

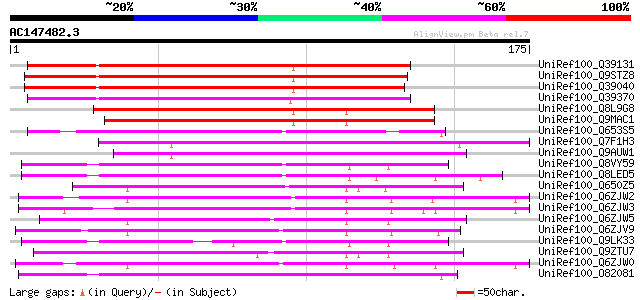
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147482.3 - phase: 0
(175 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q39131 Lamin [Arabidopsis thaliana] 122 5e-27
UniRef100_Q9STZ8 Blue copper-binding protein, 15K [Arabidopsis t... 114 8e-25
UniRef100_Q39040 Lamin [Arabidopsis thaliana] 112 3e-24
UniRef100_Q39370 Lamin [Brassica oleracea] 110 2e-23
UniRef100_Q8L9G8 Putative lamin [Arabidopsis thaliana] 103 2e-21
UniRef100_Q9MAC1 Putative lamin [Arabidopsis thaliana] 102 3e-21
UniRef100_Q653S5 Hypothetical protein OJ1065_E04.1 [Oryza sativa] 102 5e-21
UniRef100_Q7F1H3 Blue copper-binding protein-like [Oryza sativa] 90 2e-17
UniRef100_Q9AUW1 Putative blue copper-binding protein [Oryza sat... 86 4e-16
UniRef100_Q8VY59 Blue copper protein, putative [Arabidopsis thal... 74 1e-12
UniRef100_Q8LED5 Blue copper protein, putative [Arabidopsis thal... 74 1e-12
UniRef100_Q650Z5 Blue copper-binding protein-like [Oryza sativa] 74 1e-12
UniRef100_Q6ZJW2 Putative blue copper binding protein [Oryza sat... 73 3e-12
UniRef100_Q6ZJW3 Putative blue copper binding protein [Oryza sat... 72 5e-12
UniRef100_Q6ZJW5 Putative blue copper binding protein [Oryza sat... 72 6e-12
UniRef100_Q6ZJV9 Putative blue copper binding protein [Oryza sat... 72 8e-12
UniRef100_Q9LK33 Arabidopsis thaliana genomic DNA, chromosome 3,... 70 2e-11
UniRef100_Q9ZTU7 Blue copper-binding protein homolog [Triticum a... 69 7e-11
UniRef100_Q6ZJW0 Putative blue copper binding protein [Oryza sat... 68 9e-11
UniRef100_O82081 Uclacyanin I [Arabidopsis thaliana] 68 1e-10
>UniRef100_Q39131 Lamin [Arabidopsis thaliana]
Length = 172
Score = 122 bits (305), Expect = 5e-27
Identities = 62/130 (47%), Positives = 84/130 (63%), Gaps = 2/130 (1%)
Query: 7 LLMVSVVLGVLSLWPMVVMGGPKLHKVGGSKGWKENVNYTTWSSQEHVYVGDWLKFVFDK 66
+L+ +VVL L PM + K + VG +K W N+NYT W+ +H Y+GDWL FVFD+
Sbjct: 6 VLITAVVLAFLMAAPMPGVTAKK-YTVGENKFWNPNINYTIWAQGKHFYLGDWLYFVFDR 64
Query: 67 RYYNVLEVNKTGYDYCIDMTFIRNLTRG-GRDVVQLTEAKTYYFITGGGYCFHGMKVAVD 125
+N+LEVNKT Y+ CI IRN TRG GRD+V L + K YY + G G C+ GMK++V
Sbjct: 65 NQHNILEVNKTDYEGCIADHPIRNWTRGAGRDIVTLNQTKHYYLLDGKGGCYGGMKLSVK 124
Query: 126 VQEHPTPAPS 135
V++ P P S
Sbjct: 125 VEKLPPPPKS 134
>UniRef100_Q9STZ8 Blue copper-binding protein, 15K [Arabidopsis thaliana]
Length = 141
Score = 114 bits (286), Expect = 8e-25
Identities = 60/130 (46%), Positives = 79/130 (60%), Gaps = 2/130 (1%)
Query: 6 VLLMVSVVLGVLSLWPMVVMGGPKLHKVGGSKGWKENVNYTTWSSQEHVYVGDWLKFVFD 65
++L+ +VVL L P+ + K + VG K W N+NYT W+ +H YVGDWL FVF
Sbjct: 5 MVLISAVVLAFLVAAPIPEVTAKK-YLVGDKKFWNPNINYTLWAQGKHFYVGDWLYFVFY 63
Query: 66 KRYYNVLEVNKTGYDYCIDMTFIRNLTRG-GRDVVQLTEAKTYYFITGGGYCFHGMKVAV 124
+ +N+LEVNK Y+ CI IRN TRG GRD+V L E + YY + G G C GMK+ V
Sbjct: 64 RDQHNILEVNKADYEKCISNRPIRNYTRGAGRDIVPLYETRRYYLLDGRGGCVQGMKLDV 123
Query: 125 DVQEHPTPAP 134
V+ P P P
Sbjct: 124 LVETPPPPPP 133
>UniRef100_Q39040 Lamin [Arabidopsis thaliana]
Length = 134
Score = 112 bits (281), Expect = 3e-24
Identities = 59/129 (45%), Positives = 79/129 (60%), Gaps = 2/129 (1%)
Query: 6 VLLMVSVVLGVLSLWPMVVMGGPKLHKVGGSKGWKENVNYTTWSSQEHVYVGDWLKFVFD 65
++L+ +VVL L P+ + K + VG K W N+NYT W+ +H YVGDWL FVF
Sbjct: 5 MVLISAVVLAFLVAAPIPEVTAKK-YLVGDKKFWNPNINYTLWAQGKHFYVGDWLYFVFY 63
Query: 66 KRYYNVLEVNKTGYDYCIDMTFIRNLTRG-GRDVVQLTEAKTYYFITGGGYCFHGMKVAV 124
+ +N+LEVNK Y+ CI IRN TRG GRD+V L E + YY + G G C GMK+ V
Sbjct: 64 RDQHNILEVNKADYEKCISNLPIRNYTRGAGRDIVPLYETRRYYLLDGRGGCVQGMKLDV 123
Query: 125 DVQEHPTPA 133
V+ P P+
Sbjct: 124 LVETPPPPS 132
>UniRef100_Q39370 Lamin [Brassica oleracea]
Length = 165
Score = 110 bits (275), Expect = 2e-23
Identities = 58/130 (44%), Positives = 78/130 (59%), Gaps = 2/130 (1%)
Query: 7 LLMVSVVLGVLSLWPMVVMGGPKLHKVGGSKGWKENVNYTTWSSQEHVYVGDWLKFVFDK 66
+L+ + VL L P+ + K + VG K W ++NY TW +H Y+GDWL FV+ +
Sbjct: 6 VLIAAPVLAFLVAAPVPEVTAKK-YLVGDKKFWNPDINYDTWVQGKHFYLGDWLYFVYYR 64
Query: 67 RYYNVLEVNKTGYDYCIDMTFIRNLTR-GGRDVVQLTEAKTYYFITGGGYCFHGMKVAVD 125
+N+LEVNKT Y+ CI IRN TR GGRD+V L K YY + G G CF GMK+ V
Sbjct: 65 DQHNILEVNKTDYEGCISDHPIRNYTRGGGRDIVPLNVTKQYYLLDGRGGCFKGMKLTVT 124
Query: 126 VQEHPTPAPS 135
V++ P P S
Sbjct: 125 VEKLPPPPKS 134
>UniRef100_Q8L9G8 Putative lamin [Arabidopsis thaliana]
Length = 167
Score = 103 bits (257), Expect = 2e-21
Identities = 51/117 (43%), Positives = 73/117 (61%), Gaps = 2/117 (1%)
Query: 29 KLHKVGGSKGWKENVNYTTWSSQEHVYVGDWLKFVFDKRYYNVLEVNKTGYDYCIDMTFI 88
K VG +K W N+NYT W+ +H Y+ DWL FV+++ YNV+EVN+T Y C I
Sbjct: 25 KRRTVGDNKFWNPNINYTIWAQDKHFYLDDWLYFVYERNQYNVIEVNETNYISCNPNNPI 84
Query: 89 RNLTRG-GRDVVQLTEAKTYYFITG-GGYCFHGMKVAVDVQEHPTPAPSPSLSDTAK 143
N +RG GRD+V L + YY I+G GG C+ GMK+AV V++ P P + ++A+
Sbjct: 85 ANWSRGAGRDLVHLNVTRHYYLISGNGGGCYGGMKLAVLVEKPPPPPAAAPNKNSAR 141
>UniRef100_Q9MAC1 Putative lamin [Arabidopsis thaliana]
Length = 167
Score = 102 bits (255), Expect = 3e-21
Identities = 50/113 (44%), Positives = 72/113 (63%), Gaps = 2/113 (1%)
Query: 33 VGGSKGWKENVNYTTWSSQEHVYVGDWLKFVFDKRYYNVLEVNKTGYDYCIDMTFIRNLT 92
VG +K W N+NYT W+ +H Y+ DWL FV+++ YNV+EVN+T Y C I N +
Sbjct: 29 VGDNKFWNPNINYTIWAQDKHFYLDDWLYFVYERNQYNVIEVNETNYISCNPNNPIANWS 88
Query: 93 RG-GRDVVQLTEAKTYYFITG-GGYCFHGMKVAVDVQEHPTPAPSPSLSDTAK 143
RG GRD+V L + YY I+G GG C+ GMK+AV V++ P P + ++A+
Sbjct: 89 RGAGRDLVHLNVTRHYYLISGNGGGCYGGMKLAVLVEKPPPPPAAAPNKNSAR 141
>UniRef100_Q653S5 Hypothetical protein OJ1065_E04.1 [Oryza sativa]
Length = 168
Score = 102 bits (253), Expect = 5e-21
Identities = 58/142 (40%), Positives = 79/142 (54%), Gaps = 11/142 (7%)
Query: 7 LLMVSVVLGVLSLWPMVVMGGPKLHKVGGSKGWKENVNYTTWSSQEHVYVGDWLKFVFDK 66
LL+V+VV G V + G H VG + GW N++Y+ WS + YVGD + F + K
Sbjct: 5 LLLVAVVAGFA-----VSLAGATDHIVGANHGWNPNIDYSLWSGNQTFYVGDLISFRYQK 59
Query: 67 RYYNVLEVNKTGYDYCIDMTFIRNLTRGGRDVVQLTEAKTYYFITGGGYCFHGMKVAVDV 126
+NV EVN+TGYD C N T G+D + L +++ YYFI G G+C GMKVA+ V
Sbjct: 60 GTHNVFEVNQTGYDNCTMAGVAGNWT-SGKDFIPLNDSRRYYFICGNGFCQAGMKVAITV 118
Query: 127 QEHPTPAPSPSLSDTAKS-GGD 147
P + D AK+ GGD
Sbjct: 119 H----PLKHNATGDGAKNHGGD 136
>UniRef100_Q7F1H3 Blue copper-binding protein-like [Oryza sativa]
Length = 178
Score = 90.1 bits (222), Expect = 2e-17
Identities = 50/148 (33%), Positives = 75/148 (49%), Gaps = 3/148 (2%)
Query: 31 HKVGGSKGWKENVNYTTWSSQEH-VYVGDWLKFVFDKRYYNVLEVNKTGYDYCIDMTFIR 89
+ VG KGW +V+YT W + Y GDWL F + +V++V++ GYD C I
Sbjct: 26 YTVGDEKGWNPDVDYTAWVKKHRPFYKGDWLLFEYQNGRSDVVQVDEVGYDNCDKANAIS 85
Query: 90 NLTRGGRDVVQLTEAKTYYFITGGGYCFHGMKVAVDVQEHPTPAPSPSLSDTAKSGGDSI 149
+ ++G QL EAK YYFI GYC+ GMK+AV ++ + S D++ S
Sbjct: 86 SYSKGHSYAFQLKEAKDYYFICSYGYCYKGMKLAVTAKKGSASSSSSGSGDSSSSSKSDT 145
Query: 150 L--PSMYTCFGIIVANVVYVSLVLVGIL 175
S + +AN Y +L+ V I+
Sbjct: 146 ASSKSKSSAAASSLANPSYAALLAVAII 173
>UniRef100_Q9AUW1 Putative blue copper-binding protein [Oryza sativa]
Length = 165
Score = 85.9 bits (211), Expect = 4e-16
Identities = 41/120 (34%), Positives = 64/120 (53%), Gaps = 1/120 (0%)
Query: 36 SKGWKENVNYTTWSSQEH-VYVGDWLKFVFDKRYYNVLEVNKTGYDYCIDMTFIRNLTRG 94
S WK N NY+ W +Q Y GDWL F + +V++V+ GY+ C I N ++G
Sbjct: 31 SINWKPNTNYSDWPAQHGPFYKGDWLVFYYTAGQADVIQVDAAGYNTCDATNAISNYSKG 90
Query: 95 GRDVVQLTEAKTYYFITGGGYCFHGMKVAVDVQEHPTPAPSPSLSDTAKSGGDSILPSMY 154
+L E KTYYFI GYCF GM++ + ++ P P+P + D + + + S++
Sbjct: 91 RTYAFELNETKTYYFICSYGYCFGGMRLQIKTEKLPPPSPPAAAKDKSAAAFTASRASLF 150
>UniRef100_Q8VY59 Blue copper protein, putative [Arabidopsis thaliana]
Length = 174
Score = 74.3 bits (181), Expect = 1e-12
Identities = 48/150 (32%), Positives = 83/150 (55%), Gaps = 11/150 (7%)
Query: 5 RVLLMVSVVLGVLSLWPMVVMGGPKLHKVGGSKGWKENVNYTTWSSQEHVYVGDWLKFVF 64
+ +L++ V G+LS+ + H +GGS+GW+++V++ +WSS + VGD + F +
Sbjct: 4 QAVLVILVFSGLLSVKTALAAR----HVIGGSQGWEQSVDFDSWSSDQSFKVGDQIVFKY 59
Query: 65 DKRYYNVLEVNKTGYDYCIDMTFIRNLTRGGRDVVQLTEAKTYYFITGG-GYCFHGMKVA 123
+ + V ++T Y C T + +L+ G DVV+L++ T YF G G+C GMK+
Sbjct: 60 SELHSVVELGSETAYKSCDLGTSVNSLS-SGNDVVKLSKTGTRYFACGTVGHCEQGMKIK 118
Query: 124 VDV-----QEHPTPAPSPSLSDTAKSGGDS 148
V+V + +P+ S S SD+ G S
Sbjct: 119 VNVVSSDSKSASSPSGSGSGSDSGSGSGSS 148
>UniRef100_Q8LED5 Blue copper protein, putative [Arabidopsis thaliana]
Length = 172
Score = 74.3 bits (181), Expect = 1e-12
Identities = 53/172 (30%), Positives = 93/172 (53%), Gaps = 15/172 (8%)
Query: 5 RVLLMVSVVLGVLSLWPMVVMGGPKLHKVGGSKGWKENVNYTTWSSQEHVYVGDWLKFVF 64
+ +L++ V G+LS+ + H +GGS+GW+++V++ +WSS + VGD + F +
Sbjct: 4 QAVLVILVFSGLLSVKTALAAQ----HVIGGSQGWEQSVDFDSWSSDQSFKVGDQIVFKY 59
Query: 65 DKRYYNVLEVNKTGYDYCIDMTFIRNLTRGGRDVVQLTEAKTYYFITGG-GYCFHGMKV- 122
+ V ++T Y C T + +L+ G DVV+L++ T YF+ G G+C GMK+
Sbjct: 60 SGLHSVVELGSETAYKSCDLGTSVNSLS-SGNDVVKLSKTGTRYFVCGTVGHCEQGMKIK 118
Query: 123 --AVDVQEHPTPAPSPSLSDTA----KSGGDSILPSMYTCF--GIIVANVVY 166
VD + +P+ S S SD+ S GD + S+ F G +V +++
Sbjct: 119 VNVVDSKSGSSPSGSGSGSDSGSGSKSSSGDGLRASIGYMFVVGSLVIGLIW 170
>UniRef100_Q650Z5 Blue copper-binding protein-like [Oryza sativa]
Length = 159
Score = 74.3 bits (181), Expect = 1e-12
Identities = 50/139 (35%), Positives = 69/139 (48%), Gaps = 8/139 (5%)
Query: 22 MVVMGGPKLHKVGGSKG-WKENVNYTTWSSQEHVYVGDWLKFVFDKRYYNVLEVNKTGYD 80
+V M + VG G W NYT W +Q+ + GD + F + + ++V+EVNK GYD
Sbjct: 5 LVGMASAATYNVGEPGGAWDLTTNYTNWVAQKRFHPGDQIVFKYSAQRHDVVEVNKAGYD 64
Query: 81 YCIDMTFIRNLTRGGRDVVQLTEAKTYYFITG-GGYC----FHGMKVAVD-VQEHPTPAP 134
C T I T G DV+ LT T YFI G G+C MK+ +D VQ + AP
Sbjct: 65 SCSTSTSIATHTT-GNDVIPLTSTGTRYFICGFPGHCTTTGTGNMKIQIDVVQADSSSAP 123
Query: 135 SPSLSDTAKSGGDSILPSM 153
+P + T S S S+
Sbjct: 124 APVATTTPPSPPSSAATSL 142
>UniRef100_Q6ZJW2 Putative blue copper binding protein [Oryza sativa]
Length = 188
Score = 73.2 bits (178), Expect = 3e-12
Identities = 62/190 (32%), Positives = 86/190 (44%), Gaps = 24/190 (12%)
Query: 4 SRVLLMVSVVLGVLSLWPMVVMGGPKLHKVGGSKG-WKENVNYTTWSSQEHVYVGDWLKF 62
+R LL+V++ VL G + VG G W NY W S VGD L F
Sbjct: 3 ARALLVVAMAAAVLG-----TAMGVTTYTVGAPAGSWDTRTNYAQWVSAITFRVGDQLVF 57
Query: 63 VFDKRYYNVLEVNKTGYDYCIDMTFIRNLTRGGRDVVQLTEAKTYYFITG-GGYCFHGMK 121
+ ++V+EVNK YD C + I G D + L T YFI G G+C GMK
Sbjct: 58 KYSPAAHDVVEVNKADYDSCSSSSPISTFNSGD-DTIPLAAIGTRYFICGFPGHCTAGMK 116
Query: 122 VAVDVQ----EHPTPAPSPSLSDT----------AKSGGDSILPSMYTCFGIIVANVVYV 167
VAV V+ +PTP+P L T +GG PS + VA++V +
Sbjct: 117 VAVKVEAATGSNPTPSPLAPLPRTPTVMAPNAMPPTNGGRPTPPSSSASKPVGVASLVGL 176
Query: 168 SL--VLVGIL 175
SL ++ G++
Sbjct: 177 SLSAIVAGLM 186
>UniRef100_Q6ZJW3 Putative blue copper binding protein [Oryza sativa]
Length = 187
Score = 72.4 bits (176), Expect = 5e-12
Identities = 63/191 (32%), Positives = 89/191 (45%), Gaps = 27/191 (14%)
Query: 4 SRVLLMVSVVLGVL--SLWPMVVMGGPKLHKVGGSKGWKENVNYTTWSSQEHVYVGDWLK 61
+R LL+V++ VL +L +G P S W NY W S + GD +
Sbjct: 3 ARALLVVAMAAAVLGTALGATYTVGAP-------SGSWDLRTNYDQWVSNINFRAGDQIV 55
Query: 62 FVFDKRYYNVLEVNKTGYDYCIDMTFIRNLTRGGRDVVQLTEAKTYYFITG-GGYCFHGM 120
F + ++V+EVNK YD C + I G D + LT T YFI G G+C GM
Sbjct: 56 FKYSPAAHDVVEVNKADYDSCSSSSPIATFNSGD-DTIPLTATGTRYFICGFNGHCTGGM 114
Query: 121 KVAVDVQ--EHPTPAPSPSL----SDTA--------KSGGDSILPSMYTCFGIIVANVVY 166
KVAV V+ PAPSP + TA +GG + PS VA++V
Sbjct: 115 KVAVKVEAATGSNPAPSPMTPRPRTPTAMAPNAMPPTAGGRPVPPSNSASQPAGVASLVG 174
Query: 167 VSL--VLVGIL 175
+SL ++VG++
Sbjct: 175 LSLGAIVVGLM 185
>UniRef100_Q6ZJW5 Putative blue copper binding protein [Oryza sativa]
Length = 166
Score = 72.0 bits (175), Expect = 6e-12
Identities = 48/154 (31%), Positives = 71/154 (45%), Gaps = 11/154 (7%)
Query: 11 SVVLGVLSLWPMVVMGGPKLHKVGGSKG-WKENVNYTTWSSQEHVYVGDWLKFVFDKRYY 69
+VV+ L + G + VG G W +Y W ++ + GD L F + + +
Sbjct: 8 AVVMVAAVLAGQAMAAGATTYTVGAPDGLWDMETDYKEWVARRTFHPGDKLTFTYSRELH 67
Query: 70 NVLEVNKTGYDYCIDMTFIRNLTRGGRDVVQLTEAKTYYFITG-GGYCFHGMKVAVDV-- 126
+V+EV K GYD C + I + R G D+V LT T YF+ G G+C GMK+ +DV
Sbjct: 68 DVVEVTKAGYDACSNANNI-SAFRSGNDLVALTAVGTRYFLCGLTGHCGSGMKIRIDVVA 126
Query: 127 ------QEHPTPAPSPSLSDTAKSGGDSILPSMY 154
P PS S A +G +L +Y
Sbjct: 127 AASSGPAAAAAPLPSTSSVTAAVAGSRLVLVLLY 160
>UniRef100_Q6ZJV9 Putative blue copper binding protein [Oryza sativa]
Length = 190
Score = 71.6 bits (174), Expect = 8e-12
Identities = 55/161 (34%), Positives = 74/161 (45%), Gaps = 14/161 (8%)
Query: 3 NSRVLLMVSVVLGVLSLWPMVVMGGPKLHKVGGSKG-WKENVNYTTWSSQEHVYVGDWLK 61
++R LL+V+V L + G + VG G W NY W+S GD L
Sbjct: 4 SARALLVVAVAAAAAVLATTAM--GATTYTVGAPAGSWDTRTNYAQWASAATFRAGDRLV 61
Query: 62 FVFDKRYYNVLEVNKTGYDYCIDMTFIRNLTRGGRDVVQLTEAKTYYFITG-GGYCFHGM 120
F + ++V+EV K GYD C + I G D V L T YFI G G+C GM
Sbjct: 62 FRYSPAAHDVVEVTKAGYDACSAASPIATF-NSGDDTVPLAAVGTRYFICGFPGHCAAGM 120
Query: 121 KVAVDVQ--------EHPTPAPSPSLSDTAK-SGGDSILPS 152
K+AV V+ TP+PSPS + +GG + PS
Sbjct: 121 KLAVKVEAAAAAPGGSSTTPSPSPSPAALPPVNGGRPVTPS 161
>UniRef100_Q9LK33 Arabidopsis thaliana genomic DNA, chromosome 3, TAC clone:K17E12
[Arabidopsis thaliana]
Length = 169
Score = 70.5 bits (171), Expect = 2e-11
Identities = 48/151 (31%), Positives = 85/151 (55%), Gaps = 18/151 (11%)
Query: 5 RVLLMVSVVLGVLSLWPMVVMGGPKLHKVGGSKGWKENVNYTTWSSQEHVYVGDWLKFVF 64
+ +L++ V G+LS+ + H +GGS+GW+++V++ +WSS + VGD ++
Sbjct: 4 QAVLVILVFSGLLSVKTALAAR----HVIGGSQGWEQSVDFDSWSSDQSFKVGDQIEL-- 57
Query: 65 DKRYYNVLEV-NKTGYDYCIDMTFIRNLTRGGRDVVQLTEAKTYYFITGG-GYCFHGMKV 122
++V+E+ ++T Y C T + +L+ G DVV+L++ T YF G G+C GMK+
Sbjct: 58 ----HSVVELGSETAYKSCDLGTSVNSLS-SGNDVVKLSKTGTRYFACGTVGHCEQGMKI 112
Query: 123 AVDV-----QEHPTPAPSPSLSDTAKSGGDS 148
V+V + +P+ S S SD+ G S
Sbjct: 113 KVNVVSSDSKSASSPSGSGSGSDSGSGSGSS 143
>UniRef100_Q9ZTU7 Blue copper-binding protein homolog [Triticum aestivum]
Length = 176
Score = 68.6 bits (166), Expect = 7e-11
Identities = 49/157 (31%), Positives = 74/157 (46%), Gaps = 14/157 (8%)
Query: 9 MVSVVLGVLSLWPMVVMGGPKLHKVGGSKGWKENVNYTTWSSQEHVYVGDWLKFVFDKRY 68
M +L V ++ ++ + VG GW N +Y++W S + VGD + F +
Sbjct: 4 MKITLLAVAAMAALLGTASAVTYNVGEQGGWTLNTDYSSWVSGKKFNVGDEIVFKYSSAA 63
Query: 69 YNVLEVNKTGYDYC-IDMTFIRNLTRGGRDVVQLTEAKTYYFITG-GGYC----FHGMKV 122
++V+EV+K GYD C ID N + G DV+ L+ T YFI G G+C MKV
Sbjct: 64 HDVVEVSKAGYDSCSIDGAI--NTFKSGNDVIPLSATGTRYFICGITGHCSPTAAASMKV 121
Query: 123 AVDV------QEHPTPAPSPSLSDTAKSGGDSILPSM 153
+DV P PA P S++ + S S+
Sbjct: 122 MIDVASGSSSPSSPMPAAGPGASNSPPAPPSSAATSV 158
>UniRef100_Q6ZJW0 Putative blue copper binding protein [Oryza sativa]
Length = 195
Score = 68.2 bits (165), Expect = 9e-11
Identities = 58/194 (29%), Positives = 84/194 (42%), Gaps = 26/194 (13%)
Query: 3 NSRVLLMVSVVLGVLSLWPMVVMGGPKLHKVGGSKG-WKENVNYTTWSSQEHVYVGDWLK 61
N LL+V++ VL+ G + VG G W NY W S VGD L
Sbjct: 5 NRSALLVVAMAAAVLT----TTATGATTYTVGAPAGSWDTRTNYAQWVSAVTFRVGDQLV 60
Query: 62 FVFDKRYYNVLEVNKTGYDYCIDMTFIRNLTRGGRDVVQLTEAKTYYFITG-GGYCFHGM 120
F + ++V+EV K GYD C + G D V LT T YF+ G G+C GM
Sbjct: 61 FKYSPAAHDVVEVTKAGYDSCSSSGPVATF-NSGDDTVPLTATGTRYFMCGFPGHCAAGM 119
Query: 121 KVAVDVQE--------------HPTPAPSPSLSDTA---KSGGDSILPSMYTCFGIIVAN 163
K+AV V+ P P +++ A +GG + PS VA+
Sbjct: 120 KIAVKVEAATATGGSGTALSPMAPRPRTPTAMAPNAMPPMAGGRPVSPSSSASKSTGVAS 179
Query: 164 VVYVSL--VLVGIL 175
+V +SL ++ G++
Sbjct: 180 LVGLSLGAIVAGLM 193
>UniRef100_O82081 Uclacyanin I [Arabidopsis thaliana]
Length = 261
Score = 67.8 bits (164), Expect = 1e-10
Identities = 45/152 (29%), Positives = 69/152 (44%), Gaps = 8/152 (5%)
Query: 4 SRVLLMVSVVLGVLSLWPMVVMGGPKLHKVGGSKGWKENVNYTTWSSQEHVYVGDWLKFV 63
SR +L++ VL + V H +GG GW + TW++ + VGD L F
Sbjct: 3 SREMLIIISVLATTLIGLTVATD----HTIGGPSGWTVGASLRTWAAGQTFAVGDNLVFS 58
Query: 64 FDKRYYNVLEVNKTGYDYCIDMTFIRNLTRGGRDVVQLTEAKTYYFITGGGYCFHGMKVA 123
+ +++V+EV K +D C + + G V T K Y+ G+C GMK+
Sbjct: 59 YPAAFHDVVEVTKPEFDSCQAVKPLITFANGNSLVPLTTPGKRYFICGMPGHCSQGMKLE 118
Query: 124 VDVQEHPTPAPSPSLSDTAKS----GGDSILP 151
V+V T AP+ L +T S S+LP
Sbjct: 119 VNVVPTATVAPTAPLPNTVPSLNAPSPSSVLP 150
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.322 0.140 0.435
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 321,743,453
Number of Sequences: 2790947
Number of extensions: 13787435
Number of successful extensions: 32728
Number of sequences better than 10.0: 148
Number of HSP's better than 10.0 without gapping: 91
Number of HSP's successfully gapped in prelim test: 57
Number of HSP's that attempted gapping in prelim test: 32532
Number of HSP's gapped (non-prelim): 161
length of query: 175
length of database: 848,049,833
effective HSP length: 119
effective length of query: 56
effective length of database: 515,927,140
effective search space: 28891919840
effective search space used: 28891919840
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 70 (31.6 bits)
Medicago: description of AC147482.3


