/181 (89%), Gaps = 1/181 (0%)
Query: 1 IYNEQITDLLDPSQRNLQIREDVKSG-VYVENLTEEQVSTMKDVTQLLLKGLSNRRIGAT 59
IYNEQITDLLDPS ++LQIREDV++ VYVE+LT+E V T KDVTQLL+KGLSNRR GAT
Sbjct: 223 IYNEQITDLLDPSPKSLQIREDVRTACVYVESLTKELVFTTKDVTQLLVKGLSNRRTGAT 282
Query: 60 SINSESSRSHTVFTCVVESRCKSAADGVSRFKTSRINLVDLAGSERQKSTGAAGERLKEA 119
S N++SSRSH VFTCV++S K+ DG + +TSRINLVDLAGSERQK T A G+RLKEA
Sbjct: 283 SANADSSRSHCVFTCVIKSESKNLEDGSNSTRTSRINLVDLAGSERQKLTHAFGDRLKEA 342
Query: 120 GNINRSLSQLGNLINILAEVSQTGKQRHIPYRDSRLTFLLQESLGGNAKLAMVCAISPAQ 179
GNINRSLSQLGNLINILAE+SQ+GKQRH+PYRDS+LTFLLQESLGGNAKLAM+CA+SP+Q
Sbjct: 343 GNINRSLSQLGNLINILAEISQSGKQRHVPYRDSKLTFLLQESLGGNAKLAMICAVSPSQ 402
Query: 180 S 180
S
Sbjct: 403 S 403
>UniRef100_Q6K765 Putative phragmoplast-associated kinesin-related protein 1 [Oryza
sativa]
Length = 1106
Score = 257 bits (656), Expect = 1e-66
Identities = 141/194 (72%), Positives = 159/194 (81%), Gaps = 14/194 (7%)
Query: 1 IYNEQITDLLDPSQRNLQ------------IREDV-KSGVYVENLTEEQVSTMKDVTQLL 47
IYNEQITDLLDP QRNLQ IREDV S VYVE+LT+E V T+ DVTQLL
Sbjct: 240 IYNEQITDLLDPVQRNLQAASVLIRLVHFQIREDVGTSSVYVESLTKESVFTINDVTQLL 299
Query: 48 LKGLSNRRIGATSINSESSRSHTVFTCVVESRCKSAADGVSRFKTSRINLVDLAGSERQK 107
KGL+NRR AT+ N+ESSRSH VFTC ++S K+ DG + +TSRINLVDLAGSERQK
Sbjct: 300 EKGLANRRTEATTANAESSRSHCVFTCFIKSESKNMEDGSNFTRTSRINLVDLAGSERQK 359
Query: 108 STGAAGERLKEAGNINRSLSQLGNLINILAEVSQTGKQR-HIPYRDSRLTFLLQESLGGN 166
T AAG+RLKEAGNINRSLSQLGNLINILAEVSQ+GKQR HIPYRDS+LTFLLQESLGGN
Sbjct: 360 LTNAAGDRLKEAGNINRSLSQLGNLINILAEVSQSGKQRHHIPYRDSKLTFLLQESLGGN 419
Query: 167 AKLAMVCAISPAQS 180
AKLAM+CA+SP+Q+
Sbjct: 420 AKLAMICAVSPSQN 433
>UniRef100_Q9LJ60 Kinesin (Centromeric protein)-like protein [Arabidopsis thaliana]
Length = 2756
Score = 222 bits (566), Expect = 4e-56
Identities = 112/180 (62%), Positives = 147/180 (81%), Gaps = 3/180 (1%)
Query: 1 IYNEQITDLLDPSQRNLQIREDVKSGVYVENLTEEQVSTMKDVTQLLLKGLSNRRIGATS 60
IYNEQITDLL+PS NLQ+RED+KSGVYVENLTE +V +++D+ L+ +G NRR+GAT+
Sbjct: 336 IYNEQITDLLEPSSTNLQLREDIKSGVYVENLTECEVQSVQDILGLITQGSLNRRVGATN 395
Query: 61 INSESSRSHTVFTCVVESRCKSAADGVSRFKTSRINLVDLAGSERQKSTGAAGERLKEAG 120
+N ESSRSH+VFTCV+ESR + D + + +R+NLVDLAGSERQK++GA G+RLKEA
Sbjct: 396 MNRESSRSHSVFTCVIESRWEK--DSTANMRFARLNLVDLAGSERQKTSGAEGDRLKEAA 453
Query: 121 NINRSLSQLGNLINILAEVSQTGKQRHIPYRDSRLTFLLQESLGGNAKLAMVCAISPAQS 180
+IN+SLS LG++I +L +V+ GK RHIPYRDSRLTFLLQ+SLGGN+K ++ SP+ S
Sbjct: 454 SINKSLSTLGHVIMVLVDVA-NGKPRHIPYRDSRLTFLLQDSLGGNSKTMIIANASPSVS 512
>UniRef100_Q9LUT5 Kinesin-related centromere protein-like [Arabidopsis thaliana]
Length = 2158
Score = 217 bits (553), Expect = 1e-54
Identities = 111/178 (62%), Positives = 142/178 (79%), Gaps = 3/178 (1%)
Query: 1 IYNEQITDLLDPSQRNLQIREDVKSGVYVENLTEEQVSTMKDVTQLLLKGLSNRRIGATS 60
IYNEQITDLL+PS NLQ+RED+ GVYVENL E V T+ DV +LLL+G +NR+I AT
Sbjct: 311 IYNEQITDLLEPSSTNLQLREDLGKGVYVENLVEHNVRTVSDVLKLLLQGATNRKIAATR 370
Query: 61 INSESSRSHTVFTCVVESRCKSAADGVSRFKTSRINLVDLAGSERQKSTGAAGERLKEAG 120
+NSESSRSH+VFTC +ES + D ++R + +R+NLVDLAGSERQKS+GA G+RLKEA
Sbjct: 371 MNSESSRSHSVFTCTIESLWEK--DSLTRSRFARLNLVDLAGSERQKSSGAEGDRLKEAA 428
Query: 121 NINRSLSQLGNLINILAEVSQTGKQRHIPYRDSRLTFLLQESLGGNAKLAMVCAISPA 178
NIN+SLS LG +I L +++ GK RH+PYRDSRLTFLLQ+SLGGN+K ++ +SP+
Sbjct: 429 NINKSLSTLGLVIMSLVDLAH-GKHRHVPYRDSRLTFLLQDSLGGNSKTMIIANVSPS 485
>UniRef100_Q9LXV6 Kinesin-like protein [Arabidopsis thaliana]
Length = 1229
Score = 211 bits (538), Expect = 7e-53
Identities = 110/180 (61%), Positives = 141/180 (78%), Gaps = 3/180 (1%)
Query: 1 IYNEQITDLLDPSQRNLQIREDVKSGVYVENLTEEQVSTMKDVTQLLLKGLSNRRIGATS 60
IYNEQI DLLDPS NLQ+RED K G++VENL E +VS+ +DV Q L++G +NR++ AT+
Sbjct: 236 IYNEQILDLLDPSSYNLQLREDHKKGIHVENLKEIEVSSARDVIQQLMQGAANRKVAATN 295
Query: 61 INSESSRSHTVFTCVVESRCKSAADGVSRFKTSRINLVDLAGSERQKSTGAAGERLKEAG 120
+N SSRSH+VFTC++ES K + GV+ + +R+NLVDLAGSERQKS+GA GERLKEA
Sbjct: 296 MNRASSRSHSVFTCIIES--KWVSQGVTHHRFARLNLVDLAGSERQKSSGAEGERLKEAT 353
Query: 121 NINRSLSQLGNLINILAEVSQTGKQRHIPYRDSRLTFLLQESLGGNAKLAMVCAISPAQS 180
NIN+SLS LG +I L VS GK H+PYRDS+LTFLLQ+SLGGN+K ++ ISP+ S
Sbjct: 354 NINKSLSTLGLVIMNLVSVS-NGKSVHVPYRDSKLTFLLQDSLGGNSKTIIIANISPSSS 412
>UniRef100_Q75LL2 Kinesin-like protein [Oryza sativa]
Length = 1266
Score = 208 bits (529), Expect = 8e-52
Identities = 107/178 (60%), Positives = 139/178 (77%), Gaps = 3/178 (1%)
Query: 1 IYNEQITDLLDPSQRNLQIREDVKSGVYVENLTEEQVSTMKDVTQLLLKGLSNRRIGATS 60
IYNEQI DLL+P+ NLQIRED K GV+VENLTE +VS ++ Q L++G +NR++ AT+
Sbjct: 175 IYNEQILDLLNPNSVNLQIREDAKKGVHVENLTEHEVSNAREAMQQLVEGAANRKVAATN 234
Query: 61 INSESSRSHTVFTCVVESRCKSAADGVSRFKTSRINLVDLAGSERQKSTGAAGERLKEAG 120
+N SSRSH+VFTC++ES+ +S G++ + SR+NLVDLAGSERQKS+GA GERLKEA
Sbjct: 235 MNRASSRSHSVFTCLIESKWES--QGINHHRFSRLNLVDLAGSERQKSSGAEGERLKEAT 292
Query: 121 NINRSLSQLGNLINILAEVSQTGKQRHIPYRDSRLTFLLQESLGGNAKLAMVCAISPA 178
NIN+SLS LG +I L VS K H+PYRDS+LTFLLQ+SLGGN+K ++ ISP+
Sbjct: 293 NINKSLSTLGLVITNLIAVSNK-KSHHVPYRDSKLTFLLQDSLGGNSKTTIIANISPS 349
>UniRef100_Q9LLF1 Phragmoplast-associated kinesin-related protein 1 [Oryza sativa]
Length = 817
Score = 197 bits (502), Expect = 1e-48
Identities = 98/126 (77%), Positives = 113/126 (88%)
Query: 55 RIGATSINSESSRSHTVFTCVVESRCKSAADGVSRFKTSRINLVDLAGSERQKSTGAAGE 114
R GATS N++SSRSH VFTCV++S K+ DG + +TSRINLVDLAGSERQK T A G+
Sbjct: 1 RTGATSANADSSRSHCVFTCVIKSESKNLEDGSNSTRTSRINLVDLAGSERQKLTHAFGD 60
Query: 115 RLKEAGNINRSLSQLGNLINILAEVSQTGKQRHIPYRDSRLTFLLQESLGGNAKLAMVCA 174
RLKEAGNINRSLSQLGNLINILAE+SQ+GKQRH+PYRDS+LTFLLQESLGGNAKLAM+CA
Sbjct: 61 RLKEAGNINRSLSQLGNLINILAEISQSGKQRHVPYRDSKLTFLLQESLGGNAKLAMICA 120
Query: 175 ISPAQS 180
+SP+QS
Sbjct: 121 VSPSQS 126
>UniRef100_Q8GZU1 Kinesin related protein [Lycopersicon esculentum]
Length = 1191
Score = 191 bits (484), Expect = 1e-46
Identities = 98/178 (55%), Positives = 135/178 (75%), Gaps = 1/178 (0%)
Query: 1 IYNEQITDLLDPSQRNLQIREDVKSGVYVENLTEEQVSTMKDVTQLLLKGLSNRRIGATS 60
IY+E I DLLDP+QRNL+I +D + G YVEN+TEE VST +DV+Q+L+KGLS+R++G+TS
Sbjct: 234 IYDEHIGDLLDPTQRNLKIMDDPRVGFYVENITEEYVSTYEDVSQMLIKGLSSRKVGSTS 293
Query: 61 INSESSRSHTVFTCVVESRCK-SAADGVSRFKTSRINLVDLAGSERQKSTGAAGERLKEA 119
INS+SSRSH VFTCV+ES CK S++ K SR++LVDLAG ++ A + +KE
Sbjct: 294 INSKSSRSHIVFTCVIESWCKESSSTCFGSSKMSRMSLVDLAGFDKNIPDDAGKQLVKEG 353
Query: 120 GNINRSLSQLGNLINILAEVSQTGKQRHIPYRDSRLTFLLQESLGGNAKLAMVCAISP 177
+ +S S LG+L+N+L+E SQ+ K + Y S LT L++ESLGGNAKL+++CAISP
Sbjct: 354 KYVKKSTSLLGHLVNVLSERSQSRKLEDVSYSSSTLTHLMRESLGGNAKLSVICAISP 411
>UniRef100_Q9GYZ0 Kinesin-like protein KRP180 [Strongylocentrotus purpuratus]
Length = 1463
Score = 189 bits (479), Expect = 5e-46
Identities = 99/177 (55%), Positives = 131/177 (73%), Gaps = 3/177 (1%)
Query: 1 IYNEQITDLLDPSQRNLQIREDVKSGVYVENLTEEQVSTMKDVTQLLLKGLSNRRIGATS 60
IYNEQI DLLDP+ L +RE++K GV+V+ L E V++ + +L G NRR+ ATS
Sbjct: 160 IYNEQIYDLLDPASLGLHLRENMKKGVFVDGLIERAVASASEAYGVLQAGWHNRRVAATS 219
Query: 61 INSESSRSHTVFTCVVESRCKSAADGVSRFKTSRINLVDLAGSERQKSTGAAGERLKEAG 120
+N ESSRSH VFT +ES+ K A GVS + S+++LVDLAGSERQK T A G RLKEAG
Sbjct: 220 MNRESSRSHAVFTVSIESKEKKA--GVSNIRVSQLHLVDLAGSERQKDTKAIGVRLKEAG 277
Query: 121 NINRSLSQLGNLINILAEVSQTGKQRHIPYRDSRLTFLLQESLGGNAKLAMVCAISP 177
+IN+SLS LGN+I L +++ GKQRH+PYRDS+L+FLL++SLGGNAK ++ + P
Sbjct: 278 SINKSLSILGNVIMALVDIAH-GKQRHVPYRDSKLSFLLRDSLGGNAKTYIIANVHP 333
>UniRef100_Q93XF4 Kinesin heavy chain [Zea mays]
Length = 857
Score = 185 bits (469), Expect = 7e-45
Identities = 90/176 (51%), Positives = 135/176 (76%), Gaps = 2/176 (1%)
Query: 1 IYNEQITDLLDPSQRNLQIREDVKSGVYVENLTEEQVSTMKDVTQLLLKGLSNRRIGATS 60
++NEQI DLL+PSQR+LQIRE+ +G++VENLT+E VST D++Q+L+KGLSNR++G TS
Sbjct: 6 VHNEQINDLLEPSQRDLQIRENASNGIHVENLTDEYVSTADDISQILMKGLSNRKVGTTS 65
Query: 61 INSESSRSHTVFTCVVESRCKSAADGVSRFKTSRINLVDLAGSERQKSTGAAGERLKEAG 120
+N +SSRSH +FTCV+E+ K +++G S +TSRI VDLAG + + GAA KE
Sbjct: 66 MNLKSSRSHVIFTCVIEAWSKGSSNGFSSSRTSRITFVDLAGPDTDE-LGAAKHSTKEER 124
Query: 121 NINRSLSQLGNLINILAEVSQTGKQRHIPYRDSRLTFLLQESLGGNAKLAMVCAIS 176
++ +SLS+LG L+N+L+E ++ K +PY SRLT +L+++LGGN+++ +C+IS
Sbjct: 125 HLKKSLSRLGKLVNVLSETPESHKV-DLPYEQSRLTHVLKDTLGGNSRVIFLCSIS 179
>UniRef100_Q70MX5 Kinesin family member 15 [Mus musculus]
Length = 1387
Score = 182 bits (463), Expect = 3e-44
Identities = 99/177 (55%), Positives = 125/177 (69%), Gaps = 3/177 (1%)
Query: 1 IYNEQITDLLDPSQRNLQIREDVKSGVYVENLTEEQVSTMKDVTQLLLKGLSNRRIGATS 60
+YNEQI DLLD + L +RE +K GV+V E+ V++ + Q+L +G NRR+ +TS
Sbjct: 169 VYNEQIYDLLDSASVGLYLREHIKKGVFVVGAVEQAVTSAAETYQVLSRGWRNRRVASTS 228
Query: 61 INSESSRSHTVFTCVVESRCKSAADGVSRFKTSRINLVDLAGSERQKSTGAAGERLKEAG 120
+N ESSRSH VFT +ES KS+ +TS +NLVDLAGSERQK T A G RLKEAG
Sbjct: 229 MNRESSRSHAVFTITIESMEKSSE--TVNIRTSLLNLVDLAGSERQKDTHAEGMRLKEAG 286
Query: 121 NINRSLSQLGNLINILAEVSQTGKQRHIPYRDSRLTFLLQESLGGNAKLAMVCAISP 177
NINRSLS LG +I L +V GKQRHI YRDS+LTFLL++SLGGNAK A++ + P
Sbjct: 287 NINRSLSCLGQVITALVDVG-NGKQRHICYRDSKLTFLLRDSLGGNAKTAIIANVHP 342
>UniRef100_Q6P9L6 Kinesin-like 7 [Mus musculus]
Length = 1387
Score = 182 bits (463), Expect = 3e-44
Identities = 99/177 (55%), Positives = 125/177 (69%), Gaps = 3/177 (1%)
Query: 1 IYNEQITDLLDPSQRNLQIREDVKSGVYVENLTEEQVSTMKDVTQLLLKGLSNRRIGATS 60
+YNEQI DLLD + L +RE +K GV+V E+ V++ + Q+L +G NRR+ +TS
Sbjct: 169 VYNEQIYDLLDSASVGLYLREHIKKGVFVVGAVEQAVTSAAETYQVLSRGWRNRRVASTS 228
Query: 61 INSESSRSHTVFTCVVESRCKSAADGVSRFKTSRINLVDLAGSERQKSTGAAGERLKEAG 120
+N ESSRSH VFT +ES KS+ +TS +NLVDLAGSERQK T A G RLKEAG
Sbjct: 229 MNRESSRSHAVFTITIESMEKSSE--TVNIRTSLLNLVDLAGSERQKDTHAEGMRLKEAG 286
Query: 121 NINRSLSQLGNLINILAEVSQTGKQRHIPYRDSRLTFLLQESLGGNAKLAMVCAISP 177
NINRSLS LG +I L +V GKQRHI YRDS+LTFLL++SLGGNAK A++ + P
Sbjct: 287 NINRSLSCLGQVITALVDVG-NGKQRHICYRDSKLTFLLRDSLGGNAKTAIIANVHP 342
>UniRef100_UPI000036B4A8 UPI000036B4A8 UniRef100 entry
Length = 974
Score = 182 bits (462), Expect = 4e-44
Identities = 99/177 (55%), Positives = 125/177 (69%), Gaps = 3/177 (1%)
Query: 1 IYNEQITDLLDPSQRNLQIREDVKSGVYVENLTEEQVSTMKDVTQLLLKGLSNRRIGATS 60
IYNEQI DLLD + L +RE +K GV+V E+ V++ + Q+L G NRR+ +TS
Sbjct: 162 IYNEQIYDLLDSASAGLYLREHIKKGVFVVGAVEQVVTSAAEAYQVLSGGWRNRRVASTS 221
Query: 61 INSESSRSHTVFTCVVESRCKSAADGVSRFKTSRINLVDLAGSERQKSTGAAGERLKEAG 120
+N ESSRSH VFT +ES KS + + +TS +NLVDLAGSERQK T A G RLKEAG
Sbjct: 222 MNRESSRSHAVFTITIESMEKS--NEIVNIRTSLLNLVDLAGSERQKDTHAEGMRLKEAG 279
Query: 121 NINRSLSQLGNLINILAEVSQTGKQRHIPYRDSRLTFLLQESLGGNAKLAMVCAISP 177
NINRSLS LG +I L +V GKQRH+ YRDS+LTFLL++SLGGNAK A++ + P
Sbjct: 280 NINRSLSCLGQVITALVDVG-NGKQRHVCYRDSKLTFLLRDSLGGNAKTAIIANVHP 335
>UniRef100_Q9NS87 Kinesin-like protein 2 [Homo sapiens]
Length = 1388
Score = 182 bits (462), Expect = 4e-44
Identities = 99/177 (55%), Positives = 125/177 (69%), Gaps = 3/177 (1%)
Query: 1 IYNEQITDLLDPSQRNLQIREDVKSGVYVENLTEEQVSTMKDVTQLLLKGLSNRRIGATS 60
IYNEQI DLLD + L +RE +K GV+V E+ V++ + Q+L G NRR+ +TS
Sbjct: 169 IYNEQIYDLLDSASAGLYLREHIKKGVFVVGAVEQVVTSAAEAYQVLSGGWRNRRVASTS 228
Query: 61 INSESSRSHTVFTCVVESRCKSAADGVSRFKTSRINLVDLAGSERQKSTGAAGERLKEAG 120
+N ESSRSH VFT +ES KS + + +TS +NLVDLAGSERQK T A G RLKEAG
Sbjct: 229 MNRESSRSHAVFTITIESMEKS--NEIVNIRTSLLNLVDLAGSERQKDTHAEGMRLKEAG 286
Query: 121 NINRSLSQLGNLINILAEVSQTGKQRHIPYRDSRLTFLLQESLGGNAKLAMVCAISP 177
NINRSLS LG +I L +V GKQRH+ YRDS+LTFLL++SLGGNAK A++ + P
Sbjct: 287 NINRSLSCLGQVITALVDVG-NGKQRHVCYRDSKLTFLLRDSLGGNAKTAIIANVHP 342
>UniRef100_Q7TSP2 Kinesin-like protein KIF15 [Rattus norvegicus]
Length = 1385
Score = 182 bits (461), Expect = 6e-44
Identities = 98/177 (55%), Positives = 125/177 (70%), Gaps = 3/177 (1%)
Query: 1 IYNEQITDLLDPSQRNLQIREDVKSGVYVENLTEEQVSTMKDVTQLLLKGLSNRRIGATS 60
+YNEQI DLLD + L +RE +K GV+V E+ V++ + Q+L +G NRR+ +TS
Sbjct: 169 VYNEQIYDLLDSASVGLYLREHIKKGVFVVGAVEQVVASAAEAYQVLSRGWRNRRVASTS 228
Query: 61 INSESSRSHTVFTCVVESRCKSAADGVSRFKTSRINLVDLAGSERQKSTGAAGERLKEAG 120
+N ESSRSH VFT +ES KS+ +TS +NLVDLAGSERQK T A G RLKEAG
Sbjct: 229 MNRESSRSHAVFTITIESMEKSSE--AVNIRTSLLNLVDLAGSERQKDTHAEGMRLKEAG 286
Query: 121 NINRSLSQLGNLINILAEVSQTGKQRHIPYRDSRLTFLLQESLGGNAKLAMVCAISP 177
NINRSLS LG +I L +V GKQRH+ YRDS+LTFLL++SLGGNAK A++ + P
Sbjct: 287 NINRSLSCLGQVITALVDVG-NGKQRHVCYRDSKLTFLLRDSLGGNAKTAIIANVHP 342
>UniRef100_Q7TN17 Kinesin-like protein KIF15 [Rattus norvegicus]
Length = 1385
Score = 182 bits (461), Expect = 6e-44
Identities = 98/177 (55%), Positives = 125/177 (70%), Gaps = 3/177 (1%)
Query: 1 IYNEQITDLLDPSQRNLQIREDVKSGVYVENLTEEQVSTMKDVTQLLLKGLSNRRIGATS 60
+YNEQI DLLD + L +RE +K GV+V E+ V++ + Q+L +G NRR+ +TS
Sbjct: 169 VYNEQIYDLLDSASVGLYLREHIKKGVFVVGAVEQVVASAAEAYQVLSRGWRNRRVASTS 228
Query: 61 INSESSRSHTVFTCVVESRCKSAADGVSRFKTSRINLVDLAGSERQKSTGAAGERLKEAG 120
+N ESSRSH VFT +ES KS+ +TS +NLVDLAGSERQK T A G RLKEAG
Sbjct: 229 MNRESSRSHAVFTITIESMEKSSE--AVNIRTSLLNLVDLAGSERQKDTHAEGMRLKEAG 286
Query: 121 NINRSLSQLGNLINILAEVSQTGKQRHIPYRDSRLTFLLQESLGGNAKLAMVCAISP 177
NINRSLS LG +I L +V GKQRH+ YRDS+LTFLL++SLGGNAK A++ + P
Sbjct: 287 NINRSLSCLGQVITALVDVG-NGKQRHVCYRDSKLTFLLRDSLGGNAKTAIIANVHP 342
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.375 0.168 0.667
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,174,443,991
Number of Sequences: 2790947
Number of extensions: 37639830
Number of successful extensions: 239271
Number of sequences better than 10.0: 1426
Number of HSP's better than 10.0 without gapping: 1355
Number of HSP's successfully gapped in prelim test: 71
Number of HSP's that attempted gapping in prelim test: 234275
Number of HSP's gapped (non-prelim): 1911
length of query: 1011
length of database: 848,049,833
effective HSP length: 137
effective length of query: 874
effective length of database: 465,690,094
effective search space: 407013142156
effective search space used: 407013142156
T: 11
A: 40
X1: 13 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 35 (21.5 bits)
S2: 80 (35.4 bits)
Medicago: description of AC147484.10


