

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147179.8 + phase: 0
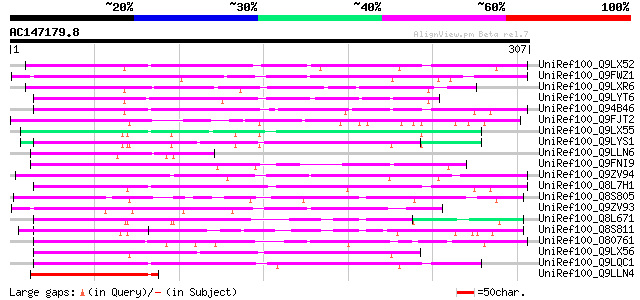
(307 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9LX52 Hypothetical protein F25L23_50 [Arabidopsis tha... 75 3e-12
UniRef100_Q9FWZ1 Hypothetical protein F24J1.28 [Arabidopsis thal... 75 3e-12
UniRef100_Q9LXR6 Hypothetical protein T20N10_210 [Arabidopsis th... 72 2e-11
UniRef100_Q9LYT6 Hypothetical protein F17J16_50 [Arabidopsis tha... 72 2e-11
UniRef100_Q94B46 Hypothetical protein At4g14100 [Arabidopsis tha... 71 3e-11
UniRef100_Q9FJT2 Emb|CAB62008.1 [Arabidopsis thaliana] 71 4e-11
UniRef100_Q9LX55 Hypothetical protein F25L23_20 [Arabidopsis tha... 70 7e-11
UniRef100_Q9LYS1 Hypothetical protein F17J16_200 [Arabidopsis th... 70 7e-11
UniRef100_Q9LLN6 Hypothetical protein DUPR11.17 [Oryza sativa] 70 9e-11
UniRef100_Q9FNI9 Arabidopsis thaliana genomic DNA, chromosome 5,... 70 9e-11
UniRef100_Q9ZV94 F9K20.20 protein [Arabidopsis thaliana] 69 2e-10
UniRef100_Q8L7H1 Hypothetical protein At4g14103 [Arabidopsis tha... 69 2e-10
UniRef100_Q8S805 Putative copia-type polyprotein [Oryza sativa] 68 4e-10
UniRef100_Q9ZV93 F9K20.21 protein [Arabidopsis thaliana] 67 5e-10
UniRef100_Q8L671 Hypothetical protein OSJNBb0072F04.2 [Oryza sat... 67 8e-10
UniRef100_Q8S811 Hypothetical protein OSJNBa0020E23.13 [Oryza sa... 66 1e-09
UniRef100_O80761 T13D8.27 protein [Arabidopsis thaliana] 66 1e-09
UniRef100_Q9LX56 Hypothetical protein F25L23_10 [Arabidopsis tha... 65 2e-09
UniRef100_Q9LQC1 F19C14.7 protein [Arabidopsis thaliana] 65 2e-09
UniRef100_Q9LLN4 Hypothetical protein DUPR11.22 [Oryza sativa] 65 3e-09
>UniRef100_Q9LX52 Hypothetical protein F25L23_50 [Arabidopsis thaliana]
Length = 514
Score = 74.7 bits (182), Expect = 3e-12
Identities = 83/329 (25%), Positives = 145/329 (43%), Gaps = 44/329 (13%)
Query: 10 DYVKEDRLSDLPNCVLLRILSLLDADEAVKTCTISSRWRHLWKHLPTLILCSSHFMR--- 66
D +D +S+LP+ +L +LS L EA T ++ RWR L +P L L + + R
Sbjct: 2 DSGSKDIISNLPDALLCHVLSFLPTTEAASTSVLAKRWRFLLAFVPNLDLDNMIYDRPKM 61
Query: 67 --------IKSFNRFVSRILCLRDVSTPLHAVDFLCRGVVDPRLLKKVVTYAVSHNVEKL 118
KSF FV R++ L+ + PL C+ DP + V + VE+L
Sbjct: 62 GRRKRLELRKSFKLFVDRVMALQG-NAPLKKFSLRCKIGSDPSRVNGWVLKVLDRGVEEL 120
Query: 119 RVHACCDFQH-FSSCFFSCHTLTSLDLCVGPPRTDERLSFPKSLNLPALTTLSLESFAFC 177
++ ++++ TL SL + TDE LP L TL L + +F
Sbjct: 121 DLYIASEYEYPLPPKVLMTKTLVSLKV----SGTDEFTIDVGEFFLPKLKTLHLSAISF- 175
Query: 178 VGDDG---CAEPFSALNSLKNLMILYCNVLDAESLCISSVTLTNLTIVGDPVYYS--TVE 232
GD+G A+ SA ++L+ L+++ E +SS +L ++I + + + +V
Sbjct: 176 -GDEGGPPFAKLISACHALEELVMIKMMWDYWEFCSVSSPSLKRVSIDCENIDENPKSVS 234
Query: 233 LSTPSLFTFDFVCYEGIPVLKLCRSKINLSSVKHVNIEVRMWTDYADASLVLLN------ 286
TP+L +F + + K+N S+ +I +RM D + L+N
Sbjct: 235 FDTPNLVYLEFT-----DTVAVKYPKVNFDSLVEASIGLRMTPDQVFDARDLVNRHHGYK 289
Query: 287 ---------WLAELANIKSLTLNHTALKV 306
++ + N+K++ L+ AL+V
Sbjct: 290 RCKGANAADFMMGVCNVKTMYLSSEALEV 318
>UniRef100_Q9FWZ1 Hypothetical protein F24J1.28 [Arabidopsis thaliana]
Length = 451
Score = 74.7 bits (182), Expect = 3e-12
Identities = 88/325 (27%), Positives = 135/325 (41%), Gaps = 36/325 (11%)
Query: 2 KRGRKKHNDYVKEDRLSDLPNCVLLRILSLLDADEAVKTCTISSRWRHLWKHLPTLILCS 61
K R K +D V D +S LP+C+L +L L + VKT +S RWR+LWKH+P L L +
Sbjct: 7 KHVRAKGSDEV--DWISKLPDCLLCEVLLNLPTKDVVKTSVLSRRWRNLWKHVPGLDLDN 64
Query: 62 SHFMRIKSFNRFVSRILCLRDVS-TPLHAVDFLCRGVVDP------RLLKKVVTYAVSHN 114
+ F +F FV L S + + C DP R + +VT V H
Sbjct: 65 TDFQEFNTFLSFVDSFLDFNSESFLQKFILKYDCDDEYDPDIFLIGRWINTIVTRKVQHI 124
Query: 115 VEKLRVHACCDFQHFSSCFFSCHTLTSLDLCVGPPRTDERLSFPKSLNLPALTTLSLESF 174
+ + Q S ++C +L SL LC L+ P+ ++LP+L + L
Sbjct: 125 DVLDDSYGSWEVQ-LPSSIYTCESLVSLKLC------GLTLASPEFVSLPSLKVMDLIIT 177
Query: 175 AFCVGDDGCAEPFSALNSLKNLMILYCNVLDAESLCISSVTLTNLTIVGDP----VYYST 230
F D G + L++L I + E L + S +L T V D V
Sbjct: 178 KF-ADDMGLETLITKCPVLESLTIERSFCDEIEVLRVRSQSLLRFTHVADSDEGVVEDLV 236
Query: 231 VELSTPSLFTFDFVCYEGIPVL-----KLCRSKI----NLSSVKHVNIEVRMWTDYADAS 281
V + P L + + KL ++ I NLSSV N D
Sbjct: 237 VSIDAPKLEYLRLSDHRVASFILNKPGKLVKADIDIVFNLSSVNKFN------PDDLPKR 290
Query: 282 LVLLNWLAELANIKSLTLNHTALKV 306
++ N+L ++ IK + + + L+V
Sbjct: 291 TMIRNFLLGISTIKDMIIFSSTLEV 315
>UniRef100_Q9LXR6 Hypothetical protein T20N10_210 [Arabidopsis thaliana]
Length = 457
Score = 72.0 bits (175), Expect = 2e-11
Identities = 78/283 (27%), Positives = 120/283 (41%), Gaps = 33/283 (11%)
Query: 10 DYVKEDRLSDLPNCVLLRILSLLDADEAVKTCTISSRWRHLWKHLPTLILCSSHFMR--- 66
D K D S LP+ V+ ILS L EA T ++ +WR+L+ +P+L S F+
Sbjct: 2 DSEKMDLFSKLPDEVISHILSSLPTKEAASTSVLAKKWRYLFAFVPSLDFNDSDFLHPQE 61
Query: 67 --------IKSFNRFVSRILCLRDVSTPLHAVDFLCRGVVDPRLLKKVVTYAVSHNVEKL 118
++SF FV R+L L+ S P+ + VDP + + + + V L
Sbjct: 62 GKREKDGILRSFMDFVDRVLALQGAS-PIKKFSLNVKTGVDPDRVDRWICNVLQRGVSHL 120
Query: 119 RVHACCDFQHFSSCFFS---CHTLTSLDLCVGPPRTDERLSFPKSLNLPALTTLSLESFA 175
+ DF+ S + TL L G + + + LP L TL LES
Sbjct: 121 ALF--MDFEEEYSLPYEISVSKTLVELKTGYGV----DLYLWDDDMFLPMLKTLVLESVE 174
Query: 176 FCVGDDGCAEPFSALNSLKNLMILYCNVLDAESLCISSVTLTNLTIVGDPVYYSTVELST 235
F G P A L+ LM+L D ++ +SS +L NL I + T+ T
Sbjct: 175 FGRGQFQTLLP--ACPVLEELMLLNMEWKD-RNVILSSSSLKNLKITSEDGCLGTLSFDT 231
Query: 236 PSLFTFDFVCY--EGIPVLKLCRSKINLSSVKHVNIEVRMWTD 276
P+L D+ Y E P+ +NL ++ V I + + D
Sbjct: 232 PNLVFLDYYDYVAEDYPI-------VNLKNLVEVGINLVLTAD 267
>UniRef100_Q9LYT6 Hypothetical protein F17J16_50 [Arabidopsis thaliana]
Length = 461
Score = 71.6 bits (174), Expect = 2e-11
Identities = 77/258 (29%), Positives = 116/258 (44%), Gaps = 27/258 (10%)
Query: 15 DRLSDLPNCVLLRILSLLDADEAVKTCTISSRWRHLWKHLPTLILCSSHFMR-------- 66
DR+ LP+ +L ILS L EA T +S RWR+L +P L F+
Sbjct: 2 DRVGSLPDELLSHILSFLTTKEAALTSLLSKRWRYLIAFVPNLAFDDIVFLHPEEGKPER 61
Query: 67 ---IKSFNRFVSRILCLRDVSTPLHAVDFLCRGVVDPRLLKKVVTYAVSHNVEKLRVHAC 123
+SF FV R+L L+ +P+ CR VD + ++ ++ V +L +
Sbjct: 62 DEIRQSFMDFVDRVLALQ-AESPIKKFSLKCRIGVDSDRVDGWISNVLNRGVSELDLLII 120
Query: 124 C-----DFQHFSSCFFSCHTLTSLDLCVGPPRTDERLSFPKSLNLPALTTLSLESFAFCV 178
D S F+ TL L++ G + S+ LP L TL L+S F V
Sbjct: 121 LGMTMEDSYRLSPKGFASKTLVKLEIGCG----IDISWVAGSIFLPMLKTLVLDSVWFYV 176
Query: 179 GDDGCAEPFSALNSLKNLMILYCNVLDAESLCISSVTLTNLTIVGDPVYYSTVELSTPSL 238
D AL +L+ L+++ N LD++ + IS+ +L LTI D + T TPSL
Sbjct: 177 --DKFETLLLALPALEELVLVDVNWLDSD-VTISNASLKTLTIDSDG-HLGTFSFDTPSL 232
Query: 239 FTFDFVCY--EGIPVLKL 254
F + Y E PV+K+
Sbjct: 233 VYFCYSDYAAEDYPVVKM 250
>UniRef100_Q94B46 Hypothetical protein At4g14100 [Arabidopsis thaliana]
Length = 468
Score = 71.2 bits (173), Expect = 3e-11
Identities = 82/309 (26%), Positives = 137/309 (43%), Gaps = 31/309 (10%)
Query: 15 DRLSDLPNCVLLRILSLLDADEAVKTCTISSRWRHLWKHLPTLILCSSHFMR-------I 67
D +S LP + ILS L EA T +S +WR+L+ +P L L S ++
Sbjct: 8 DIISSLPEAISCHILSFLPTKEAASTSVLSKKWRYLFAFVPNLDLDESVYLNPENETEVS 67
Query: 68 KSFNRFVSRILCLRDVSTPLHAVDFLCRGVVDPRLLKKVVTYAVSHNVEKLRVHACCDFQ 127
SF FV R+L L+ ++PLH V+P + + + V L +H + +
Sbjct: 68 SSFMDFVDRVLALQG-NSPLHKFSLKIGDGVEPDRIIPWINNVLERGVSDLDLHVYMETE 126
Query: 128 H-FSSCFFSCHTLTSLDLCVGPPRTDERLSFPKSLNLPALTTLSLESFAFCVGDDGCAEP 186
F S F TL L L + P L F + + LP L TL ++S F G +
Sbjct: 127 FVFPSEMFLSKTLVRLKLMLYP-----LLEF-EDVYLPKLKTLYIDSCYFEKYGIGLTKL 180
Query: 187 FSALNSLKNLM---ILYCNVLDAESLCISSV-TLTNLTIVGDPVYYSTVELSTPSLFTFD 242
S L++L+ I +C D S+ + ++ LT T V D + +V + TP+L
Sbjct: 181 LSGCPILEDLVLDDIPWC-TWDFASVSVPTLKRLTFSTQVRDE-FPKSVSIDTPNLVYLK 238
Query: 243 FVCYEGIPVLKLCRSKINLSSVKHVNIEVRM---WTDYADASLV--LLNWLAELANIKSL 297
F + K+N S+ +I++R+ Y + +V +++ + N+K+L
Sbjct: 239 FT-----DTVAGKYPKVNFDSLVEAHIDLRLLQGHQGYGENDMVGNATDFIMRICNVKTL 293
Query: 298 TLNHTALKV 306
L+ L+V
Sbjct: 294 YLSSNTLQV 302
>UniRef100_Q9FJT2 Emb|CAB62008.1 [Arabidopsis thaliana]
Length = 435
Score = 70.9 bits (172), Expect = 4e-11
Identities = 98/372 (26%), Positives = 152/372 (40%), Gaps = 100/372 (26%)
Query: 1 MKRGRKKHNDYVKEDRLSDLPNCVLLRILSLLDADEAVKTCTISSRWRHLWKHLPTLILC 60
M+ R D DR+S LPN +L RILSL+ +A+ T +S RW+ +WK LPTL+
Sbjct: 1 MEAARSGIEDVTSPDRISQLPNDLLFRILSLIPVSDAMSTSLLSKRWKSVWKMLPTLVYN 60
Query: 61 SSHFMRIKS--FNRFVSRILCLRDVSTPLHAVDFLCRGVVDPRLLKKVVTYAVSHNVEKL 118
+ I S F++F R L L + L K +T + + L
Sbjct: 61 ENSCSNIGSLGFDQFCGRSLQLHEAP------------------LLKTLTLELRKQTDSL 102
Query: 119 RVHACCDFQHFSSCFFSCHTLTSLDLCV---GPPRTDERLSFPKSLNL-PALTTLSLESF 174
SS F + H+ T L+ + G P +SFP +L++ L L L+
Sbjct: 103 D----------SSIFPNIHS-TLLEFSIKSTGYPVYYSTISFPNNLDVFQTLVVLKLQG- 150
Query: 175 AFCVGDDGCAEPFSALNSLK------------NLMILYCNVLD---AESLC--------I 211
C+ F +L SL + ++ C VL+ + LC I
Sbjct: 151 NICLDVVDSPVCFQSLKSLYLTCVNFENEESFSKLLSACPVLEDLFLQRLCSVGRFLFSI 210
Query: 212 SSVTLTNLTIVGDPVYYST----VELSTPS---LFTFD----FVCYEGIPVLKLCRSKIN 260
S +L LT + YYS +E++ PS L FD F E +P L ++
Sbjct: 211 SVPSLQRLTYTKEQAYYSNDEAILEITAPSLKHLNIFDRVGVFSFIEDMPKLVEASVRVK 270
Query: 261 LS----------SVKHVNIE------------------VRMWTDYAD--ASLVLLNWLAE 290
LS SV+H++++ + + D D S +LL+ L +
Sbjct: 271 LSKNEKLPKVLTSVEHLSLDLYPSMVFHLDDRFISKQLLHLKLDIYDNFQSNLLLSLLKD 330
Query: 291 LANIKSLTLNHT 302
L N++SL LNH+
Sbjct: 331 LPNLQSLKLNHS 342
>UniRef100_Q9LX55 Hypothetical protein F25L23_20 [Arabidopsis thaliana]
Length = 464
Score = 70.1 bits (170), Expect = 7e-11
Identities = 83/312 (26%), Positives = 124/312 (39%), Gaps = 48/312 (15%)
Query: 7 KHNDYVKEDRLSDLPNCVLLRILSLLDADEAVKTCTISSRWRHLWKHLPTLILCSSHFM- 65
K D +D ++DLP+ +L +LS L EA T +S RWR+L +P L S ++
Sbjct: 5 KRMDSGSKDMINDLPDALLCHVLSYLTTKEAASTSLLSRRWRYLLAFVPNLEFDDSAYLH 64
Query: 66 ---RIK------------------------SFNRFVSRILCLRDVSTPLHAVDFLC---R 95
R+K SF FV RIL L+ ++PL
Sbjct: 65 RDKRVKNPLHEKGLVGFVLTVDDKRKKLSTSFPDFVDRILDLQG-NSPLDKFSLKMVDDH 123
Query: 96 GVVDPRLLKKVVTYAVSHNVEKLRVHACCDFQHFSSC---FFSCHTLTSLDLCV--GPPR 150
VDP + + + V L H D ++S F TL L L + GPP
Sbjct: 124 DPVDPDCVAPWIHKVLVRGVSDL--HLVIDMNEWTSLPAKIFLTETLVKLTLKIRDGPPI 181
Query: 151 TDERLSFPKSLNLPALTTLSLESFAFCVGDDGCAEPFSALNSLKNLMILYCNVLDAESLC 210
K ++LP L TL LES F D G ++ S L+ L++ + S
Sbjct: 182 D------VKHVHLPKLKTLHLESVMFDEEDIGFSKLLSGCPELEELVLHHIWSCVWTSCS 235
Query: 211 ISSVTLTNLTIVGDPVYYSTVELSTPSLFTFD---FVCYEGIPVLKLCRSKINLSSVKHV 267
+S TL LT + + + + P+ +FD V E V+ K+N S+
Sbjct: 236 VSVATLKRLTFCCNNMKFCGMHEENPNNVSFDTPNLVYLEYAEVIANNYPKVNFDSLVEA 295
Query: 268 NIEVRMWTDYAD 279
I++ M D D
Sbjct: 296 KIDIWMTNDQLD 307
>UniRef100_Q9LYS1 Hypothetical protein F17J16_200 [Arabidopsis thaliana]
Length = 827
Score = 70.1 bits (170), Expect = 7e-11
Identities = 83/312 (26%), Positives = 124/312 (39%), Gaps = 48/312 (15%)
Query: 7 KHNDYVKEDRLSDLPNCVLLRILSLLDADEAVKTCTISSRWRHLWKHLPTLILCSSHFM- 65
K D +D ++DLP+ +L +LS L EA T +S RWR+L +P L S ++
Sbjct: 487 KRMDSGSKDMINDLPDALLCHVLSYLTTKEAASTSLLSRRWRYLLAFVPNLEFDDSAYLH 546
Query: 66 ---RIK------------------------SFNRFVSRILCLRDVSTPLHAVDFLC---R 95
R+K SF FV RIL L+ ++PL
Sbjct: 547 RDKRVKNPLHEKGLVGFVLTVDDKRKKLSTSFPDFVDRILDLQG-NSPLDKFSLKMVDDH 605
Query: 96 GVVDPRLLKKVVTYAVSHNVEKLRVHACCDFQHFSSC---FFSCHTLTSLDLCV--GPPR 150
VDP + + + V L H D ++S F TL L L + GPP
Sbjct: 606 DPVDPDCVAPWIHKVLVRGVSDL--HLVIDMNEWTSLPAKIFLTETLVKLTLKIRDGPPI 663
Query: 151 TDERLSFPKSLNLPALTTLSLESFAFCVGDDGCAEPFSALNSLKNLMILYCNVLDAESLC 210
K ++LP L TL LES F D G ++ S L+ L++ + S
Sbjct: 664 D------VKHVHLPKLKTLHLESVMFDEEDIGFSKLLSGCPELEELVLHHIWSCVWTSCS 717
Query: 211 ISSVTLTNLTIVGDPVYYSTVELSTPSLFTFD---FVCYEGIPVLKLCRSKINLSSVKHV 267
+S TL LT + + + + P+ +FD V E V+ K+N S+
Sbjct: 718 VSVATLKRLTFCCNNMKFCGMHEENPNNVSFDTPNLVYLEYAEVIANNYPKVNFDSLVEA 777
Query: 268 NIEVRMWTDYAD 279
I++ M D D
Sbjct: 778 KIDIWMTNDQLD 789
Score = 65.1 bits (157), Expect = 2e-09
Identities = 75/243 (30%), Positives = 107/243 (43%), Gaps = 18/243 (7%)
Query: 15 DRLSDLPNCVLLRILSLLDADEAVKTCTISSRWRHLWKHLPTLILCSSHFMRIKS----- 69
D +S+LPN +L RILS L EA T +S RW +L +P L S ++ +
Sbjct: 13 DVISNLPNDLLCRILSYLSTKEAALTSILSKRWSNLLLSIPILDFDDSVLLKPQKGQRKN 72
Query: 70 --FNRFVSRILCLR-DVSTPLHAVDFLCR-GVVDPRLLKKVVTYAVSHNVEKLRVHACCD 125
F FV R+L R + S+ + V CR G V+P + K + V ++ L + C D
Sbjct: 73 VFFKAFVDRLLSQRVETSSHVQRVSLKCRQGGVEPDCVIKWILTTV-RDLGVLDLSLCID 131
Query: 126 FQHFSSCFFSCHTLTSLDLCVGPPRTDERLS-FPKSLNLPALTTLSLESFAFCVGDD-GC 183
F F F + T + L +G T RLS FP + P L +L L+ F GD
Sbjct: 132 FGIFHLPFNVFRSKTLVKLRIG---TMIRLSQFPSDVVSPTLNSLVLDLVEFRDGDKVEF 188
Query: 184 AEPFSALNSLKNLMILYCNVLDAESLCISSVTLTNLTIVGD---PVYYSTVELSTPSLFT 240
+ A SL++L + N + SS TL +L D V TPSL
Sbjct: 189 RQILLAFPSLQSLRVHESNKWKFWNGSASSRTLKSLVYRSDDDSSAPKPCVSFDTPSLVY 248
Query: 241 FDF 243
D+
Sbjct: 249 LDY 251
>UniRef100_Q9LLN6 Hypothetical protein DUPR11.17 [Oryza sativa]
Length = 518
Score = 69.7 bits (169), Expect = 9e-11
Identities = 49/122 (40%), Positives = 68/122 (55%), Gaps = 14/122 (11%)
Query: 13 KEDRLSDLPNCVLLRILSLLDADEAVKTCTISSRWRHLWKHLPTLILCSS--HFMRIKSF 70
+E + DLP+ +L +ILSLL ADEAVKTC +S RWRHLWK L + S + F
Sbjct: 99 EECGIDDLPDELLQQILSLLSADEAVKTCVLSRRWRHLWKSTDILRVAYSTDRWKSSDEF 158
Query: 71 NRFVSRILCLRDVSTPLHAVD--FLCR--------GVVDP-RLLKKVVTYAVSHNVEKLR 119
+FV+ ++ LR +S PL +D F R G DP + + V YAV V+ L+
Sbjct: 159 KKFVNHLVLLRGIS-PLRELDLRFNARRYEDVVHDGGSDPYQCVMLWVMYAVMCRVQVLK 217
Query: 120 VH 121
+H
Sbjct: 218 IH 219
>UniRef100_Q9FNI9 Arabidopsis thaliana genomic DNA, chromosome 5, P1 clone:MDJ22
[Arabidopsis thaliana]
Length = 422
Score = 69.7 bits (169), Expect = 9e-11
Identities = 73/270 (27%), Positives = 116/270 (42%), Gaps = 47/270 (17%)
Query: 13 KEDRLSDLPNCVLLRILSLLDADEAVKTCTISSRWRHLWKHLPTLILCSSHFMRIKSFNR 72
KED +S LP+ ++ +IL L +AV T +S RWR LW P L+L S+ F +F
Sbjct: 21 KEDLISQLPDSLITQILFYLQTKKAVTTSVLSKRWRSLWLSTPGLVLISNDFTDYNAFVS 80
Query: 73 FVSRILCL-RDVSTPLHAVDFLCR-GVVDPRLLKKVVTYAVSHNVEKLRVH----ACCDF 126
FV + L R+ LH + R G D + + + + + ++ L V C F
Sbjct: 81 FVDKFLGFSREQKLCLHKLKLSIRKGENDQDCVTRWIDFVATPKLKHLDVEIGPTRCECF 140
Query: 127 QHFSSCFFSCHTLTSLDL---CVGPPRTDERLSFPKSLNLPALTTLSLESFAFCVGDDGC 183
+ +SC +L L L C+G +S++LP L T+SLE
Sbjct: 141 EVIPLSLYSCESLLYLRLNHVCLGK---------FESVSLPCLKTMSLE----------- 180
Query: 184 AEPFSALNSLKNLMILYCNVLDAESLCISSVTLTNLTIVGDPVYYSTVELSTPSLFTFDF 243
+Y N D ESL + L +L+ V Y V + T
Sbjct: 181 -------------QNIYANEADLESLISTCPVLEDLSFVSGA--YDKVNVLRVQSQTLTS 225
Query: 244 VCYEG-IPVLKLCRSK--INLSSVKHVNIE 270
+ EG + L L +S+ I+ + +K++N+E
Sbjct: 226 LNIEGCVEYLDLDKSEVLIDATRLKYLNLE 255
>UniRef100_Q9ZV94 F9K20.20 protein [Arabidopsis thaliana]
Length = 452
Score = 68.6 bits (166), Expect = 2e-10
Identities = 76/318 (23%), Positives = 143/318 (44%), Gaps = 28/318 (8%)
Query: 4 GRKKHNDYVKEDRLSDLPNCVLLRILSLLDADEAVKTCTISSRWRHLWKHLPTLILCSSH 63
G +K +EDRLS+LP ++ +I+ + + VK+ +S RWR+LW+++P L + +
Sbjct: 5 GDEKRARRSEEDRLSNLPESLICQIMLNIPTKDVVKSSVLSKRWRNLWRYVPGLNVEYNQ 64
Query: 64 FMRIKSFNRFVSRILCL-RDVSTPLHAVDFLCRGVVDPRLLKKVVTYAVSHNVEKLRVHA 122
F+ +F FV R L L R+ + + C + R + V + +KL+V
Sbjct: 65 FLDYNAFVSFVDRFLALDRESCFQSFRLRYDCD--EEERTISNVKRWINIVVDQKLKVLD 122
Query: 123 CCDFQ------HFSSCFFSCHTLTSLDLCVGPPRTDERLSFPKSLNLPALTTLSLESFAF 176
D+ ++C +L SL LC + L PK ++LP + + L+ F
Sbjct: 123 VLDYTWGNEEVQIPPSVYTCESLVSLKLC------NVILPNPKVISLPLVKVIELDIVKF 176
Query: 177 CVGDDGCAEPFSALNSLKNLMILYCNVLDAESLCISSVTLTNLTIVG---DPVYYSTVEL 233
+ S+ ++L++L+I +V D L +SS +L + +G D V +
Sbjct: 177 S-NALVLEKIISSCSALESLIISRSSVDDINVLRVSSRSLLSFKHIGNCSDGWDELEVAI 235
Query: 234 STPSLFTFDFVCYEGIPVL-----KLCRSKINLSSVKHVNIEVRMWTDYADASLVLLNWL 288
P L + + L +KIN+ N+E + ++ ++L
Sbjct: 236 DAPKLEYLNISDHSTAKFKMKNSGSLVEAKINII----FNMEELPHPNDRPKRKMIQDFL 291
Query: 289 AELANIKSLTLNHTALKV 306
AE++++K L ++ L+V
Sbjct: 292 AEISSVKKLFISSHTLEV 309
>UniRef100_Q8L7H1 Hypothetical protein At4g14103 [Arabidopsis thaliana]
Length = 381
Score = 68.6 bits (166), Expect = 2e-10
Identities = 75/312 (24%), Positives = 136/312 (43%), Gaps = 30/312 (9%)
Query: 15 DRLSDLPNCVLLRILSLLDADEAVKTCTISSRWRHLWKHLPTLILCSSHFMRIK------ 68
D +S LP+ + ILS L EA T +S +WR+L+ +P L L S ++ +
Sbjct: 8 DVISSLPDDISSHILSFLPTKEAASTSVLSKKWRYLFAFVPNLDLDDSVYLNPENETEIS 67
Query: 69 -SFNRFVSRILCLRDVSTPLHAVDFLCRGVVDPRLLKKVVTYAVSHNVEKLRVHACCDFQ 127
SF FV R+L L+ ++PLH +DP + + + V L +H + +
Sbjct: 68 TSFMDFVDRVLALQG-NSPLHKFSLKIGDGIDPVRIIPWINNVLERGVSDLDLHLNLESE 126
Query: 128 H-FSSCFFSCHTLTSLDLCVGPPRTDERLSFPKSLNLPALTTLSLESFAFCVGDDGCAEP 186
S + C TL L L G T + + ++LP L TL +E+ F G +
Sbjct: 127 FLLPSQVYLCKTLVWLKLRFGLYPTID----VEDVHLPKLKTLYIEATHFEEHGVGLTKL 182
Query: 187 FSALNSLKNLMI--LYCNVLDAESLCISSVTLTNLTIVGDPVYYSTVELSTPSLFTFDFV 244
S L++L++ + + D S+ + ++ + + +V L TP+L F
Sbjct: 183 LSGCPMLEDLVLDDISWFIWDFASVSVPTLKRLRFSWQERDEFPKSVLLDTPNLVYLKFT 242
Query: 245 CYEGIPVLKLCRSKINLSSVKHVNIEVRM--------WTDYADASLV--LLNWLAELANI 294
+ K+NL S+ +I++R+ Y + +V +++ + N+
Sbjct: 243 -----DTVAGKYPKVNLDSLVEAHIDLRLLKPLLINYHQGYGENDMVGNATDFIMRICNV 297
Query: 295 KSLTLNHTALKV 306
K+L L+ L+V
Sbjct: 298 KTLYLSANTLQV 309
>UniRef100_Q8S805 Putative copia-type polyprotein [Oryza sativa]
Length = 1803
Score = 67.8 bits (164), Expect = 4e-10
Identities = 91/350 (26%), Positives = 145/350 (41%), Gaps = 89/350 (25%)
Query: 3 RGRKKHNDYVKEDRLSDLPNCVLLRILSLLDADEAVKTCTISSRWRHLWKHLPTLILCSS 62
R R D DRLSDLP+ +L RI+S L+A +AV+TC +S RWR+LW +P + ++
Sbjct: 1412 RARPPMEDAAGRDRLSDLPDEILHRIMSFLNARQAVQTCVLSRRWRNLWHTVPCI---NA 1468
Query: 63 HFMRIKS---------FNRFVSRILCLRDVSTPLHAVDFLCRGVVDPRLLKKVVTYAVSH 113
F+ S F RFV+R+L RD ++ V+D LLK YA+
Sbjct: 1469 DFVEFDSIGYQGPVVPFKRFVNRLLEFRDPAS-----------VIDTFLLK----YAMPD 1513
Query: 114 NVEKLRVHACCDFQHFSSCFFSCHTLTS----LDLCVGPPRTDERLSFPKSLNLPALTTL 169
++ + + + + H L L++ V FP L+ T+
Sbjct: 1514 RLDGYKA------SNEEANRWIGHALQKQARILEVAV--------FFFPLDLDHSVFTSF 1559
Query: 170 SLE--SFAFCVGDDGCAEPF-SALNSLKNLMILYCNVLDAESLCISSVTLTNLTIVGDPV 226
L F+ G E + L++L++ C + D E ISS TL LT+ +
Sbjct: 1560 YLRRIEFSHVYLRKGFFEQIETGCPLLEDLLLHQCFIWDGE---ISSQTLKVLTVDATEL 1616
Query: 227 Y-YSTVELSTPSL---------------------------FTFDFVCYEGIPVLKLCRSK 258
Y + +STP+L TFD + ++G R
Sbjct: 1617 YTVKEMSISTPNLTSLTLSGLEYPKAVLKDMPLLVTASVSVTFDALNFDGYYDANDLRQY 1676
Query: 259 I-NLSSVKHVNIEVRMWTDYADASLVLLN---WLAELANIKSLTLNHTAL 304
+ LS+V+++ Y A L++ N W E ++ +LTL L
Sbjct: 1677 LWGLSAVRNLEFH------YEGAELMIANNSQWCPEFVDVVNLTLGEWCL 1720
>UniRef100_Q9ZV93 F9K20.21 protein [Arabidopsis thaliana]
Length = 458
Score = 67.4 bits (163), Expect = 5e-10
Identities = 78/269 (28%), Positives = 114/269 (41%), Gaps = 34/269 (12%)
Query: 2 KRGRKKHNDYVKEDRLSDLPNCVLLRILSLLDADEAVKTCTISSRWRHLWKHLP--TLIL 59
KR R K + V D +S+LP +L ++L L + VK+ +SSRWR+LWK++P L
Sbjct: 7 KRVRAKGSREV--DWISNLPETLLCQVLFYLPTKDVVKSSVLSSRWRNLWKYVPGFNLSY 64
Query: 60 CSSHFMRIKSFN-----RFVSRILCLRDVS-TPLHAVDFLCRGVVDPR--LLKKVVTYAV 111
C H S++ RFV + S +++ G +P+ L+++ + V
Sbjct: 65 CDFHVRNTFSYDHNTFLRFVDSFMGFNSQSCLQSFRLEYDSSGYGEPKLALIRRWINSVV 124
Query: 112 SHNVEKLRV-HACCDFQHFS--SCFFSCHTLTSLDLCVGPPRTDERLSFPKSLNLPALTT 168
S V+ L V CD F ++C TL L L L+ PK ++LP+L
Sbjct: 125 SRKVKYLGVLDDSCDNYEFEMPPTLYTCETLVYLTL------DGLSLASPKFVSLPSLKE 178
Query: 169 LSLESFAFCVGDDGCAEP-FSALNSLKNLMILYCNVLDAESLCISSVTLTNLTIVGDPVY 227
L L F D E S L+NL I D E C+ S +L + T V D
Sbjct: 179 LHLSIVKF--ADHMALETLISQCPVLENLNINRSFCDDFEFTCVRSQSLLSFTHVAD--- 233
Query: 228 YSTVELSTPSLFTFDFVCYEGIPVLKLCR 256
T + D V P LK R
Sbjct: 234 -------TDEMLNEDLVVAIDAPKLKYLR 255
>UniRef100_Q8L671 Hypothetical protein OSJNBb0072F04.2 [Oryza sativa]
Length = 960
Score = 66.6 bits (161), Expect = 8e-10
Identities = 86/333 (25%), Positives = 125/333 (36%), Gaps = 81/333 (24%)
Query: 15 DRLSDLPNCVLLRILSLLDADEAVKTCTISSRWRHLWKHLPTLILCSSHFMRI------- 67
D SD+P+ V+L I+S L +AV+TC +S RW +LW+ +P + F R
Sbjct: 583 DWFSDVPDDVILNIMSFLTTRQAVQTCVLSRRWLNLWRSVPCINADVGEFQRSDTEWEEY 642
Query: 68 -----KSFNRFVSRILCLRDVSTPLHAVDFLC------RGVVDPRLLKKVVTYAVSHNVE 116
+F F+ R+L LR+ + P+ F C G D + + +T+A+
Sbjct: 643 DQERESAFKMFMDRVLELRNPAAPIRTFRFRCCRLDGFEGTSDEADMNRWITHAMQKQPW 702
Query: 117 KLRVHACCDFQHFSSCFFSCHTLTSLDLCVGPPRTDERLSFPKSLNLPALTTLSLESFAF 176
L + D F+C LT R+ F L +P F
Sbjct: 703 VLDILVLYDALKLDHSAFTCRYLT-------------RIKFINVLMMPGF---------F 740
Query: 177 CVGDDGCAEPFSALNSLKNLMILYCNVLDAESLCISSVTLTNLTIVGDPVYYS-TVELST 235
+ GC L+NL + V D E ISS TL LTI Y +ST
Sbjct: 741 QQLEMGCP-------VLENLFLDESIVADVE---ISSRTLKVLTIKSTQFSYKFRTTIST 790
Query: 236 PS---------------------LFTFDFVCYEGIPVLKLCRSKINLSSVKHVNIEVRMW 274
PS L T V Y+ C++ +LS+ K R+
Sbjct: 791 PSVTYLKLWRPVNGIYVFNDMPLLVTSILVLYDVQDSSDFCQNLRSLSAAK------RLE 844
Query: 275 TDYADASLVLLNWL---AELANIKSLTLNHTAL 304
DY L + N L + N+ SLTL L
Sbjct: 845 FDYFGRKLTMENNLQLYPKFNNLVSLTLGQWCL 877
Score = 66.2 bits (160), Expect = 1e-09
Identities = 61/244 (25%), Positives = 99/244 (40%), Gaps = 52/244 (21%)
Query: 15 DRLSDLPNCVLLRILSLLDADEAVKTCTISSRWRHLWKHLPTLILCSSHFMRIK------ 68
D SD+P+ ++L I+S L +AV+TC +S RWR+LW+ +P + F R
Sbjct: 21 DWFSDVPDDIILNIMSFLTTRQAVQTCVLSRRWRNLWRSVPCINSDIDEFTRDSDSEGYY 80
Query: 69 ------SFNRFVSRILCLRDVSTPLHAVDFLCR--------GVVDPRLLKKVVTYAVSHN 114
+F F+ R++ LRD + + F C+ + DP + +++AV
Sbjct: 81 DEKTELAFIMFMERVMELRDPAALISTFQFRCKFELDEGFDDISDPEDINAWISHAVQKQ 140
Query: 115 VEKLRVHACCDFQHFSSCFFSCHTLTSLDLCVGPPRTDERLSFPKSLNLPALTTLSLESF 174
L + CD + F+ LT ++ T++ L
Sbjct: 141 ARVLDIVVLCDKLYLDHSEFASRYLTRIE----------------------FTSVVLMEG 178
Query: 175 AFCVGDDGCAEPFSALNSLKNLMILYCNVLDAESLCISSVTLTNLTIVGDPVYYSTVELS 234
F + GC + ++L + C V D E ISS TL LTI +S
Sbjct: 179 FFKQLEMGCP-------AWESLFLDECAVNDVE---ISSQTLKVLTIKNTLFSSDKTTIS 228
Query: 235 TPSL 238
TPS+
Sbjct: 229 TPSV 232
>UniRef100_Q8S811 Hypothetical protein OSJNBa0020E23.13 [Oryza sativa]
Length = 949
Score = 66.2 bits (160), Expect = 1e-09
Identities = 85/326 (26%), Positives = 134/326 (41%), Gaps = 65/326 (19%)
Query: 15 DRLSDLPNCVLLRILSLLDADEAVKTCTISSRWRHLWKHLPTLILCSSHF------MRIK 68
D +SDLP+ +L RI+S L+A +AV+TC +S RWR+LW+ +P + F
Sbjct: 549 DWISDLPDEILHRIMSFLNARQAVQTCVLSRRWRNLWRTVPCINADCKEFDFFGFRRSEV 608
Query: 69 SFNRFVSRILCLRDVSTPLHAVDFLCRGVVDPRLLKKVVTYAVSHNVEKLRVHACCDFQH 128
F RFV+R+L LRD + A F + +SH ++K +
Sbjct: 609 EFKRFVNRLLELRDPIAMMDAFWFRYHKLDTDTTSSADTNRWISHALQK-------QARV 661
Query: 129 FSSCFFSCHTLTSLDLCVGPPRTDERLSFPKSLNLPALTTLSLESFAFCVGDDGCAEPFS 188
+ + CH L LD R R+ F + + L+ F + GC
Sbjct: 662 LEAVMYPCH-LLDLDHSSFTSRYLRRIGF---------SGVRLDQGFFKQLEAGCP---- 707
Query: 189 ALNSLKNLMILYCNVLDAESLCISSVTLTNLTIVGDPVYY---------STVELSTPSLF 239
+L++L + +C + D + ISS TL LTI D Y+ +S PS+
Sbjct: 708 ---ALEDLFLHHCTIEDDK---ISSQTLKVLTI--DRTYFLIAINATDVQKKSISAPSVT 759
Query: 240 TFDFVCYEG-IPVLKLCRSKINLS---SVKHVNIEVR-----MWT---------DYADAS 281
+ EG + +LK S + S S V+ + +W+ +Y
Sbjct: 760 SLTMYSPEGSLHILKDMTSLVTASVSFSEFRVHFDANDFYQYLWSLSGVTNLEFNYQGPK 819
Query: 282 LVL---LNWLAELANIKSLTLNHTAL 304
L + L W E N+ +LTL L
Sbjct: 820 LKIENNLQWCPEFVNVVNLTLGKWCL 845
Score = 56.6 bits (135), Expect = 8e-07
Identities = 32/89 (35%), Positives = 49/89 (54%), Gaps = 12/89 (13%)
Query: 6 KKHNDYVKEDRLSDLPNCVLLRILSLLDADEAVKTCTISSRWRHLWKHLPTLILCSSHFM 65
K H + DR+S LP+ +L I+S L+ +AV+TC +S RWR+LW+ +P + F
Sbjct: 81 KAHVRAARMDRISGLPDKILHHIMSFLNTRQAVQTCVLSRRWRNLWRTMPCINADFDEFD 140
Query: 66 RIK------------SFNRFVSRILCLRD 82
I +F RFV+++L LRD
Sbjct: 141 FIAYQGDDEDYNDEVAFKRFVNQMLELRD 169
>UniRef100_O80761 T13D8.27 protein [Arabidopsis thaliana]
Length = 403
Score = 65.9 bits (159), Expect = 1e-09
Identities = 76/318 (23%), Positives = 133/318 (40%), Gaps = 54/318 (16%)
Query: 15 DRLSDLPNCVLLRILSLLDADEAVKTCTISSRWRHLWKHLPTLILCSSHFMRIKSFNRFV 74
DRLS LP +L RILS L ++V+T +S WR+LW H+P L L +S F F F+
Sbjct: 14 DRLSALPEHLLCRILSELSTKDSVRTSVLSKHWRNLWLHVPVLELETSDFPDNLVFREFI 73
Query: 75 SRILCLRDVSTPLHAVD--------------FLCRGVVDPRLLKKVVT---YAVSHNVEK 117
R + D L + D ++ VV R+ +VT Y V+ + K
Sbjct: 74 DRFVGF-DKEIDLKSFDIFYDVNVLWYDDFLWMIDDVVKRRVCDLMVTNNPYVVNEKLVK 132
Query: 118 LRV--HACCDFQHFSSCFFSCHTLTSLDLCVGPPRTDERLSFPKSLNLPALTTLSLESFA 175
+ + ++C + + F + + L S +C LP + TL L
Sbjct: 133 MPISLYSCATLVNLNLSFVAMNNLPSESVC-----------------LPRVKTLYLHGVK 175
Query: 176 FCVGDDGCAEPFSALNSLKNLMIL-----YCNVLDAESLCISSVTLTNLTIVGDPVYYST 230
GD S+ + L++L ++ Y V+ S + S T+ + DP
Sbjct: 176 L-DGDSILGTLVSSCSVLEDLTVVTHPGDYEKVVCFRSQSVKSFTIESQCEYKDP----N 230
Query: 231 VELSTPSLFTFDFVCYEGIPVLKLCRSKINLSSVKHVNIEVRMWTDYAD--ASLVLLNWL 288
VE+ P L +++C I V++++ +Y D A ++ N+L
Sbjct: 231 VEIDCPRL---EYMCIREYQSESFVVHSI--GPYAKVDVDIFFEVEYEDPLAISMIRNFL 285
Query: 289 AELANIKSLTLNHTALKV 306
++ ++ +T++ L+V
Sbjct: 286 TGISKVREMTISSRTLEV 303
>UniRef100_Q9LX56 Hypothetical protein F25L23_10 [Arabidopsis thaliana]
Length = 475
Score = 65.1 bits (157), Expect = 2e-09
Identities = 75/243 (30%), Positives = 107/243 (43%), Gaps = 18/243 (7%)
Query: 15 DRLSDLPNCVLLRILSLLDADEAVKTCTISSRWRHLWKHLPTLILCSSHFMRIKS----- 69
D +S+LPN +L RILS L EA T +S RW +L +P L S ++ +
Sbjct: 13 DVISNLPNDLLCRILSYLSTKEAALTSILSKRWSNLLLSIPILDFDDSVLLKPQKGQRKN 72
Query: 70 --FNRFVSRILCLR-DVSTPLHAVDFLCR-GVVDPRLLKKVVTYAVSHNVEKLRVHACCD 125
F FV R+L R + S+ + V CR G V+P + K + V ++ L + C D
Sbjct: 73 VFFKAFVDRLLSQRVETSSHVQRVSLKCRQGGVEPDCVIKWILTTV-RDLGVLDLSLCID 131
Query: 126 FQHFSSCFFSCHTLTSLDLCVGPPRTDERLS-FPKSLNLPALTTLSLESFAFCVGDD-GC 183
F F F + T + L +G T RLS FP + P L +L L+ F GD
Sbjct: 132 FGIFHLPFNVFRSKTLVKLRIG---TMIRLSQFPSDVVSPTLNSLVLDLVEFRDGDKVEF 188
Query: 184 AEPFSALNSLKNLMILYCNVLDAESLCISSVTLTNLTIVGD---PVYYSTVELSTPSLFT 240
+ A SL++L + N + SS TL +L D V TPSL
Sbjct: 189 RQILLAFPSLQSLRVHESNKWKFWNGSASSRTLKSLVYRSDDDSSAPKPCVSFDTPSLVY 248
Query: 241 FDF 243
D+
Sbjct: 249 LDY 251
>UniRef100_Q9LQC1 F19C14.7 protein [Arabidopsis thaliana]
Length = 482
Score = 65.1 bits (157), Expect = 2e-09
Identities = 72/272 (26%), Positives = 119/272 (43%), Gaps = 19/272 (6%)
Query: 15 DRLSDLPNCVLLRILSLLDADEAVKTCTISSRWRHLWKHLPTLILCSSHFMRIKSFNRFV 74
D +S LP+ +L ILS L+ EA T ++ +WR+L+ +P L S +R+ N V
Sbjct: 8 DIISGLPDSLLCHILSFLNTKEAASTSVLAKKWRYLFASVPNLDFDDSVHLRLGKRNPAV 67
Query: 75 S-RILCLRDVSTPLHAVDFLCRGVVDPRLLKKVVTYAVSHNVEKLRVHACCDFQ-HFSSC 132
S ++L L+D ++PLH R VD + + + V L + ++ S
Sbjct: 68 SDQVLRLQD-NSPLHKFSLKIRDCVDIVRIICWILKVLERGVSDLELDMHLKWKSSLPSK 126
Query: 133 FFSCHTLTSLDLCVGPPRTDERLSF--PKSLNLPALTTLSLESFAFCVGDDGCAEPFSAL 190
F TL L L V ER F + ++LP L TL + S F G + S
Sbjct: 127 IFLSETLVRLKLSV------ERGPFIDVEDVHLPKLKTLHIVSVKFEKHGIGLNKLLSGC 180
Query: 191 NSLKNLMILYCNVLDAESLCISSVTLTNLTIVGDPVYYS---TVELSTPSLFTFDFVCYE 247
+ L+ L + Y + + + +S TL LT G+ + +V +TP+L F
Sbjct: 181 HILEELNLEYISWCLWDFVSVSLTTLKRLTFCGEVMQDENPISVSFNTPNLVYLMFT--- 237
Query: 248 GIPVLKLCRSKINLSSVKHVNIEVRMWTDYAD 279
+ S +N S+ +I ++M D +
Sbjct: 238 --DAIADEYSTVNFDSLVEAHINLQMSEDQVE 267
>UniRef100_Q9LLN4 Hypothetical protein DUPR11.22 [Oryza sativa]
Length = 431
Score = 64.7 bits (156), Expect = 3e-09
Identities = 34/77 (44%), Positives = 49/77 (63%), Gaps = 2/77 (2%)
Query: 13 KEDRLSDLPNCVLLRILSLLDADEAVKTCTISSRWRHLWKHLPTLILCSSHFMRI-KSFN 71
+ED + LP+ +L ILS L A++AVKTC +S RWRHLWK P L + ++ R K F
Sbjct: 6 EEDGIDVLPDALLQHILSFLPAEDAVKTCVLSRRWRHLWKLTPILCITNTERWRSPKDFI 65
Query: 72 RFVSRILCLRDVSTPLH 88
+ V+ ++ R S+PLH
Sbjct: 66 KLVNHLVLFRG-SSPLH 81
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.326 0.138 0.431
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 491,008,709
Number of Sequences: 2790947
Number of extensions: 18772541
Number of successful extensions: 47663
Number of sequences better than 10.0: 359
Number of HSP's better than 10.0 without gapping: 297
Number of HSP's successfully gapped in prelim test: 62
Number of HSP's that attempted gapping in prelim test: 47234
Number of HSP's gapped (non-prelim): 427
length of query: 307
length of database: 848,049,833
effective HSP length: 127
effective length of query: 180
effective length of database: 493,599,564
effective search space: 88847921520
effective search space used: 88847921520
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 74 (33.1 bits)
Medicago: description of AC147179.8


