

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147435.10 + phase: 0 /pseudo
(612 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
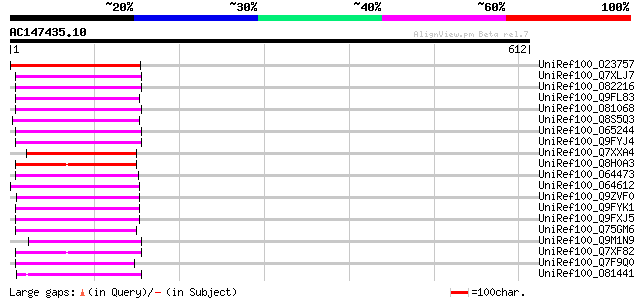
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_O23757 Reverse transcriptase [Beta vulgaris subsp. vul... 135 4e-30
UniRef100_Q7XLJ7 OSJNBa0009K15.13 protein [Oryza sativa] 106 2e-21
UniRef100_O82216 Putative non-LTR retroelement reverse transcrip... 102 3e-20
UniRef100_Q9FL83 Non-LTR retroelement reverse transcriptase-like... 100 1e-19
UniRef100_O81068 Putative non-LTR retroelement reverse transcrip... 99 3e-19
UniRef100_Q8S5Q3 Putative non-LTR retroelement reverse transcrip... 99 4e-19
UniRef100_O65244 F21E10.5 protein [Arabidopsis thaliana] 98 6e-19
UniRef100_Q9FYJ4 F17F8.5 [Arabidopsis thaliana] 96 3e-18
UniRef100_Q7XXA4 OSJNBa0019G23.12 protein [Oryza sativa] 95 7e-18
UniRef100_Q8H0A3 Putative reverse transcriptase [Oryza sativa] 94 1e-17
UniRef100_O64473 Putative non-LTR retroelement reverse transcrip... 94 2e-17
UniRef100_O64612 Putative reverse transcriptase [Arabidopsis tha... 92 6e-17
UniRef100_Q9ZVF0 Putative non-LTR retroelement reverse transcrip... 91 1e-16
UniRef100_Q9FYK1 F21J9.30 [Arabidopsis thaliana] 91 1e-16
UniRef100_Q9FXJ5 F5A9.24 [Arabidopsis thaliana] 91 1e-16
UniRef100_Q75GM6 Putative non-LTR retroelement reverse transcrip... 90 2e-16
UniRef100_Q9M1N9 Hypothetical protein T18B22.50 [Arabidopsis tha... 90 2e-16
UniRef100_Q7XF82 Putative retroelement [Oryza sativa] 90 2e-16
UniRef100_Q7F9Q0 OSJNBa0045O17.3 protein [Oryza sativa] 90 2e-16
UniRef100_O81441 T24H24.17 protein [Arabidopsis thaliana] 90 2e-16
>UniRef100_O23757 Reverse transcriptase [Beta vulgaris subsp. vulgaris]
Length = 507
Score = 135 bits (340), Expect = 4e-30
Identities = 64/154 (41%), Positives = 100/154 (64%)
Query: 1 DGVNFNRLSEVDNDLLVAPFHMEEIEVVVRESDGNKSPGADEYNFAFVKEFWYLMKNEVR 60
+G+NFN+LSE + L+APF EEI+ V D K+PG D +NF F+K W ++K ++
Sbjct: 279 NGLNFNKLSESECSSLIAPFTHEEIDEAVDSCDAQKAPGPDGFNFKFIKSSWEVIKKDIY 338
Query: 61 IMFDQIHANELLPKSMLAYFVALIPKVSSPLELKDYRTISLLGSLYKLLAKVLARRLAGV 120
+ + A+ LP + F+ALIPK P ++R IS++G LYK++AK+LARRL V
Sbjct: 339 KIVEDFWASGSLPHGCNSAFIALIPKTQHPRGFNEFRPISMVGCLYKIIAKILARRLQRV 398
Query: 121 LSSVISTTQSAFLKGTNLVDGVLVVNELVDYAKK 154
+ +I QS+F+KG ++DGVL+ +EL+D ++
Sbjct: 399 MDHLIGPYQSSFIKGRQILDGVLIASELIDSCRR 432
>UniRef100_Q7XLJ7 OSJNBa0009K15.13 protein [Oryza sativa]
Length = 779
Score = 106 bits (265), Expect = 2e-21
Identities = 57/149 (38%), Positives = 85/149 (56%)
Query: 7 RLSEVDNDLLVAPFHMEEIEVVVRESDGNKSPGADEYNFAFVKEFWYLMKNEVRIMFDQI 66
R+ + DND LV PF MEEIE + E N +PG D F KEFW +K++++ M D +
Sbjct: 141 RVDQEDNDFLVRPFSMEEIERALAEMKTNTAPGPDGLPVCFYKEFWEQLKDQIKEMLDSL 200
Query: 67 HANELLPKSMLAYFVALIPKVSSPLELKDYRTISLLGSLYKLLAKVLARRLAGVLSSVIS 126
L + + LIPK+ +K +R I LL +KLL KVL R+ V + VI
Sbjct: 201 FEGRLDLWRLNYGVITLIPKIKEANNIKAFRPICLLNVCFKLLTKVLTMRITHVANKVIG 260
Query: 127 TTQSAFLKGTNLVDGVLVVNELVDYAKKS 155
+Q+AFL G ++DGV++++E++ K S
Sbjct: 261 ESQTAFLPGRFILDGVVILHEVLHELKNS 289
>UniRef100_O82216 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1216
Score = 102 bits (254), Expect = 3e-20
Identities = 63/150 (42%), Positives = 82/150 (54%), Gaps = 1/150 (0%)
Query: 7 RLSEVDNDLLVAPFHMEEIEVVVRESDGNKSPGADEYNFAFVKEFWYLMKNEVRIMFDQI 66
R SE D+ LL EEI+ V+ +KSPG D Y F K W ++ +EV I
Sbjct: 161 RCSEDDHRLLTRVVTGEEIKKVIFSMPKDKSPGPDGYTSEFYKASWEIIGDEVIIAIQSF 220
Query: 67 HANELLPKSMLAYFVALIPKVSSPLELKDYRTISLLGSLYKLLAKVLARRLAGVLSSVIS 126
A LPK + + +ALIPK E+KDYR IS LYK ++K+LA RL +L I
Sbjct: 221 FAKGFLPKGVNSTILALIPKKKEAREIKDYRPISCCNVLYKAISKILANRLKRILPKFIV 280
Query: 127 TTQSAFLKGTNLVDGVLVVNELV-DYAKKS 155
QSAF+K L++ VL+ ELV DY K S
Sbjct: 281 GNQSAFVKDRLLIENVLLATELVKDYHKDS 310
>UniRef100_Q9FL83 Non-LTR retroelement reverse transcriptase-like protein
[Arabidopsis thaliana]
Length = 1223
Score = 100 bits (249), Expect = 1e-19
Identities = 57/148 (38%), Positives = 81/148 (54%), Gaps = 1/148 (0%)
Query: 7 RLSEVDNDLLVAPFHMEEIEVVVRESDGNKSPGADEYNFAFVKEFWYLMKNEVRIMFDQI 66
R S+ D L+ P EEI V+ +KSPG D Y F K W ++ +E +
Sbjct: 440 RCSDADQQSLIRPVTAEEIRKVLFRMPSDKSPGPDGYTSEFFKATWEIIGDEFTLAVQSF 499
Query: 67 HANELLPKSMLAYFVALIPKVSSPLELKDYRTISLLGSLYKLLAKVLARRLAGVLSSVIS 126
LPK + + +ALIPK + E+KDYR IS LYK+++K++A RL VL I+
Sbjct: 500 FTKGFLPKGINSTILALIPKKTEAREMKDYRPISCCNVLYKVISKIIANRLKLVLPKFIA 559
Query: 127 TTQSAFLKGTNLVDGVLVVNELV-DYAK 153
QSAF+K L++ +L+ ELV DY K
Sbjct: 560 GNQSAFVKDRLLIENLLLATELVKDYHK 587
>UniRef100_O81068 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1529
Score = 99.4 bits (246), Expect = 3e-19
Identities = 61/150 (40%), Positives = 80/150 (52%), Gaps = 1/150 (0%)
Query: 7 RLSEVDNDLLVAPFHMEEIEVVVRESDGNKSPGADEYNFAFVKEFWYLMKNEVRIMFDQI 66
R S D ++L EEI+ V+ NKSPG D Y F K W L +
Sbjct: 737 RCSVTDQNILTREVTGEEIQKVLFAMPNNKSPGPDGYTSEFFKATWSLTGPDFIAAIQSF 796
Query: 67 HANELLPKSMLAYFVALIPKVSSPLELKDYRTISLLGSLYKLLAKVLARRLAGVLSSVIS 126
LPK + A +ALIPK +E+KDYR IS LYK+++K+LA RL +L S I
Sbjct: 797 FVKGFLPKGLNATILALIPKKDEAIEMKDYRPISCCNVLYKVISKILANRLKLLLPSFIL 856
Query: 127 TTQSAFLKGTNLVDGVLVVNELV-DYAKKS 155
QSAF+K L++ VL+ ELV DY K+S
Sbjct: 857 QNQSAFVKERLLMENVLLATELVKDYHKES 886
>UniRef100_Q8S5Q3 Putative non-LTR retroelement reverse transcriptase [Oryza sativa]
Length = 1228
Score = 99.0 bits (245), Expect = 4e-19
Identities = 49/150 (32%), Positives = 88/150 (58%)
Query: 4 NFNRLSEVDNDLLVAPFHMEEIEVVVRESDGNKSPGADEYNFAFVKEFWYLMKNEVRIMF 63
N LS+ + L PF M E++ V+ E+ N +PG D ++ F + FW +KN++ M
Sbjct: 940 NHLNLSDASKENLTRPFVMAELDKVINEAKLNTAPGPDGFSIPFYRAFWPQVKNDLFEML 999
Query: 64 DQIHANELLPKSMLAYFVALIPKVSSPLELKDYRTISLLGSLYKLLAKVLARRLAGVLSS 123
+H +L K + ++LIPK ++P ++K +R I +L +K L+KV+ RL V
Sbjct: 1000 LMLHNEDLDLKRLNFGVISLIPKNNNPTDIKQFRPICVLNDCFKFLSKVVTNRLTEVADE 1059
Query: 124 VISTTQSAFLKGTNLVDGVLVVNELVDYAK 153
V+S TQ+AF+ G +++G ++++E++ K
Sbjct: 1060 VVSQTQTAFIPGRFILEGCVIIHEVLHELK 1089
>UniRef100_O65244 F21E10.5 protein [Arabidopsis thaliana]
Length = 928
Score = 98.2 bits (243), Expect = 6e-19
Identities = 59/150 (39%), Positives = 80/150 (53%), Gaps = 1/150 (0%)
Query: 7 RLSEVDNDLLVAPFHMEEIEVVVRESDGNKSPGADEYNFAFVKEFWYLMKNEVRIMFDQI 66
R S+ D ++L EEI VV +KSPG D Y F K W ++ E +
Sbjct: 328 RCSDSDKEMLTNHVSAEEIHKVVFSMPNDKSPGPDGYTAEFYKGAWNIIGAEFILAIQSF 387
Query: 67 HANELLPKSMLAYFVALIPKVSSPLELKDYRTISLLGSLYKLLAKVLARRLAGVLSSVIS 126
A LPK + + +ALIPK E+KDYR IS LYK+++K++A RL VL I
Sbjct: 388 FAKGFLPKGINSTILALIPKKKEAKEMKDYRPISCCNVLYKVISKIIANRLKLVLPKFIV 447
Query: 127 TTQSAFLKGTNLVDGVLVVNELV-DYAKKS 155
QSAF+K L++ VL+ E+V DY K S
Sbjct: 448 GNQSAFVKDRLLIENVLLATEIVKDYHKDS 477
>UniRef100_Q9FYJ4 F17F8.5 [Arabidopsis thaliana]
Length = 872
Score = 95.9 bits (237), Expect = 3e-18
Identities = 55/150 (36%), Positives = 82/150 (54%), Gaps = 1/150 (0%)
Query: 7 RLSEVDNDLLVAPFHMEEIEVVVRESDGNKSPGADEYNFAFVKEFWYLMKNEVRIMFDQI 66
R + DN++L EEI+ V+ +KSPG D Y F K W ++ E +
Sbjct: 87 RCTNSDNEMLTREVSSEEIKTVLFSMPKDKSPGPDGYTSEFYKATWDIIGQEFTLPVQSF 146
Query: 67 HANELLPKSMLAYFVALIPKVSSPLELKDYRTISLLGSLYKLLAKVLARRLAGVLSSVIS 126
LPK + + +ALIPK + E++DYR IS LYK+++K++A RL +L I+
Sbjct: 147 FQKGFLPKGINSIILALIPKKLAAKEMRDYRPISCCNVLYKVISKIIANRLKLLLPRFIA 206
Query: 127 TTQSAFLKGTNLVDGVLVVNELV-DYAKKS 155
QSAF+K L++ +L+ ELV DY K S
Sbjct: 207 ENQSAFVKDRLLIENLLLATELVKDYHKDS 236
>UniRef100_Q7XXA4 OSJNBa0019G23.12 protein [Oryza sativa]
Length = 1140
Score = 94.7 bits (234), Expect = 7e-18
Identities = 43/129 (33%), Positives = 78/129 (60%)
Query: 21 HMEEIEVVVRESDGNKSPGADEYNFAFVKEFWYLMKNEVRIMFDQIHANELLPKSMLAYF 80
H+ EI+ + D NK+PG D + F ++FW ++K+++ +F++ H+ L +
Sbjct: 665 HLREIKEAIFAMDHNKAPGPDGFPVEFYQKFWEVIKHDLLNLFNEFHSGSLPIFGLNFGV 724
Query: 81 VALIPKVSSPLELKDYRTISLLGSLYKLLAKVLARRLAGVLSSVISTTQSAFLKGTNLVD 140
+ LIPKV ++ YR I LL YK+ KV R++ V ++++ TQ+AF++G N++D
Sbjct: 725 ITLIPKVEEANRIQQYRPICLLNVSYKIFTKVATNRISSVADNLVNPTQTAFMRGRNILD 784
Query: 141 GVLVVNELV 149
GV +++E V
Sbjct: 785 GVAIIHETV 793
>UniRef100_Q8H0A3 Putative reverse transcriptase [Oryza sativa]
Length = 563
Score = 94.0 bits (232), Expect = 1e-17
Identities = 47/144 (32%), Positives = 88/144 (60%), Gaps = 2/144 (1%)
Query: 7 RLSEVDNDLLVAPFHMEEIEVVVRESDGNKSPGADEYNFAFVKEFWYLMKNEVRIMFDQI 66
++S+++N+ L F +EI+ + + + NK+PG D + F + FW ++K+++ +F +
Sbjct: 188 QVSQLENEALTQEFTEDEIKKAIFQMEHNKAPGPDGFPAEFYQVFWNVIKSDLLDLFIEF 247
Query: 67 HANELLPKSMLAYF-VALIPKVSSPLELKDYRTISLLGSLYKLLAKVLARRLAGVLSSVI 125
H N LP L + + L+PK ++++ YR I LL +K+ KV R+ GV VI
Sbjct: 248 H-NGTLPLFSLNFGPIILLPKCMEAMKIQQYRPICLLNVSFKIFTKVATNRIIGVAQKVI 306
Query: 126 STTQSAFLKGTNLVDGVLVVNELV 149
S TQ+AF+ G N+++GV++++E +
Sbjct: 307 SPTQTAFIPGRNIMEGVVILHETI 330
>UniRef100_O64473 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1447
Score = 93.6 bits (231), Expect = 2e-17
Identities = 51/146 (34%), Positives = 78/146 (52%)
Query: 7 RLSEVDNDLLVAPFHMEEIEVVVRESDGNKSPGADEYNFAFVKEFWYLMKNEVRIMFDQI 66
R SE +++LL EEI+ V+ +KSPG D + F K W ++ NE +
Sbjct: 848 RCSETEHELLTRVVTAEEIKKVLFSMPNDKSPGPDGFTSEFFKATWEILGNEFILAIQSF 907
Query: 67 HANELLPKSMLAYFVALIPKVSSPLELKDYRTISLLGSLYKLLAKVLARRLAGVLSSVIS 126
A LPK + +ALIPK E+KDYR IS +YK+++K++A RL V I+
Sbjct: 908 FAKGFLPKGINTTILALIPKKKEAKEMKDYRPISCCNVIYKVISKIIANRLKLVPPKFIA 967
Query: 127 TTQSAFLKGTNLVDGVLVVNELVDYA 152
QSAF+K L++ VL+ + ++
Sbjct: 968 GNQSAFVKDRLLIENVLLATDTASFS 993
>UniRef100_O64612 Putative reverse transcriptase [Arabidopsis thaliana]
Length = 1412
Score = 91.7 bits (226), Expect = 6e-17
Identities = 58/153 (37%), Positives = 82/153 (52%), Gaps = 1/153 (0%)
Query: 2 GVNFNRLSEVDNDLLVAPFHMEEIEVVVRESDGNKSPGADEYNFAFVKEFWYLMKNEVRI 61
G+ R S + +LLVA E+ V NKSPG D Y F +E W ++ EV +
Sbjct: 701 GILQYRYSLHEQNLLVAEITEAEVMKVFFSIPLNKSPGPDGYTVEFFRETWSVIGQEVTM 760
Query: 62 MFDQIHANELLPKSMLAYFVALIPKVSSPLELKDYRTISLLGSLYKLLAKVLARRLAGVL 121
LPK + + +ALIPK + E+KDYR IS LYK ++K+LA RL +L
Sbjct: 761 AIKSFFTYGFLPKGLNSTILALIPKRTYAKEMKDYRPISCCNVLYKAISKLLANRLKCLL 820
Query: 122 SSVISTTQSAFLKGTNLVDGVLVVNELV-DYAK 153
I+ QSAF+ L++ +L+ +ELV DY K
Sbjct: 821 PEFIAPNQSAFISDRLLMENLLLASELVKDYHK 853
>UniRef100_Q9ZVF0 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1449
Score = 90.9 bits (224), Expect = 1e-16
Identities = 53/146 (36%), Positives = 77/146 (52%), Gaps = 1/146 (0%)
Query: 9 SEVDNDLLVAPFHMEEIEVVVRESDGNKSPGADEYNFAFVKEFWYLMKNEVRIMFDQIHA 68
S + D+L A +EI + +KSPG D Y F K W ++ E +
Sbjct: 857 SPAEKDMLTASVSAKEIRGALFSMPNDKSPGPDGYTSEFYKRAWDIIGAEFVLAVKSFFE 916
Query: 69 NELLPKSMLAYFVALIPKVSSPLELKDYRTISLLGSLYKLLAKVLARRLAGVLSSVISTT 128
LPK + +ALIPK E+KDYR IS +YK+++K++A RL VL + I+
Sbjct: 917 KGFLPKGVNTTILALIPKKLEAKEMKDYRPISCCNVIYKVISKIIANRLKHVLPNFIAGN 976
Query: 129 QSAFLKGTNLVDGVLVVNELV-DYAK 153
QSAF+K L++ +L+ ELV DY K
Sbjct: 977 QSAFVKDRLLIENLLLATELVKDYHK 1002
>UniRef100_Q9FYK1 F21J9.30 [Arabidopsis thaliana]
Length = 1270
Score = 90.5 bits (223), Expect = 1e-16
Identities = 51/147 (34%), Positives = 79/147 (53%)
Query: 7 RLSEVDNDLLVAPFHMEEIEVVVRESDGNKSPGADEYNFAFVKEFWYLMKNEVRIMFDQI 66
R+SEV N+ LV +EI+ V +PG D + F + +W + N+V +
Sbjct: 378 RVSEVMNESLVGEVSAQEIKEAVFSIKPASAPGPDGMSALFFQHYWSTVGNQVTSEVKKF 437
Query: 67 HANELLPKSMLAYFVALIPKVSSPLELKDYRTISLLGSLYKLLAKVLARRLAGVLSSVIS 126
A+ ++P + LIPK P E+ D R ISL LYK+++K++A+RL L ++S
Sbjct: 438 FADGIMPAEWNYTHLCLIPKTQHPTEMVDLRPISLCSVLYKIISKIMAKRLQPWLPEIVS 497
Query: 127 TTQSAFLKGTNLVDGVLVVNELVDYAK 153
TQSAF+ + D +LV +ELV K
Sbjct: 498 DTQSAFVSERLITDNILVAHELVHSLK 524
>UniRef100_Q9FXJ5 F5A9.24 [Arabidopsis thaliana]
Length = 1254
Score = 90.5 bits (223), Expect = 1e-16
Identities = 51/147 (34%), Positives = 79/147 (53%)
Query: 7 RLSEVDNDLLVAPFHMEEIEVVVRESDGNKSPGADEYNFAFVKEFWYLMKNEVRIMFDQI 66
R+SEV N+ LV +EI+ V +PG D + F + +W + N+V +
Sbjct: 381 RVSEVMNESLVGEVSAQEIKEAVFSIKPASAPGPDGMSALFFQHYWSTVGNQVTSEVKKF 440
Query: 67 HANELLPKSMLAYFVALIPKVSSPLELKDYRTISLLGSLYKLLAKVLARRLAGVLSSVIS 126
A+ ++P + LIPK P E+ D R ISL LYK+++K++A+RL L ++S
Sbjct: 441 FADGIMPAEWNYTHLCLIPKTQHPTEMVDLRPISLCSVLYKIISKIMAKRLQPWLPEIVS 500
Query: 127 TTQSAFLKGTNLVDGVLVVNELVDYAK 153
TQSAF+ + D +LV +ELV K
Sbjct: 501 DTQSAFVSERLITDNILVAHELVHSLK 527
>UniRef100_Q75GM6 Putative non-LTR retroelement reverse transcriptase [Oryza sativa]
Length = 1614
Score = 90.1 bits (222), Expect = 2e-16
Identities = 45/142 (31%), Positives = 85/142 (59%)
Query: 8 LSEVDNDLLVAPFHMEEIEVVVRESDGNKSPGADEYNFAFVKEFWYLMKNEVRIMFDQIH 67
L E D + L+ PF +EE+ V+RE+ N + G D ++ F + FW ++ ++ M ++
Sbjct: 1046 LKEEDKNTLMRPFTIEELYKVLREAKPNTAFGPDGFSIPFYRAFWPQLRPDLFEMLLMLY 1105
Query: 68 ANELLPKSMLAYFVALIPKVSSPLELKDYRTISLLGSLYKLLAKVLARRLAGVLSSVIST 127
EL K + ++LIPK S+P ++K +R I +L +K ++K + RL + VIS
Sbjct: 1106 NEELDLKRLNFGVISLIPKNSNPTDIKQFRPICVLNDCFKFISKCVCNRLTEIARDVISP 1165
Query: 128 TQSAFLKGTNLVDGVLVVNELV 149
TQ+AF+ G +++G ++++E++
Sbjct: 1166 TQTAFIPGRFILEGCVIIHEVL 1187
>UniRef100_Q9M1N9 Hypothetical protein T18B22.50 [Arabidopsis thaliana]
Length = 762
Score = 89.7 bits (221), Expect = 2e-16
Identities = 52/134 (38%), Positives = 76/134 (55%), Gaps = 1/134 (0%)
Query: 23 EEIEVVVRESDGNKSPGADEYNFAFVKEFWYLMKNEVRIMFDQIHANELLPKSMLAYFVA 82
EEI+ V+ +KSPG D + F KE W ++ E + A LPK + + +A
Sbjct: 9 EEIKKVLFSMPNDKSPGPDGFTSEFFKESWEILGPEFILAIQSFFALGFLPKGVNSTILA 68
Query: 83 LIPKVSSPLELKDYRTISLLGSLYKLLAKVLARRLAGVLSSVISTTQSAFLKGTNLVDGV 142
LIPK E+KDYR IS +YK+++K+LA RL +L I+ QS+F+K L++ V
Sbjct: 69 LIPKKLESKEMKDYRPISCCNVMYKVISKILANRLKLLLPQFIAGNQSSFVKDRLLIENV 128
Query: 143 LVVNELV-DYAKKS 155
L+ +LV DY K S
Sbjct: 129 LLATDLVKDYHKDS 142
>UniRef100_Q7XF82 Putative retroelement [Oryza sativa]
Length = 586
Score = 89.7 bits (221), Expect = 2e-16
Identities = 51/152 (33%), Positives = 90/152 (58%), Gaps = 4/152 (2%)
Query: 7 RLSEVDNDLLVAPFHMEEIEVVVRESDGNKSPGADEYNFAFVKEFWYLMKNEVRIMFDQI 66
++S+ D L PF +EE++ VV E N++PG D + F +FW ++K E+ + +
Sbjct: 6 QVSKEDKVELTKPFSLEEVKKVVFEMRQNRAPGPDGFPAEFYVKFWGIIKFELLDLINDF 65
Query: 67 HANELLPKSMLAY-FVALIPKVSSPLELKDYRTISLLGSLYKLLAKVLARRLAGVLSSVI 125
H N L L Y V L+PK +++ +R I LL +K++ KVL RL +++ +I
Sbjct: 66 H-NGFLEVERLNYGIVTLVPKTKDAEQIQKFRPICLLNVSFKIITKVLMNRLDRIMTYII 124
Query: 126 STTQSAFLKGTNLVDGVLVVNELVD--YAKKS 155
S Q+AFLK +++G+++++EL++ + KKS
Sbjct: 125 SKNQTAFLKNRFIMEGIVILHELLNSLHTKKS 156
>UniRef100_Q7F9Q0 OSJNBa0045O17.3 protein [Oryza sativa]
Length = 492
Score = 89.7 bits (221), Expect = 2e-16
Identities = 45/141 (31%), Positives = 82/141 (57%)
Query: 7 RLSEVDNDLLVAPFHMEEIEVVVRESDGNKSPGADEYNFAFVKEFWYLMKNEVRIMFDQI 66
++S ++N+ L F EI+ + + + NK+PG D + F + FW ++KN++ +F +
Sbjct: 199 QVSHLENEALTQEFIEMEIKEAIFQMEHNKAPGPDGFPAEFYQVFWSVIKNDLFDLFTEF 258
Query: 67 HANELLPKSMLAYFVALIPKVSSPLELKDYRTISLLGSLYKLLAKVLARRLAGVLSSVIS 126
H L S+ + L+PK ++++ YR I LL +K+ KV R+ V VIS
Sbjct: 259 HNGSLPLFSLNFGTIILLPKCVEAMKIQQYRPICLLNVSFKIFTKVATNRIMCVAQKVIS 318
Query: 127 TTQSAFLKGTNLVDGVLVVNE 147
TQ+AF+ G N+++GV++++E
Sbjct: 319 PTQTAFIPGRNIMEGVVILHE 339
>UniRef100_O81441 T24H24.17 protein [Arabidopsis thaliana]
Length = 1077
Score = 89.7 bits (221), Expect = 2e-16
Identities = 49/147 (33%), Positives = 83/147 (56%), Gaps = 1/147 (0%)
Query: 9 SEVDNDLLVAPFHMEEIEVVVRESDGNKSPGADEYNFAFVKEFWYLMKNEVRIMFDQIHA 68
++++N L+ P + EI+ +K+PG D ++ +F + W + E+
Sbjct: 394 TDMNNTLISLPT-VAEIKQACLSIHPDKAPGRDGFSASFFQSNWSTVSEEITAEVQAFFI 452
Query: 69 NELLPKSMLAYFVALIPKVSSPLELKDYRTISLLGSLYKLLAKVLARRLAGVLSSVISTT 128
+E+LP+ + V LIPK+ +P ++ DYR I+L YK++AK+LA+RL +L S IS
Sbjct: 453 SEILPQKINHTHVRLIPKIPAPQKVSDYRPIALCSVYYKIIAKLLAKRLQPILHSCISEN 512
Query: 129 QSAFLKGTNLVDGVLVVNELVDYAKKS 155
QSAF+ + D VL+ +E + Y K S
Sbjct: 513 QSAFVPQRAISDNVLITHEALHYLKNS 539
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.358 0.165 0.605
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 873,270,529
Number of Sequences: 2790947
Number of extensions: 32817549
Number of successful extensions: 150975
Number of sequences better than 10.0: 492
Number of HSP's better than 10.0 without gapping: 343
Number of HSP's successfully gapped in prelim test: 149
Number of HSP's that attempted gapping in prelim test: 150198
Number of HSP's gapped (non-prelim): 602
length of query: 612
length of database: 848,049,833
effective HSP length: 133
effective length of query: 479
effective length of database: 476,853,882
effective search space: 228413009478
effective search space used: 228413009478
T: 11
A: 40
X1: 14 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (21.7 bits)
S2: 78 (34.7 bits)
Medicago: description of AC147435.10


