

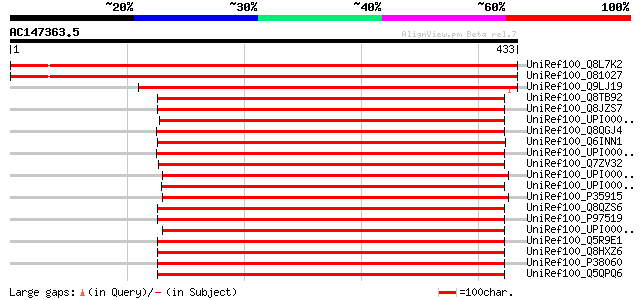
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147363.5 - phase: 0
(433 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8L7K2 Putative hydroxymethylglutaryl-CoA lyase [Arabi... 629 e-179
UniRef100_O81027 Putative hydroxymethylglutaryl-CoA lyase [Arabi... 629 e-179
UniRef100_Q9LJ19 ESTs C97134 [Oryza sativa] 493 e-138
UniRef100_Q8TB92 HMGCLL1 protein [Homo sapiens] 404 e-111
UniRef100_Q8JZS7 3-hydroxymethyl-3-methylglutaryl-Coenzyme A lya... 402 e-110
UniRef100_UPI00003AD0F1 UPI00003AD0F1 UniRef100 entry 402 e-110
UniRef100_Q8QGJ4 Hydroxymethylglutaryl-CoA lyase [Fugu rubripes] 402 e-110
UniRef100_Q6INN1 MGC82338 protein [Xenopus laevis] 401 e-110
UniRef100_UPI0000360D1C UPI0000360D1C UniRef100 entry 401 e-110
UniRef100_Q7ZV32 Similar to 3-hydroxymethyl-3-methylglutaryl-Coe... 401 e-110
UniRef100_UPI00003AC63A UPI00003AC63A UniRef100 entry 400 e-110
UniRef100_UPI0000318F4C UPI0000318F4C UniRef100 entry 400 e-110
UniRef100_P35915 Hydroxymethylglutaryl-CoA lyase [Gallus gallus] 398 e-109
UniRef100_Q8QZS6 Hmgcl protein [Mus musculus] 395 e-108
UniRef100_P97519 Hydroxymethylglutaryl-CoA lyase, mitochondrial ... 395 e-108
UniRef100_UPI00003624B7 UPI00003624B7 UniRef100 entry 392 e-107
UniRef100_Q5R9E1 Hypothetical protein DKFZp469I2412 [Pongo pygma... 390 e-107
UniRef100_Q8HXZ6 3-hydroxymethyl-3-methylglutaryl-Coenzyme A lya... 390 e-107
UniRef100_P38060 Hydroxymethylglutaryl-CoA lyase, mitochondrial ... 389 e-106
UniRef100_Q5QPQ6 OTTHUMP00000044830 [Homo sapiens] 387 e-106
>UniRef100_Q8L7K2 Putative hydroxymethylglutaryl-CoA lyase [Arabidopsis thaliana]
Length = 433
Score = 629 bits (1621), Expect = e-179
Identities = 316/434 (72%), Positives = 363/434 (82%), Gaps = 2/434 (0%)
Query: 1 MSSLEEPLGHDKLPSMSTMDRVQRFSSGCCRPQVDNLGMGNCFIEGRSCSTSNSCNEDNE 60
MSSLEEPL DKLPSMSTMDR+QRFSSG CRP+ D++GMG+ +IEGR C+TSNSC +D++
Sbjct: 1 MSSLEEPLSFDKLPSMSTMDRIQRFSSGACRPR-DDVGMGHRWIEGRDCTTSNSCIDDDK 59
Query: 61 DYTAETYPWKRQTRDMSRGDSFSPRTMTTGRNTLKSGIVDNSFYTSDYQYSQKRNNKDMQ 120
+ E++PW+R TR +S G+ GR + SG + S + +YS N
Sbjct: 60 SFAKESFPWRRHTRKLSEGEHMFRNISFAGRTSTVSGTLRESKSFKEQKYSTFSNENGTS 119
Query: 121 DMAYKFMKGMPEFVKIVEVGPRDGLQNEKNIVPTDVKIELIHRLASTGLSVIEATSFVSP 180
++ K KG+P+FVKIVEVGPRDGLQNEKNIVPT VK+ELI RL S+GL V+EATSFVSP
Sbjct: 120 HISNKISKGIPKFVKIVEVGPRDGLQNEKNIVPTSVKVELIQRLVSSGLPVVEATSFVSP 179
Query: 181 KWVPQLADAKDVMQAVHNLRGIRLPVLTPNLKGFEAAVAAGAREVAVFASASESFSKSNI 240
KWVPQLADAKDVM AV+ L G RLPVLTPNLKGF+AAV+AGA+EVA+FASASESFS SNI
Sbjct: 180 KWVPQLADAKDVMDAVNTLDGARLPVLTPNLKGFQAAVSAGAKEVAIFASASESFSLSNI 239
Query: 241 NCSIEESLSRYRAVTRAAKELSIPVRGYVSCVVGCPVEGPVPPSKVAYVAKALYDMGCFE 300
NC+IEESL RYR V AAKE S+PVRGYVSCVVGCPVEGPV PSKVAYV K LYDMGCFE
Sbjct: 240 NCTIEESLLRYRVVATAAKEHSVPVRGYVSCVVGCPVEGPVLPSKVAYVVKELYDMGCFE 299
Query: 301 ISLGDTIGVGTPGTVVPMLLAVMAIVPTEKLAVHFHDTYGQSLPNILVSLQMGISAVDSS 360
ISLGDTIG+GTPG+VVPML AVMA+VP +KLAVHFHDTYGQ+L NILVSLQMGIS VDSS
Sbjct: 300 ISLGDTIGIGTPGSVVPMLEAVMAVVPADKLAVHFHDTYGQALANILVSLQMGISIVDSS 359
Query: 361 VAGLGGCPYAKGASGNVATEDVVYMLNGLGIKTNVDIGKLMSAGDFIGKQLGRPSGSKTA 420
+AGLGGCPYAKGASGNVATEDVVYMLNGLG+ TNVD+GKL++AGDFI K LGRP+GSK A
Sbjct: 360 IAGLGGCPYAKGASGNVATEDVVYMLNGLGVHTNVDLGKLIAAGDFISKHLGRPNGSKAA 419
Query: 421 IALN-RVTADASKI 433
+ALN R+TADASKI
Sbjct: 420 VALNRRITADASKI 433
>UniRef100_O81027 Putative hydroxymethylglutaryl-CoA lyase [Arabidopsis thaliana]
Length = 468
Score = 629 bits (1621), Expect = e-179
Identities = 316/434 (72%), Positives = 363/434 (82%), Gaps = 2/434 (0%)
Query: 1 MSSLEEPLGHDKLPSMSTMDRVQRFSSGCCRPQVDNLGMGNCFIEGRSCSTSNSCNEDNE 60
MSSLEEPL DKLPSMSTMDR+QRFSSG CRP+ D++GMG+ +IEGR C+TSNSC +D++
Sbjct: 36 MSSLEEPLSFDKLPSMSTMDRIQRFSSGACRPR-DDVGMGHRWIEGRDCTTSNSCIDDDK 94
Query: 61 DYTAETYPWKRQTRDMSRGDSFSPRTMTTGRNTLKSGIVDNSFYTSDYQYSQKRNNKDMQ 120
+ E++PW+R TR +S G+ GR + SG + S + +YS N
Sbjct: 95 SFAKESFPWRRHTRKLSEGEHMFRNISFAGRTSTVSGTLRESKSFKEQKYSTFSNENGTS 154
Query: 121 DMAYKFMKGMPEFVKIVEVGPRDGLQNEKNIVPTDVKIELIHRLASTGLSVIEATSFVSP 180
++ K KG+P+FVKIVEVGPRDGLQNEKNIVPT VK+ELI RL S+GL V+EATSFVSP
Sbjct: 155 HISNKISKGIPKFVKIVEVGPRDGLQNEKNIVPTSVKVELIQRLVSSGLPVVEATSFVSP 214
Query: 181 KWVPQLADAKDVMQAVHNLRGIRLPVLTPNLKGFEAAVAAGAREVAVFASASESFSKSNI 240
KWVPQLADAKDVM AV+ L G RLPVLTPNLKGF+AAV+AGA+EVA+FASASESFS SNI
Sbjct: 215 KWVPQLADAKDVMDAVNTLDGARLPVLTPNLKGFQAAVSAGAKEVAIFASASESFSLSNI 274
Query: 241 NCSIEESLSRYRAVTRAAKELSIPVRGYVSCVVGCPVEGPVPPSKVAYVAKALYDMGCFE 300
NC+IEESL RYR V AAKE S+PVRGYVSCVVGCPVEGPV PSKVAYV K LYDMGCFE
Sbjct: 275 NCTIEESLLRYRVVATAAKEHSVPVRGYVSCVVGCPVEGPVLPSKVAYVVKELYDMGCFE 334
Query: 301 ISLGDTIGVGTPGTVVPMLLAVMAIVPTEKLAVHFHDTYGQSLPNILVSLQMGISAVDSS 360
ISLGDTIG+GTPG+VVPML AVMA+VP +KLAVHFHDTYGQ+L NILVSLQMGIS VDSS
Sbjct: 335 ISLGDTIGIGTPGSVVPMLEAVMAVVPADKLAVHFHDTYGQALANILVSLQMGISIVDSS 394
Query: 361 VAGLGGCPYAKGASGNVATEDVVYMLNGLGIKTNVDIGKLMSAGDFIGKQLGRPSGSKTA 420
+AGLGGCPYAKGASGNVATEDVVYMLNGLG+ TNVD+GKL++AGDFI K LGRP+GSK A
Sbjct: 395 IAGLGGCPYAKGASGNVATEDVVYMLNGLGVHTNVDLGKLIAAGDFISKHLGRPNGSKAA 454
Query: 421 IALN-RVTADASKI 433
+ALN R+TADASKI
Sbjct: 455 VALNRRITADASKI 468
>UniRef100_Q9LJ19 ESTs C97134 [Oryza sativa]
Length = 407
Score = 493 bits (1268), Expect = e-138
Identities = 252/325 (77%), Positives = 279/325 (85%), Gaps = 2/325 (0%)
Query: 111 SQKRNNKDMQDMAYKFMKGMPEFVKIVEVGPRDGLQNEKNIVPTDVKIELIHRLASTGLS 170
S +N + K + +P VKIVEVGPRDGLQNEKNIVPT VKIELI RLA++GLS
Sbjct: 83 SSSSDNPASIGIGNKIIHDLPRSVKIVEVGPRDGLQNEKNIVPTHVKIELIQRLATSGLS 142
Query: 171 VIEATSFVSPKWVPQLADAKDVMQAVHNLRGIRLPVLTPNLKGFEAAVAAGAREVAVFAS 230
V+EATSFVSPKWVPQLADAKDVM V N+ G+ LPVLTPNLKGFEAAVAAGA+EVAVFAS
Sbjct: 143 VVEATSFVSPKWVPQLADAKDVMDVVRNIEGVSLPVLTPNLKGFEAAVAAGAKEVAVFAS 202
Query: 231 ASESFSKSNINCSIEESLSRYRAVTRAAKELSIPVRGYVSCVVGCPVEGPVPPSKVAYVA 290
ASE+FSKSNINC+I+ESL+RY+ V AAKEL IP+RGYVSCVVGCPVEG VPPS VA+VA
Sbjct: 203 ASEAFSKSNINCTIKESLARYKDVALAAKELKIPMRGYVSCVVGCPVEGYVPPSNVAHVA 262
Query: 291 KALYDMGCFEISLGDTIGVGTPGTVVPMLLAVMAIVPTEKLAVHFHDTYGQSLPNILVSL 350
K LYDMGC+E+SLGDTIGVGTPGTVVPML AVM VP EKLAVHFHDTYGQSL NIL+SL
Sbjct: 263 KELYDMGCYEVSLGDTIGVGTPGTVVPMLEAVMFFVPKEKLAVHFHDTYGQSLSNILISL 322
Query: 351 QMGISAVDSSVAGLGGCPYAKGASGNVATEDVVYMLNGLGIKTNVDIGKLMSAGDFIGKQ 410
QMG+S VDSSVAGLGGCPYAKGASGNVATEDVVYMLNGLGI TNVD+GK+M+AG+FI
Sbjct: 323 QMGVSVVDSSVAGLGGCPYAKGASGNVATEDVVYMLNGLGISTNVDLGKVMAAGEFICNH 382
Query: 411 LGRPSGSKTAIALNR--VTADASKI 433
LGR SGSK AIAL TA+ASK+
Sbjct: 383 LGRQSGSKAAIALGSKVATANASKL 407
>UniRef100_Q8TB92 HMGCLL1 protein [Homo sapiens]
Length = 340
Score = 404 bits (1038), Expect = e-111
Identities = 195/296 (65%), Positives = 240/296 (80%)
Query: 127 MKGMPEFVKIVEVGPRDGLQNEKNIVPTDVKIELIHRLASTGLSVIEATSFVSPKWVPQL 186
+ G+PEFVKIVEVGPRDGLQNEK IVPTD+KIE I+RL+ TGLSVIE TSFVS +WVPQ+
Sbjct: 41 LSGLPEFVKIVEVGPRDGLQNEKVIVPTDIKIEFINRLSQTGLSVIEVTSFVSSRWVPQM 100
Query: 187 ADAKDVMQAVHNLRGIRLPVLTPNLKGFEAAVAAGAREVAVFASASESFSKSNINCSIEE 246
AD +VM+ +H G+R PVLTPNL+GF AVAAGA E++VF +ASESFSK NINCSIEE
Sbjct: 101 ADHTEVMKGIHQYPGVRYPVLTPNLQGFHHAVAAGATEISVFGAASESFSKKNINCSIEE 160
Query: 247 SLSRYRAVTRAAKELSIPVRGYVSCVVGCPVEGPVPPSKVAYVAKALYDMGCFEISLGDT 306
S+ ++ V ++A+ ++IP RGYVSC +GCP EG + P KV V+K LY MGC+EISLGDT
Sbjct: 161 SMGKFEEVVKSARHMNIPARGYVSCALGCPYEGSITPQKVTEVSKRLYGMGCYEISLGDT 220
Query: 307 IGVGTPGTVVPMLLAVMAIVPTEKLAVHFHDTYGQSLPNILVSLQMGISAVDSSVAGLGG 366
IGVGTPG++ ML +VM +P LAVH HDTYGQ+L NIL +LQMGI+ VDS+V+GLGG
Sbjct: 221 IGVGTPGSMKRMLESVMKEIPPGALAVHCHDTYGQALANILTALQMGINVVDSAVSGLGG 280
Query: 367 CPYAKGASGNVATEDVVYMLNGLGIKTNVDIGKLMSAGDFIGKQLGRPSGSKTAIA 422
CPYAKGASGNVATED++YMLNGLG+ T V++ K+M AGDFI K + + + SK A A
Sbjct: 281 CPYAKGASGNVATEDLIYMLNGLGLNTGVNLYKVMEAGDFICKAVNKTTNSKVAQA 336
>UniRef100_Q8JZS7 3-hydroxymethyl-3-methylglutaryl-Coenzyme A lyase-like 1 [Mus
musculus]
Length = 343
Score = 402 bits (1033), Expect = e-110
Identities = 195/296 (65%), Positives = 241/296 (80%)
Query: 127 MKGMPEFVKIVEVGPRDGLQNEKNIVPTDVKIELIHRLASTGLSVIEATSFVSPKWVPQL 186
+ G+PE+VKIVEVGPRDGLQNEK IVPTD+KIELI++L+ TGLSVIE TSFVS +WVPQ+
Sbjct: 41 LPGLPEYVKIVEVGPRDGLQNEKVIVPTDIKIELINQLSQTGLSVIEVTSFVSSRWVPQM 100
Query: 187 ADAKDVMQAVHNLRGIRLPVLTPNLKGFEAAVAAGAREVAVFASASESFSKSNINCSIEE 246
AD +VM+ + G+R PVLTPNL+GF+ AVAAGA E+AVF +ASESFSK NINCSIEE
Sbjct: 101 ADHAEVMRGIRQYPGVRYPVLTPNLQGFQHAVAAGATEIAVFGAASESFSKKNINCSIEE 160
Query: 247 SLSRYRAVTRAAKELSIPVRGYVSCVVGCPVEGPVPPSKVAYVAKALYDMGCFEISLGDT 306
S+ R++ V +A+ + IPVRGYVSC +GCP EG + P KV V+K LY MGC+EISLGDT
Sbjct: 161 SMGRFQEVISSARHMDIPVRGYVSCALGCPYEGSITPQKVTEVSKRLYGMGCYEISLGDT 220
Query: 307 IGVGTPGTVVPMLLAVMAIVPTEKLAVHFHDTYGQSLPNILVSLQMGISAVDSSVAGLGG 366
IGVGTPG++ ML +VM +P LAVH HDTYGQ+L NIL +LQMGI+ VDS+V+GLGG
Sbjct: 221 IGVGTPGSMKMMLESVMKEIPPGALAVHCHDTYGQALANILTALQMGINVVDSAVSGLGG 280
Query: 367 CPYAKGASGNVATEDVVYMLNGLGIKTNVDIGKLMSAGDFIGKQLGRPSGSKTAIA 422
CPYAKGASGNVATED++YMLNG+G+ T VD+ K+M AG+FI K + + + SK A A
Sbjct: 281 CPYAKGASGNVATEDLIYMLNGMGLNTGVDLYKVMEAGEFICKAVNKTTNSKVAQA 336
>UniRef100_UPI00003AD0F1 UPI00003AD0F1 UniRef100 entry
Length = 337
Score = 402 bits (1032), Expect = e-110
Identities = 196/294 (66%), Positives = 239/294 (80%)
Query: 129 GMPEFVKIVEVGPRDGLQNEKNIVPTDVKIELIHRLASTGLSVIEATSFVSPKWVPQLAD 188
G+PE+VKIVEVGPRDGLQNEK IVPTD KIELI+RL+ TGL IE TSFVS KWVPQ+AD
Sbjct: 41 GLPEYVKIVEVGPRDGLQNEKVIVPTDTKIELINRLSKTGLPAIEVTSFVSSKWVPQMAD 100
Query: 189 AKDVMQAVHNLRGIRLPVLTPNLKGFEAAVAAGAREVAVFASASESFSKSNINCSIEESL 248
K+VM+ + G++ PVLTPNLKGF +A+AAGA EV+VF +ASESFSK NINCSIEES+
Sbjct: 101 HKEVMRGIERHPGVQYPVLTPNLKGFHSAIAAGATEVSVFGAASESFSKMNINCSIEESI 160
Query: 249 SRYRAVTRAAKELSIPVRGYVSCVVGCPVEGPVPPSKVAYVAKALYDMGCFEISLGDTIG 308
++ V ++A+ ++IPVRGYVSC +GCP EG + P+KVA V+K LY MGC+EISLGDTIG
Sbjct: 161 EKFEEVAKSARNMNIPVRGYVSCALGCPYEGDITPAKVAEVSKRLYSMGCYEISLGDTIG 220
Query: 309 VGTPGTVVPMLLAVMAIVPTEKLAVHFHDTYGQSLPNILVSLQMGISAVDSSVAGLGGCP 368
VGTPG++ ML AVM +P LAVH HDTYGQ+L NIL ++QMG++ VDSSVAGLGGCP
Sbjct: 221 VGTPGSMKRMLEAVMKEIPLSALAVHCHDTYGQALANILTAIQMGVAVVDSSVAGLGGCP 280
Query: 369 YAKGASGNVATEDVVYMLNGLGIKTNVDIGKLMSAGDFIGKQLGRPSGSKTAIA 422
YAKGA+GNVATEDV+YMLNGLGI T V++ +M AG+FI L + + SK A A
Sbjct: 281 YAKGATGNVATEDVIYMLNGLGINTGVNLYTVMEAGNFICTALNKKTNSKVAQA 334
>UniRef100_Q8QGJ4 Hydroxymethylglutaryl-CoA lyase [Fugu rubripes]
Length = 325
Score = 402 bits (1032), Expect = e-110
Identities = 202/297 (68%), Positives = 236/297 (79%)
Query: 126 FMKGMPEFVKIVEVGPRDGLQNEKNIVPTDVKIELIHRLASTGLSVIEATSFVSPKWVPQ 185
F + VKIVEVGPRDGLQNEK IVP + KI LI L+++GL VIEATSFVSPKWVPQ
Sbjct: 25 FSSAKLQSVKIVEVGPRDGLQNEKTIVPLETKIHLIDMLSASGLPVIEATSFVSPKWVPQ 84
Query: 186 LADAKDVMQAVHNLRGIRLPVLTPNLKGFEAAVAAGAREVAVFASASESFSKSNINCSIE 245
+AD +VM+ + G+ PVLTPNLKGF+AAV+AGA EVA+F +ASE FSK NINCS++
Sbjct: 85 MADQVEVMKGISRKPGVSYPVLTPNLKGFQAAVSAGASEVAIFGAASELFSKKNINCSVD 144
Query: 246 ESLSRYRAVTRAAKELSIPVRGYVSCVVGCPVEGPVPPSKVAYVAKALYDMGCFEISLGD 305
ESL R+ V +AAKE +PVRGYVSCV+GCP EG V P+KVA+VAK LY MGC+EISLGD
Sbjct: 145 ESLQRFDEVMKAAKEAGVPVRGYVSCVLGCPYEGKVEPAKVAHVAKRLYSMGCYEISLGD 204
Query: 306 TIGVGTPGTVVPMLLAVMAIVPTEKLAVHFHDTYGQSLPNILVSLQMGISAVDSSVAGLG 365
TIGVGTPG++ ML V VP LAVH HDTYGQ+L NILV+LQMG+S VDSSVAGLG
Sbjct: 205 TIGVGTPGSMCEMLKVVSREVPVSALAVHCHDTYGQALANILVALQMGVSVVDSSVAGLG 264
Query: 366 GCPYAKGASGNVATEDVVYMLNGLGIKTNVDIGKLMSAGDFIGKQLGRPSGSKTAIA 422
GCPYA+GASGNVATEDVVYML+GLGI+T VD+ K+M AG FI + L R S SK A A
Sbjct: 265 GCPYAQGASGNVATEDVVYMLHGLGIQTGVDLSKVMDAGAFICRSLNRKSNSKVAQA 321
>UniRef100_Q6INN1 MGC82338 protein [Xenopus laevis]
Length = 328
Score = 401 bits (1031), Expect = e-110
Identities = 201/297 (67%), Positives = 238/297 (79%)
Query: 127 MKGMPEFVKIVEVGPRDGLQNEKNIVPTDVKIELIHRLASTGLSVIEATSFVSPKWVPQL 186
++ P+ VKIVEVGPRDGLQNEK +VPTDVKI LI+ L+ GL IEATSFVSPKWVPQ+
Sbjct: 29 IQSFPKEVKIVEVGPRDGLQNEKTVVPTDVKIHLINMLSEAGLQAIEATSFVSPKWVPQM 88
Query: 187 ADAKDVMQAVHNLRGIRLPVLTPNLKGFEAAVAAGAREVAVFASASESFSKSNINCSIEE 246
AD K+VMQ + I PVLTPNL GF+AAV GA+EVA+F +ASE FSK NINCSI+E
Sbjct: 89 ADHKNVMQGIKKYPNISYPVLTPNLTGFQAAVECGAKEVAIFGAASELFSKKNINCSIDE 148
Query: 247 SLSRYRAVTRAAKELSIPVRGYVSCVVGCPVEGPVPPSKVAYVAKALYDMGCFEISLGDT 306
SL R++AV AKE +IPVRGYVSCV+GCP EG V PSKVA VA ++ MGC+EISLGDT
Sbjct: 149 SLQRFKAVITEAKEANIPVRGYVSCVLGCPYEGKVAPSKVAEVAYKMFSMGCYEISLGDT 208
Query: 307 IGVGTPGTVVPMLLAVMAIVPTEKLAVHFHDTYGQSLPNILVSLQMGISAVDSSVAGLGG 366
IGVGTPG + ML AV+ +VP + LAVH HDTYGQ+L NILV+LQMG+ VD+SVAGLGG
Sbjct: 209 IGVGTPGGMRDMLSAVLDVVPAKALAVHCHDTYGQALANILVALQMGVQVVDASVAGLGG 268
Query: 367 CPYAKGASGNVATEDVVYMLNGLGIKTNVDIGKLMSAGDFIGKQLGRPSGSKTAIAL 423
CPYA+GASGNVATEDVVYML+GLGI+T +D+ KL AG FI K LG+ S SK + A+
Sbjct: 269 CPYAQGASGNVATEDVVYMLHGLGIQTGIDLKKLTEAGAFICKALGKKSHSKVSQAI 325
>UniRef100_UPI0000360D1C UPI0000360D1C UniRef100 entry
Length = 325
Score = 401 bits (1030), Expect = e-110
Identities = 202/297 (68%), Positives = 235/297 (79%)
Query: 126 FMKGMPEFVKIVEVGPRDGLQNEKNIVPTDVKIELIHRLASTGLSVIEATSFVSPKWVPQ 185
F + VKIVEVGPRDGLQNEK IVP + KI LI L+++GL VIEATSFVSPKWVPQ
Sbjct: 25 FSSAKLQSVKIVEVGPRDGLQNEKTIVPLETKIHLIDMLSASGLPVIEATSFVSPKWVPQ 84
Query: 186 LADAKDVMQAVHNLRGIRLPVLTPNLKGFEAAVAAGAREVAVFASASESFSKSNINCSIE 245
+AD +VM+ + G+ PVLTPNLKGF+AAV AGA EVA+F +ASE FSK NINCS++
Sbjct: 85 MADQVEVMKGISRKPGVSYPVLTPNLKGFQAAVKAGASEVAIFGAASELFSKKNINCSVD 144
Query: 246 ESLSRYRAVTRAAKELSIPVRGYVSCVVGCPVEGPVPPSKVAYVAKALYDMGCFEISLGD 305
ESL R+ V +AAKE +PVRGYVSCV+GCP EG V P+KVA+VAK LY MGC+EISLGD
Sbjct: 145 ESLQRFDEVMKAAKEAGVPVRGYVSCVLGCPYEGKVEPAKVAHVAKRLYSMGCYEISLGD 204
Query: 306 TIGVGTPGTVVPMLLAVMAIVPTEKLAVHFHDTYGQSLPNILVSLQMGISAVDSSVAGLG 365
TIGVGTPG++ ML V VP LAVH HDTYGQ+L NILV+LQMG+S VDSSVAGLG
Sbjct: 205 TIGVGTPGSMCEMLKVVSREVPVSALAVHCHDTYGQALANILVALQMGVSVVDSSVAGLG 264
Query: 366 GCPYAKGASGNVATEDVVYMLNGLGIKTNVDIGKLMSAGDFIGKQLGRPSGSKTAIA 422
GCPYA+GASGNVATEDVVYML+GLGI+T VD+ K+M AG FI + L R S SK A A
Sbjct: 265 GCPYAQGASGNVATEDVVYMLHGLGIQTGVDLSKVMDAGAFICRSLNRKSNSKVAQA 321
>UniRef100_Q7ZV32 Similar to 3-hydroxymethyl-3-methylglutaryl-Coenzyme A lyase
[Brachydanio rerio]
Length = 340
Score = 401 bits (1030), Expect = e-110
Identities = 202/295 (68%), Positives = 235/295 (79%)
Query: 128 KGMPEFVKIVEVGPRDGLQNEKNIVPTDVKIELIHRLASTGLSVIEATSFVSPKWVPQLA 187
+ +PE VKIVEVGPRDGLQNEK IVPT+VKI LI L+ GL VIEATSFVSPKWVPQ+A
Sbjct: 42 QALPERVKIVEVGPRDGLQNEKTIVPTEVKIRLIDMLSEAGLPVIEATSFVSPKWVPQMA 101
Query: 188 DAKDVMQAVHNLRGIRLPVLTPNLKGFEAAVAAGAREVAVFASASESFSKSNINCSIEES 247
D ++VM+ +H G+ PVLTPNLKGF+AAV AGA+EVA+F +ASE FSK NINCS+EES
Sbjct: 102 DQEEVMRGLHKKPGVNYPVLTPNLKGFQAAVKAGAKEVAIFGAASELFSKKNINCSVEES 161
Query: 248 LSRYRAVTRAAKELSIPVRGYVSCVVGCPVEGPVPPSKVAYVAKALYDMGCFEISLGDTI 307
L R+ V AAK+ + VRGYVSCV+GCP EG V PSKVA VAK LY MGC+E+SLGDTI
Sbjct: 162 LVRFEEVMTAAKQEGVSVRGYVSCVLGCPYEGKVSPSKVAEVAKRLYSMGCYEVSLGDTI 221
Query: 308 GVGTPGTVVPMLLAVMAIVPTEKLAVHFHDTYGQSLPNILVSLQMGISAVDSSVAGLGGC 367
GVGTPG + ML AV +P E LAVH HDTYGQ+L NILV+LQ G+S VDSSVAGLGGC
Sbjct: 222 GVGTPGGMTEMLNAVKKELPVEALAVHCHDTYGQALANILVALQNGVSVVDSSVAGLGGC 281
Query: 368 PYAKGASGNVATEDVVYMLNGLGIKTNVDIGKLMSAGDFIGKQLGRPSGSKTAIA 422
PYA+GASGNVATEDVVYML+GLGI T VD+ +L+ AG FI + R + SK A A
Sbjct: 282 PYAQGASGNVATEDVVYMLHGLGIHTGVDLPRLLDAGSFICHSINRRTNSKVAQA 336
>UniRef100_UPI00003AC63A UPI00003AC63A UniRef100 entry
Length = 298
Score = 400 bits (1029), Expect = e-110
Identities = 200/296 (67%), Positives = 238/296 (79%)
Query: 131 PEFVKIVEVGPRDGLQNEKNIVPTDVKIELIHRLASTGLSVIEATSFVSPKWVPQLADAK 190
P+ VK+VEVGPRDGLQNEK++VPT VKI LI L+ TGL VIEATSFVSP+WVPQ+AD
Sbjct: 3 PQRVKVVEVGPRDGLQNEKSVVPTPVKIRLIDMLSETGLPVIEATSFVSPRWVPQMADHA 62
Query: 191 DVMQAVHNLRGIRLPVLTPNLKGFEAAVAAGAREVAVFASASESFSKSNINCSIEESLSR 250
+VMQ ++ L G+ PVLTPNLKGF+AAVAAGA+EV++F +ASE F+K NINCSIEESL R
Sbjct: 63 EVMQGINKLPGVSYPVLTPNLKGFQAAVAAGAKEVSIFGAASELFTKKNINCSIEESLER 122
Query: 251 YRAVTRAAKELSIPVRGYVSCVVGCPVEGPVPPSKVAYVAKALYDMGCFEISLGDTIGVG 310
+ V AA+ SIPVRGYVSCV+GCP EG + +KVA V+K +Y MGC+EISLGDTIG+G
Sbjct: 123 FSEVMNAARAASIPVRGYVSCVLGCPYEGNISAAKVAEVSKKMYSMGCYEISLGDTIGIG 182
Query: 311 TPGTVVPMLLAVMAIVPTEKLAVHFHDTYGQSLPNILVSLQMGISAVDSSVAGLGGCPYA 370
TPG++ ML AVM VP LAVH HDTYGQ+L NILV+LQMG+S VD+SVAGLGGCPYA
Sbjct: 183 TPGSMKEMLAAVMKEVPVGALAVHCHDTYGQALANILVALQMGVSVVDASVAGLGGCPYA 242
Query: 371 KGASGNVATEDVVYMLNGLGIKTNVDIGKLMSAGDFIGKQLGRPSGSKTAIALNRV 426
+GASGNVATED+VYMLNGLGI T VD+ KLM G FI L R + SK + A R+
Sbjct: 243 QGASGNVATEDLVYMLNGLGIHTGVDLQKLMDTGTFICNALNRRTNSKVSQAACRL 298
>UniRef100_UPI0000318F4C UPI0000318F4C UniRef100 entry
Length = 337
Score = 400 bits (1029), Expect = e-110
Identities = 202/293 (68%), Positives = 236/293 (79%)
Query: 130 MPEFVKIVEVGPRDGLQNEKNIVPTDVKIELIHRLASTGLSVIEATSFVSPKWVPQLADA 189
+PE VKIVEVGPRDGLQNEK IVP++ KI LI L+++GL VIEATSFVSPKWVPQ+AD
Sbjct: 41 LPEKVKIVEVGPRDGLQNEKTIVPSETKIHLIDLLSASGLPVIEATSFVSPKWVPQMADQ 100
Query: 190 KDVMQAVHNLRGIRLPVLTPNLKGFEAAVAAGAREVAVFASASESFSKSNINCSIEESLS 249
+VM+ + G+ PVLTPNLKGF+AAV AGA EVA+F +ASE FSK NINCS++ESL
Sbjct: 101 VEVMKGISRRPGVSYPVLTPNLKGFQAAVQAGASEVAIFGAASELFSKKNINCSVDESLQ 160
Query: 250 RYRAVTRAAKELSIPVRGYVSCVVGCPVEGPVPPSKVAYVAKALYDMGCFEISLGDTIGV 309
R+ V +AAKE +PVRGYVSCV+GCP EG V P KVA+VAK LY MGC+EISLGDTIGV
Sbjct: 161 RFDEVMKAAKEAGVPVRGYVSCVLGCPYEGKVEPVKVAHVAKRLYAMGCYEISLGDTIGV 220
Query: 310 GTPGTVVPMLLAVMAIVPTEKLAVHFHDTYGQSLPNILVSLQMGISAVDSSVAGLGGCPY 369
GTPG++ ML V VP LAVH HDTYGQ+L NILV+LQMG+S VDSSVAGLGGCPY
Sbjct: 221 GTPGSMRDMLEVVSREVPVSALAVHCHDTYGQALANILVALQMGVSVVDSSVAGLGGCPY 280
Query: 370 AKGASGNVATEDVVYMLNGLGIKTNVDIGKLMSAGDFIGKQLGRPSGSKTAIA 422
A+GASGNVATEDVVYML+GLGI+T VD+ K+M AG FI + L R + SK A A
Sbjct: 281 AQGASGNVATEDVVYMLHGLGIQTGVDLSKVMDAGAFICRSLNRRTNSKVAQA 333
>UniRef100_P35915 Hydroxymethylglutaryl-CoA lyase [Gallus gallus]
Length = 298
Score = 398 bits (1023), Expect = e-109
Identities = 199/296 (67%), Positives = 237/296 (79%)
Query: 131 PEFVKIVEVGPRDGLQNEKNIVPTDVKIELIHRLASTGLSVIEATSFVSPKWVPQLADAK 190
P+ VK+VEVGPRDGLQNEK++VPT VKI LI L+ TGL VIEATSFVSP+WVPQ+AD
Sbjct: 3 PQRVKVVEVGPRDGLQNEKSVVPTPVKIRLIDMLSETGLPVIEATSFVSPRWVPQMADHA 62
Query: 191 DVMQAVHNLRGIRLPVLTPNLKGFEAAVAAGAREVAVFASASESFSKSNINCSIEESLSR 250
+VMQ ++ L G+ PVLTPNLKGF+AAVAAGA+EV++F +ASE F+K NINCSIEESL R
Sbjct: 63 EVMQGINKLPGVSYPVLTPNLKGFQAAVAAGAKEVSIFGAASELFTKKNINCSIEESLER 122
Query: 251 YRAVTRAAKELSIPVRGYVSCVVGCPVEGPVPPSKVAYVAKALYDMGCFEISLGDTIGVG 310
+ V AA+ SIPVRGYVSCV+GCP EG + +KVA V+K +Y MGC+EISLGD IG+G
Sbjct: 123 FSEVMNAARAASIPVRGYVSCVLGCPYEGNISAAKVAEVSKKMYSMGCYEISLGDRIGIG 182
Query: 311 TPGTVVPMLLAVMAIVPTEKLAVHFHDTYGQSLPNILVSLQMGISAVDSSVAGLGGCPYA 370
TPG++ ML AVM VP LAVH HDTYGQ+L NILV+LQMG+S VD+SVAGLGGCPYA
Sbjct: 183 TPGSMKEMLAAVMKEVPVGALAVHCHDTYGQALANILVALQMGVSVVDASVAGLGGCPYA 242
Query: 371 KGASGNVATEDVVYMLNGLGIKTNVDIGKLMSAGDFIGKQLGRPSGSKTAIALNRV 426
+GASGNVATED+VYMLNGLGI T VD+ KLM G FI L R + SK + A R+
Sbjct: 243 QGASGNVATEDLVYMLNGLGIHTGVDLQKLMDTGTFICNALNRRTNSKVSQAACRL 298
>UniRef100_Q8QZS6 Hmgcl protein [Mus musculus]
Length = 325
Score = 395 bits (1015), Expect = e-108
Identities = 201/296 (67%), Positives = 232/296 (77%)
Query: 127 MKGMPEFVKIVEVGPRDGLQNEKNIVPTDVKIELIHRLASTGLSVIEATSFVSPKWVPQL 186
M +P+ VKIVEVGPRDGLQNEK+IVPT VKI LI L+ GL VIEATSFVSPKWVPQ+
Sbjct: 26 MGTLPKQVKIVEVGPRDGLQNEKSIVPTPVKIRLIDMLSEAGLPVIEATSFVSPKWVPQM 85
Query: 187 ADAKDVMQAVHNLRGIRLPVLTPNLKGFEAAVAAGAREVAVFASASESFSKSNINCSIEE 246
AD DV++ + GI PVLTPN+KGFE AVAAGA+EV+VF + SE F++ N NCSIEE
Sbjct: 86 ADHSDVLKGIQKFPGINYPVLTPNMKGFEEAVAAGAKEVSVFGAVSELFTRKNANCSIEE 145
Query: 247 SLSRYRAVTRAAKELSIPVRGYVSCVVGCPVEGPVPPSKVAYVAKALYDMGCFEISLGDT 306
S R+ V +AA+ SI VRGYVSC +GCP EG V P+KVA VAK LY MGC+EISLGDT
Sbjct: 146 SFQRFAGVMQAAQAASISVRGYVSCALGCPYEGKVSPAKVAEVAKKLYSMGCYEISLGDT 205
Query: 307 IGVGTPGTVVPMLLAVMAIVPTEKLAVHFHDTYGQSLPNILVSLQMGISAVDSSVAGLGG 366
IGVGTPG + ML AVM VP LAVH HDTYGQ+L N LV+LQMG+S VDSSVAGLGG
Sbjct: 206 IGVGTPGLMKDMLTAVMHEVPVTALAVHCHDTYGQALANTLVALQMGVSVVDSSVAGLGG 265
Query: 367 CPYAKGASGNVATEDVVYMLNGLGIKTNVDIGKLMSAGDFIGKQLGRPSGSKTAIA 422
CPYAKGASGN+ATED+VYMLNGLGI T V++ KL+ AGDFI + L R + SK A A
Sbjct: 266 CPYAKGASGNLATEDLVYMLNGLGIHTGVNLQKLLEAGDFICQALNRKTSSKVAQA 321
>UniRef100_P97519 Hydroxymethylglutaryl-CoA lyase, mitochondrial precursor [Rattus
norvegicus]
Length = 325
Score = 395 bits (1014), Expect = e-108
Identities = 199/296 (67%), Positives = 234/296 (78%)
Query: 127 MKGMPEFVKIVEVGPRDGLQNEKNIVPTDVKIELIHRLASTGLSVIEATSFVSPKWVPQL 186
M +P+ VKIVEVGPRDGLQNEK+IVPT VKI+LI L+ GL VIEATSFVSPKWVPQ+
Sbjct: 26 MGTLPKRVKIVEVGPRDGLQNEKSIVPTPVKIKLIDMLSEAGLPVIEATSFVSPKWVPQM 85
Query: 187 ADAKDVMQAVHNLRGIRLPVLTPNLKGFEAAVAAGAREVAVFASASESFSKSNINCSIEE 246
AD DV++ + GI PVLTPN+KGFE AVAAGA+EV++F +ASE F++ N+NCSIEE
Sbjct: 86 ADHSDVLKGIQKFPGINYPVLTPNMKGFEEAVAAGAKEVSIFGAASELFTRKNVNCSIEE 145
Query: 247 SLSRYRAVTRAAKELSIPVRGYVSCVVGCPVEGPVPPSKVAYVAKALYDMGCFEISLGDT 306
S R+ V +AA+ SI VRGYVSC +GCP EG V P+KVA VAK LY MGC+EISLGDT
Sbjct: 146 SFQRFDGVMQAARAASISVRGYVSCALGCPYEGKVSPAKVAEVAKKLYSMGCYEISLGDT 205
Query: 307 IGVGTPGTVVPMLLAVMAIVPTEKLAVHFHDTYGQSLPNILVSLQMGISAVDSSVAGLGG 366
IGVGTPG + ML AV+ VP LAVH HDTYGQ+L N LV+LQMG+S VDSSVAGLGG
Sbjct: 206 IGVGTPGLMKDMLTAVLHEVPVAALAVHCHDTYGQALANTLVALQMGVSVVDSSVAGLGG 265
Query: 367 CPYAKGASGNVATEDVVYMLNGLGIKTNVDIGKLMSAGDFIGKQLGRPSGSKTAIA 422
CPYAKGASGN+ATED+VYML GLGI T V++ KL+ AGDFI + L R + SK A A
Sbjct: 266 CPYAKGASGNLATEDLVYMLTGLGIHTGVNLQKLLEAGDFICQALNRKTSSKVAQA 321
>UniRef100_UPI00003624B7 UPI00003624B7 UniRef100 entry
Length = 331
Score = 392 bits (1006), Expect = e-107
Identities = 194/292 (66%), Positives = 236/292 (80%)
Query: 131 PEFVKIVEVGPRDGLQNEKNIVPTDVKIELIHRLASTGLSVIEATSFVSPKWVPQLADAK 190
P+FVK+VEVGPRDGLQNEK I+PT VKI+LI L+ +GL VIEATSFVS KWVPQ+AD
Sbjct: 40 PKFVKVVEVGPRDGLQNEKEILPTGVKIQLIDMLSGSGLPVIEATSFVSSKWVPQMADHT 99
Query: 191 DVMQAVHNLRGIRLPVLTPNLKGFEAAVAAGAREVAVFASASESFSKSNINCSIEESLSR 250
DV++ + ++ PVLTPN++GF+ AVAAGA EVAVF S SE+FSK NINC+I+ES+ R
Sbjct: 100 DVLRGIQRAPHVQYPVLTPNIQGFQDAVAAGASEVAVFGSVSETFSKKNINCTIDESMLR 159
Query: 251 YRAVTRAAKELSIPVRGYVSCVVGCPVEGPVPPSKVAYVAKALYDMGCFEISLGDTIGVG 310
++ V AAKE IPVRGY+SC +GCP EG + PSKVA VAK LY MGC+EISLGDTIGVG
Sbjct: 160 FQDVINAAKEQKIPVRGYISCALGCPYEGHMEPSKVAEVAKRLYGMGCYEISLGDTIGVG 219
Query: 311 TPGTVVPMLLAVMAIVPTEKLAVHFHDTYGQSLPNILVSLQMGISAVDSSVAGLGGCPYA 370
TPG+++ ML +VM VP LAVH HDTYGQ+LPNIL +LQMGI VDS+VAGLGGCPYA
Sbjct: 220 TPGSMLKMLQSVMKEVPANALAVHCHDTYGQALPNILTALQMGICVVDSAVAGLGGCPYA 279
Query: 371 KGASGNVATEDVVYMLNGLGIKTNVDIGKLMSAGDFIGKQLGRPSGSKTAIA 422
+G+SGNVATEDV+YMLNG+GI+T V++ K++ AGDFI L R + SK A A
Sbjct: 280 QGSSGNVATEDVLYMLNGMGIETGVNLAKVIEAGDFICCALQRKTNSKVAQA 331
>UniRef100_Q5R9E1 Hypothetical protein DKFZp469I2412 [Pongo pygmaeus]
Length = 325
Score = 390 bits (1002), Expect = e-107
Identities = 197/296 (66%), Positives = 233/296 (78%)
Query: 127 MKGMPEFVKIVEVGPRDGLQNEKNIVPTDVKIELIHRLASTGLSVIEATSFVSPKWVPQL 186
M +P+ VKIVEVGPRDGLQNEKNIV T VKI+LI L+ GLSVIE TSFVSPKWVPQ+
Sbjct: 26 MGTLPKRVKIVEVGPRDGLQNEKNIVSTPVKIKLIDMLSEAGLSVIETTSFVSPKWVPQM 85
Query: 187 ADAKDVMQAVHNLRGIRLPVLTPNLKGFEAAVAAGAREVAVFASASESFSKSNINCSIEE 246
D +V++ + GI PVLTPNLKGFEAAVAAGA+EVA+F +ASE F+K NINCSIEE
Sbjct: 86 GDHTEVLKGIQKFPGINYPVLTPNLKGFEAAVAAGAKEVAIFGAASELFTKKNINCSIEE 145
Query: 247 SLSRYRAVTRAAKELSIPVRGYVSCVVGCPVEGPVPPSKVAYVAKALYDMGCFEISLGDT 306
S R+ A+ +AA+ +I VRGYVSC +GCP EG + P+KVA V K LY MGC+EISLGDT
Sbjct: 146 SFQRFDAILKAAQSANISVRGYVSCALGCPYEGKISPAKVAEVTKKLYSMGCYEISLGDT 205
Query: 307 IGVGTPGTVVPMLLAVMAIVPTEKLAVHFHDTYGQSLPNILVSLQMGISAVDSSVAGLGG 366
IGVGTPG + ML AVM VP LAVH HDTYGQ+L N L++LQMG+S VDSSVAGLGG
Sbjct: 206 IGVGTPGIMKGMLSAVMQEVPLAALAVHCHDTYGQALANTLMALQMGVSVVDSSVAGLGG 265
Query: 367 CPYAKGASGNVATEDVVYMLNGLGIKTNVDIGKLMSAGDFIGKQLGRPSGSKTAIA 422
CPYA+GASGN+ATED+VYML GLGI T V++ KL+ AG+FI + L R + SK A A
Sbjct: 266 CPYAQGASGNLATEDLVYMLEGLGIHTGVNLQKLLEAGNFICQALNRKTSSKVAQA 321
>UniRef100_Q8HXZ6 3-hydroxymethyl-3-methylglutaryl-Coenzyme A lyase [Macaca
fascicularis]
Length = 325
Score = 390 bits (1001), Expect = e-107
Identities = 196/296 (66%), Positives = 234/296 (78%)
Query: 127 MKGMPEFVKIVEVGPRDGLQNEKNIVPTDVKIELIHRLASTGLSVIEATSFVSPKWVPQL 186
M +P+ VKIVEVGPRDGLQNEKNIV T VKI+LI L+ GLSVIEATSFVSPKWVPQ+
Sbjct: 26 MDTLPKQVKIVEVGPRDGLQNEKNIVSTPVKIKLIDMLSEAGLSVIEATSFVSPKWVPQM 85
Query: 187 ADAKDVMQAVHNLRGIRLPVLTPNLKGFEAAVAAGAREVAVFASASESFSKSNINCSIEE 246
AD +V++ + GI PVL PNLKGFEAAVAAGA+EV++F +ASE F+K N+NCSIEE
Sbjct: 86 ADHAEVLKGIQKFPGITYPVLIPNLKGFEAAVAAGAKEVSIFGAASELFTKKNVNCSIEE 145
Query: 247 SLSRYRAVTRAAKELSIPVRGYVSCVVGCPVEGPVPPSKVAYVAKALYDMGCFEISLGDT 306
S R+ A+ +AA+ +I VRGYVSCV+GCP EG + P+KVA V K Y MGC+EISLGDT
Sbjct: 146 SFQRFDAILKAAQSANISVRGYVSCVLGCPYEGKISPAKVAEVTKKFYSMGCYEISLGDT 205
Query: 307 IGVGTPGTVVPMLLAVMAIVPTEKLAVHFHDTYGQSLPNILVSLQMGISAVDSSVAGLGG 366
IGVGTPG + ML AVM VP LAVH HDTYGQ+L N L++LQMG+S VDSSVAGLGG
Sbjct: 206 IGVGTPGIMKDMLSAVMQEVPPAALAVHCHDTYGQALANTLMALQMGVSVVDSSVAGLGG 265
Query: 367 CPYAKGASGNVATEDVVYMLNGLGIKTNVDIGKLMSAGDFIGKQLGRPSGSKTAIA 422
CPYA+GASGN+ATED+VYML GLGI T V++ KL+ AG+FI + L R + SK A A
Sbjct: 266 CPYAQGASGNLATEDLVYMLEGLGIHTGVNLQKLLEAGNFICQALNRKTSSKVAQA 321
>UniRef100_P38060 Hydroxymethylglutaryl-CoA lyase, mitochondrial precursor [Mus
musculus]
Length = 325
Score = 389 bits (998), Expect = e-106
Identities = 198/296 (66%), Positives = 229/296 (76%)
Query: 127 MKGMPEFVKIVEVGPRDGLQNEKNIVPTDVKIELIHRLASTGLSVIEATSFVSPKWVPQL 186
M +P+ VKIVEVGPRDGLQNEK+IVPT VKI LI L+ GL VIEATSFVSP WVPQ+
Sbjct: 26 MGTLPKQVKIVEVGPRDGLQNEKSIVPTPVKIRLIDMLSEAGLPVIEATSFVSPNWVPQM 85
Query: 187 ADAKDVMQAVHNLRGIRLPVLTPNLKGFEAAVAAGAREVAVFASASESFSKSNINCSIEE 246
AD DV++ + GI PVLTPN+KGFE AVAAGA+EV+VF + SE F++ N NCSIEE
Sbjct: 86 ADHSDVLKGIQKFPGINYPVLTPNMKGFEEAVAAGAKEVSVFGAVSELFTRKNANCSIEE 145
Query: 247 SLSRYRAVTRAAKELSIPVRGYVSCVVGCPVEGPVPPSKVAYVAKALYDMGCFEISLGDT 306
S R+ V +AA+ SI VRGYVSC +GCP EG V P+KVA VAK LY MGC+EISLGDT
Sbjct: 146 SFQRFAGVMQAAQAASISVRGYVSCALGCPYEGKVSPAKVAEVAKKLYSMGCYEISLGDT 205
Query: 307 IGVGTPGTVVPMLLAVMAIVPTEKLAVHFHDTYGQSLPNILVSLQMGISAVDSSVAGLGG 366
IGVGTPG + ML AVM VP L VH HDT GQ+L N LV+LQMG+S VDSSVAGLGG
Sbjct: 206 IGVGTPGLMKDMLTAVMHEVPVTALGVHCHDTIGQALANTLVALQMGVSVVDSSVAGLGG 265
Query: 367 CPYAKGASGNVATEDVVYMLNGLGIKTNVDIGKLMSAGDFIGKQLGRPSGSKTAIA 422
CPYAKGASGN+ATED+VYMLNGLGI T V++ KL+ AGDFI + L R + SK A A
Sbjct: 266 CPYAKGASGNLATEDLVYMLNGLGIHTGVNLQKLLEAGDFICQALNRKTSSKVAQA 321
>UniRef100_Q5QPQ6 OTTHUMP00000044830 [Homo sapiens]
Length = 300
Score = 387 bits (995), Expect = e-106
Identities = 195/296 (65%), Positives = 231/296 (77%)
Query: 127 MKGMPEFVKIVEVGPRDGLQNEKNIVPTDVKIELIHRLASTGLSVIEATSFVSPKWVPQL 186
M +P+ VKIVEVGPRDGLQNEKNIV T VKI+LI L+ GLSVIE TSFVSPKWVPQ+
Sbjct: 1 MGTLPKRVKIVEVGPRDGLQNEKNIVSTPVKIKLIDMLSEAGLSVIETTSFVSPKWVPQM 60
Query: 187 ADAKDVMQAVHNLRGIRLPVLTPNLKGFEAAVAAGAREVAVFASASESFSKSNINCSIEE 246
D +V++ + GI PVLTPNLKGFEAAVAAGA+EV +F +ASE F+K NINCSIEE
Sbjct: 61 GDHTEVLKGIQKFPGINYPVLTPNLKGFEAAVAAGAKEVVIFGAASELFTKKNINCSIEE 120
Query: 247 SLSRYRAVTRAAKELSIPVRGYVSCVVGCPVEGPVPPSKVAYVAKALYDMGCFEISLGDT 306
S R+ A+ +AA+ +I VRGYVSC +GCP EG + P+KVA V K Y MGC+EISLGDT
Sbjct: 121 SFQRFDAILKAAQSANISVRGYVSCALGCPYEGKISPAKVAEVTKKFYSMGCYEISLGDT 180
Query: 307 IGVGTPGTVVPMLLAVMAIVPTEKLAVHFHDTYGQSLPNILVSLQMGISAVDSSVAGLGG 366
IGVGTPG + ML AVM VP LAVH HDTYGQ+L N L++LQMG+S VDSSVAGLGG
Sbjct: 181 IGVGTPGIMKDMLSAVMQEVPLAALAVHCHDTYGQALANTLMALQMGVSVVDSSVAGLGG 240
Query: 367 CPYAKGASGNVATEDVVYMLNGLGIKTNVDIGKLMSAGDFIGKQLGRPSGSKTAIA 422
CPYA+GASGN+ATED+VYML GLGI T V++ KL+ AG+FI + L R + SK A A
Sbjct: 241 CPYAQGASGNLATEDLVYMLEGLGIHTGVNLQKLLEAGNFICQALNRKTSSKVAQA 296
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.316 0.133 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 682,620,548
Number of Sequences: 2790947
Number of extensions: 28268392
Number of successful extensions: 85592
Number of sequences better than 10.0: 1154
Number of HSP's better than 10.0 without gapping: 486
Number of HSP's successfully gapped in prelim test: 668
Number of HSP's that attempted gapping in prelim test: 84446
Number of HSP's gapped (non-prelim): 1262
length of query: 433
length of database: 848,049,833
effective HSP length: 130
effective length of query: 303
effective length of database: 485,226,723
effective search space: 147023697069
effective search space used: 147023697069
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 76 (33.9 bits)
Medicago: description of AC147363.5


