

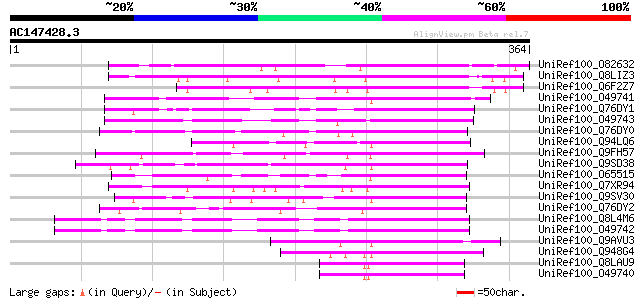
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147428.3 - phase: 0
(364 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_O82632 Hypothetical protein AT4g32890 [Arabidopsis tha... 264 3e-69
UniRef100_Q8LIZ3 OSJNBa0014K08.18 protein [Oryza sativa] 255 1e-66
UniRef100_Q6F2Z7 Hypothetical protein P0483D07.7 [Oryza sativa] 246 7e-64
UniRef100_O49741 Homologous to GATA-binding transcription factor... 207 4e-52
UniRef100_Q76DY1 AG-motif binding protein-3 [Nicotiana tabacum] 191 3e-47
UniRef100_O49743 Homologous to GATA-binding transcription factor... 189 1e-46
UniRef100_Q76DY0 AG-motif binding protein-4 [Nicotiana tabacum] 176 1e-42
UniRef100_Q94LQ6 Putative transcription factor [Oryza sativa] 170 5e-41
UniRef100_Q9FH57 GATA-binding transcription factor-like protein ... 166 7e-40
UniRef100_Q9SD38 Putative transcription factor [Arabidopsis thal... 154 5e-36
UniRef100_O65515 Hypothetical protein F23E13.130 [Arabidopsis th... 147 3e-34
UniRef100_Q7XR94 OSJNBa0011L07.7 protein [Oryza sativa] 139 9e-32
UniRef100_Q9SV30 Hypothetical protein F28P10.210 [Arabidopsis th... 135 1e-30
UniRef100_Q76DY2 AG-motif binding protein-2 [Nicotiana tabacum] 134 3e-30
UniRef100_Q8L4M6 GATA transcription factor 3 [Arabidopsis thaliana] 134 4e-30
UniRef100_O49742 AtGATA-3 protein [Arabidopsis thaliana] 134 4e-30
UniRef100_Q9AVU3 GATA-1 zinc finger protein [Nicotiana tabacum] 132 1e-29
UniRef100_Q948G4 Putative GATA-1 zinc finger protein [Oryza sativa] 129 1e-28
UniRef100_Q8LAU9 GATA transcription factor 1 [Arabidopsis thaliana] 125 1e-27
UniRef100_O49740 Protein homologous to GATA-binding transcriptio... 125 1e-27
>UniRef100_O82632 Hypothetical protein AT4g32890 [Arabidopsis thaliana]
Length = 308
Score = 264 bits (674), Expect = 3e-69
Identities = 156/318 (49%), Positives = 189/318 (59%), Gaps = 48/318 (15%)
Query: 70 DHFIVEDLFDFSNEDVAIEDPTFEESPPTNSNDSPPLETNPTSNFFTDNSCQNSADGPFS 129
D F+V+DL DFSN+D ++D DS L T ++ + NS DG
Sbjct: 16 DSFVVDDLLDFSNDDGEVDDGL------NTLPDSSTLSTGTLTD--SSNSSSLFTDGTGF 67
Query: 130 GELSVPYDDLAELEWVSKFAEESFSSEDLHKLQLISGLKAPNNVAS------KPYEESNP 183
+L +P DD+AELEW+S F EESF+ ED KL L SGLK P S KP E +
Sbjct: 68 SDLYIPNDDIAELEWLSNFVEESFAGEDQDKLHLFSGLKNPQTTGSTLTHLIKPEPELDH 127
Query: 184 TV----HSQVSVPAKARSKRSRVPPCNWTSRLLVLSPTTTTTTTTTTSSHSDTMAPPKKP 239
S V+VPAKARSKRSR W SRLL L+ + T PKK
Sbjct: 128 QFIDIDESNVAVPAKARSKRSRSAASTWASRLLSLADSDETN--------------PKKK 173
Query: 240 SPRKR----------DPNDGGEGRKCLHCATDKTPQWRTGPLGPKTLCNACGVRYKSGRL 289
R + D + G GR+CLHCAT+KTPQWRTGP+GPKTLCNACGVRYKSGRL
Sbjct: 174 QRRVKEQDFAGDMDVDCGESGGGRRCLHCATEKTPQWRTGPMGPKTLCNACGVRYKSGRL 233
Query: 290 VPEYRPAASPTFVLTKHSNSHRKVQELRRQKEMMRAQQHQLLQLQHHHSIMFEGPSNGDD 349
VPEYRPA+SPTFV+ +HSNSHRKV ELRRQKEM +H L QL+ + +M + SNG+D
Sbjct: 234 VPEYRPASSPTFVMARHSNSHRKVMELRRQKEM--RDEHLLSQLRCENLLM-DIRSNGED 290
Query: 350 YLIH---QHVGPDFTHLI 364
+L+H HV PDF HLI
Sbjct: 291 FLMHNNTNHVAPDFRHLI 308
>UniRef100_Q8LIZ3 OSJNBa0014K08.18 protein [Oryza sativa]
Length = 387
Score = 255 bits (652), Expect = 1e-66
Identities = 162/365 (44%), Positives = 201/365 (54%), Gaps = 84/365 (23%)
Query: 70 DHFIVEDLFDFSNEDVAIEDPTFEESPPTNSNDSPPLETNPTSNFFTD---NSCQNS--- 123
DHF V+DL + ED T E T + N +++ T +SC NS
Sbjct: 29 DHFAVDDLLVLPYGE---EDETTREGEATGGKEEAAGFGNASADSSTITALDSCSNSFGL 85
Query: 124 ADGPFSGELSVPYDDLAELEWVSKFAEE---SFSSEDLHKLQLISGLKAPN-NVASKPYE 179
ADG F GEL PYD LAELEW+S + E +F++EDL KLQLISG+ + + AS P
Sbjct: 86 ADGDFPGELCEPYDQLAELEWLSNYMNEGDDAFATEDLQKLQLISGIPSGGFSTASVPSA 145
Query: 180 ESNPTVHS------------QVSVPAKARSKRSRVPPCNWTSRLLVLSPTTTTTTTTTT- 226
++ + + VPAKARSKRSR P NW+SRLLVL P + + +
Sbjct: 146 QAQAASAAASMAVQPGGFLPEAPVPAKARSKRSRAAPGNWSSRLLVLPPPPASPPSPASM 205
Query: 227 ---------SSHSDTMAPPKKPSPRKRDPND---------------------GGEGRKCL 256
S+H+ + P KP+ +K P GEGR+CL
Sbjct: 206 AISPAESGVSAHAFPIKKPSKPAKKKDAPAPPAQAQLSSVPVHSGGSAPAAAAGEGRRCL 265
Query: 257 HCATDKTPQWRTGPLGPKTLCNACGVRYKSGRLVPEYRPAASPTFVLTKHSNSHRKVQEL 316
HC TDKTPQWRTGP+GPKTLCNACGVRYKSGRLVPEYRPAASPTF+++KHSNSHRKV EL
Sbjct: 266 HCETDKTPQWRTGPMGPKTLCNACGVRYKSGRLVPEYRPAASPTFMVSKHSNSHRKVLEL 325
Query: 317 RRQKEMMRAQQHQLLQLQHHH------------------SIMFEGPS---NGDDYLIHQH 355
RRQKEM HQ Q HHH S++F+G S +GDD+LIH H
Sbjct: 326 RRQKEM-----HQ--QTPHHHQPQVAAAGGVGSLMHMQSSMLFDGVSPVVSGDDFLIHHH 378
Query: 356 VGPDF 360
+ DF
Sbjct: 379 LRTDF 383
>UniRef100_Q6F2Z7 Hypothetical protein P0483D07.7 [Oryza sativa]
Length = 386
Score = 246 bits (628), Expect = 7e-64
Identities = 145/305 (47%), Positives = 179/305 (58%), Gaps = 71/305 (23%)
Query: 118 NSCQNS----ADGPFSGELSVPYDDLAELEWVSKF-AEESFSSEDLHKLQLISGL----K 168
+SC NS ADG FSG L PY+ LAELEWVS + EE+ +EDL KLQLISG+ +
Sbjct: 86 DSCSNSFSGLADGDFSGGLCEPYEQLAELEWVSTYMGEETLPTEDLRKLQLISGIPAAPR 145
Query: 169 APNNVASKPYE---ESNPTVHSQVSVPAKARSKRSRVPPCNWTSRLLVLSPTTTTTTTTT 225
AP +A + + ++ VP KARSKRSRV PC+W+SRL+VL P + +
Sbjct: 146 APPALAVSAVQLPAGGAGALPTEAPVPGKARSKRSRVAPCSWSSRLMVLPPPPASPPSPA 205
Query: 226 TS----SHSDTMAP--PKKPSPRKRDPNDG--------------GEGRKCLHCATDKTPQ 265
++ S S T AP P K + + DG EGR+CLHC TDKTPQ
Sbjct: 206 SAVISPSESGTAAPAFPAKKAAKSAKKKDGPSPAPAPNAAAQAAAEGRRCLHCETDKTPQ 265
Query: 266 WRTGPLGPKTLCNACGVRYKSGRLVPEYRPAASPTFVLTKHSNSHRKVQELRRQKEMMRA 325
WRTGP+GPKTLCNACGVRYKSGRLVPEYRPAASPTFV++KHSNSHRKV ELRRQKEM
Sbjct: 266 WRTGPMGPKTLCNACGVRYKSGRLVPEYRPAASPTFVVSKHSNSHRKVVELRRQKEM--- 322
Query: 326 QQHQLLQLQHHHS-----------------------IMFEGPSN-------GDDYLIHQH 355
QL HHH ++F+GP++ D++LIH
Sbjct: 323 ------QLLHHHQQPPPHVGAGGGGAAGGLLHVTSPLLFDGPTSSAPLFAGADEFLIHNR 376
Query: 356 VGPDF 360
+ PD+
Sbjct: 377 ISPDY 381
>UniRef100_O49741 Homologous to GATA-binding transcription factors [Arabidopsis
thaliana]
Length = 264
Score = 207 bits (527), Expect = 4e-52
Identities = 122/289 (42%), Positives = 159/289 (54%), Gaps = 52/289 (17%)
Query: 67 AASDHFIVEDLFDFSNEDVAIEDPTFEESPPTNSNDSPPLETNPTSNFFTDNSCQNSADG 126
++ D ++DL DFSNED+ + + T+S+ PP P + F + +SAD
Sbjct: 7 SSPDLLRIDDLLDFSNEDIFSASSSGGSTAATSSSSFPP----PQNPSFHHHHLPSSADH 62
Query: 127 -PFSGELSVPYDDLAELEWVSKFAEESFSSEDLHKLQLISGLKAPNNVASKPYEESNPTV 185
F ++ VP DD A LEW+S+F ++SF+ + + P + +V
Sbjct: 63 HSFLHDICVPSDDAAHLEWLSQFVDDSFA-----------------DFPANPLGGTMTSV 105
Query: 186 HSQVSVPAKARSKRSRVPPCNWTSRLLVLSPTTTTTTTTTTSSHSDTMAPPKKPSPRKRD 245
++ S P K RSKRSR P +P T + S + K P+K
Sbjct: 106 KTETSFPGKPRSKRSRAP-----------APFAGTWSPMPLESEHQQLHSAAKFKPKKEQ 154
Query: 246 PNDGGEG-----------------RKCLHCATDKTPQWRTGPLGPKTLCNACGVRYKSGR 288
GG G R+C HCA++KTPQWRTGPLGPKTLCNACGVR+KSGR
Sbjct: 155 SGGGGGGGGRHQSSSSETTEGGGMRRCTHCASEKTPQWRTGPLGPKTLCNACGVRFKSGR 214
Query: 289 LVPEYRPAASPTFVLTKHSNSHRKVQELRRQKEMMRAQQHQLLQLQHHH 337
LVPEYRPA+SPTFVLT+HSNSHRKV ELRRQKE+MR Q Q +QL HHH
Sbjct: 215 LVPEYRPASSPTFVLTQHSNSHRKVMELRRQKEVMR--QPQQVQLHHHH 261
>UniRef100_Q76DY1 AG-motif binding protein-3 [Nicotiana tabacum]
Length = 256
Score = 191 bits (485), Expect = 3e-47
Identities = 120/272 (44%), Positives = 153/272 (56%), Gaps = 50/272 (18%)
Query: 67 AASDHFIVEDLFDFSNEDV-----------AIEDPTFEESPPTNSNDSPPLETNPTSNFF 115
+A D F ++DL DFSN+++ A D P + N S T+N++
Sbjct: 7 SAPDLFRIDDLLDFSNDEIFSINSNSSSTTATPDSQHHHHQPHSDNSSAA-----TANYY 61
Query: 116 TDNSCQNSADGPFSGELSVPYDDLAELEWVSKFAEESFSSEDLHKLQLISGLKAPNNVAS 175
D N +D F+ L VP DD+AELEW+S F E+SFS+ + + L +
Sbjct: 62 -DALLPNCSDD-FTDNLCVPSDDVAELEWLSNFVEDSFSNFPTNSITGTMNLSS------ 113
Query: 176 KPYEESNPTVHSQVSVPAKARSKRSRVPPCNWTSRLLVLSPTTTTTTTTTTSSHSDTMAP 235
+S S +++RSKRSR +WTS L TT S H+
Sbjct: 114 ----------NSTASFHSRSRSKRSR-STSSWTSSL---QNPNTTMKNKEISVHT----- 154
Query: 236 PKKPSPRKRDPN-DGGEGRKCLHCATDKTPQWRTGPLGPKTLCNACGVRYKSGRLVPEYR 294
R+R + D R+C HCA++KTPQWRTGPLGPKTLCNACGVR+KSGRLVPEYR
Sbjct: 155 ------RERSSSMDDDVPRRCTHCASEKTPQWRTGPLGPKTLCNACGVRFKSGRLVPEYR 208
Query: 295 PAASPTFVLTKHSNSHRKVQELRRQKEMMRAQ 326
PAASPTFVLT+HSNSHRKV ELRRQKEM+ Q
Sbjct: 209 PAASPTFVLTQHSNSHRKVMELRRQKEMVHQQ 240
>UniRef100_O49743 Homologous to GATA-binding transcription factors [Arabidopsis
thaliana]
Length = 240
Score = 189 bits (479), Expect = 1e-46
Identities = 110/262 (41%), Positives = 146/262 (54%), Gaps = 41/262 (15%)
Query: 67 AASDHFIVEDLFDFSNEDVAIEDPTFEESPPTNSNDSPPLETNPTSNFFTDNSCQNSADG 126
++ D ++DL DFSN+++ T S +++ S + P+S + + +
Sbjct: 7 SSPDLLRIDDLLDFSNDEIFSSSSTVTSSAASSAASSENPFSFPSSTYTSPTLLTD---- 62
Query: 127 PFSGELSVPYDDLAELEWVSKFAEESFSSEDLHKLQLISGLKAPNNVASKPYEESNPTVH 186
F+ +L VP DD A LEW+S+F ++SFS + L + TV
Sbjct: 63 -FTHDLCVPSDDAAHLEWLSRFVDDSFSDFPANPLTM--------------------TVR 101
Query: 187 SQVSVPAKARSKRSRVPPCNWTSRLLVLSPTTTTTTTTTTSS---HSDTMAPPKKPSPRK 243
++S K RS+RSR P +P+ T + S HS PKK +
Sbjct: 102 PEISFTGKPRSRRSRAP-----------APSVAGTWAPMSESELCHSVAKPKPKKVYNAE 150
Query: 244 RDPNDGGEGRKCLHCATDKTPQWRTGPLGPKTLCNACGVRYKSGRLVPEYRPAASPTFVL 303
DG R+C HCA++KTPQWRTGPLGPKTLCNACGVRYKSGRLVPEYRPA+SPTFVL
Sbjct: 151 SVTADGA--RRCTHCASEKTPQWRTGPLGPKTLCNACGVRYKSGRLVPEYRPASSPTFVL 208
Query: 304 TKHSNSHRKVQELRRQKEMMRA 325
T+HSNSHRKV ELRRQKE +
Sbjct: 209 TQHSNSHRKVMELRRQKEQQES 230
>UniRef100_Q76DY0 AG-motif binding protein-4 [Nicotiana tabacum]
Length = 326
Score = 176 bits (445), Expect = 1e-42
Identities = 113/288 (39%), Positives = 144/288 (49%), Gaps = 43/288 (14%)
Query: 64 VNTAASDHFIVEDLFDFSNEDVAIEDPTFEESPPTNSNDSPPLETNPTSNFFTDNSCQNS 123
+N SD F V+DL DFS++D + + +E + DS + ++ ++ SC +S
Sbjct: 36 INNVPSDDFSVDDLLDFSDKDFK-DGQSLQELHEDDEKDSFSGSSQHRNSQVSNFSCMDS 94
Query: 124 ADGPFSGELSVPYDDLAELEWVSKFAEESFSSEDLHKLQLISGLKAPNNVASKPYEESNP 183
FSGEL VP D+L LEW+S+F ++S S L L + S P
Sbjct: 95 ----FSGELPVPVDELENLEWLSQFVDDSTSEFSL----LCPAGSFKDKTGGFQVSRSEP 146
Query: 184 TVHSQVS----------VPAKARSKRSRVPPCNWTSRLLVLSPTTTTTTTTTTSSH---- 229
V V V K R+ RSR W+ SPT + + + TSS
Sbjct: 147 VVRPVVQKLKVPCFPLPVVQKPRTYRSRPAGRKWSFS----SPTVSADSCSPTSSSYGSS 202
Query: 230 -------------SDTMAPPKKP---SPRKRDPNDGGEGRKCLHCATDKTPQWRTGPLGP 273
D +KP P+K + G GR+C HC KTPQWR GPLGP
Sbjct: 203 PFPSVLFSNPVLDGDLFCSVEKPPLKKPKKLSTAETGSGRRCTHCQVQKTPQWRAGPLGP 262
Query: 274 KTLCNACGVRYKSGRLVPEYRPAASPTFVLTKHSNSHRKVQELRRQKE 321
KTLCNACGVRYKSGRL PEYRPA SPTF HSNSHRKV E+RR+KE
Sbjct: 263 KTLCNACGVRYKSGRLFPEYRPACSPTFSQEVHSNSHRKVLEMRRKKE 310
>UniRef100_Q94LQ6 Putative transcription factor [Oryza sativa]
Length = 387
Score = 170 bits (431), Expect = 5e-41
Identities = 100/225 (44%), Positives = 126/225 (55%), Gaps = 38/225 (16%)
Query: 128 FSGELSVPYDDLAELEWVSKFAEESFSS-----EDLHKLQLISGLKAPNNVASKPYEESN 182
F+ + +P +D AELEW+SKF ++S+S H + A NN + +
Sbjct: 131 FADDFYIPTEDAAELEWLSKFVDDSYSDMPNYQSSAHAAMAAAAASAANNGGGSSAGQDS 190
Query: 183 PTVHSQVSVPAK-ARSKRSRVPPCN---WTSRLLVLSPTTTTTTTTTTSSHSDTMAPPKK 238
+ P + ARSKRSR W S L P + ++ ++S S K
Sbjct: 191 ----CLTAAPGRGARSKRSRATAAAAAAWHS----LVPRPPSQSSPSSSCSSSDFPSSNK 242
Query: 239 PSPRKRDPNDGGEG--------------------RKCLHCATDKTPQWRTGPLGPKTLCN 278
PS R PN G G R+C HCA++KTPQWRTGPLGPKTLCN
Sbjct: 243 PSGTAR-PNGSGGGSRGKKSPGPAGAEVGMEAGVRRCTHCASEKTPQWRTGPLGPKTLCN 301
Query: 279 ACGVRYKSGRLVPEYRPAASPTFVLTKHSNSHRKVQELRRQKEMM 323
ACGVR+KSGRL+PEYRPAASPTFVLT+HSNSHRKV ELRRQKE++
Sbjct: 302 ACGVRFKSGRLMPEYRPAASPTFVLTQHSNSHRKVMELRRQKELL 346
>UniRef100_Q9FH57 GATA-binding transcription factor-like protein [Arabidopsis
thaliana]
Length = 339
Score = 166 bits (421), Expect = 7e-40
Identities = 118/311 (37%), Positives = 151/311 (47%), Gaps = 49/311 (15%)
Query: 61 TNNVNTAASDHFIVEDLFDFSNEDVAIEDPT---FEESPPTNSNDSPPLETNPTSNFFTD 117
T N + D F V+DL D SN+DV ++ T + S++ P + +
Sbjct: 30 TTAQNGFSVDDFSVDDLLDLSNDDVFADEETDLKAQHEMVRVSSEEPNDDGDALRRSSDF 89
Query: 118 NSCQNSADGPFSGELSVPYDDLAELEWVSKFAEESFSSEDLHKLQLISGLKAPNNVASKP 177
+ C + P S ELS+P DDLA LEW+S F E+SF+ SG KP
Sbjct: 90 SGCDDFGSLPTS-ELSLPADDLANLEWLSHFVEDSFTE--------YSGPNLTGTPTEKP 140
Query: 178 YEESNPTVHSQVSV----------PAKARSKRSRVPPCNWTSRLLVLSPTTTTTTTTTTS 227
+ H +V PAKARSKR+R W+ L S + +++ +T+S
Sbjct: 141 AWLTGDRKHPVTAVTEETCFKSPVPAKARSKRNRNGLKVWS--LGSSSSSGPSSSGSTSS 198
Query: 228 SHSDTMAP-------------------PKKPSPRKRDPNDGGE------GRKCLHCATDK 262
S S +P PKK R + GE RKC HC K
Sbjct: 199 SSSGPSSPWFSGAELLEPVVTSERPPFPKKHKKRSAESVFSGELQQLQPQRKCSHCGVQK 258
Query: 263 TPQWRTGPLGPKTLCNACGVRYKSGRLVPEYRPAASPTFVLTKHSNSHRKVQELRRQKEM 322
TPQWR GP+G KTLCNACGVRYKSGRL+PEYRPA SPTF HSN HRKV E+RR+KE
Sbjct: 259 TPQWRAGPMGAKTLCNACGVRYKSGRLLPEYRPACSPTFSSELHSNHHRKVIEMRRKKEP 318
Query: 323 MRAQQHQLLQL 333
+ L QL
Sbjct: 319 TSDNETGLNQL 329
>UniRef100_Q9SD38 Putative transcription factor [Arabidopsis thaliana]
Length = 312
Score = 154 bits (388), Expect = 5e-36
Identities = 111/300 (37%), Positives = 146/300 (48%), Gaps = 36/300 (12%)
Query: 47 MEAQEFFQSDNNSNTNNVNTAAS--DHFIVEDLFDFSNE----DVAIEDPTFEESPPTNS 100
ME+ E ++N + A D F V+DL DFS E DV +ED E
Sbjct: 1 MESVELTLKNSNMKDKTLTGGAQNGDDFSVDDLLDFSKEEEDDDVLVEDEA--ELKVQRK 58
Query: 101 NDSPPLETNPTSNFFTDNSCQNSADGPFSGELSVPYDDLAELEWVSKFAEESFSSEDLHK 160
T SN F+ +AD SG LSVP DD+AELEW+S F ++S +
Sbjct: 59 RGVSDENTLHRSNDFS------TADFHTSG-LSVPMDDIAELEWLSNFVDDSSFTPYSAP 111
Query: 161 LQLISGLKAPNNVASKPYEESNPTVHSQVSVPAKARSKRSRVPPCNWTSRLLVLSPTTTT 220
L +P +E T K R KR+R W+ L+ ++++
Sbjct: 112 TNKPVWLTGNRRHLVQPVKEE--TCFKSQHPAVKTRPKRARTGVRVWSHGSQSLTDSSSS 169
Query: 221 TTTTTTSSHSDTMA-----------PPKKPSPRKRDPNDGGEG--------RKCLHCATD 261
+TT+++SS + P K +K+ + G+ R+C HC
Sbjct: 170 STTSSSSSPRPSSPLWLASGQFLDEPMTKTQKKKKVWKNAGQTQTQTQTQTRQCGHCGVQ 229
Query: 262 KTPQWRTGPLGPKTLCNACGVRYKSGRLVPEYRPAASPTFVLTKHSNSHRKVQELRRQKE 321
KTPQWR GPLG KTLCNACGVRYKSGRL+PEYRPA SPTF HSN H KV E+RR+KE
Sbjct: 230 KTPQWRAGPLGAKTLCNACGVRYKSGRLLPEYRPACSPTFSSELHSNHHSKVIEMRRKKE 289
>UniRef100_O65515 Hypothetical protein F23E13.130 [Arabidopsis thaliana]
Length = 238
Score = 147 bits (372), Expect = 3e-34
Identities = 107/259 (41%), Positives = 124/259 (47%), Gaps = 48/259 (18%)
Query: 72 FIVEDLFDFSNEDVAIEDPTFEESPPTNSNDSPPLETNPTSNFFTDNSCQNSADGPFSGE 131
F V+DL D SN D ++E S+ S E F S Q++ P
Sbjct: 11 FSVDDLLDLSNADTSLE-----------SSSSQRKEDEQEREKFKSFSDQSTRLSPPEDL 59
Query: 132 LSVPYD----DLAELEWVSKFAEESFSSEDLHKLQLISGLKAPNNVASKPYEESNPTVHS 187
LS P D DL +LEW+S F E+SFS IS N VAS
Sbjct: 60 LSFPGDAPVGDLEDLEWLSNFVEDSFSES------YISSDFPVNPVASVEVRRQ------ 107
Query: 188 QVSVPAKARSKRSRVPPCNWTSRLLVLSPTTTTTTTTTTSSHSDTMAPPKKPSPRKRDPN 247
VP K RSKR R W+ SP+ +T +A KK +K D +
Sbjct: 108 --CVPVKPRSKRRRTNGRIWSME----SPSPLLSTA---------VARRKKRGRQKVDAS 152
Query: 248 DGGE------GRKCLHCATDKTPQWRTGPLGPKTLCNACGVRYKSGRLVPEYRPAASPTF 301
GG R C HC KTPQWR GPLG KTLCNACGVR+KSGRL+PEYRPA SPTF
Sbjct: 153 YGGVVQQQQLRRCCSHCGVQKTPQWRMGPLGAKTLCNACGVRFKSGRLLPEYRPACSPTF 212
Query: 302 VLTKHSNSHRKVQELRRQK 320
HSNSHRKV ELR K
Sbjct: 213 TNEIHSNSHRKVLELRLMK 231
>UniRef100_Q7XR94 OSJNBa0011L07.7 protein [Oryza sativa]
Length = 392
Score = 139 bits (351), Expect = 9e-32
Identities = 115/299 (38%), Positives = 148/299 (49%), Gaps = 54/299 (18%)
Query: 70 DHFIVEDLFDFSNEDVAIEDPTFEESPPTNSNDSPPLETNPTSNFFTDNSCQNS------ 123
D F VEDL D A +D E + + L P D+S +S
Sbjct: 84 DGFSVEDLLDLEEFCEAEKDAAEE------NEQALALVAAPEEEKSKDDSQPSSVVTYEL 137
Query: 124 -ADGPFSGEL-SVPYDDLAELEWVSKFAEESFSS---EDLHKLQLISGLKA----PNNVA 174
A P E+ +P D+ ELEWVS+ ++S S +++ L A P +
Sbjct: 138 VAPPPPPPEIVDLPAHDVEELEWVSRIMDDSLSELPPPPQPPASVVASLAARPPQPRQLQ 197
Query: 175 SKP------------YEESNPTV---HSQVSVPAKA-RSKRSRVPPCNWT-SRLLVLSPT 217
+P Y PT+ ++ VP KA RSKRSR W+ S S +
Sbjct: 198 RRPQDGAYRALPPASYPVRTPTICALSTEALVPVKAKRSKRSRATA--WSLSGAPPFSDS 255
Query: 218 TTTTTTTTTSSHSDT-----MAPPKK----PSPRKRD-PNDG----GEGRKCLHCATDKT 263
T++++TTTTSS S + +P K P D P+D GE + HC KT
Sbjct: 256 TSSSSTTTTSSCSSSASFSSFSPLLKFEWHPLGGTSDLPDDHLLPPGEEVQARHCGVQKT 315
Query: 264 PQWRTGPLGPKTLCNACGVRYKSGRLVPEYRPAASPTFVLTKHSNSHRKVQELRRQKEM 322
PQWR GP G KTLCNACGVRYKSGRL+PEYRPA SPTFV HSNSHRKV E+RR+KE+
Sbjct: 316 PQWRAGPEGAKTLCNACGVRYKSGRLLPEYRPACSPTFVSAIHSNSHRKVLEMRRKKEV 374
>UniRef100_Q9SV30 Hypothetical protein F28P10.210 [Arabidopsis thaliana]
Length = 322
Score = 135 bits (341), Expect = 1e-30
Identities = 100/293 (34%), Positives = 133/293 (45%), Gaps = 61/293 (20%)
Query: 74 VEDLFDFSNEDV----AIEDPTFEESPPTNSNDSPPLETNPTSNFFTDNSCQNSADGPFS 129
++DL DF D+ I D + T +D+ P ++P F+ N+ +S S
Sbjct: 19 MDDLMDFPGGDIDVGFGIGDSDSFPTIWTTHHDTWPAASDP---LFSSNTNSDS-----S 70
Query: 130 GELSVPYDDLAELEWVSKFAEESF--SSEDLHKLQLISGLK-------APNNVASKPYEE 180
EL VP++D+ ++E F EE+ ED S +P +V
Sbjct: 71 PELYVPFEDIVKVERPPSFVEETLVEKKEDSFSTNTDSSSSHSQFRSSSPVSVLESSSSS 130
Query: 181 SNPTVHSQVSVPAK---ARSKRSRVPP----------CNWTSRLLVLSPTTTTTTTTTTS 227
S T + + +P K R+KR R P C SRL++ P
Sbjct: 131 SQTTNTTSLVLPGKHGRPRTKRPRPPVQDKDRVKDNVCGGDSRLIIRIPKQFL------- 183
Query: 228 SHSDTMAPPKKPSPRKRDPNDGGEG--------------------RKCLHCATDKTPQWR 267
S + M KK K + G RKC+HC KTPQWR
Sbjct: 184 SDHNKMINKKKKKKAKITSSSSSSGIDLEVNGNNVDSYSSEQYPLRKCMHCEVTKTPQWR 243
Query: 268 TGPLGPKTLCNACGVRYKSGRLVPEYRPAASPTFVLTKHSNSHRKVQELRRQK 320
GP+GPKTLCNACGVRYKSGRL PEYRPAASPTF HSNSH+KV E+R ++
Sbjct: 244 LGPMGPKTLCNACGVRYKSGRLFPEYRPAASPTFTPALHSNSHKKVAEMRNKR 296
>UniRef100_Q76DY2 AG-motif binding protein-2 [Nicotiana tabacum]
Length = 289
Score = 134 bits (338), Expect = 3e-30
Identities = 100/292 (34%), Positives = 140/292 (47%), Gaps = 60/292 (20%)
Query: 64 VNTAASDHFIVE---DLFDFSNEDVAIEDPTFEESPPTNSNDSPPLETNPTSNFFTDN-- 118
++ +A+ F+V+ DL +FS ED + D E++ + + PL ++ +S+ + N
Sbjct: 6 LDPSAASCFMVDVDDDLLNFSLEDETVFDDD-EKTTKSITKHKHPLSSSYSSSLDSSNPV 64
Query: 119 -SCQNSADGPFSGELSVPYDDLAELEWVSKFAEESFSSEDLHKLQLISGLKAPNNVASKP 177
S S P E ELEW+S +++F + + L + ++
Sbjct: 65 LSLLPSQQHPECVE--------EELEWLSN--KDAFPAVEFGILADNPSIVFDHHSPVSV 114
Query: 178 YEESNPTVHSQ--------------------VSVPAKARSKRSRVPPCNWTSRLLVLSPT 217
E S+ T +S V+ P +ARSKR R
Sbjct: 115 LENSSSTCNSSGNGSANANAYMSCCASLKVPVNYPVRARSKRRR--------------RR 160
Query: 218 TTTTTTTTTSSHSDTMAPPKKPSPRKRDP---------NDGGEGRKCLHCATDKTPQWRT 268
+ S H ++ P S ++R+P GR+C HC DKTPQWR
Sbjct: 161 QRGSFADLPSEHCMSVNKPSFKSVKQREPLLSLPLNSAKSASIGRRCQHCGADKTPQWRA 220
Query: 269 GPLGPKTLCNACGVRYKSGRLVPEYRPAASPTFVLTKHSNSHRKVQELRRQK 320
GPLGPKTLCNACGVRYKSGRL+PEYRPA SPTF T HSNSHRKV E+R+QK
Sbjct: 221 GPLGPKTLCNACGVRYKSGRLLPEYRPANSPTFSPTVHSNSHRKVLEMRKQK 272
>UniRef100_Q8L4M6 GATA transcription factor 3 [Arabidopsis thaliana]
Length = 269
Score = 134 bits (337), Expect = 4e-30
Identities = 98/291 (33%), Positives = 135/291 (45%), Gaps = 48/291 (16%)
Query: 32 ASTSKPNNNNNLTQPMEAQEFFQSDNNSNTNNVNTAASDHFIVEDLFDFSNEDVAIEDPT 91
A K + T ++ + S++ S T+++ + F VE DFS E
Sbjct: 7 ARALKASLRGESTISLKHHQVIVSEDLSQTSSL----PEDFSVECFLDFS------EGQK 56
Query: 92 FEESPPTNSNDSPPLETNPTSNFFTDNSCQNSADGPFSGELSVPYDDLAELEWVSKFAEE 151
EE + + S E F+ C F S+P +D+ ELEWVS+ ++
Sbjct: 57 EEEEEVVSVSSSQEQEEQEHDCVFSSQPCI------FDQLPSLPDEDVEELEWVSRVVDD 110
Query: 152 SFSSEDLHKLQLISGLKAPNNVASKPYEESNPTVHSQVSVPAKARSKRSRVPPCNWTSRL 211
S E S +++ T S +P K R+KRSR
Sbjct: 111 CSSPE-----------------VSLLLTQTHKTKPSFSRIPVKPRTKRSRNS-------- 145
Query: 212 LVLSPTTTTTTTTTTSSHSDTMAPPKKPSPRKRDPNDGGEGRKCLHCATDKTPQWRTGPL 271
L+ + +T H+ T + +K+ R+C HC T+ TPQWRTGP+
Sbjct: 146 --LTGSRVWPLVSTNHQHAAT-----EQLRKKKQETVLVFQRRCSHCGTNNTPQWRTGPV 198
Query: 272 GPKTLCNACGVRYKSGRLVPEYRPAASPTFVLTKHSNSHRKVQELRRQKEM 322
GPKTLCNACGVR+KSGRL PEYRPA SPTF HSN HRKV ELR+ KE+
Sbjct: 199 GPKTLCNACGVRFKSGRLCPEYRPADSPTFSNEIHSNLHRKVLELRKSKEL 249
>UniRef100_O49742 AtGATA-3 protein [Arabidopsis thaliana]
Length = 269
Score = 134 bits (337), Expect = 4e-30
Identities = 98/291 (33%), Positives = 135/291 (45%), Gaps = 48/291 (16%)
Query: 32 ASTSKPNNNNNLTQPMEAQEFFQSDNNSNTNNVNTAASDHFIVEDLFDFSNEDVAIEDPT 91
A K + T ++ + S++ S T+++ + F VE DFS E
Sbjct: 7 ARALKASLRGESTISLKHHQVIVSEDLSRTSSL----PEDFSVECFLDFS------EGQK 56
Query: 92 FEESPPTNSNDSPPLETNPTSNFFTDNSCQNSADGPFSGELSVPYDDLAELEWVSKFAEE 151
EE + + S E F+ C F S+P +D+ ELEWVS+ ++
Sbjct: 57 EEEEEVVSVSSSQEQEEQEHDCVFSSQPCI------FDQLPSLPDEDVEELEWVSRVVDD 110
Query: 152 SFSSEDLHKLQLISGLKAPNNVASKPYEESNPTVHSQVSVPAKARSKRSRVPPCNWTSRL 211
S E S +++ T S +P K R+KRSR
Sbjct: 111 CSSPE-----------------VSLLLTQTHKTKPSFSRIPVKPRTKRSRNS-------- 145
Query: 212 LVLSPTTTTTTTTTTSSHSDTMAPPKKPSPRKRDPNDGGEGRKCLHCATDKTPQWRTGPL 271
L+ + +T H+ T + +K+ R+C HC T+ TPQWRTGP+
Sbjct: 146 --LTGSRVWPLVSTNHQHAAT-----EQLRKKKQETVLVFQRRCSHCGTNNTPQWRTGPV 198
Query: 272 GPKTLCNACGVRYKSGRLVPEYRPAASPTFVLTKHSNSHRKVQELRRQKEM 322
GPKTLCNACGVR+KSGRL PEYRPA SPTF HSN HRKV ELR+ KE+
Sbjct: 199 GPKTLCNACGVRFKSGRLCPEYRPADSPTFSNEIHSNLHRKVLELRKSKEL 249
>UniRef100_Q9AVU3 GATA-1 zinc finger protein [Nicotiana tabacum]
Length = 305
Score = 132 bits (332), Expect = 1e-29
Identities = 72/170 (42%), Positives = 97/170 (56%), Gaps = 14/170 (8%)
Query: 184 TVHSQVSVPAKARSKRSRVPPCNWTSRLLVLSPTTTTTTTTTTSSHS-------DTMAPP 236
++ +++P + RSKR R N + +S T + T + ++ P
Sbjct: 122 SIKHDIAIPVRPRSKRPRSSALNPWILMPPISSTRFASKKTCDARKGKEKKRKMSLLSVP 181
Query: 237 KKPSPRKRDPNDGGEG--RKCLHCATDKTPQWRTGPLGPKTLCNACGVRYKSGRLVPEYR 294
+ K+ G + +KC HC KTPQWR GPLGPKTLCNACGVRY+SGRL PEYR
Sbjct: 182 QIADVTKKKTTSGQQFSFKKCTHCQVTKTPQWREGPLGPKTLCNACGVRYRSGRLFPEYR 241
Query: 295 PAASPTFVLTKHSNSHRKVQELRRQKEMMRAQQHQLLQLQHHHSIMFEGP 344
PAASPTFV T HSNSHRKV E+R+ +A + L+ H+++ EGP
Sbjct: 242 PAASPTFVPTLHSNSHRKVVEMRK-----KAIYGETSALEEPHNVIVEGP 286
>UniRef100_Q948G4 Putative GATA-1 zinc finger protein [Oryza sativa]
Length = 418
Score = 129 bits (324), Expect = 1e-28
Identities = 79/180 (43%), Positives = 95/180 (51%), Gaps = 38/180 (21%)
Query: 191 VPAKARSKRSRVPPCNWTSRLLVLSPTTTTTTT---TTTSSHSDTMA--------PPKKP 239
+PA+ARSKRSR + TT T ++TSSHSD + PP K
Sbjct: 233 IPARARSKRSRPSAFPAVRGAPAATETTILVPTPMYSSTSSHSDPESIAESNPHPPPMKK 292
Query: 240 SPRKRDPN-----------------DGGEG----------RKCLHCATDKTPQWRTGPLG 272
+ + P D EG R+C HC +KTPQWR GPLG
Sbjct: 293 KKKAKKPAAPAAASDAEADADAADADYEEGGALALPPGTVRRCTHCQIEKTPQWRAGPLG 352
Query: 273 PKTLCNACGVRYKSGRLVPEYRPAASPTFVLTKHSNSHRKVQELRRQKEMMRAQQHQLLQ 332
PKTLCNACGVRYKSGRL PEYRPAASPTF+ + HSNSH+KV E+R++ LLQ
Sbjct: 353 PKTLCNACGVRYKSGRLFPEYRPAASPTFMPSIHSNSHKKVVEMRQKATRTADPSCDLLQ 412
>UniRef100_Q8LAU9 GATA transcription factor 1 [Arabidopsis thaliana]
Length = 268
Score = 125 bits (315), Expect = 1e-27
Identities = 68/128 (53%), Positives = 73/128 (56%), Gaps = 26/128 (20%)
Query: 218 TTTTTTTTTSSHSDTMAPPKKPSPRKRDPN-----------DGG---------------E 251
TTTTT T S AP K S R+R GG
Sbjct: 127 TTTTTPTIMSCCVGFKAPAKARSKRRRTGRRDLRVLWTGNEQGGIQKKKTMTVAAAALIM 186
Query: 252 GRKCLHCATDKTPQWRTGPLGPKTLCNACGVRYKSGRLVPEYRPAASPTFVLTKHSNSHR 311
GRKC HC +KTPQWR GP GPKTLCNACGVRYKSGRLVPEYRPA SPTF HSNSHR
Sbjct: 187 GRKCQHCGAEKTPQWRAGPAGPKTLCNACGVRYKSGRLVPEYRPANSPTFTAELHSNSHR 246
Query: 312 KVQELRRQ 319
K+ E+R+Q
Sbjct: 247 KIVEMRKQ 254
>UniRef100_O49740 Protein homologous to GATA-binding transcription factors
[Arabidopsis thaliana]
Length = 274
Score = 125 bits (315), Expect = 1e-27
Identities = 68/128 (53%), Positives = 73/128 (56%), Gaps = 26/128 (20%)
Query: 218 TTTTTTTTTSSHSDTMAPPKKPSPRKRDPN-----------DGG---------------E 251
TTTTT T S AP K S R+R GG
Sbjct: 133 TTTTTPTIMSCCVGFKAPAKARSKRRRTGRRDLRVLWTGNEQGGIQKKKTMTVAAAALIM 192
Query: 252 GRKCLHCATDKTPQWRTGPLGPKTLCNACGVRYKSGRLVPEYRPAASPTFVLTKHSNSHR 311
GRKC HC +KTPQWR GP GPKTLCNACGVRYKSGRLVPEYRPA SPTF HSNSHR
Sbjct: 193 GRKCQHCGAEKTPQWRAGPAGPKTLCNACGVRYKSGRLVPEYRPANSPTFTAELHSNSHR 252
Query: 312 KVQELRRQ 319
K+ E+R+Q
Sbjct: 253 KIVEMRKQ 260
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.313 0.129 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 678,889,348
Number of Sequences: 2790947
Number of extensions: 30676423
Number of successful extensions: 183690
Number of sequences better than 10.0: 1188
Number of HSP's better than 10.0 without gapping: 323
Number of HSP's successfully gapped in prelim test: 907
Number of HSP's that attempted gapping in prelim test: 168829
Number of HSP's gapped (non-prelim): 6821
length of query: 364
length of database: 848,049,833
effective HSP length: 129
effective length of query: 235
effective length of database: 488,017,670
effective search space: 114684152450
effective search space used: 114684152450
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 75 (33.5 bits)
Medicago: description of AC147428.3


