

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147471.4 + phase: 0
(274 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
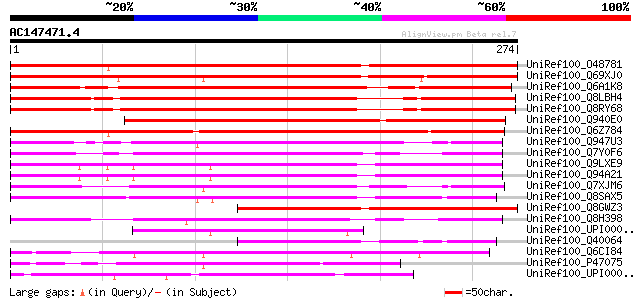
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_O48781 Hypothetical protein At2g26660 [Arabidopsis tha... 298 1e-79
UniRef100_Q69XJ0 Putative ids-4 protein [Oryza sativa] 274 2e-72
UniRef100_Q6A1K8 IDS4-like protein [Lycopersicon esculentum] 274 2e-72
UniRef100_Q8LBH4 Ids4-like protein [Arabidopsis thaliana] 270 4e-71
UniRef100_Q8RY68 AT5g20150/F5O24_40 [Arabidopsis thaliana] 268 1e-70
UniRef100_Q940E0 IDS-4-like protein [Castanea sativa] 248 9e-65
UniRef100_Q6Z784 Putative SPX (SYG1/Pho81/XPR1) domain-containin... 240 3e-62
UniRef100_Q947U3 Hypothetical protein OSJNBa0068A07.13 [Oryza sa... 197 2e-49
UniRef100_Q7Y0F6 SPX domain containing protein [Oryza sativa] 189 9e-47
UniRef100_Q9LXE9 Ids-4 protein-like [Arabidopsis thaliana] 183 4e-45
UniRef100_Q94A21 AT5g15330/F8M21_220 [Arabidopsis thaliana] 183 4e-45
UniRef100_Q7XJM6 At2g45130 protein [Arabidopsis thaliana] 176 8e-43
UniRef100_Q8SAX5 Putative signal transduction protein [Oryza sat... 165 1e-39
UniRef100_Q8GWZ3 Hypothetical protein At2g26660 [Arabidopsis tha... 157 2e-37
UniRef100_Q8H398 Putative ids-4 protein [Oryza sativa] 148 2e-34
UniRef100_UPI00002C8327 UPI00002C8327 UniRef100 entry 99 2e-19
UniRef100_Q40064 Iron-deficiency specific clone No.4 [Hordeum vu... 91 2e-17
UniRef100_Q6CI84 Similar to sp|Q01317 Neurospora crassa Ankyrin ... 72 1e-11
UniRef100_P47075 VTC4 protein [Saccharomyces cerevisiae] 71 3e-11
UniRef100_UPI000046B7EB UPI000046B7EB UniRef100 entry 70 6e-11
>UniRef100_O48781 Hypothetical protein At2g26660 [Arabidopsis thaliana]
Length = 287
Score = 298 bits (763), Expect = 1e-79
Identities = 158/277 (57%), Positives = 200/277 (72%), Gaps = 7/277 (2%)
Query: 1 MKFWKILNNQIEQTLPDWRDKFLSYKDLKKQLKLIVPKEIDSSCSKRRRLDD---DGGAE 57
MKF K L+NQIE+TLP+WRDKFLSYK+LKK+LKL+ P+ +++ +KR R D D
Sbjct: 1 MKFGKSLSNQIEETLPEWRDKFLSYKELKKKLKLMEPRSVENRPNKRSRSDSNSVDTDPT 60
Query: 58 GEVTKEVKDFLRLLEVEIEKFNGFFVEKEEEYVIKWKELQDKVAWAKSSDIELMTVGREI 117
+TKE DF+ LLE E+EKFN FFVE+EEEY+I+ KEL+D+VA AK+S+ E++ + +EI
Sbjct: 61 VGMTKEELDFISLLEDELEKFNSFFVEQEEEYIIRLKELKDQVAKAKNSNEEMINIKKEI 120
Query: 118 VDFHGEMVLLENYSALNYTGLVKIIKKYDKRTGALLRLPFIQDVLNQPFFKIDVLNKLVK 177
VDFHGEMVLL NYSALNYTGL KI+KKYDKRTGAL+RLPFIQ VL +PFF D+LN VK
Sbjct: 121 VDFHGEMVLLMNYSALNYTGLAKILKKYDKRTGALIRLPFIQKVLQEPFFTTDLLNTFVK 180
Query: 178 ECEVMLSIIFPKSGPLGQSLSTSEVFEEVARETTTANETKETLDHVPKEFSEIQNMENIF 237
ECE ML +FP + +S + E E L VPKE SEI+ ME+++
Sbjct: 181 ECEAMLDRLFPSN----KSRNLDEEGEPTTSGMVKTGTDDSELLRVPKELSEIEYMESLY 236
Query: 238 IKLTTSALDTLKEIRGGSSTVSIYSLPPLHSETLVED 274
+K T SAL LKEIR GSSTVS++SLPPL + L +D
Sbjct: 237 MKSTVSALKVLKEIRSGSSTVSVFSLPPLPASGLEDD 273
>UniRef100_Q69XJ0 Putative ids-4 protein [Oryza sativa]
Length = 295
Score = 274 bits (701), Expect = 2e-72
Identities = 158/283 (55%), Positives = 201/283 (70%), Gaps = 13/283 (4%)
Query: 1 MKFWKILNNQIEQTLPDWRDKFLSYKDLKKQLKLIVPKEI-DSSCSKRRRLDDDGGAE-- 57
MKF K L++QI +TLP+WRDKFLSYKDLKK+LKLI + +KR R+ DGG E
Sbjct: 1 MKFGKSLSSQIVETLPEWRDKFLSYKDLKKRLKLIGGGGGGEERQAKRARVAADGGEEEA 60
Query: 58 --GEVTKEVKDFLRLLEVEIEKFNGFFVEKEEEYVIKWKELQDKVAWA--KSSDIELMTV 113
+T E F+RLLE E++KFN FFVEKEEEY+I+ KELQD+VA A + S ELM V
Sbjct: 61 AAAAMTPEEAGFMRLLEAELDKFNSFFVEKEEEYIIRQKELQDRVARAAGRESKEELMRV 120
Query: 114 GREIVDFHGEMVLLENYSALNYTGLVKIIKKYDKRTGALLRLPFIQDVLNQPFFKIDVLN 173
+EIVDFHGEMVLLENYSALNYTGLVKI+KKYDKRTGAL+RLPFIQ VL QPFF D+L
Sbjct: 121 RKEIVDFHGEMVLLENYSALNYTGLVKILKKYDKRTGALIRLPFIQKVLQQPFFTTDLLY 180
Query: 174 KLVKECEVMLSIIFPKSGPLGQSLSTSEVFEEVARETTTANETKETLD--HVPKEFSEIQ 231
KLVK+CE ML + P + S+S+ + + E +N + ++ +P E EI+
Sbjct: 181 KLVKQCEAMLDQLLPSN---ELSVSSEDGRGDSTNEDKPSNPSSSLVNGGTIP-ELDEIE 236
Query: 232 NMENIFIKLTTSALDTLKEIRGGSSTVSIYSLPPLHSETLVED 274
ME++++K T +AL +LKEIR GSSTVS +SLPPL ++ E+
Sbjct: 237 YMESMYMKGTVAALRSLKEIRSGSSTVSAFSLPPLQGDSSPEE 279
>UniRef100_Q6A1K8 IDS4-like protein [Lycopersicon esculentum]
Length = 266
Score = 274 bits (700), Expect = 2e-72
Identities = 149/271 (54%), Positives = 197/271 (71%), Gaps = 20/271 (7%)
Query: 1 MKFWKILNNQIEQTLPDWRDKFLSYKDLKKQLKLIVPKEIDSSCSKRRRLDDDGGAEGEV 60
MKFWKIL + IE+TLP+W+DKFLSYKDLKK+LKLI P+ D K++RL++D E+
Sbjct: 1 MKFWKILKSHIEETLPEWQDKFLSYKDLKKELKLIYPQ--DDRPIKKQRLNND-----EL 53
Query: 61 TKEVKDFLRLLEVEIEKFNGFFVEKEEEYVIKWKELQDKVAWAKSSDIELMTVGREIVDF 120
KEV DF++LLE EI+KFN FFVEKEE+Y+I K L+++VA S+ E+ +GR+IVD
Sbjct: 54 AKEVNDFVKLLEEEIDKFNTFFVEKEEDYIIHLKVLKERVAEMGKSNEEVNRLGRDIVDL 113
Query: 121 HGEMVLLENYSALNYTGLVKIIKKYDKRTGALLRLPFIQDVLNQPFFKIDVLNKLVKECE 180
HGEMVLLENYSALNYTG+VKI+KKYDK +G LLRLPF VL +PFF+ +VLNKLVKEC+
Sbjct: 114 HGEMVLLENYSALNYTGVVKILKKYDKLSGELLRLPFHPKVLAEPFFETEVLNKLVKECD 173
Query: 181 VMLSIIFPKSGPLGQSLSTSEVFEEVARETTTANETKETLDHVPKEFSEIQNMENIFIKL 240
+LS + ++ PL +VA + VP+E +EI+NMEN++++L
Sbjct: 174 TLLSHLLYQTEPL-----------KVAGGGGGGGGERPV--KVPQELAEIKNMENMYLRL 220
Query: 241 TTSALDTLKEIRGGSSTVSIYSLPPLHSETL 271
T SAL L+E+R GSSTVSI+SLPP+ + L
Sbjct: 221 TYSALRVLQEMRSGSSTVSIFSLPPMKTNAL 251
>UniRef100_Q8LBH4 Ids4-like protein [Arabidopsis thaliana]
Length = 256
Score = 270 bits (689), Expect = 4e-71
Identities = 154/273 (56%), Positives = 189/273 (68%), Gaps = 30/273 (10%)
Query: 1 MKFWKILNNQIEQTLPDWRDKFLSYKDLKKQLKLIVPKEIDSSCSKRRRLDDDGGAEGEV 60
MKF K L+NQIEQTLP+W+DKFLSYK+LKK+LKLI K D KR RLD+ +
Sbjct: 1 MKFGKSLSNQIEQTLPEWQDKFLSYKELKKRLKLIGSKTADRPV-KRLRLDEFSVG---I 56
Query: 61 TKEVKDFLRLLEVEIEKFNGFFVEKEEEYVIKWKELQDKVAWAKSSDIELMTVGREIVDF 120
+KE DF++LLE E+EKFN FFVEKEEEY+I+ KE +D++A AK S +++ + +EIVDF
Sbjct: 57 SKEEIDFIQLLEDELEKFNNFFVEKEEEYIIRLKEFRDRIAKAKDSMEKMIKIRKEIVDF 116
Query: 121 HGEMVLLENYSALNYTGLVKIIKKYDKRTGALLRLPFIQDVLNQPFFKIDVLNKLVKECE 180
HGEMVLLENYSALNYTGLVKI+KKYDKRTG L+RLPFIQ VL QPF+ D+L KLVKE E
Sbjct: 117 HGEMVLLENYSALNYTGLVKILKKYDKRTGDLMRLPFIQKVLQQPFYTTDLLFKLVKESE 176
Query: 181 VMLSIIFPKSGPLGQSLSTSEVFEEVARETTTANETKETLDHVPKEFSEIQNMENIFIKL 240
ML IFP ANET+ + + E SE + ME++ +K
Sbjct: 177 AMLDQIFP------------------------ANETESEI--IQAELSEHKFMESLHMKS 210
Query: 241 TTSALDTLKEIRGGSSTVSIYSLPPLHSETLVE 273
T +AL LKEIR GSSTVS++SLPPL L E
Sbjct: 211 TIAALRVLKEIRSGSSTVSVFSLPPLQLNGLDE 243
>UniRef100_Q8RY68 AT5g20150/F5O24_40 [Arabidopsis thaliana]
Length = 256
Score = 268 bits (684), Expect = 1e-70
Identities = 153/273 (56%), Positives = 189/273 (69%), Gaps = 30/273 (10%)
Query: 1 MKFWKILNNQIEQTLPDWRDKFLSYKDLKKQLKLIVPKEIDSSCSKRRRLDDDGGAEGEV 60
MKF K L+NQIEQTLP+W+DKFLSYK+LKK+LKLI K D KR RLD+ +
Sbjct: 1 MKFGKSLSNQIEQTLPEWQDKFLSYKELKKRLKLIGSKTADRPV-KRLRLDEFSVG---I 56
Query: 61 TKEVKDFLRLLEVEIEKFNGFFVEKEEEYVIKWKELQDKVAWAKSSDIELMTVGREIVDF 120
+KE +F++LLE E+EKFN FFVEKEEEY+I+ KE +D++A AK S +++ + +EIVDF
Sbjct: 57 SKEEINFIQLLEDELEKFNNFFVEKEEEYIIRLKEFRDRIAKAKDSMEKMIKIRKEIVDF 116
Query: 121 HGEMVLLENYSALNYTGLVKIIKKYDKRTGALLRLPFIQDVLNQPFFKIDVLNKLVKECE 180
HGEMVLLENYSALNYTGLVKI+KKYDKRTG L+RLPFIQ VL QPF+ D+L KLVKE E
Sbjct: 117 HGEMVLLENYSALNYTGLVKILKKYDKRTGDLMRLPFIQKVLQQPFYTTDLLFKLVKESE 176
Query: 181 VMLSIIFPKSGPLGQSLSTSEVFEEVARETTTANETKETLDHVPKEFSEIQNMENIFIKL 240
ML IFP ANET+ + + E SE + ME++ +K
Sbjct: 177 AMLDQIFP------------------------ANETESEI--IQAELSEHKFMESLHMKS 210
Query: 241 TTSALDTLKEIRGGSSTVSIYSLPPLHSETLVE 273
T +AL LKEIR GSSTVS++SLPPL L E
Sbjct: 211 TIAALRVLKEIRSGSSTVSVFSLPPLQLNGLDE 243
>UniRef100_Q940E0 IDS-4-like protein [Castanea sativa]
Length = 224
Score = 248 bits (634), Expect = 9e-65
Identities = 129/207 (62%), Positives = 160/207 (76%), Gaps = 4/207 (1%)
Query: 63 EVKDFLRLLEVEIEKFNGFFVEKEEEYVIKWKELQDKVAWAKSSDIELMTVGREIVDFHG 122
E+ DFLRLLEVEI+KFN FFV+KEEEYVI+WKELQD +A AK S EL+ VG+E+VDFHG
Sbjct: 1 ELVDFLRLLEVEIDKFNAFFVDKEEEYVIRWKELQDSIAKAKDSSEELIEVGKEVVDFHG 60
Query: 123 EMVLLENYSALNYTGLVKIIKKYDKRTGALLRLPFIQDVLNQPFFKIDVLNKLVKECEVM 182
EM+LLENYSALNYTGLVKI+KKYDKR+GAL+RLPFIQ VL +PFF DVLN LVKECE +
Sbjct: 61 EMILLENYSALNYTGLVKILKKYDKRSGALVRLPFIQKVLQEPFFSTDVLNNLVKECECV 120
Query: 183 LSIIFPKS-GPLGQSLSTSEVFEEVARETTTANETKETLDHVPKEFSEIQNMENIFIKLT 241
L +F K+ P G +T+ +E + E+K+ VPKE +EI+ ME++++KLT
Sbjct: 121 LDNLFSKNDDPSGCPEATN---KEEGNDPKAVTESKQKQLKVPKELAEIEKMESMYMKLT 177
Query: 242 TSALDTLKEIRGGSSTVSIYSLPPLHS 268
SAL +KEI GSSTVS +SLPPL +
Sbjct: 178 LSALRAVKEISSGSSTVSEFSLPPLQN 204
>UniRef100_Q6Z784 Putative SPX (SYG1/Pho81/XPR1) domain-containing protein [Oryza
sativa]
Length = 280
Score = 240 bits (613), Expect = 3e-62
Identities = 140/274 (51%), Positives = 181/274 (65%), Gaps = 11/274 (4%)
Query: 1 MKFWKILNNQIEQTLPDWRDKFLSYKDLKKQLKLIVPKEIDSSCSKRRRLDD------DG 54
MKF K L++QI + P+WRD FLSYKDLKK+L LI SKRRR+
Sbjct: 1 MKFGKSLSSQIVEMQPEWRDNFLSYKDLKKRLNLISGGAAGERASKRRRVGGATAVTVTA 60
Query: 55 GAEGEVTKEVKDFLRLLEVEIEKFNGFFVEKEEEYVIKWKELQDKVAWAKSSDIELMTVG 114
A G +T E F+ LL+ E++KFN FF+EKEEEYVIK KEL+++ +S E+M V
Sbjct: 61 AAAGGMTLEQAGFVGLLDAELDKFNFFFLEKEEEYVIKQKELRER---KMASAEEVMRVR 117
Query: 115 REIVDFHGEMVLLENYSALNYTGLVKIIKKYDKRTGALLRLPFIQDVLNQPFFKIDVLNK 174
+EIVD HGEMVLLENYSALNYTGLVKI+KKYDKRTG+++RLPF+Q VL QPFF D+L K
Sbjct: 118 KEIVDLHGEMVLLENYSALNYTGLVKILKKYDKRTGSMIRLPFVQKVLQQPFFTTDLLYK 177
Query: 175 LVKECEVMLSIIFPKSGPLGQSLSTSEVFEEVARETTTANETKETLDHVPKEFSEIQNME 234
LVKECE ML + P + S + E + + ++ + VP E +E ++
Sbjct: 178 LVKECEEMLDQLMPTNEHSVASEDGKDDSEGEEKGSKPSSSSSANGGAVPGE-AEAEDER 236
Query: 235 NIFIKLT-TSALDTLKEIRGGSSTVSIYSLPPLH 267
+ +K T T+AL L+EIR GSSTVS++SLPPLH
Sbjct: 237 STDMKSTVTAALRALREIRSGSSTVSVFSLPPLH 270
>UniRef100_Q947U3 Hypothetical protein OSJNBa0068A07.13 [Oryza sativa]
Length = 277
Score = 197 bits (502), Expect = 2e-49
Identities = 117/274 (42%), Positives = 165/274 (59%), Gaps = 39/274 (14%)
Query: 1 MKFWKILNNQIEQTLPDWRDKFLSYKDLKKQLKLIVPKEIDSSCSKRRRLDDDGGAEGEV 60
MKF K L Q+E++LP+WRDKFL+YK LKK ++L+ SS S D GG GE
Sbjct: 1 MKFGKRLKKQVEESLPEWRDKFLAYKRLKKLVRLV------SSSSG----DVGGGGGGEA 50
Query: 61 TKEVKDFLRLLEVEIEKFNGFFVEKEEEYVIKWKELQDKV--------AWAKSSDIELMT 112
+F+RLL+ E+++ N FF+E+EEE+VI+ +ELQ+ V + + E+
Sbjct: 51 -----EFVRLLDGEVDRINAFFLEQEEEFVIRQRELQETVEKVAGGGGGGRRPAAAEMRR 105
Query: 113 VGREIVDFHGEMVLLENYSALNYTGLVKIIKKYDKRTGALLRLPFIQDVLNQPFFKIDVL 172
V +EIVD HGEMVLL NYSA+NYTGL KI+KKYDKRTG LLRLPFI+ VL QPFF +++
Sbjct: 106 VRKEIVDLHGEMVLLLNYSAVNYTGLAKILKKYDKRTGRLLRLPFIEKVLRQPFFTTELI 165
Query: 173 NKLVKECEVMLSIIFPKSGPLGQSLSTSEVFEEVARETTTANETKETLDHVPKEFSEIQN 232
++LV++CE + IF S ++ + + A + +
Sbjct: 166 SRLVRDCEATMEAIFTSSVATTAMAGDRRTWKGCSGDAGMA---------------PMAD 210
Query: 233 MENIFIKLTTSALDTLKEIRGGSSTVSIYSLPPL 266
+ IF + T +AL T+KE+R GSST +SLPP+
Sbjct: 211 QQGIF-RNTVAALATMKELRSGSSTYGRFSLPPM 243
>UniRef100_Q7Y0F6 SPX domain containing protein [Oryza sativa]
Length = 247
Score = 189 bits (479), Expect = 9e-47
Identities = 110/267 (41%), Positives = 152/267 (56%), Gaps = 50/267 (18%)
Query: 1 MKFWKILNNQIEQTLPDWRDKFLSYKDLKKQLKLIVPKEIDSSCSKRRRLDDDGGAEGEV 60
MKF K L QIE++LP+WRD FL+YK+LK++L + D AE
Sbjct: 1 MKFGKRLKRQIEESLPEWRDHFLNYKELKRRLNAVS--------------SPDPAAEAR- 45
Query: 61 TKEVKDFLRLLEVEIEKFNGFFVEKEEEYVIKWKELQDKVAWAKSSDIELM-TVGREIVD 119
FL LL E++KFN FF+E+EE++VI+ +ELQ+++ + S+ E+ V RE+VD
Sbjct: 46 ------FLALLHAEVDKFNAFFLEQEEDFVIRQRELQERIQSSSSAAAEMEGRVRREVVD 99
Query: 120 FHGEMVLLENYSALNYTGLVKIIKKYDKRTGALLRLPFIQDVLNQPFFKIDVLNKLVKEC 179
HGEMVLL NYS++NYTGL KI+KKYDKRTG +LRLP I VL QPF+ D+L+ LV++C
Sbjct: 100 LHGEMVLLLNYSSINYTGLAKILKKYDKRTGGVLRLPVIAGVLRQPFYATDLLSSLVRDC 159
Query: 180 EVMLSIIFPKSGPLGQSLSTSEVFEEVARETTTANETKETLDHVPKEFSEIQNMENIFIK 239
E ++ +FP S S AR E +
Sbjct: 160 EAIMDAVFPSLP------SPSAAAAAAARAAA----------------------EQAIFR 191
Query: 240 LTTSALDTLKEIRGGSSTVSIYSLPPL 266
T +AL T++E+R GSST +SLPP+
Sbjct: 192 NTVAALLTMQEVRSGSSTYGHFSLPPM 218
>UniRef100_Q9LXE9 Ids-4 protein-like [Arabidopsis thaliana]
Length = 387
Score = 183 bits (465), Expect = 4e-45
Identities = 116/295 (39%), Positives = 163/295 (54%), Gaps = 38/295 (12%)
Query: 1 MKFWKILNNQIEQTLPDWRDKFLSYKDLKKQLKLIV-------PKEIDSSCSKRRRLD-- 51
MKF K +E+TLP+WRDKFL YK LKK LK P D + S+ D
Sbjct: 70 MKFGKEFRTHLEETLPEWRDKFLCYKPLKKLLKYYPYYSADFGPANSDHNDSRPVFADTT 129
Query: 52 ------DDGGAEGEVTKEVK---DFLRLLEVEIEKFNGFFVEKEEEYVIKWKELQDKVAW 102
DDGG V F+R+L E+EKFN F+V+KEE++VI+ +EL++++
Sbjct: 130 NISSAADDGGVVPGVRPSEDLQGSFVRILNDELEKFNDFYVDKEEDFVIRLQELKERIEQ 189
Query: 103 AKSSD----------IELMTVGREIVDFHGEMVLLENYSALNYTGLVKIIKKYDKRTGAL 152
K + E+M + R++V HGEMVLL+NYS+LN+ GLVKI+KKYDKRTG L
Sbjct: 190 VKEKNGEFASESEFSEEMMDIRRDLVTIHGEMVLLKNYSSLNFAGLVKILKKYDKRTGGL 249
Query: 153 LRLPFIQDVLNQPFFKIDVLNKLVKECEVMLSIIFPKSGPLGQSLSTSEVFEEVARETTT 212
LRLPF Q VL+QPFF + L +LV+ECE L ++FP S +EV E +
Sbjct: 250 LRLPFTQLVLHQPFFTTEPLTRLVRECEANLELLFP---------SEAEVVESSSAVQAH 300
Query: 213 ANETKETLDHVPKEFSEIQNMENIFI-KLTTSALDTLKEIRGGSSTVSIYSLPPL 266
++ + + E S EN+ I K T +A+ ++ ++ SST + S L
Sbjct: 301 SSSHQHNSPRISAETSSTLGNENLDIYKSTLAAMRAIRGLQKASSTYNPLSFSSL 355
>UniRef100_Q94A21 AT5g15330/F8M21_220 [Arabidopsis thaliana]
Length = 318
Score = 183 bits (465), Expect = 4e-45
Identities = 116/295 (39%), Positives = 163/295 (54%), Gaps = 38/295 (12%)
Query: 1 MKFWKILNNQIEQTLPDWRDKFLSYKDLKKQLKLIV-------PKEIDSSCSKRRRLD-- 51
MKF K +E+TLP+WRDKFL YK LKK LK P D + S+ D
Sbjct: 1 MKFGKEFRTHLEETLPEWRDKFLCYKPLKKLLKYYPYYSADFGPANSDHNDSRPVFADTT 60
Query: 52 ------DDGGAEGEVTKEVK---DFLRLLEVEIEKFNGFFVEKEEEYVIKWKELQDKVAW 102
DDGG V F+R+L E+EKFN F+V+KEE++VI+ +EL++++
Sbjct: 61 NISSAADDGGVVPGVRPSEDLQGSFVRILNDELEKFNDFYVDKEEDFVIRLQELKERIEQ 120
Query: 103 AKSSD----------IELMTVGREIVDFHGEMVLLENYSALNYTGLVKIIKKYDKRTGAL 152
K + E+M + R++V HGEMVLL+NYS+LN+ GLVKI+KKYDKRTG L
Sbjct: 121 VKEKNGEFASESEFSEEMMDIRRDLVTIHGEMVLLKNYSSLNFAGLVKILKKYDKRTGGL 180
Query: 153 LRLPFIQDVLNQPFFKIDVLNKLVKECEVMLSIIFPKSGPLGQSLSTSEVFEEVARETTT 212
LRLPF Q VL+QPFF + L +LV+ECE L ++FP S +EV E +
Sbjct: 181 LRLPFTQLVLHQPFFTTEPLTRLVRECEANLELLFP---------SEAEVVESSSAVQAH 231
Query: 213 ANETKETLDHVPKEFSEIQNMENIFI-KLTTSALDTLKEIRGGSSTVSIYSLPPL 266
++ + + E S EN+ I K T +A+ ++ ++ SST + S L
Sbjct: 232 SSSHQHNSPRISAETSSTLGNENLDIYKSTLAAMRAIRGLQKASSTYNPLSFSSL 286
>UniRef100_Q7XJM6 At2g45130 protein [Arabidopsis thaliana]
Length = 245
Score = 176 bits (445), Expect = 8e-43
Identities = 111/275 (40%), Positives = 153/275 (55%), Gaps = 58/275 (21%)
Query: 1 MKFWKILNNQIEQTLPDWRDKFLSYKDLKKQLKLIVPKEIDSSCSKRRRLDDDGGAEGEV 60
MKF K + QI+++LP+WRDKFL YK+LK + P E
Sbjct: 1 MKFGKRIKEQIQESLPEWRDKFLRYKELKNLISSPAPVE--------------------- 39
Query: 61 TKEVKDFLRLLEVEIEKFNGFFVEKEEEYVIKWKELQDKVAWA--------KSSDIELMT 112
F+ LL EI+KFN FFVE+EE+++I KELQ ++ + S +
Sbjct: 40 ----SIFVGLLNAEIDKFNAFFVEQEEDFIIHHKELQYRIQRLVEKCGHNDEMSRENISE 95
Query: 113 VGREIVDFHGEMVLLENYSALNYTGLVKIIKKYDKRTGALLRLPFIQDVLNQPFFKIDVL 172
+ ++IV+FHGEMVLL NYS +NYTGL KI+KKYDKRT LR PFIQ VL+QPFFK D++
Sbjct: 96 IRKDIVNFHGEMVLLVNYSNINYTGLAKILKKYDKRTRGGLRSPFIQKVLHQPFFKTDLV 155
Query: 173 NKLVKECEVMLSIIFPKSGPLGQSLSTSEVFEEVARETTTANETKETLDHVPKEFSEIQN 232
++LV+E E + + P ++ +E +E A T+ A
Sbjct: 156 SRLVREWETTMDAVDP------VKVAEAEGYERCAAVTSAAAG----------------- 192
Query: 233 MENIFIKLTTSALDTLKEIRGGSSTVSIYSLPPLH 267
E IF + T +AL T+KE+R GSST S +SLPPL+
Sbjct: 193 -EGIF-RNTVAALLTMKEMRRGSSTYSAFSLPPLN 225
>UniRef100_Q8SAX5 Putative signal transduction protein [Oryza sativa]
Length = 320
Score = 165 bits (417), Expect = 1e-39
Identities = 96/275 (34%), Positives = 153/275 (54%), Gaps = 24/275 (8%)
Query: 1 MKFWKILNNQIEQTLPDWRDKFLSYKDLKKQLKLIVPKEIDSSCSKRRRLD-DDGGAEGE 59
MKF K + +E+TLP WRDK+L+YK LKK +K + P + + DG +G
Sbjct: 1 MKFGKDFRSHLEETLPAWRDKYLAYKSLKKLIKNLPPDGDPPPVAAAAEVPAGDGDGDGG 60
Query: 60 VTKEVKDFLRLLEVEIEKFNGFFVEKEEEYVIKWKELQDKV---------AWAKSSDI-- 108
+ F R+L++E++K N F++E+EE YVI+ + L++++ A+ S+
Sbjct: 61 IALG-NWFARVLDMELQKLNDFYIEREEWYVIRLQVLKERIERVKAKKNGAFTSKSEFTE 119
Query: 109 ELMTVGREIVDFHGEMVLLENYSALNYTGLVKIIKKYDKRTGALLRLPFIQDVLNQPFFK 168
E++ + + V HGEM+LL+ YS+LN+ GLVKI+KKYDKRTG LL LPF Q +QPFF
Sbjct: 120 EMLEIRKAFVIIHGEMILLQTYSSLNFAGLVKILKKYDKRTGGLLSLPFTQRARHQPFFT 179
Query: 169 IDVLNKLVKECEVMLSIIFPKSGPLGQSLSTSEVFEEVARETTTANETKETLDHVPKEFS 228
+ L +LV+ECE L ++FP +EV E + + + H P
Sbjct: 180 TEPLTRLVRECEANLELLFP---------IEAEVLESASSSAKLQPQNDDAASHDPASSV 230
Query: 229 EIQNMENIFIKLTTSALDTLKEIRGGSSTVSIYSL 263
+++ + + T +A+ ++ +R SST + SL
Sbjct: 231 DVETSD--VYRSTLAAMKAIQGLRKASSTYNPLSL 263
>UniRef100_Q8GWZ3 Hypothetical protein At2g26660 [Arabidopsis thaliana]
Length = 161
Score = 157 bits (398), Expect = 2e-37
Identities = 85/151 (56%), Positives = 102/151 (67%), Gaps = 4/151 (2%)
Query: 124 MVLLENYSALNYTGLVKIIKKYDKRTGALLRLPFIQDVLNQPFFKIDVLNKLVKECEVML 183
MVLL NYSALNYTGL KI+KKYDKRTGAL+RLPFIQ VL +PFF D+LN VKECE ML
Sbjct: 1 MVLLMNYSALNYTGLAKILKKYDKRTGALIRLPFIQKVLQEPFFTTDLLNTFVKECEAML 60
Query: 184 SIIFPKSGPLGQSLSTSEVFEEVARETTTANETKETLDHVPKEFSEIQNMENIFIKLTTS 243
+FP + +S + E E L VPKE SEI+ ME++++K T S
Sbjct: 61 DRLFPSN----KSRNLDEEGEPTTSGMVKTGTDDSELLRVPKELSEIEYMESLYMKSTVS 116
Query: 244 ALDTLKEIRGGSSTVSIYSLPPLHSETLVED 274
AL LKEIR GSSTVS++SLPPL + L +D
Sbjct: 117 ALKVLKEIRSGSSTVSVFSLPPLPASGLEDD 147
>UniRef100_Q8H398 Putative ids-4 protein [Oryza sativa]
Length = 244
Score = 148 bits (373), Expect = 2e-34
Identities = 97/277 (35%), Positives = 145/277 (52%), Gaps = 64/277 (23%)
Query: 1 MKFWKILNNQIEQTLPDWRDKFLSYKDLKKQLKLIVPKEIDSSCSKRRRLDDDGGAEGEV 60
MKF K+L QIEQ+LP+WRDKF+SYK+LK+ + I D +
Sbjct: 1 MKFGKLLKRQIEQSLPEWRDKFVSYKELKRIVASISGSPADEAA---------------- 44
Query: 61 TKEVKDFLRLLEVEIEKFNGFFVEKEEEYVIKWK----------ELQDKVAWAKSSDIEL 110
F+ L +I+K + FF+E+EEE+VI+ + ELQ+ + A + E+
Sbjct: 45 ------FVAALAADIDKIDSFFLEQEEEFVIRHRARTPIRFNSFELQEAIKKAAEAAAEV 98
Query: 111 MTVGREIVDFHGEMVLLENYSALNYTGLVKIIKKYDKRTGALLRLPFIQDVL-NQPFFKI 169
+ REIVDFHGEMVLL +YS++NY G+ KI+KK+DKRTG L P + V + FFK
Sbjct: 99 AGIRREIVDFHGEMVLLLSYSSINYIGVGKILKKHDKRTGGALAAPVAEAVRERRHFFKT 158
Query: 170 DVLNKLVKECEVMLSIIFPKSGPLGQSLSTSEVFEEVARETTTANETKETLDHVPKEFSE 229
+ ++++V+ECE M++ + + E A E A
Sbjct: 159 ETVSRMVRECEAMMA-------------EAAVLPAEAAPEALAA---------------- 189
Query: 230 IQNMENIFIKLTTSALDTLKEIRGGSSTVSIYSLPPL 266
E+ + T +AL T++++R GSST +SLPPL
Sbjct: 190 --AAEHGIFRNTVAALLTMEDVRRGSSTHGRHSLPPL 224
>UniRef100_UPI00002C8327 UPI00002C8327 UniRef100 entry
Length = 220
Score = 98.6 bits (244), Expect = 2e-19
Identities = 53/137 (38%), Positives = 82/137 (59%), Gaps = 12/137 (8%)
Query: 67 FLRLLEVEIEKFNGFFVEKEEEYVIKWKELQDKVAWAKSSD---------IELMTVGREI 117
F++ L E++KFN FF++ EE+ VIK L+ + D + +E
Sbjct: 4 FVKTLNAELQKFNKFFMDAEEDLVIKDSLLEQAYREVVNEDGKRAESFSMKKYRKTCQEF 63
Query: 118 VDFHGEMVLLENYSALNYTGLVKIIKKYDKRTGALLRLPFIQDVLNQPFFKIDVLNKLVK 177
DFHGE+VL+E++ LNYT LVKI+KK+DKR+ LR PF+ VL QPF++ +VL++L+
Sbjct: 64 ADFHGELVLMEHWVGLNYTALVKILKKHDKRSNLSLRSPFLVSVLQQPFYRTEVLSQLIT 123
Query: 178 ECEV---MLSIIFPKSG 191
+ E L+ + P+ G
Sbjct: 124 KTETSFRKLNALLPEGG 140
>UniRef100_Q40064 Iron-deficiency specific clone No.4 [Hordeum vulgare]
Length = 189
Score = 91.3 bits (225), Expect = 2e-17
Identities = 54/140 (38%), Positives = 79/140 (55%), Gaps = 13/140 (9%)
Query: 124 MVLLENYSALNYTGLVKIIKKYDKRTGALLRLPFIQDVLNQPFFKIDVLNKLVKECEVML 183
M+LL+ YS+LN+ GLVKI+KKYDKRTG +L LPF Q +QPFF + L +LV+ECE L
Sbjct: 1 MILLQTYSSLNFAGLVKILKKYDKRTGGVLSLPFTQRARHQPFFTTEPLTRLVRECEANL 60
Query: 184 SIIFPKSGPLGQSLSTSEVFEEVARETTTANETKETLDHVPKEFSEIQNMENIFIKLTTS 243
I+FP EV E + A+ + D P+ S+ + E + T +
Sbjct: 61 EILFPVE---------DEVLESGSSSKHQAHNDAASRD--PESSSDAETSE--VYRSTLA 107
Query: 244 ALDTLKEIRGGSSTVSIYSL 263
A+ ++ ++ SST + SL
Sbjct: 108 AMKAIEGLKKASSTYNALSL 127
>UniRef100_Q6CI84 Similar to sp|Q01317 Neurospora crassa Ankyrin repeat protein nuc-
2 [Yarrowia lipolytica]
Length = 985
Score = 72.4 bits (176), Expect = 1e-11
Identities = 73/286 (25%), Positives = 125/286 (43%), Gaps = 44/286 (15%)
Query: 1 MKFWKIL-NNQIEQTLPDWRDKFLSYKDLKKQLKLIVPKEIDSSCSKRRRLDDDGGAEGE 59
MKF K L Q+E +P++ + F++YK LKK +K L + A G
Sbjct: 1 MKFGKYLAKRQLE--VPEYGNYFINYKALKKLIK---------------SLSNQAAAGGN 43
Query: 60 VTKEVKD----FLRLLEVEIEKFNGFFVEKEEEYVIKWKELQDKVAWA-------KSSDI 108
V + ++D F LE E+EK N F+++KE E ++ L +K A A S+ +
Sbjct: 44 VEQALRDNKATFFFRLERELEKVNSFYLQKEAELKLRIDILMEKKADAYASGSLTSSTSV 103
Query: 109 ELMTVGREIVDFHGEMVLLENYSALNYTGLVKIIKKYDKRTGALLRLPFIQDVLN-QPFF 167
+++ F ++ LE + LN TG K++KK+DKR+ + ++ + QP F
Sbjct: 104 SYISLYEGFQRFRRDLSKLEQFIELNATGFSKVLKKWDKRSKQQTKELYLSRAVEVQPVF 163
Query: 168 KIDVLNKLVKECEVML---------SIIFPKSGPLGQSLSTSEVFEEVARETTTANETKE 218
D+L +L + + +IF S + + + A TA
Sbjct: 164 HRDILARLSDQASSSILELEAWADGDVIFDDSRRGSEIQAATAPATAPATAPVTAPVAPV 223
Query: 219 TL-----DHVPKEFSEIQNMENIFIKLTTSALDTLKEIRGGSSTVS 259
T D + EF E + EN+ + ++ LK++R S V+
Sbjct: 224 TAPGAKDDDLYYEFVETASAENVTTADISEWVEVLKKVRDAKSRVT 269
>UniRef100_P47075 VTC4 protein [Saccharomyces cerevisiae]
Length = 721
Score = 71.2 bits (173), Expect = 3e-11
Identities = 61/220 (27%), Positives = 105/220 (47%), Gaps = 30/220 (13%)
Query: 1 MKFWKILNNQIEQTLPDWRDKFLSYKDLKKQLKLIVPKEIDSSCSKRRRLDDDGGAEGEV 60
MKF + L+ + + + ++SY DLK E++ + SK G+
Sbjct: 1 MKFGEHLSKSL---IRQYSYYYISYDDLKT--------ELEDNLSKNN---------GQW 40
Query: 61 TKEVK-DFLRLLEVEIEKFNGFFVEKEEEYVIKWKELQDKVAWA--------KSSDIELM 111
T+E++ DFL LE+E++K F K E + KE+Q++V + ++
Sbjct: 41 TQELETDFLESLEIELDKVYTFCKVKHSEVFRRVKEVQEQVQHTVRLLDSNNPPTQLDFE 100
Query: 112 TVGREIVDFHGEMVLLENYSALNYTGLVKIIKKYDKRTGALLRLPFIQDVLNQPFFKIDV 171
+ E+ D ++ L +S LNYTG KIIKK+DK+TG +L+ F + ++PFFK +
Sbjct: 101 ILEEELSDIIADVHDLAKFSRLNYTGFQKIIKKHDKKTGFILKPVFQVRLDSKPFFK-EN 159
Query: 172 LNKLVKECEVMLSIIFPKSGPLGQSLSTSEVFEEVARETT 211
++LV + + I P+ S + R+TT
Sbjct: 160 YDELVVKISQLYDIARTSGRPIKGDSSAGGKQQNFVRQTT 199
>UniRef100_UPI000046B7EB UPI000046B7EB UniRef100 entry
Length = 857
Score = 70.1 bits (170), Expect = 6e-11
Identities = 63/228 (27%), Positives = 102/228 (44%), Gaps = 21/228 (9%)
Query: 1 MKFWKILNNQIEQTLPDWRDKFLSYKDLKKQLKLIVPKEI-DSSCSKRRRLDDDGG---- 55
MKF K L+ E+ P +RD ++SYK+LK +KLI I D+S + + + G
Sbjct: 1 MKFRKKLH---EEAHPKYRDHYISYKELKNVIKLITGNNINDTSTYTIKEITTNFGNIRA 57
Query: 56 -AEGEVTKEVKDFLRLLEVEIEKFNGFFV----EKEEEYVIKWKELQDKVAWAKSSDIEL 110
E F +L E++K N F V + +E I +KEL+ I++
Sbjct: 58 LTGAEYKSPESRFQDILNGELDKINKFSVVIIKQWFKEAEIYYKELKR----GNEESIDI 113
Query: 111 MTVGREIVDFHGEMVLLENYSALNYTGLVKIIKKYDKRTGALLRLPFIQDVLNQPFFKID 170
+ + +++ + ++ LE Y +N+ G KI KK+DK G + F +V+ + FF
Sbjct: 114 LNIEKKLNELGNTLIFLEKYKHINFIGFRKITKKFDKHNGKTVSSSFYINVVIKSFFMTF 173
Query: 171 VLNKLVKECEVMLSIIFPKSGPLGQSLSTSEVFEEVARETTTANETKE 218
+N LV +LSI + + EV E E N E
Sbjct: 174 DINFLV----YILSICYKYYRDIKNKNKIIEVKENTKDEQYLLNNNVE 217
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.316 0.136 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 442,885,143
Number of Sequences: 2790947
Number of extensions: 18174811
Number of successful extensions: 64886
Number of sequences better than 10.0: 266
Number of HSP's better than 10.0 without gapping: 123
Number of HSP's successfully gapped in prelim test: 145
Number of HSP's that attempted gapping in prelim test: 64519
Number of HSP's gapped (non-prelim): 397
length of query: 274
length of database: 848,049,833
effective HSP length: 125
effective length of query: 149
effective length of database: 499,181,458
effective search space: 74378037242
effective search space used: 74378037242
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 74 (33.1 bits)
Medicago: description of AC147471.4


