

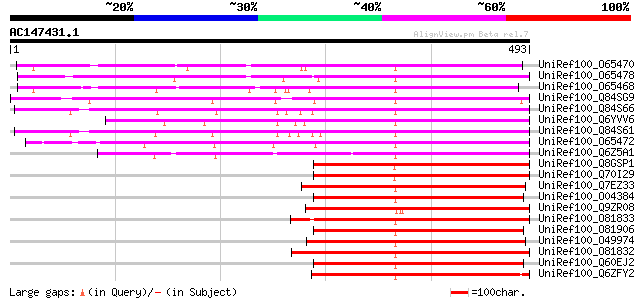
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147431.1 + phase: 0 /pseudo
(493 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_O65470 Serine/threonine kinase-like protein [Arabidops... 351 3e-95
UniRef100_O65478 Serine/threonine kinase [Arabidopsis thaliana] 328 2e-88
UniRef100_O65468 Hypothetical protein F21P8.50 [Arabidopsis thal... 313 9e-84
UniRef100_Q84SG9 Serine/threonine kinase receptor-like protein [... 293 6e-78
UniRef100_Q84S66 Putative serine/threonine kinase protein [Oryza... 290 9e-77
UniRef100_Q6YVV6 Putative serine/threonine-specific protein kina... 289 1e-76
UniRef100_Q84S61 Putative serine/threonine kinase protein [Oryza... 283 6e-75
UniRef100_O65472 Serine /threonine kinase-like protein [Arabidop... 277 6e-73
UniRef100_Q6Z5A1 Putative serine/threonine kinase [Oryza sativa] 271 2e-71
UniRef100_Q8GSP1 Similar to S-receptor kinase [Lotus japonicus] 265 2e-69
UniRef100_Q70I29 S-receptor kinase-like protein 2 [Lotus japonicus] 265 2e-69
UniRef100_Q7EZ33 Putative S-receptor kinase KIK1 [Oryza sativa] 263 7e-69
UniRef100_O04384 Serine/threonine kinase [Brassica oleracea] 263 7e-69
UniRef100_Q9ZR08 Putative receptor kinase [Arabidopsis thaliana] 261 3e-68
UniRef100_O81833 Putative receptor protein kinase [Arabidopsis t... 261 4e-68
UniRef100_O81906 Serine/threonine kinase-like protein [Arabidops... 260 6e-68
UniRef100_O49974 KI domain interacting kinase 1 [Zea mays] 259 9e-68
UniRef100_O81832 Putative receptor like kinase [Arabidopsis thal... 259 1e-67
UniRef100_Q60EJ2 Putative receptor-like protein kinase [Oryza sa... 256 8e-67
UniRef100_Q6ZFY2 Receptor protein kinase-like [Oryza sativa] 255 2e-66
>UniRef100_O65470 Serine/threonine kinase-like protein [Arabidopsis thaliana]
Length = 633
Score = 351 bits (900), Expect = 3e-95
Identities = 206/528 (39%), Positives = 291/528 (55%), Gaps = 58/528 (10%)
Query: 7 FLSFILLHIFLFTLT---NAQSPYYRLNFCQNSTEKTNNTSYRSNVRNLLLWINTDSATG 63
+ SF L +F F + +AQ P Y + CQN+ T+N++Y +N++ LL +++ +A+
Sbjct: 4 YSSFFFLFLFSFLTSFRVSAQDPTYVYHTCQNTANYTSNSTYNNNLKTLLASLSSRNASY 63
Query: 64 SVSNHIAINSNKTNSNDNDGDVVYGFYDCRGDVTGSFCQFCINAAVRAIAQCCPNGVSAM 123
S A D V G ++CRGDV+ C+ C++ AV CPN A
Sbjct: 64 STGFQNATVGQAP-------DRVTGLFNCRGDVSTEVCRRCVSFAVNDTLTRCPNQKEAT 116
Query: 124 IWYDICVIGYSDHNTSGKISVTPSWNLTGTKNTKDSTELEKAVD---DMRNLIGRVTAEA 180
++YD CV+ YS+ N + T L T+N S +L+ D N V +
Sbjct: 117 LYYDECVLRYSNQNILSTLITTGGVILVNTRNVT-SNQLDLLSDLVLPTLNQAATVALNS 175
Query: 181 NSNWAVGEFDWSDTEKRYGWVQCNSDLTKDGCRYCLETMLDKVPQCCGTKVKWAVVSPSC 240
+ + + +++ + YG VQC DLT+ C CL+ +++++P ++ +++PSC
Sbjct: 176 SKKFGTRKNNFTALQSFYGLVQCTPDLTRQDCSRCLQLVINQIPT---DRIGARIINPSC 232
Query: 241 GMEIDDNKFYNFQTESPPSLPNPGGLLLRTITPKS-------------FRDH--VPREDS 285
+ FY PP P + P+S F+D V +D
Sbjct: 233 TSRYEIYAFYTESAVPPPPPPPSISTPPVSAPPRSGQSSVFFLSCFSCFKDFHCVTGDDI 292
Query: 286 FNGDLPTIPLTVIQQSTDNFSESFKLGEGGFGPVYKGTLPDGTEIAAKRLSETSGQGLEE 345
D + IQ +TD+F ES K+G+GGFG VYKGTL DGTE+A KRLS++SGQG E
Sbjct: 293 TTADSLQLDYRTIQTATDDFVESNKIGQGGFGEVYKGTLSDGTEVAVKRLSKSSGQGEVE 352
Query: 346 FKNEVIFIAKLQHRNLVKLL--------------------------DEEKHKHLDWKLRL 379
FKNEV+ +AKLQHRNLV+LL D K LDW R
Sbjct: 353 FKNEVVLVAKLQHRNLVRLLGFCLDGEERVLVYEYVPNKSLDYFLFDPAKKGQLDWTRRY 412
Query: 380 SIIKGIARGLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKISDFGLARAFEKDQCHTKTK 439
II G+ARG+LYLH+DS L +IHRDLKASN+LLD +MNPKI+DFG+AR F DQ T
Sbjct: 413 KIIGGVARGILYLHQDSRLTIIHRDLKASNILLDADMNPKIADFGMARIFGLDQTEENTS 472
Query: 440 RVIGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSE 487
R++GTYGYM+PEYAM G +S+KSDV+SFGVLVLEII GK+N F+ ++
Sbjct: 473 RIVGTYGYMSPEYAMHGQYSMKSDVYSFGVLVLEIISGKKNSSFYQTD 520
>UniRef100_O65478 Serine/threonine kinase [Arabidopsis thaliana]
Length = 581
Score = 328 bits (842), Expect = 2e-88
Identities = 194/521 (37%), Positives = 278/521 (53%), Gaps = 45/521 (8%)
Query: 8 LSFILLHIFLFTLTNAQSPYYRLNFCQNSTEKTNNTSYRSNVRNLLLWINTDSATGSVSN 67
+SFI L +F +AQ+ +Y + C +T ++N++Y +N++ LL S+ S++
Sbjct: 5 ISFIFLFLFSSITASAQNTFYLYHNCSVTTTFSSNSTYSTNLKTLL------SSLSSLNA 58
Query: 68 HIAINSNKTNSNDNDGDVVYGFYDCRGDVTGSFCQFCINAAVRAIAQCCPNGVSAMIWYD 127
+T + D V G + CR DV+ C+ C+ AV CP + +Y+
Sbjct: 59 SSYSTGFQTATAGQAPDRVTGLFLCRVDVSSEVCRSCVTFAVNETLTRCPKDKEGVFYYE 118
Query: 128 ICVIGYSDHNTSGKISVTPSWNLTGTKN--TKDSTELEKAVDDMRNLIGRVTAEANSNWA 185
C++ YS+ N ++ + +N + + V NL A + WA
Sbjct: 119 QCLLRYSNRNIVATLNTDGGMFMQSARNPLSVKQDQFRDLVLTPMNLAAVEAARSFKKWA 178
Query: 186 VGEFDWSDTEKRYGWVQCNSDLTKDGCRYCLETMLDKVPQCCGTKVKWAVVSPSCGMEID 245
V + D + ++ YG V+C DL + C CL+ +++V K+ ++ PSC D
Sbjct: 179 VRKIDLNASQSLYGMVRCTPDLREQDCLDCLKIGINQVTY---DKIGGRILLPSCASRYD 235
Query: 246 DNKFYNFQTESPP--SLPNPGGLLLRTITPKSFRDHVPREDSFNGDLPT-----IPLTVI 298
+ FYN P S P P +R ++ + P + N D+ T I
Sbjct: 236 NYAFYNESNVGTPQDSSPRPAVFSVRAKNKRTLNEKEPVAEDGN-DITTAGSLQFDFKAI 294
Query: 299 QQSTDNFSESFKLGEGGFGPVYKGTLPDGTEIAAKRLSETSGQGLEEFKNEVIFIAKLQH 358
+ +T+ F KLG+GGFG VYKGTL G ++A KRLS+TSGQG +EF+NEV+ +AKLQH
Sbjct: 295 EAATNCFLPINKLGQGGFGEVYKGTLSSGLQVAVKRLSKTSGQGEKEFENEVVVVAKLQH 354
Query: 359 RNLVKLL--------------------------DEEKHKHLDWKLRLSIIKGIARGLLYL 392
RNLVKLL D LDW R II GIARG+LYL
Sbjct: 355 RNLVKLLGYCLEGEEKILVYEFVPNKSLDHFLFDSTMKMKLDWTRRYKIIGGIARGILYL 414
Query: 393 HEDSPLRVIHRDLKASNVLLDDEMNPKISDFGLARAFEKDQCHTKTKRVIGTYGYMAPEY 452
H+DS L +IHRDLKA N+LLDD+MNPKI+DFG+AR F DQ T+RV+GTYGYM+PEY
Sbjct: 415 HQDSRLTIIHRDLKAGNILLDDDMNPKIADFGMARIFGMDQTEAMTRRVVGTYGYMSPEY 474
Query: 453 AMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSEHNLETL 493
AM G FS+KSDV+SFGVLVLEII G +N + + ++ L
Sbjct: 475 AMYGQFSMKSDVYSFGVLVLEIISGMKNSSLYQMDESVGNL 515
>UniRef100_O65468 Hypothetical protein F21P8.50 [Arabidopsis thaliana]
Length = 1240
Score = 313 bits (801), Expect = 9e-84
Identities = 200/533 (37%), Positives = 282/533 (52%), Gaps = 70/533 (13%)
Query: 8 LSFILLHIFLFTLT---NAQSPYYRLNFCQNSTEKTNNTSYRSNVRNLLLWINTDSATGS 64
+ FI L +F F + +AQ+P+Y + C N T ++N++Y +N++ LL + +A+ S
Sbjct: 602 ICFIFLFLFSFLTSFKASAQNPFYLNHDCPNRTTYSSNSTYSTNLKTLLSSFASRNASYS 661
Query: 65 VSNHIAINSNKTNSNDNDGDVVYGFYDCRGDVTGSFCQFCINAAVRAIAQCCPNGVSAMI 124
I + +T D V G + CRGD++ C C+ +V CPN A+
Sbjct: 662 TGFQ-NIRAGQTP------DRVTGLFLCRGDLSPEVCSNCVAFSVNESLTRCPNQREAVF 714
Query: 125 WYDICVIGYSDHNT------SGKISVTPSWNLTGTKNTKDSTELEKAVDDMRNLIGRVTA 178
+Y+ C++ YS N G++ + N++ +N +D + V N A
Sbjct: 715 YYEECILRYSHKNFLSTVTYEGELIMRNPNNISSIQNQRD--QFIDLVQSNMNQAANEAA 772
Query: 179 EANSNWAVGEFDWSDTEKRYGWVQCNSDLTKDGCRYCLETMLDKVPQC--CGTKVKWAVV 236
++ ++ + + + + YG VQC DL + C CL + ++++ G + W
Sbjct: 773 NSSRKFSTIKTELTSLQTLYGLVQCTPDLARQDCFSCLTSSINRMMPLFRIGARQFW--- 829
Query: 237 SPSCGMEIDDNKFYN---FQTESPPSL--------PNP------GGLLLRTITPKSFRDH 279
PSC + FYN T SPP L +P G L T K+F
Sbjct: 830 -PSCNSRYELYAFYNETAIGTPSPPPLFPGSTPPLTSPSIPALVGYCFLAQRTKKTFDTA 888
Query: 280 VPRE---DSFNGDLPTIPLTVIQQSTDNFSESFKLGEGGFGPVYKGTLPDGTEIAAKRLS 336
E D D + IQ +T++F+ES K+G GGFG VYKGT +G E+A KRLS
Sbjct: 889 SASEVGDDMATADSLQLDYRTIQTATNDFAESNKIGRGGFGEVYKGTFSNGKEVAVKRLS 948
Query: 337 ETSGQGLEEFKNEVIFIAKLQHRNLVKLL--------------------------DEEKH 370
+ S QG EFK EV+ +AKLQHRNLV+LL D K
Sbjct: 949 KNSRQGEAEFKTEVVVVAKLQHRNLVRLLGFSLQGEERILVYEYMPNKSLDCLLFDPTKQ 1008
Query: 371 KHLDWKLRLSIIKGIARGLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKISDFGLARAFE 430
LDW R +II GIARG+LYLH+DS L +IHRDLKASN+LLD ++NPKI+DFG+AR F
Sbjct: 1009 TQLDWMQRYNIIGGIARGILYLHQDSRLTIIHRDLKASNILLDADINPKIADFGMARIFG 1068
Query: 431 KDQCHTKTKRVIGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDF 483
DQ T R++GTYGYMAPEYAM G FS+KSDV+SFGVLVLEII G++N F
Sbjct: 1069 LDQTQDNTSRIVGTYGYMAPEYAMHGQFSMKSDVYSFGVLVLEIISGRKNSSF 1121
>UniRef100_Q84SG9 Serine/threonine kinase receptor-like protein [Oryza sativa]
Length = 624
Score = 293 bits (751), Expect = 6e-78
Identities = 196/541 (36%), Positives = 270/541 (49%), Gaps = 66/541 (12%)
Query: 1 MTCTARFLSFILLHIFLFTLTNAQSPYYRLNFCQNSTEKTNNTSYRSNVRNLLLWINTDS 60
M A LS +L + + Y C + T N++Y++N+ D+
Sbjct: 1 MAHRAGCLSMLLTAAVVLLALAPRGAAYPWQVCGTTGNFTANSTYQANL---------DA 51
Query: 61 ATGSVSNHIAINSN--KTNSNDNDGDVVYGFYDCRGDVTGSFCQFCINAAVRAIAQCCPN 118
++ +I+ + + T + V CRGD + C C+ A + + C
Sbjct: 52 VAAALPRNISSSPDLFATAMVGAVPEQVSALALCRGDANATECSGCLATAFQDVQNMCAY 111
Query: 119 GVSAMIWYDICVIGYSDHNTSGKISVTPSWNLTGTKN-TKDSTELEKAVDDMRNLIGRVT 177
A I+YD C++ YS+ + S + +N T D V + N
Sbjct: 112 DKDAAIYYDPCILYYSNVPFLSSVDNAASTSRVNLQNVTSDPGRFNGMVAALVNATADYA 171
Query: 178 AEANSN-WAVGEFDW---SDTEKRYGWVQCNSDLTKDGCRYCLETMLDKVPQCCGTKVKW 233
A ++ +A GE S+ K Y W QC DLT C CL ++ K+P+ ++
Sbjct: 172 AHNSTRRYASGEAVLDRESEFPKVYSWAQCTPDLTPAQCGDCLAAIIAKLPRLFTNRIGG 231
Query: 234 AVVSPSCGMEIDDNKFYN----FQTESPPSLPNPGGLLLRTITPKSFRDHVPREDSFNG- 288
V+ C + N F N +PP + T +P + EDS N
Sbjct: 232 RVLGVRCSYRYEVNPFLNGLVMVHLTAPP---------IPTASPPAAAAAAAGEDSSNSA 282
Query: 289 --------DLPTIPLTVIQQSTDNFSESFKLGEGGFGPVYKGTLPDGTEIAAKRLSETSG 340
+ I ++ ++ +T F+E KLGEGGFG VYKGTLPDG EIA KRLS++S
Sbjct: 283 ESENISSVESMLIDISTLRAATGCFAERNKLGEGGFGAVYKGTLPDGDEIAVKRLSKSSA 342
Query: 341 QGLEEFKNEVIFIAKLQHRNLVKLL--------------------------DEEKHKHLD 374
QG+ E KNE+ +AKLQH+NLV+L+ D +K + LD
Sbjct: 343 QGVGELKNELALVAKLQHKNLVRLVGVCLEQEERLLVYEFVPNRSLDQILFDADKRQQLD 402
Query: 375 WKLRLSIIKGIARGLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKISDFGLARAFEKDQC 434
W R II GIARGL YLHEDS L+V+HRDLKASN+LLD MNPKISDFGLAR F +DQ
Sbjct: 403 WGKRYKIINGIARGLQYLHEDSQLKVVHRDLKASNILLDMNMNPKISDFGLARLFGRDQT 462
Query: 435 HTKTKRVIGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFF--LSEHNLET 492
T VIGTYGYM+PEYAM G +S+KSDVFSFGV+VLEI+ GK+N D + L +L T
Sbjct: 463 QGVTNLVIGTYGYMSPEYAMRGNYSLKSDVFSFGVMVLEIVTGKKNNDCYNSLQSEDLLT 522
Query: 493 L 493
L
Sbjct: 523 L 523
>UniRef100_Q84S66 Putative serine/threonine kinase protein [Oryza sativa]
Length = 682
Score = 290 bits (741), Expect = 9e-77
Identities = 195/574 (33%), Positives = 287/574 (49%), Gaps = 94/574 (16%)
Query: 5 ARFLSFILLHIFLFTLTNAQSPYYRLNFCQNSTEKTNNTSYRSNVRNLLLWI--NTDSAT 62
A FL +LLH+ L + P+Y + S + N+++++NV L + NT S+
Sbjct: 15 AAFLLAVLLHVHLAAGEDEPPPWYLCDPYSASGRYSENSTFQANVNRLSATLPRNTSSSP 74
Query: 63 GSVSNHIAINSNKTNSNDNDGDVVYGFYDCRGDVTGSF-CQFCINAAVRAIAQCCPNGVS 121
+ T + + D VYG+ CRGDV + C+ C+ AA+R + CP
Sbjct: 75 AMYA---------TGAAGDVPDKVYGYALCRGDVADAHACERCVAAALRDAPRVCPLVKD 125
Query: 122 AMIWYDICVIGYSDHNTS-------GKISVTPSWNLTGTKNTKDSTELEKAVDDMRNLIG 174
A++++D+C + YS+ N S+ S L + AV + N
Sbjct: 126 ALVFHDLCQLRYSNRNFLLDDDYYVATYSLQRSSRLVSAPAPAAVAAFDAAVAMLANATA 185
Query: 175 RVTAEANSNWAVGEFDWSDTE--------KRYGWVQCNSDLTKDGCRYCLETMLD-KVPQ 225
A AN++ G + + + Y QC D D CR CL T+ ++P+
Sbjct: 186 EYAAAANTSRRYGTAEEEGVDGDGDSGRPRMYALAQCTPDKAADVCRACLTTLTTVQLPK 245
Query: 226 C-CGTKVKWAVVSPSCGMEIDDNKFYNFQ--------TESPPSLP------------NPG 264
G + V C + + F++ + E+PP N
Sbjct: 246 LYSGGRTGGGVFGVWCNLRYEVFPFFSGRPLLHLPAFVEAPPPATSAAATRRGEKKRNKT 305
Query: 265 GLLLRTITPK-------------SFRDHVPREDSF------NGDLPTIPLTVIQQSTDNF 305
G++L + P +R P E +F + D + L+ ++ +TD+F
Sbjct: 306 GIVLAIVMPTIAAMLLIVVAYFCCWRRRRPEEQTFLPYDIQSIDSLLLDLSTLRAATDDF 365
Query: 306 SESFKLGEGGFGPVYKGTLPDGTEIAAKRLSETSGQGLEEFKNEVIFIAKLQHRNLVKLL 365
+E+ +G GGFG VYKG LP+G E+A KRL ++SGQG+EE K+E++ +AKL H+NLV+L+
Sbjct: 366 AETKMIGRGGFGMVYKGVLPEGQEVAVKRLCQSSGQGIEELKSELVLVAKLYHKNLVRLI 425
Query: 366 --------------------------DEEKHKHLDWKLRLSIIKGIARGLLYLHEDSPLR 399
D +K+ LDW R II GIA+GL YLHEDS L+
Sbjct: 426 GVCLEQQEKILVYEYMSNKSLDTILFDIDKNIELDWGKRFKIINGIAQGLQYLHEDSRLK 485
Query: 400 VIHRDLKASNVLLDDEMNPKISDFGLARAFEKDQCHTKTKRVIGTYGYMAPEYAMAGLFS 459
++HRDLKASN+LLD + NPKISDFGLA+ F+ DQ T R+ GTYGYMAPEYAM G +S
Sbjct: 486 IVHRDLKASNILLDFDYNPKISDFGLAKIFDGDQSKDITHRIAGTYGYMAPEYAMHGHYS 545
Query: 460 VKSDVFSFGVLVLEIIYGKRNGDFFLSEHNLETL 493
VK DVFSFGVLVLEI+ G+RN + S +L+ L
Sbjct: 546 VKLDVFSFGVLVLEIVTGRRNSGSYDSGQDLDLL 579
>UniRef100_Q6YVV6 Putative serine/threonine-specific protein kinase [Oryza sativa]
Length = 638
Score = 289 bits (739), Expect = 1e-76
Identities = 181/451 (40%), Positives = 238/451 (52%), Gaps = 49/451 (10%)
Query: 92 CRGDVTGSFCQFCINAAVRAIAQCCPNGVSAMIWYDICVIGYSDHN-TSGKISVT--PSW 148
CRGD S C C+ A + C A I+YD C++ YS+ +G S P +
Sbjct: 90 CRGDANASTCLACLTQAFLDLPNACAYHKVAAIFYDSCLLAYSNATIAAGDFSSEKIPIY 149
Query: 149 NLTGTKN-TKDSTELEKAVDDMRNLIGRVTAEANSN--WAVGEFDWS-DTEKRYGWVQCN 204
N T + + V + N A ++ +A GE D++ + K Y W QC
Sbjct: 150 GFYSNANATTEQARFNRLVAALVNATADYAARNSTRRRYASGEADFNAEFPKVYSWAQCT 209
Query: 205 SDLTKDGCRYCLETMLDKVPQCCGTKVKWAVVSPSCGMEIDDNKFYNFQ-------TESP 257
DLT CR CL ++ +V V + C + F + T
Sbjct: 210 PDLTPASCRSCLAQIIGTYIGFFENRVGGFVRAVWCSFQYSTTPFLDGPMLVRLQGTSGA 269
Query: 258 PSLPNPGGLLLRT------ITPKSFRD---HVPREDSFNGDLPTIPLTVIQQSTDNFSES 308
P+P ++ TP D ED N D I +++++ +T +F+ES
Sbjct: 270 SPAPSPAAVVPAVNQTPPAATPTLEGDANYSTEAEDIENLDSMLIDISILRSATGDFAES 329
Query: 309 FKLGEGGFGPVYKGTLPDGTEIAAKRLSETSGQGLEEFKNEVIFIAKLQHRNLVKLL--- 365
KLGEGGFG VYKG LPDG EIA KRLS++S QG+EE KNE+ +AKL+H+NLV L+
Sbjct: 330 NKLGEGGFGAVYKGVLPDGYEIAVKRLSKSSTQGVEELKNELALVAKLKHKNLVSLVGVC 389
Query: 366 -----------------------DEEKHKHLDWKLRLSIIKGIARGLLYLHEDSPLRVIH 402
D EK + LDW+ R II GIARGL YLHEDS L+V+H
Sbjct: 390 LEQQERLLVYEFVPNRSLDLILFDTEKSEQLDWEKRYKIINGIARGLQYLHEDSQLKVVH 449
Query: 403 RDLKASNVLLDDEMNPKISDFGLARAFEKDQCHTKTKRVIGTYGYMAPEYAMAGLFSVKS 462
RDLKASN+LLD MNPKISDFGLAR F +DQ TK VIGTYGYMAPEY G +SVKS
Sbjct: 450 RDLKASNILLDVNMNPKISDFGLARIFGRDQTQAVTKNVIGTYGYMAPEYLTRGNYSVKS 509
Query: 463 DVFSFGVLVLEIIYGKRNGDFFLSEHNLETL 493
DVFSFGV+VLEI+ G++N + S+ + + L
Sbjct: 510 DVFSFGVMVLEIVTGRKNNHSYNSQQSEDLL 540
>UniRef100_Q84S61 Putative serine/threonine kinase protein [Oryza sativa]
Length = 702
Score = 283 bits (725), Expect = 6e-75
Identities = 194/578 (33%), Positives = 283/578 (48%), Gaps = 98/578 (16%)
Query: 5 ARFLSFILLHIFLFTLTNAQSPYYRLNFCQNSTEKTNNTSYRSNVRNLLLWI--NTDSAT 62
A FL +LLH L + P+ S + N +Y+ N+ L + NT S+
Sbjct: 16 ATFLLAVLLHAPLAAGEDEPPPWVLCGPYPPSGNYSKNGTYQVNLDLLSTTLPKNTSSSP 75
Query: 63 GSVSNHIAINSNKTNSNDNDGDVVYGFYDCRGDVTGSFCQFCINAAVRAIAQCCPNGVSA 122
+ T + + D VYG CRGD S C+ C+ AA+R + CP
Sbjct: 76 AMYA---------TGTVGDVPDKVYGLALCRGDANASACERCVAAALRDAPRRCPLVKDV 126
Query: 123 MIWYDICVIGYSDHN---TSGKISVTPSWNLTGTKNTKDSTELEKAVDDMRNLIG-RVTA 178
+++YD+C + YS+ + T + + + + AV + N A
Sbjct: 127 LVFYDLCQLRYSNRDFFLDDDYFVTTYTLQRSRRVGAAAAAAFDAAVAVLVNATADYAAA 186
Query: 179 EANSNWAVGEFDW----SDTEKRYGWVQCNSDLTKDGCRYCLETMLDKVPQCCGTKVKWA 234
+++ + GE + SD K Y QC D T + CR CL T++ ++P+ +
Sbjct: 187 DSSRRYGTGEEEGVDGDSDRPKIYALAQCTPDKTPEVCRTCLSTVIGQLPKEFSGRTGGG 246
Query: 235 VVSPSCGMEIDDNKFYNFQ--------TESPPSLPNPG-----------GLLLRTITP-- 273
+ C + F++ + E+PP P+P G +L + P
Sbjct: 247 MFGVWCNFRYEVFPFFSGRPLLQLPAFVETPPPPPSPSATSGEKTKNRIGTVLAIVMPAI 306
Query: 274 --------------KSFRDHVPREDSF-------------NGDLPTI-----PLTVIQQS 301
K + P E +F + D+ +I L I+ +
Sbjct: 307 AAILLMVVACFCCWKRIKKRRPEEQTFLSCKSAKNYDSVSSDDIQSIDSLILDLPTIRVA 366
Query: 302 TDNFSESFKLGEGGFGPVYKGTLPDGTEIAAKRLSETSGQGLEEFKNEVIFIAKLQHRNL 361
TD+F+++ +G+GGFG VYKG LPDG EIA KRL ++S QG+ E K+E+I +AKL H+NL
Sbjct: 367 TDDFADTKMIGQGGFGMVYKGVLPDGQEIAVKRLCQSSRQGIGELKSELILVAKLYHKNL 426
Query: 362 VKLL--------------------------DEEKHKHLDWKLRLSIIKGIARGLLYLHED 395
V+L+ D +K++ LDW R II GIARGL YLHED
Sbjct: 427 VRLIGVCLEQQEKILVYEYMPNGSLDIVLFDTDKNRELDWGKRFKIINGIARGLQYLHED 486
Query: 396 SPLRVIHRDLKASNVLLDDEMNPKISDFGLARAFEKDQCHTKTKRVIGTYGYMAPEYAMA 455
S L+++HRDLKASN+LLD + +PKISDFGLA+ F DQ T R+ GTYGYMAPEYAM
Sbjct: 487 SQLKIVHRDLKASNILLDFDYSPKISDFGLAKIFGGDQSEDVTNRIAGTYGYMAPEYAMR 546
Query: 456 GLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSEHNLETL 493
G +S+KSDVFSFGVLVLEII G+RN + S +++ L
Sbjct: 547 GNYSIKSDVFSFGVLVLEIITGRRNTGSYDSGQDVDLL 584
>UniRef100_O65472 Serine /threonine kinase-like protein [Arabidopsis thaliana]
Length = 648
Score = 277 bits (708), Expect = 6e-73
Identities = 184/541 (34%), Positives = 263/541 (48%), Gaps = 72/541 (13%)
Query: 16 FLFTLTNAQSPYYRLNFCQNSTEKTNNTSYRSNVRNLLLWINTDSATGSVSNHIAINSNK 75
F T T+ S L C N+T N++Y +N R +L + ++ V++H +
Sbjct: 8 FFLTSTSLVSALQTLP-CINTTYFIPNSTYDTNRRVILSLLPSN-----VTSHFGFFNGS 61
Query: 76 TNSNDNDGDVVYGFYDCRGDVTGSFCQFCINAAVRAIAQCCPNGVSAMIWY---DICVIG 132
N VY C C C+ +A + + C +A+IW IC+I
Sbjct: 62 IGQAPNR---VYAVGMCLPGTEEESCIGCLLSASNTLLETCLTEENALIWIANRTICMIR 118
Query: 133 YSDHNTSGKISVTPSWNLTGTKNTK-DSTELEKAVDDM-RNLIGRVTAEANSNWAVGEFD 190
YSD + G + P K + TE + + ++ ++ ++ W+ ++
Sbjct: 119 YSDTSFVGSFELEPHREFLSIHGYKTNETEFNTVWSRLTQRMVQEASSSTDATWSGAKYY 178
Query: 191 WSD------TEKRYGWVQCNSDLTKDGCRYCLETMLDKVPQCCGTKVKWAVVSPSCGMEI 244
+D ++ Y +QC DL+ C CL + CC + ++V SC
Sbjct: 179 TADVAALPDSQTLYAMMQCTPDLSPAECNLCLTESVVNYQSCCLGRQGGSIVRLSCAFRA 238
Query: 245 DDNKF---YNFQTESPPSLPNPGGLLLRTITPKSFRDHVPREDSFNGD------------ 289
+ F + T P S P P + K + FNG+
Sbjct: 239 ELYPFGGAFTVMTARPLSQPPPSLIKKGEFFAKFMSNSQEPRKVFNGNYCCNCCSHYSGR 298
Query: 290 -----------LPTIPLTVIQQSTDNFSESFKLGEGGFGPVYKGTLPDGTEIAAKRLSET 338
+ I+ +T+NF+++ KLG+GGFG VYKGTL +GTE+A KRLS+T
Sbjct: 299 YHLLAGITTLHFQQLDFKTIEVATENFAKTNKLGQGGFGEVYKGTLVNGTEVAVKRLSKT 358
Query: 339 SGQGLEEFKNEVIFIAKLQHRNLVKLL--------------------------DEEKHKH 372
S QG +EFKNEV+ +AKLQHRNLVKLL D K
Sbjct: 359 SEQGAQEFKNEVVLVAKLQHRNLVKLLGYCLEPEEKILVYEFVPNKSLDYFLFDPTKQGQ 418
Query: 373 LDWKLRLSIIKGIARGLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKISDFGLARAFEKD 432
LDW R +II GI RG+LYLH+DS L +IHRDLKASN+LLD +M PKI+DFG+AR D
Sbjct: 419 LDWTKRYNIIGGITRGILYLHQDSRLTIIHRDLKASNILLDADMIPKIADFGMARISGID 478
Query: 433 QCHTKTKRVIGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSEHNLET 492
Q TKR+ GT+GYM PEY + G FS+KSDV+SFGVL+LEII GK+N F+ ++ E
Sbjct: 479 QSVANTKRIAGTFGYMPPEYVIHGQFSMKSDVYSFGVLILEIICGKKNRSFYQADTKAEN 538
Query: 493 L 493
L
Sbjct: 539 L 539
>UniRef100_Q6Z5A1 Putative serine/threonine kinase [Oryza sativa]
Length = 609
Score = 271 bits (694), Expect = 2e-71
Identities = 177/450 (39%), Positives = 243/450 (53%), Gaps = 48/450 (10%)
Query: 84 DVVYGFYDCRGDVTGSFCQFCINAAVRAIAQCCPNGVSAMIWYDICVIGYSDH------- 136
D VYG CRGD+ S C C R C A++ Y+ C +SD
Sbjct: 75 DAVYGLLLCRGDMNPSDCFDCGTNVWRDAGPTCNRTKDAILVYNQCYAQFSDRGDFLAAT 134
Query: 137 NTSGKISVTPSW-NLTGTKNTKDSTELEKAVDDMRNLIGRVTAEANSN-WAVGEFDWSDT 194
N SG++S+ S N+T T D ++AV ++ N R E ++ +A G+ +DT
Sbjct: 135 NNSGEVSLLISGTNITST----DVAGYDRAVTELLNATVRYAVENSTKLFATGQRVGNDT 190
Query: 195 --EKRYGWVQCNSDLTKDGCRYCLETMLDKVPQCCGTKVKWA-VVSPSCGMEIDDNKFYN 251
Y QC+ DL+ CR CL+ ++ + + K A V P C + + FY
Sbjct: 191 GFSNIYSMAQCSPDLSPAQCRSCLDGLVGQWWKTFPLNGKGARVAGPRCYLRSELGPFY- 249
Query: 252 FQTESPP-SLPNPGGLLLRTITPKSFRDHVPREDSFNGDLPTIPLTVIQQSTDNFSESFK 310
T +P LP L + + ED + + L +Q +TDNF ES K
Sbjct: 250 --TGNPMVRLPVKADELTQFSQNYANLPADTNEDFESVKSTLLSLASLQVATDNFHESNK 307
Query: 311 LGEGGFGPVYKGTLPDGTEIAAKRLSETSGQGLEEFKNEVIFIAKLQHRNLVKLL----- 365
+GEGGFG VYKG L G E+A KR+++ S QGLEE KNE++ +AKL HRNLV+L+
Sbjct: 308 IGEGGFGAVYKGIL-HGQEVAVKRMAKGSNQGLEELKNELVLVAKLHHRNLVRLVGFCLD 366
Query: 366 ---------------------DEEKHKHLDWKLRLSIIKGIARGLLYLHEDSPLRVIHRD 404
D E+ + LDW +R II+GIARGL YLH+DS +++HRD
Sbjct: 367 EGERLLIYEYMSNKSLDTFLFDAEQKRKLDWAVRFKIIEGIARGLQYLHQDSQKKIVHRD 426
Query: 405 LKASNVLLDDEMNPKISDFGLARAFEKDQCHTKTKRVIGTYGYMAPEYAMAGLFSVKSDV 464
+KASN+LLD +MNPKI DFGLAR F +DQ T R+ GT+GYM PEY + G +S KSDV
Sbjct: 427 MKASNILLDADMNPKIGDFGLARLFGQDQTREVTSRIAGTFGYMPPEYVLRGQYSTKSDV 486
Query: 465 FSFGVLVLEIIYG-KRNGDFFLSEHNLETL 493
FSFG+LV+EI+ G +RN +LSE N E +
Sbjct: 487 FSFGILVIEIVTGRRRNSGPYLSEQNDEDI 516
>UniRef100_Q8GSP1 Similar to S-receptor kinase [Lotus japonicus]
Length = 862
Score = 265 bits (678), Expect = 2e-69
Identities = 142/226 (62%), Positives = 163/226 (71%), Gaps = 21/226 (9%)
Query: 289 DLPTI-PLTVIQQSTDNFSESFKLGEGGFGPVYKGTLPDGTEIAAKRLSETSGQGLEEFK 347
DL TI + I +T++FS S KLGEGGFGPVYKG L +G EIA KRLS TSGQG+EEFK
Sbjct: 538 DLATIFDFSTISSATNHFSLSNKLGEGGFGPVYKGLLANGQEIAVKRLSNTSGQGMEEFK 597
Query: 348 NEVIFIAKLQHRNLVKL--------------------LDEEKHKHLDWKLRLSIIKGIAR 387
NE+ IA+LQHRNLVKL LD + K +DW RL II GIAR
Sbjct: 598 NEIKLIARLQHRNLVKLFGCSVHQDENSHANKKMKILLDSTRSKLVDWNKRLQIIDGIAR 657
Query: 388 GLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKISDFGLARAFEKDQCHTKTKRVIGTYGY 447
GLLYLH+DS LR+IHRDLK SN+LLDDEMNPKISDFGLAR F DQ +TKRV+GTYGY
Sbjct: 658 GLLYLHQDSRLRIIHRDLKTSNILLDDEMNPKISDFGLARIFIGDQVEARTKRVMGTYGY 717
Query: 448 MAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSEHNLETL 493
M PEYA+ G FS+KSDVFSFGV+VLEII GK+ G F+ H+L L
Sbjct: 718 MPPEYAVHGSFSIKSDVFSFGVIVLEIISGKKVGRFYDPHHHLNLL 763
>UniRef100_Q70I29 S-receptor kinase-like protein 2 [Lotus japonicus]
Length = 865
Score = 265 bits (678), Expect = 2e-69
Identities = 142/226 (62%), Positives = 163/226 (71%), Gaps = 21/226 (9%)
Query: 289 DLPTI-PLTVIQQSTDNFSESFKLGEGGFGPVYKGTLPDGTEIAAKRLSETSGQGLEEFK 347
DL TI + I +T++FS S KLGEGGFGPVYKG L +G EIA KRLS TSGQG+EEFK
Sbjct: 541 DLATIFDFSTISSATNHFSLSNKLGEGGFGPVYKGLLANGQEIAVKRLSNTSGQGMEEFK 600
Query: 348 NEVIFIAKLQHRNLVKL--------------------LDEEKHKHLDWKLRLSIIKGIAR 387
NE+ IA+LQHRNLVKL LD + K +DW RL II GIAR
Sbjct: 601 NEIKLIARLQHRNLVKLFGCSVHQDENSHANKKMKILLDSTRSKLVDWNKRLQIIDGIAR 660
Query: 388 GLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKISDFGLARAFEKDQCHTKTKRVIGTYGY 447
GLLYLH+DS LR+IHRDLK SN+LLDDEMNPKISDFGLAR F DQ +TKRV+GTYGY
Sbjct: 661 GLLYLHQDSRLRIIHRDLKTSNILLDDEMNPKISDFGLARIFIGDQVEARTKRVMGTYGY 720
Query: 448 MAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSEHNLETL 493
M PEYA+ G FS+KSDVFSFGV+VLEII GK+ G F+ H+L L
Sbjct: 721 MPPEYAVHGSFSIKSDVFSFGVIVLEIISGKKIGRFYDPHHHLNLL 766
>UniRef100_Q7EZ33 Putative S-receptor kinase KIK1 [Oryza sativa]
Length = 865
Score = 263 bits (673), Expect = 7e-69
Identities = 139/239 (58%), Positives = 163/239 (68%), Gaps = 26/239 (10%)
Query: 278 DHVPREDSFNGDLPTIPLTVIQQSTDNFSESFKLGEGGFGPVYKGTLPDGTEIAAKRLSE 337
+H E+ N +LP + +TDNFS S KLGEGGFG VYKG LP G EIA KRLS
Sbjct: 517 EHEKSEEGKNCELPLFAFETLATATDNFSISNKLGEGGFGHVYKGRLPGGEEIAVKRLSR 576
Query: 338 TSGQGLEEFKNEVIFIAKLQHRNLVKLL--------------------------DEEKHK 371
+SGQGLEEFKNEVI IAKLQHRNLV+LL D E+
Sbjct: 577 SSGQGLEEFKNEVILIAKLQHRNLVRLLGCCIQGEEKILVYEYMPNKSLDAFLFDPERRG 636
Query: 372 HLDWKLRLSIIKGIARGLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKISDFGLARAFEK 431
LDW+ R II+G+ARGLLYLH DS LRV+HRDLKASN+LLD +MNPKISDFG+AR F
Sbjct: 637 LLDWRTRFQIIEGVARGLLYLHRDSRLRVVHRDLKASNILLDRDMNPKISDFGMARIFGG 696
Query: 432 DQCHTKTKRVIGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSEHNL 490
DQ T RV+GT GYM+PEYAM GLFSV+SDV+SFG+L+LEII G++N F E +L
Sbjct: 697 DQNQVNTNRVVGTLGYMSPEYAMEGLFSVRSDVYSFGILILEIITGQKNSSFHHMEGSL 755
>UniRef100_O04384 Serine/threonine kinase [Brassica oleracea]
Length = 850
Score = 263 bits (673), Expect = 7e-69
Identities = 140/226 (61%), Positives = 161/226 (70%), Gaps = 26/226 (11%)
Query: 289 DLPTIPLTVIQQSTDNFSESFKLGEGGFGPVYKGTLPDGTEIAAKRLSETSGQGLEEFKN 348
+LP L VI ++T++FS +LG GGFGPVYKG L DG EIA KRLS SGQG++EFKN
Sbjct: 513 ELPVFCLKVIVKATNDFSRENELGRGGFGPVYKGVLEDGQEIAVKRLSGKSGQGVDEFKN 572
Query: 349 EVIFIAKLQHRNLVKLL--------------------------DEEKHKHLDWKLRLSII 382
E+I IAKLQHRNLV+LL DE K + +DWKLR +II
Sbjct: 573 EIILIAKLQHRNLVRLLGCCFEGEEKMLVYEYMPNKSLDFFIFDEMKQELVDWKLRFAII 632
Query: 383 KGIARGLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKISDFGLARAFEKDQCHTKTKRVI 442
+GIARGLLYLH DS LR+IHRDLK SNVLLD EMNPKISDFG+AR F +Q T RV+
Sbjct: 633 EGIARGLLYLHRDSRLRIIHRDLKVSNVLLDGEMNPKISDFGMARIFGGNQNEANTVRVV 692
Query: 443 GTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSEH 488
GTYGYM+PEYAM GLFSVKSDV+SFGVL+LEII GKRN SEH
Sbjct: 693 GTYGYMSPEYAMEGLFSVKSDVYSFGVLLLEIISGKRNTSLRASEH 738
>UniRef100_Q9ZR08 Putative receptor kinase [Arabidopsis thaliana]
Length = 852
Score = 261 bits (667), Expect = 3e-68
Identities = 142/238 (59%), Positives = 164/238 (68%), Gaps = 26/238 (10%)
Query: 282 REDSFNGDLPTIPLTVIQQSTDNFSESFKLGEGGFGPVYKGTLPDGTEIAAKRLSETSGQ 341
++DS D+P+ L I +T NFS + KLG+GGFGPVYKG P EIA KRLS SGQ
Sbjct: 509 QDDSQGIDVPSFELETILYATSNFSNANKLGQGGFGPVYKGMFPGDQEIAVKRLSRCSGQ 568
Query: 342 GLEEFKNEVIFIAKLQHRNLVKLLD-----EEK--------HK-------------HLDW 375
GLEEFKNEV+ IAKLQHRNLV+LL EEK HK LDW
Sbjct: 569 GLEEFKNEVVLIAKLQHRNLVRLLGYCVAGEEKLLLYEYMPHKSLDFFIFDRKLCQRLDW 628
Query: 376 KLRLSIIKGIARGLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKISDFGLARAFEKDQCH 435
K+R +II GIARGLLYLH+DS LR+IHRDLK SN+LLD+EMNPKISDFGLAR F +
Sbjct: 629 KMRCNIILGIARGLLYLHQDSRLRIIHRDLKTSNILLDEEMNPKISDFGLARIFGGSETS 688
Query: 436 TKTKRVIGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSEHNLETL 493
T RV+GTYGYM+PEYA+ GLFS KSDVFSFGV+V+E I GKRN F E +L L
Sbjct: 689 ANTNRVVGTYGYMSPEYALEGLFSFKSDVFSFGVVVIETISGKRNTGFHEPEKSLSLL 746
>UniRef100_O81833 Putative receptor protein kinase [Arabidopsis thaliana]
Length = 815
Score = 261 bits (666), Expect = 4e-68
Identities = 142/253 (56%), Positives = 169/253 (66%), Gaps = 29/253 (11%)
Query: 267 LLRTITPKSFRDHVPREDSFNGDLPTIPLTVIQQSTDNFSESFKLGEGGFGPVYKGTLPD 326
+++ ++FR + ED DLP I +TD+FS LG GGFGPVYKG L D
Sbjct: 465 IMKRYRGENFRKGIEEEDL---DLPIFDRKTISIATDDFSYVNFLGRGGFGPVYKGKLED 521
Query: 327 GTEIAAKRLSETSGQGLEEFKNEVIFIAKLQHRNLVKLL--------------------- 365
G EIA KRLS SGQG+EEFKNEV IAKLQHRNLV+LL
Sbjct: 522 GQEIAVKRLSANSGQGVEEFKNEVKLIAKLQHRNLVRLLGCCIQGEECMLIYEYMPNKSL 581
Query: 366 -----DEEKHKHLDWKLRLSIIKGIARGLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKI 420
DE + LDWK R++II G+ARG+LYLH+DS LR+IHRDLKA NVLLD++MNPKI
Sbjct: 582 DFFIFDERRSTELDWKKRMNIINGVARGILYLHQDSRLRIIHRDLKAGNVLLDNDMNPKI 641
Query: 421 SDFGLARAFEKDQCHTKTKRVIGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRN 480
SDFGLA++F DQ + T RV+GTYGYM PEYA+ G FSVKSDVFSFGVLVLEII GK N
Sbjct: 642 SDFGLAKSFGGDQSESSTNRVVGTYGYMPPEYAIDGHFSVKSDVFSFGVLVLEIITGKTN 701
Query: 481 GDFFLSEHNLETL 493
F ++H+L L
Sbjct: 702 RGFRHADHDLNLL 714
>UniRef100_O81906 Serine/threonine kinase-like protein [Arabidopsis thaliana]
Length = 849
Score = 260 bits (665), Expect = 6e-68
Identities = 138/226 (61%), Positives = 158/226 (69%), Gaps = 26/226 (11%)
Query: 289 DLPTIPLTVIQQSTDNFSESFKLGEGGFGPVYKGTLPDGTEIAAKRLSETSGQGLEEFKN 348
+LP L I +T++F + +LG GGFGPVYKG L DG EIA KRLS SGQG++EFKN
Sbjct: 513 ELPVFSLNAIAIATNDFCKENELGRGGFGPVYKGVLEDGREIAVKRLSGKSGQGVDEFKN 572
Query: 349 EVIFIAKLQHRNLVKLL--------------------------DEEKHKHLDWKLRLSII 382
E+I IAKLQHRNLV+LL DE K +DWKLR SII
Sbjct: 573 EIILIAKLQHRNLVRLLGCCFEGEEKMLVYEYMPNKSLDFFLFDETKQALIDWKLRFSII 632
Query: 383 KGIARGLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKISDFGLARAFEKDQCHTKTKRVI 442
+GIARGLLYLH DS LR+IHRDLK SNVLLD EMNPKISDFG+AR F +Q T RV+
Sbjct: 633 EGIARGLLYLHRDSRLRIIHRDLKVSNVLLDAEMNPKISDFGMARIFGGNQNEANTVRVV 692
Query: 443 GTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSEH 488
GTYGYM+PEYAM GLFSVKSDV+SFGVL+LEI+ GKRN SEH
Sbjct: 693 GTYGYMSPEYAMEGLFSVKSDVYSFGVLLLEIVSGKRNTSLRSSEH 738
>UniRef100_O49974 KI domain interacting kinase 1 [Zea mays]
Length = 848
Score = 259 bits (663), Expect = 9e-68
Identities = 139/234 (59%), Positives = 160/234 (67%), Gaps = 26/234 (11%)
Query: 283 EDSFNGDLPTIPLTVIQQSTDNFSESFKLGEGGFGPVYKGTLPDGTEIAAKRLSETSGQG 342
ED + +L L I+ +T NFS+S KLGEGGFGPVY GTLP G E+A KRL SGQG
Sbjct: 508 EDGKSHELKVYSLDRIRTATSNFSDSNKLGEGGFGPVYMGTLPGGEEVAVKRLCRNSGQG 567
Query: 343 LEEFKNEVIFIAKLQHRNLVKLL--------------------------DEEKHKHLDWK 376
LEEFKNEVI IAKLQHRNLV+LL + EK + LDWK
Sbjct: 568 LEEFKNEVILIAKLQHRNLVRLLGCCIPREEKILVYEYMPNKSLDAFLFNPEKQRLLDWK 627
Query: 377 LRLSIIKGIARGLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKISDFGLARAFEKDQCHT 436
R II+GIARGLLYLH DS LRV+HRDLKASN+LLD +M PKISDFG+AR F DQ
Sbjct: 628 KRFDIIEGIARGLLYLHRDSRLRVVHRDLKASNILLDADMKPKISDFGMARMFGGDQNQF 687
Query: 437 KTKRVIGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSEHNL 490
T RV+GT+GYM+PEYAM G+FSVKSDV+ FGVL+LEII GKR F E +L
Sbjct: 688 NTNRVVGTFGYMSPEYAMEGIFSVKSDVYGFGVLILEIITGKRAVSFHCHEDSL 741
>UniRef100_O81832 Putative receptor like kinase [Arabidopsis thaliana]
Length = 772
Score = 259 bits (662), Expect = 1e-67
Identities = 137/252 (54%), Positives = 168/252 (66%), Gaps = 26/252 (10%)
Query: 268 LRTITPKSFRDHVPREDSFNGDLPTIPLTVIQQSTDNFSESFKLGEGGFGPVYKGTLPDG 327
+ T+ +S R +++ + +LP + L + ++T FS KLG+GGFGPVYKGTL G
Sbjct: 417 IETLQRESSRVSSRKQEEEDLELPFLDLDTVSEATSGFSAGNKLGQGGFGPVYKGTLACG 476
Query: 328 TEIAAKRLSETSGQGLEEFKNEVIFIAKLQHRNLVKLL---------------------- 365
E+A KRLS TS QG+EEFKNE+ IAKLQHRNLVK+L
Sbjct: 477 QEVAVKRLSRTSRQGVEEFKNEIKLIAKLQHRNLVKILGYCVDEEERMLIYEYQPNKSLD 536
Query: 366 ----DEEKHKHLDWKLRLSIIKGIARGLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKIS 421
D+E+ + LDW R+ IIKGIARG+LYLHEDS LR+IHRDLKASNVLLD +MN KIS
Sbjct: 537 SFIFDKERRRELDWPKRVEIIKGIARGMLYLHEDSRLRIIHRDLKASNVLLDSDMNAKIS 596
Query: 422 DFGLARAFEKDQCHTKTKRVIGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNG 481
DFGLAR D+ T RV+GTYGYM+PEY + G FS+KSDVFSFGVLVLEI+ G+RN
Sbjct: 597 DFGLARTLGGDETEANTTRVVGTYGYMSPEYQIDGYFSLKSDVFSFGVLVLEIVSGRRNR 656
Query: 482 DFFLSEHNLETL 493
F EH L L
Sbjct: 657 GFRNEEHKLNLL 668
>UniRef100_Q60EJ2 Putative receptor-like protein kinase [Oryza sativa]
Length = 837
Score = 256 bits (655), Expect = 8e-67
Identities = 138/227 (60%), Positives = 157/227 (68%), Gaps = 27/227 (11%)
Query: 289 DLPTIPLTVIQQSTDNFSESFKLGEGGFGPVYKGTLPDGTEIAAKRLSETSGQGLEEFKN 348
DLP L I +T+ FS KLGEGGFGPVYKGTL DG EIA K LS+TS QGL+EF+N
Sbjct: 503 DLPLFDLETIASATNGFSADNKLGEGGFGPVYKGTLEDGQEIAVKTLSKTSVQGLDEFRN 562
Query: 349 EVIFIAKLQHRNLVKLL--------------------------DEEKHKHLDWKLRLSII 382
EV+ IAKLQHRNLV+L+ D+ K K LDW+ R II
Sbjct: 563 EVMLIAKLQHRNLVQLIGYSVCGQEKMLLYEFMENKSLDCFLFDKSKSKLLDWQTRYHII 622
Query: 383 KGIARGLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKISDFGLARAFEKDQCHTKTKRVI 442
+GIARGLLYLH+DS R+IHRDLK SN+LLD EM PKISDFG+AR F D T RV+
Sbjct: 623 EGIARGLLYLHQDSRYRIIHRDLKTSNILLDKEMTPKISDFGMARMFGSDDTEINTVRVV 682
Query: 443 GTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRN-GDFFLSEH 488
GTYGYMAPEYAM G+FSVKSDVFSFGV+VLEII GKRN G + S H
Sbjct: 683 GTYGYMAPEYAMDGVFSVKSDVFSFGVIVLEIISGKRNRGVYSYSSH 729
>UniRef100_Q6ZFY2 Receptor protein kinase-like [Oryza sativa]
Length = 426
Score = 255 bits (652), Expect = 2e-66
Identities = 138/233 (59%), Positives = 162/233 (69%), Gaps = 27/233 (11%)
Query: 287 NGDLPTIPLTVIQQSTDNFSESFKLGEGGFGPVYKGTLPDGTEIAAKRLSETSGQGLEEF 346
N DLP + L+ + +T+ FS+ KLGEGGFGPVY+G L G EIA KRLS S QG EF
Sbjct: 83 NSDLPLMDLSSMYDATNQFSKENKLGEGGFGPVYRGVLGGGAEIAVKRLSARSRQGAAEF 142
Query: 347 KNEVIFIAKLQHRNLVKLL--------------------------DEEKHKHLDWKLRLS 380
+NEV IAKLQHRNLV+LL D K LDWK R S
Sbjct: 143 RNEVELIAKLQHRNLVRLLGCCVEKEEKMLIYEYLPNRSLDAFLFDSRKRAQLDWKTRQS 202
Query: 381 IIKGIARGLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKISDFGLARAFEKDQCHTKTKR 440
II GIARGLLYLHEDS L+VIHRDLKASNVLLD++MNPKISDFG+A+ FE++ T
Sbjct: 203 IILGIARGLLYLHEDSCLKVIHRDLKASNVLLDNKMNPKISDFGMAKIFEEESNEVNTGH 262
Query: 441 VIGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSEHNLETL 493
V+GTYGYMAPEYAM G+FSVKSDVFS GVLVLEI+ G+RNG +L ++N +TL
Sbjct: 263 VVGTYGYMAPEYAMEGVFSVKSDVFSLGVLVLEILSGQRNGAMYL-QNNQQTL 314
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.319 0.136 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 872,910,470
Number of Sequences: 2790947
Number of extensions: 39021617
Number of successful extensions: 156068
Number of sequences better than 10.0: 17621
Number of HSP's better than 10.0 without gapping: 9620
Number of HSP's successfully gapped in prelim test: 8002
Number of HSP's that attempted gapping in prelim test: 122538
Number of HSP's gapped (non-prelim): 23543
length of query: 493
length of database: 848,049,833
effective HSP length: 131
effective length of query: 362
effective length of database: 482,435,776
effective search space: 174641750912
effective search space used: 174641750912
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 77 (34.3 bits)
Medicago: description of AC147431.1


